Abstract
Mouse embryonic stem cells (mESCs) have a remarkable capacity to maintain normal genome stability and karyotype in culture. We previously showed that infrequent bursts of Zscan4 expression (Z4 events) are important for the maintenance of telomere length and genome stability in mESCs. However, the molecular details of Z4 events remain unclear. Here we show that Z4 events involve unexpected transcriptional derepression in heterochromatin regions that usually remain silent. During a Z4 event, we see rapid derepression and rerepression of heterochromatin leading to a burst of transcription that coincides with transient histone hyperacetylation and DNA demethylation, clustering of pericentromeric heterochromatin around the nucleolus, and accumulation of activating and repressive chromatin remodelling complexes. This heterochromatin-based transcriptional activity suggests that mESCs may maintain their extraordinary genome stability at least in part by transiently resetting their heterochromatin.
Keywords: heterochromatin, pericentromere, embryonic stem cells
1. Introduction
Mouse embryonic stem cells (mESCs) are typically derived from the inner cell mass (ICM) of blastocysts.1,2 Remarkably, mESCs can be maintained in vitro for extended periods without losing their replicative capacity (self-renewal) or ability to differentiate into nearly all cell types except trophoblasts (pluripotency).3,4 Pluripotency has been studied intensively5,6 and is now understood to have states of different degree. For example, depending on the culture condition, mESCs can be maintained as a naive pluripotent state (ICM-like) or a primed pluripotent state (epiblast-like).7–9 It has also been proposed that mESCs are a heterogenous mixture of various transient pluripotent states with distinct expression levels of Nanog and other key transcription factors:10,11 mESCs would fluctuate among these states. Furthermore, recent reports proposed that mESCs can also take on a ‘totipotent’ state by acquiring the additional capacity to differentiate into trophoblast cells: the totipotent cells can be marked and isolated by their expression of the Hhex gene12 or the expression of a MuERV-L retrotransposon.13 As mESCs in the latter state were found to express some genes specifically activated in two-cell embryos, this particular totipotent state is called the ‘two-cell (2C) state’.13
An equally remarkable though less studied feature of mESCs is their capacity to maintain genome stability and normal karyotype.14,15 We recently demonstrated that this feature involves the action of a mammalian-specific gene, Zscan4 (zinc finger and SCAN domain containing 4). Originally identified as a gene expressed specifically in the two-cell mouse embryos, Zscan4 is not expressed in mESCs most of the time, but it is occasionally expressed transiently for several hours.16,17 Such a burst of Zscan4 transcription, called hereafter ‘Zscan4 event’ or ‘Z4 event’, is accompanied by transient expression of other two-cell or preimplantation genes.16–18 It has been shown that Z4 events are accompanied by critical biological events including rapid extension of telomeres17,18 and global blockage of translation.19 Because Z4 events occur rarely, Zscan4 is expressed in only 1–5% of ES cells at any given time in culture.16,17,20 Therefore, despite the occurrence of Z4 events in all mouse ES cell lines examined thus far,21 all the previous studies on mESCs have been obtained essentially with Zscan4− mESCs. To define the molecular events occurring during the Z4 events more clearly, we have compared gene expression profiles and the genome-wide chromatin conformations in isolated Zscan4+ cells and Zscan4− cells. Our study has revealed an unexpected link between Z4 event and changes in the conformation of heterochromatin.
Mammalian genomes form two main chromatin structures: heterochromatin and euchromatin. Prototypical heterochromatin, often called ‘constitutive heterochromatin’, is in a tightly packed form and usually organizes on repetitive sequences such as centromeres, telomeres, and retrotransposons.22,23 Constitutive heterochromatin was considered transcriptionally silent, but recent studies have shown that constitutive heterochromatin can also be transcribed.24 In Schizosaccharomyces Pombe, it has been shown that constitutive heterochromatin is transcribed in S phase of every cell cycle, which is required for the timely assembly of constitutive heterochromatin.25 Another form of heterochromatin, the ‘facultative heterochromatin’, including repressed X-chromosome, is transcribed in some cell types.26–28 In this paper, we have identified a unique set of facultative heterochromatin regions that respond specifically to Z4 event (tentatively called ‘Z4 event-associated heterochromatin, zHC’). We have also provided the detailed molecular mechanism by showing that Z4 event is accompanied by transient rapid derepression and rerepression of both constitutive heterochromatin and zHC. We discuss biological significance of Z4 event.
2. Materials and methods
2.1. ES cell culture
MC1 ES cells (129S6/SvEvTac)29 were used for all experiments unless otherwise specified. Generation of MC1-ZE7 ES cells, Tet-Zscan4c ES cells, and Tet-shZscan4c ES cells has been described previously.17,30 All ES cell lines were maintained on gelatin-coated feeder-free plates in complete ES medium.17 In Tet-Zscan4c ES cells, Zscan4-Flag was repressed in the complete ES medium supplemented with 1.5 µg/ml of Dox, but induced in the absence of Dox. Tet-shZscan4c ES cells were cultured in the complete ES medium supplemented with 1.5 µg/ml of Dox to induce shRNA against Zscan4c. For immunofluorescense staining, ES cells were cultured on cover glass with mitomycin C-treated feeders.
2.2. FACS
Sorting of MC1-ZE7 ES cells was performed using a BD FACSAria II. The cells were sorted according to the fluorescent intensity of Emerald.
2.3. RNA-seq
Total RNA was extracted using TRIzol solution (Roche) from FACS-sorted MC1-ZE7 cells following the manufacturer's instruction. The quality of total RNA was checked by Nanodrop (260/280 = 1.9–2.1, 260/230 > 2.0) and BioAnalyzer 2100 (RIN > 7). Five hundred nanograms of total RNA was used to make rRNA- and mtrRNA-free cDNA libraries using TruSeq Stranded Total RNA with Ribo-Zero Gold Sample Prep Kit (Illumina). The cDNA was fragmented with alkaline-denatured fragmentation with 2 min at 94°C, which produced the fragments in 130–290 bp size range. The DNA libraries were quantified by KAPA quantification kit and sequenced by HiSeq 2500.
2.4. Antibodies
The following antibodies were used: mouse polyclonal anti-Zscan4 (Abnova H00201516- B01P), rabbit polyclonal anti-Zscan4,17 HP1α (Millipore MAB3584 for immunostaining, Millipore no. 05-689 for immunoblotting), H3K9me3 (Upstate no. 07-442), H4K20me3 (Millipore no. 07-463), H3K4me3 (Millipore no. 07-473), H3K9ac (Millipore no. 07-352), H3K14ac (Millipore no. 17-10051), H3K18ac (Epitomics no. 1766-1), H3K27ac (Active Motif no. 39-133 for immunostaining and ChIP, Abcam ab4729 for immunostaining and immunoblotting), H3K9me2 (Abcam ab1220), pan-H3 (Abcam ab1791), p300 (Millipore no. 05-257), CBP (Santa Cruz sc-583), HDAC1 (Abcam ab7028 for immunostaining, ab31263 for western blot), HDAC2 (ab7029), MTA1 (Bethyl lab. A300-911A for immunostaining, Abcam ab71153 for IP and western blot), MTA2 (Santa Cruz sc-28731), RBBP4 (Epitomics no. 2599-1), RBBP7 (Epitomics no. 3503-1), LSD1 (Abcam ab17721), BRG1 (Abcam ab4081 for immunostaining, a gift from Dr Wang for IP and western blot), KAP1 (Abcam ab22553), CREST (Antibodies Inc. no. 15-235), DIG (Roche no. 11093274910), and Flag M2 (Sigma F1804).
2.5. Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature and permeabilized in 0.25% NP-40 in phosphate-buffered saline (PBS) for 10 min. The cells were blocked for 10 min in PBS, 1% bovine serum albumin, 10% fetal bovine serum, and 0.2% saponin and incubated overnight at 4°C with the primary antibodies (1 : 500) in a blocking solution. After three washes in PBS, the cells were incubated for 1 h at room temperature with Alexa dye-conjugated secondary antibodies (1 : 500; Invitrogen) in a 1/10 blocking solution. Nuclei were counterstained with DAPI (Roche) for 5 min at room temperature. Immunofluorescence was visualized by a laser scanning confocal microscope LSM 510 (Zeiss). Images were obtained using the Zeiss LSM software. The quantitative analysis of the fluorescence signals was conducted using the NIH Image program. The procedure for Immuno-DNA-FISH is included in the Supplementary Information.
2.6. ChIP-seq
ChIP was performed using standard methods on FACS-sorted MC1-ZE7 cells (Em+/Em−). Sequencing analysis was performed on the Illumina Genome Analyzer II, and sequence reads were aligned to the mouse genome (mm9) using Bowtie (http://bowtie-bio.sourceforge.net). H3K27ac peaks were identified using HOMER (http://biowhat.ucsd.edu/homer/chipseq/). The details are described in the Supplementary Information.
2.7. Genome-wide DNA methylation analysis using the HELP tagging method
The extraction of genomic DNA and the HELP tagging assay was performed as previously described31 with several modifications (see the Supplementary Information for details).
2.8. Bisulfite sequencing
Genomic DNA was subjected to bisulfite conversion using an EZ DNA Methylation-Gold kit (Zymo Research). The bisulfite converted DNA was PCR amplified by EpiTaq HS polymerase (Takara) using the primer sets listed in Supplementary Table S1. The PCR products were subcloned into a pGEM-T Easy vector (Promega) and sequenced using a SP6 primer.
2.9. Immunoprecipitation
Nuclear extract was prepared from Tet-Zscan4c ES cells after overexpressing Flag-tagged Zscan4 and diluted 5 times with IP buffer for immunoprecipitation. Nuclear extract and antibody were incubated at 4°C overnight and then washed 4 times with the IP buffer for 10 min at room temperature. The proteins were eluted with Laemmli's sample buffer for western blotting.
3. Results
3.1. Transcriptional bursts from specific genomic regions and constitutive heterochromatin during a Z4 event
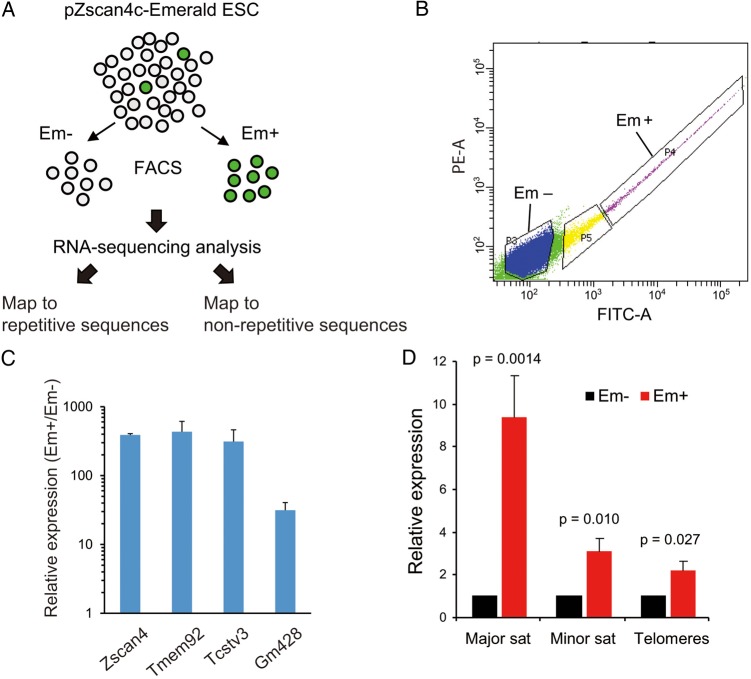
To characterize the transcriptome specific to the transient Z4 event, we carried out RNA-sequencing (RNA-seq) analyses with isolated Zscan4+ and Zscan4− cells. To separate the two kinds of cells, we FACS-sorted the MC1-ZE7 cell line,17 in which an Emerald fluorescence protein (Em) was controlled by the Zscan4 promoter, distinguishing strongly Em+ (i.e. Zscan4+) from Em− (i.e. Zscan4−) cells (Fig. 1A and B). Over 35 million reads per sample were mapped to non-repetitive regions of the mouse genome in two biological replications, detecting 24,538 genes (RefSeq). We found that among 476 differentially expressed genes (fold change > 2, FDR < 0.05), nearly all (469) were more highly expressed in Zscan4+ cells than in Zscan4− cells (Supplementary Table S2). This result indicates that a specific molecular program is activated and superimposed on the regular mESC program. The findings were consistent with previous microarray analyses.18 For brevity, we refer to genes specifically up-regulated during Z4 event as ‘Z4 event-associated genes (ZAGs).’ The ZAGs included many preimplantation embryo genes such as Zscan4, Tmem92, Gm428, and Tcstv3, whose differential expression was also confirmed by quantitative PCR (Fig. 1C). During Z4 event, essentially all Zscan4 paralogues were abundantly transcribed from their canonical transcription start sites; six newly identified copies of Tmem92 were also abundantly transcribed (Supplementary Fig. S1A), suggesting that expression of these genes is strictly controlled even though they are only expressed for a very short time.
Figure 1.
Zscan4-associated heterochromatin transcription in mouse ES cells. (A) A schematic presentation for whole transcriptome analyses of ES cells in the Zscan4+ cells and Zscan4− cells. (B) FACS sorting of ES cells into Emerald-positive cells (Em+, Zscan4+ cells) and Emerald-negative cells (Em−, Zscan4− cells). (C) Expression levels of Zscan4-related genes (Zscan4, Tmem92, Tcstv3, Gm428) in FACS-sorted Em+ cells compared with Em− cells. The expression levels were normalized by GAPDH. Error bars, S.D. (D) Abundance of sequence reads matched to major satellites, minor satellites, and telomeres in the Em+ cells relative to that in Em− cells. Error bars, S.E. See also Supplementary Fig. S1. This figure is available in black and white in print and in colour at DNA Research online.
We also mapped the RNA-seq reads to repetitive regions of the mouse genome (Fig. 1A). The analysis revealed significant increases of transcripts from repetitive sequences that are usually packed in silenced chromatin (constitutive heterochromatin), including major satellites, minor satellites, and telomeres during Z4 event (Fig. 1D). Increases of transcription from many retrotransposons were also detected (Supplementary Fig. S1B). Comparable activation of retrotransposons has already been reported in MuERV-L-marked mESCs (2C state).13
The burst of transcription from usually silent constitutive heterochromatin might be considered anomalous. However, the constitutive heterochromatin is highly transcribed in mouse preimplantation embryos, and the transcripts are required for proper embryonic development.32,33 This may indicate that a burst of constitutive heterochromatin transcription during Z4 event is also a part of normal physiological processes in mESCs.
3.2. Histone acetylation in heterochromatin increases during the Z4 event
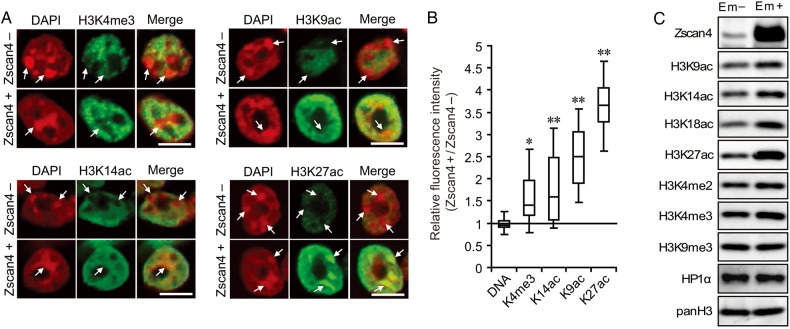
The unusual transcriptional burst from constitutive heterochromatin during Z4 event prompted us to examine the histone modifications involved in the regulation of transcriptionally active chromatin: histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 lysine 9, 14, and 27 acetylation (H3K9ac, H3K14ac, and H3K27ac). We found higher levels of these active histone modifications, especially H3K27ac, in the Zscan4+ cells compared with Zscan4– cells (Fig. 2A and Supplementary Fig. S2A). This finding was further supported by the quantification of fluorescence intensity (Fig. 2B, P < 0.001) and immunoblot analyses (Fig. 2C).
Figure 2.
Activation of heterochromatin in the Zscan4+ cells. (A) ES cells were co-immunostained for Zscan4 (not shown) and euchromatin markers—H3K4me3, H3K9ac, H3K14ac, and H3K27ac (green). DNA was counterstained with DAPI (red). Arrows indicate DNA-dense heterochromatin foci. Scale bars, 5 µm. (B) Fluorescence intensities of euchromatin markers in the Zscan4+ cells compared with those in the Zscan4− cells. n = 15 for each group. Error bars, S.D. *P < 0.01, **P < 0.001. (C) Immunoblot analyses of Zscan4, H3K9ac, H3K14ac, H3K18ac, H3K27ac, H3K4me2, H3K4me3, H3K9me3, HP1α, and pan-H3 marker. The MC1-ZE7 cells were FACS-sorted into cells with Em+ (i.e. Zscan4+ cells) and cells with Em− (i.e. Zscan4− cells) and analysed by the immunoblotting. See also Supplementary Fig. S2.
As expected, we also found that specifically in the Zscan4+ cells, histone acetylations—particularly H3K27ac—localized not only in euchromatin but also in heterochromatin—DAPI-dense regions (Fig. 2A and Supplementary Fig. S2A). This was further confirmed by colocalization with major satellite (Supplementary Fig. S2B) and heterochromatin-specific protein—HP1α (heterochromatin protein 1α) (Supplementary Fig. S2C). The association of heterochromatin with active histone modifications is consistent with a burst of constitutive heterochromatin transcription during Z4 event.
3.3. H3K27ac is enriched in constitutive heterochromatin and ZAGs during the Z4 event
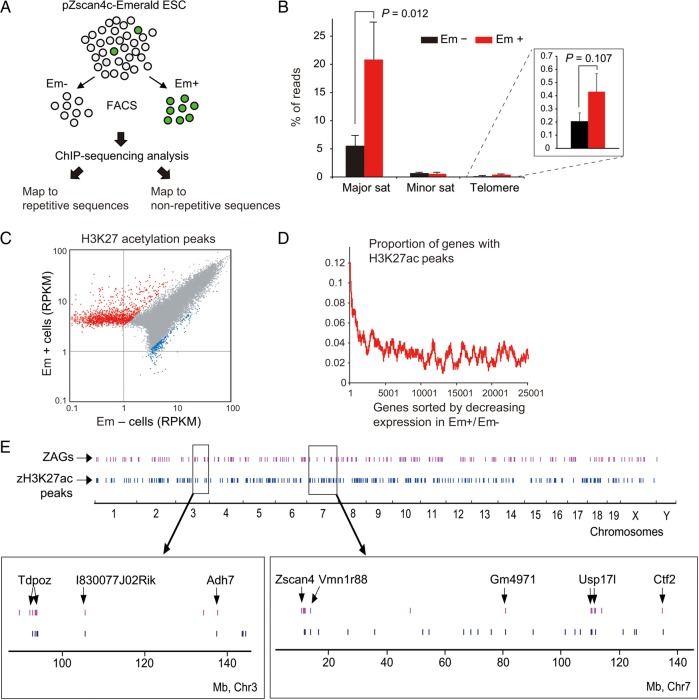
Among the histone modifications we examined thus far, H3K27ac showed the greatest up-regulation and the most specific nuclear localization in a Z4 event-specific manner. Therefore, we decided to identify the genomic localization of H3K27ac by chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq). To compare the genome-wide H3K27ac distributions reliably, we carried out ChIP-seq in duplicate using two independently FACS-sorted samples of Em+ and Em− cells (Fig. 3A). The independently replicated ChIP-seq results showed remarkable consistency, indicating the high specificity of H3K27ac ChIP-seq signals. We first analysed sequence reads that matched to repetitive sequences. Consistent with the immunostaining analyses, the number of sequence reads matched to major satellites and telomeres was 4- and 2-fold higher in the Em+ cells than in the Em− cells, respectively (Fig. 3B). Similarly, retrotransposons were more abundantly marked with H3K27ac in Em+ cells than in Em− cells (Supplementary Fig. S3A). These results further support the involvement of H3K27ac in the transcriptional burst from repetitive sequences during Z4 event.
Figure 3.
Correlation between histone hyperacetylations and gene expression in the Zscan4+ cells. (A) MC1-ZE7 cells were FACS-sorted into Em+ and Em− cells and analysed by the ChIP-seq using an anti-H3K27ac antibody. (B) Fractions (%) of sequence reads matched to major satellites, minor satellites, and telomeres in the Em+ or Em− cells. Error bars, S.E. (C) A scatterplot showing the comparison of H3K27ac peaks between the Em+ and Em− cells. Red dots (1,429), named ‘zH3K27ac peaks’, indicate H3K27ac peaks with significantly more (>3-fold) sequence reads in the Em+ cells compared with the Em− cells. (D) A plot showing the correlation between the gene expression differences in Em+ versus Em− cells (x-axis) and the proportion of genes with at least one zH3K27ac peak within 100 Kb from TSS, identified with a sliding window of 500 genes (y-axis). (E) Localizations of ZAGs (purple) and zH3K27ac peaks (blue) on mouse genomes. See also Supplementary Fig. S3.
We next analysed sequence reads matching non-repetitive sequences. H3K27ac peaks with significantly more (>3-fold) sequence reads in the Em+ cells compared with that in the Em− cells were identified, resulting in 1,429 peaks—tentatively named ‘Z4 event-associated H3K27ac peaks (zH3K27ac peaks)’ (Fig. 3C). The majority of these zH3K27ac peaks were located in intronic and intergenic regions (Supplementary Fig. S3B) and away from transcription start sites (TSSs) (Supplementary Fig. S3C). These results are consistent with H3K27ac function as a transcriptional active mark at enhancers.34 Indeed, the locations of zH3K27ac peaks were positively correlated with genes whose expression was higher in Em+ cells than in Em− cells, that is ZAGs (Fig. 3D and E), suggesting that the expression of ZAGs is also regulated by H3K27ac at their enhancers. Overlap between ZAGs and zH3K27ac was even more clearly observed when more stringent thresholds were used: 150 (59%) of 253 ZAGs (>5-fold Em+/Em− expression difference) were located within 0.5 Mb from zH3K27ac peaks (P = 6 × 10−6).
3.4. zH3K27ac peaks are located in facultative heterochromatin, silenced in Zscan4− cells, but activated during Z4 event
The analyses thus far indicated that repetitive sequences such as major satellites, telomeres, and retrotransposons, which are located in usually silenced heterochromatin, acquire H3K27ac marks and are transcribed. Are zH3K27ac peaks located in non-repetitive genomic regions also silenced and heterochromatinized in mESCs most of the time, but acquire H3K27ac marks and transcribed in a Z4 event-specific manner?
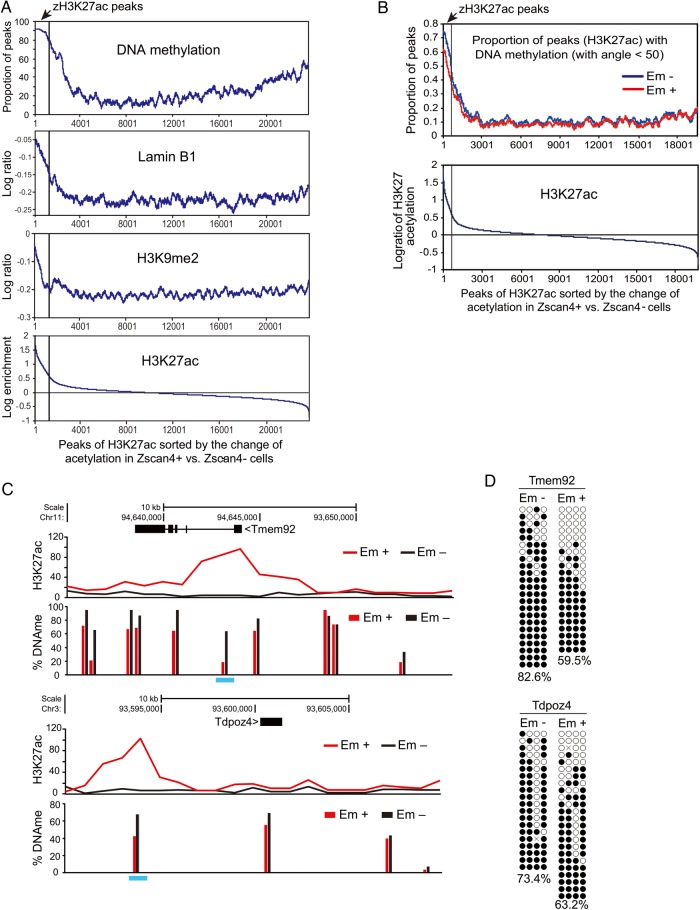
To address this question, we examined the association of zH3K27ac peaks with previously published chromatin features in mouse ES cells—essentially equivalent to Zscan4− cells (Fig. 4A and Supplementary Table S3, Fig. S4). Remarkably, zH3K27ac peaks were found to be enriched for heterochromatin features, that is high levels of DNA methylation;35 Lamin B1 binding;36 and H3K9me2 marks.37 These results suggest that not only the repetitive sequences such as major satellites (pericentromeres) and telomeres, but also zH3K27ac peaks such as ZAGs are enveloped in heterochromatin in the Zscan4− cells. In other words, pericentromeres, telomeres, retrotransposons, and unique sets of genes such as ZAGs are usually in heterochromatin in mESCs, but are transiently activated during Z4 event.
Figure 4.
DNA demethylation coupled with histone hyperacetylation in heterochromatin in Zscan4+ cells. (A) Proportion of H3K27ac peaks with the high levels of DNA methylation (>50% methylated CpGs per total CpGs by the bisulfite sequencing),35 the average log-enrichment in binding of Lamin B1,36 the average log-enrichment of H3K9me2, and the log-enrichment of H3K27ac, in a sliding window of 300 H3K27ac peaks, which were sorted by the difference in the H3K27ac between Em+ and Em− cells. (B) Comparison of DNA methylation levels with the changes in H3K27ac levels between Em+ and Em− cells. H3K27ac peaks were sorted by the decreasing log ratio of H3K27ac in the Em+ versus Em− cells (x-axis), and then the proportion of peaks with DNA methylation (y-axis) was estimated in a sliding window of 300 peaks. (C) Representative examples of genes with H3K27 hyperacetylation (H3K27ac ChIP-seq, this study) and DNA demethylation (by the HELP assay, this study) in the Em+ cells compared with the Em− cells. (D) Bisulfite sequencing analyses of Tmem92 and Tdpoz4 regions (blue bars in Fig. 4C) were performed on Em+ and Em− cells. Open and filled circles indicate unmethylated or methylated CpG sites, respectively. The percentage of methylated CpGs per total CpGs is presented below each data set. See also Supplementary Fig. S4.
To further examine the epigenetic changes specific to the Z4 event, we carried out DNA methylation analysis by the HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) assay on Em+ and Em− cells. As expected, zH3K27 peaks were enriched with high levels of DNA methylation (Fig. 4B). However, the levels of DNA methylation in the zH3K27ac peaks were slightly reduced in Em+ cells compared with Em− cells (Fig. 4B). For example, the regulatory regions of representative ZAGs such as Tmem92 and Tdpoz4 showed H3K27ac and DNA hypomethylation (Fig. 4C)—a finding confirmed independently by bisulfite sequencing of the regions (Fig. 4D). Thus, both repetitive sequences and non-repetitive ZAGs are transcriptionally silenced and heterochromatinized in Zscan4− cells (usual mESCs), but become transcriptionally active, acquiring H3K27ac—active histone modifications and DNA demethylation. For brevity, we call the subset of facultative heterochromatin where zH3K27ac and ZAGs are located ‘zHC’.
3.5. Zscan4 forms complexes with chromatin remodellers during the Z4 event
To explore which chromatin remodelling factors are related to changes in heterochromatin during Z4 event, we examined protein complexes associated with Zscan4. We used an anti-Flag antibody and a previously established ES cell line (tet-Zscan4c ES cells), in which an exogenous Zscan4 fused to a Flag-tag can be induced by switching from the Dox+ to the Dox− condition.30 Immunoblot analysis of proteins extracted from these cells (Dox− condition, i.e. Zscan4-induced condition) and fractionated by gel filtration chromatography identified a ∼70 KD Zscan4 band, which was present from ∼150 KD to ∼2,000 KD with a major peak at ∼500 KD, suggesting that Zscan4 proteins, when expressed, can form protein complexes of various sizes (Supplementary Fig. S5A).
Next, protein mixtures from each column fraction were immunoprecipitated (IP) with anti-Flag antibody and subjected to mass spectrometry analyses. After removing common contaminants often detected by Flag-tag-based IP mass spectrometry, the number of identified peptides for each protein by mass spectrometry was tabulated for each protein fraction (Supplementary Fig. S5B). The analyses revealed that Zscan4 complexes contained primarily repressing chromatin remodelling complexes (HDAC1, HDAC2, LSD1/KDM1A, NuRD, Sin3A), but also showed evidence of activating chromatin remodelling complexes (SWI/SNF). Zscan4 complexes also contained KAP1—one of the key proteins in the regulation of heterochromatin. Consistent with the mass spectrometry data, protein mixtures pulled down by Flag-IP contained Zscan4, LSD1/KDM1A, MTA1 (NuRD), BRG1 (SWI/SNF), and KAP1 (Fig. 5A); and protein mixtures pulled down by LSD1-IP, MTA1-IP, and BRG1-IP, respectively, all contained Zscan4, MTA1, BRG1, HDAC1, and KAP1. These results suggest that both activating and repressing chromatin remodelling complexes are associated with Zscan4 protein.
Figure 5.
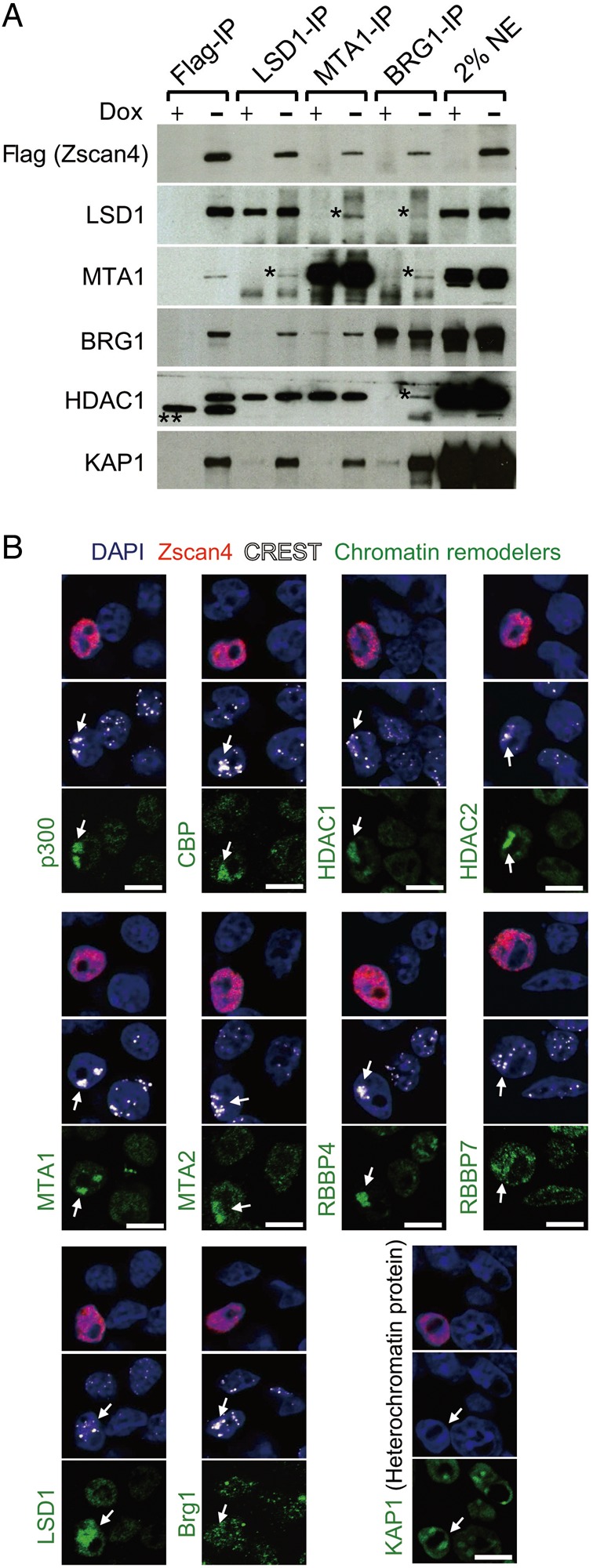
Both activating and repressing chromatin remodelling complexes localize in heterochromatin in the Zscan4+ cells. (A) Immunoblot analyses of Flag-Zscan4, Lsd1/Kdm1a, Mta1, Brg1, Hdac1, and Kap1/Trim28 proteins after immunoprecipitating the nuclear extracts of tet-Zscan4 ES cells by antibodies against Flag-tag, Lsd1/Kdm1a, Mta1, and Brg1. The tet-Zscan4 ES cells were cultured in the Dox+ (without Zscan4 overexpression) and Dox− (with Zscan4 overexpression) for 3 days. *, possible cross-reactive polypeptides. **, non-specific bands. (B) Triple immunostaining analyses of ES cells with the Zscan4 antibody (red), the CREST antibody (white, an anti-centromere protein), and various antibodies indicated (green). Blue, DAPI. Arrows indicate the clustered centromeres. Scale bars, 5 µm. See also Supplementary Fig. S5.
To further verify Zscan4-dependent colocalization of these chromatin remodelling complexes on heterochromatin, we carried out the immunostaining analyses of these proteins in mouse ES cells (Fig. 5B). For the activating chromatin remodelling complexes, the staining of BRG1 was similar for Zscan4+ and Zscan4− cells, but the histone acetyltransferases (HATs), p300 and CBP, which are known to specifically mediate the acetylation of H3K27,38 accumulated in the heterochromatin labeled with anti-centromere antibody in a Z4 event-specific manner. For the repressing chromatin remodelling complexes, HDAC1, HDAC2, MTA1, MTA2, Rbbp4, and Rbbp7, all of which are components of the NuRD complex,39 were localized in the heterochromatin in a Z4 event-specific manner. The staining of LSD1 was observed in both euchromatin and heterochromatin, but was stronger in the Zscan4+ cells than in Zscan4− cells. As a control, KAP1, heterochromatin marker, was detected in heterochromatin both in the Zscan4+ and Zscan4− cells. Interestingly, Zscan4 staining showed its localization not only in pericentromeric heterochromatin, but also in euchromatic regions, suggesting the detection of zHC, which are located in euchromatin. Alternatively, these results may indicate that Zscan4 functions not only in heterochromatin, but also in euchromatin. Overall, the heterochromatin is associated with both activating and repressing chromatin remodelling complexes during Z4 event (though a single site probably moves through a cycle of derepression and rerepression; see below).
3.6. Pericentromeric heterochromatin clusters around nucleolus during Z4 event
The results thus far indicate that heterochromatin is transiently in an activated conformation in a Z4 event-specific manner. We suspected that Z4 event-specific activation of heterochromatin has some unique features. A hint came from the immunohistochemical analysis of histone modifications during Z4 event (Fig. 2A). We noticed the clustering of heterochromatin in a Z4 event-specific manner and further investigated the distribution of heterochromatin in nuclei by costaining for Zscan4 and HP1α. In the great majority (∼95%) of ESCs, which were Zscan4−, heterochromatin recognized by HP1α appeared as discrete multiple foci scattered in the nucleoplasm and overlapping regions of high DNA density detected by DAPI (Fig. 6A and Supplementary Fig. S6A). This pattern of heterochromatin localization is commonly observed in mESCs and other cell types.40 In contrast, during Z4 event the heterochromatin appeared in larger and fewer clusters that also overlapped with the regions of high DNA density, mostly perinucleolar (Fig. 6A and Supplementary Fig. S6A). These observations were confirmed by quantitative morphometric analyses of a large number of cells: heterochromatin foci in Zscan4+ cells were significantly larger (Fig. 6B, P < 0.001) and fewer (Fig. 6C, P < 0.001) compared with those in Zscan4− cells. The clustering and relocalization of heterochromatin during Z4 event were also confirmed by immunostaining for H3K9me3 and H4K20me3—histone modifications associated with heterochromatin (Supplementary Fig. S6B and C).
Figure 6.
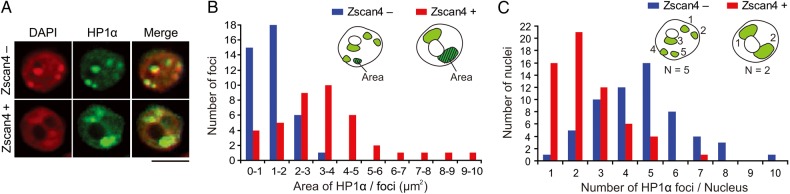
Heterochromatin clustering in the Zscan4+ cells. (A) Co-immunostaining of ES cells with an HP1α antibody (green) and a Zscan4 antibody (not shown). Red, DAPI. Scale bars, 5 µm. More examples are shown in Supplementary Fig. S2A. (B) Size distribution of nuclear foci stained with an HP1α antibody in the Zscan4+ cells (red bars) and in the Zscan4− cells (blue bars). Average areas of each focus was 3.6 and 1.5 µm2 in the Zscan4+ cells and in the Zscan4− cells, respectively. n = 40. (C) Number distribution of nuclear foci stained with an HP1α antibody in the Zscan4+ cells (red bars) and in the Zscan4− cells (blue bars). Average numbers of foci in each nucleus were 2.4 and 4.7 in the Zscan4+ cells and in the Zscan4− cells, respectively. n = 60. See also Supplementary Fig. S6.
Heterochromatin foci are known to be composed of pericentromeric regions that contain major satellite DNA repeats.41 Indeed, DNA fluorescent in situ hybridization analyses using probes specific to major satellites and telomeres showed clustering of major satellite repeats specifically during Z4 event (Supplementary Fig. S6D). In contrast, clustering of telomeres was not seen (Supplementary Fig. S6D).
As far as we know, this is the first report for heterochromatin clustering into a large cluster. Taken together, these results indicate that the unusual clustering of pericentromeric heterochromatin occurs transiently as a unique feature of Z4 event.
3.7. Zscan4 plays active roles in the epigenetic modifications of heterochromatin during Z4 event
All the dynamic changes in heterochromatin described thus far occurred specifically in Zscan4+ cells, that is a small fraction (1–5%) of ESCs in culture. Previously we have shown that Zscan4 is a causative gene for biological events occurring specifically during Z4 event (see Discussion). Therefore, we expected that Zscan4 causes rapid derepression and rerepression of heterochromatin in a Z4 event-specific manner. To test this notion, we examined heterochromatin dynamics in both loss-of-function and gain-of-function studies of Zscan4.
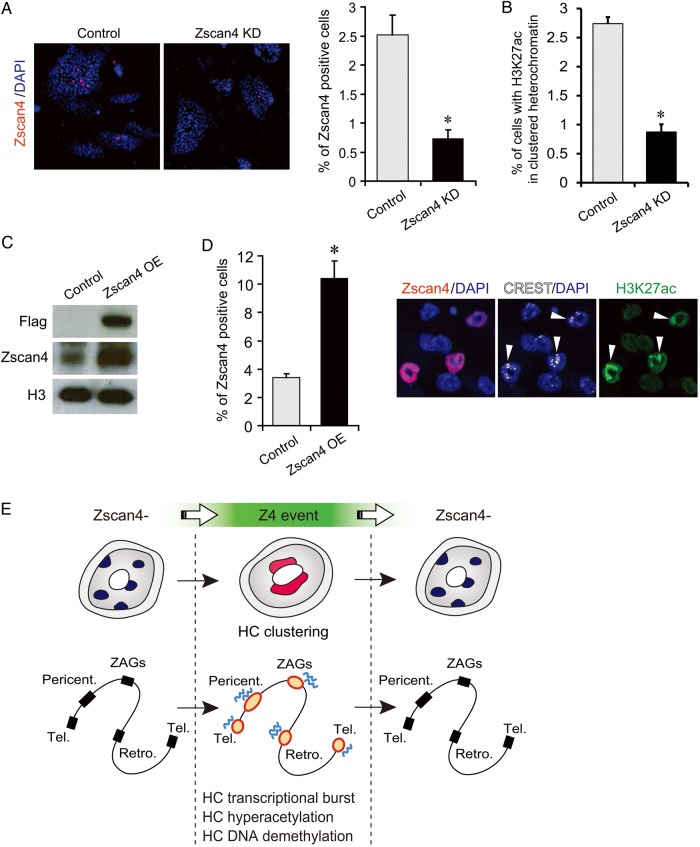
For the loss-of-function assays, we generated a mESC line carrying a Dox-inducible shRNA (short hairpin RNA) directed against Zscan4. We confirmed that Dox-induced shRNA expression for 8 days decreased the fraction of Zscan4+ mESCs (Fig. 7A). In these Zscan4 knockdown cell colonies, the number of cells displaying H3K27 acetylation in heterochromatin was significantly reduced (Fig. 7B). Furthermore, the expression of major satellites was decreased by ∼50% in Zscan4 knockdown cells (Supplementary Fig. S7A, P < 0.01).
Figure 7.
Active roles of Zscan4 in the heterochromatin regulation revealed by Zscan4 knockdown and Zscan4 overexpression experiments. (A, B) Inducible knockdown of Zscan4 expression (Zscan4 KD). (A) Left panel: Immunostaining analyses of the ES cells expressing a Dox-inducible shRNA against Zscan4, cultured for 8 days in the absence (−Dox) or the presence of 2 µg/ml Dox (+Dox), with the antibody against Zscan4 (red). DNA is counterstained with DAPI (red). Right panel: The percentage of cells stained with a Zscan4 antibody was significantly reduced by the Dox treatment (*P < 0.01, t-test). Error bars indicate S.D. (B) The percentage of the cells with H3K27 acetylation in heterochromatin was significantly reduced by the Dox treatment (*P < 0.01, t-test). Error bars indicate S.D. (C, D) Inducible overexpression of Zscan4 using tet-Zscan4 cells (Zscan4 OE). (C) Western blots using an anti-Flag antibody, anti-Zscan4 antibody, and anti-histone H3 antibody (loading control). (D) Left panel: The percentage of Zscan4+ cells in the Dox+ and Dox− conditions. *P < 0.01 versus +Dox, t-test. Right panel: Immunostaining of tet-Zscan4 ES cells with an anti-Zscan4, anti-CREST, and anti-H3K27ac. The nuclei of the Zscan4+ cells showed the hyperacetylation of H3K27 in clustered centromeric regions (arrows). (E) A schematic summary of heterochromatin dynamics during Z4 event. Tel., telomeres, Pericent., pericentromeres, Retro., retrotransposons, ZAGs, Z4 event-associated genes. See also Supplementary Fig. S7.
For the gain-of-function assays, we examined whether ectopic overexpression of Zscan4 can induce these epigenetic changes in mESCs, using the tet-Zscan4 ES cell line.30 As expected, the overexpression of Zscan4, which was confirmed by the western blot (Fig. 7C), increased the number of cells stained with Zscan4 antibodies (Fig. 7D). Zscan4+ cells were also strongly stained with the H3K27ac in the heterochromatin (Fig. 7D) and the H3K9ac (Supplementary Fig. S7B). Furthermore, ChIP-qPCR analyses showed that the overexpression of Zscan4 up-regulated H3K27ac on Tmem92, Tdpoz3/4, Zscan4c/d regions (Supplementary Fig. S7C, P < 0.05). Next, we carried out MeDIP-seq (methylated DNA immunoprecipitation followed by sequencing) analyses and found that the overexpression of Zscan4 induced the demethylation of DNAs in telomeric and major satellite regions—and to some extent in minor satellite regions (Supplementary Fig. S7D). Subsequent analyses of non-repeated genome regions revealed that the overexpression of Zscan4 also induced the demethylation of DNAs in the zH3K27ac peaks (Supplementary Fig. S7E).
4. Discussion
The Z4 event—a short burst of Zscan4 transcription—occurs very infrequently in usual mESC culture conditions, so that only 1–5% of mESCs are undergoing Z4 event at a given time.16–18 However, essentially all mESCs undergo Z4 event at least once within nine passages.17 In this paper, we have carried out detailed molecular analyses of the Z4 event, finding that it coincides with rapid and unusual molecular changes in chromatin, particularly in heterochromatin (Fig. 7E).
4.1. Heterochromatin regulation and dynamics during Z4 event
Constitutive heterochromatin is usually considered silenced for transcription. However, as summarized in Fig. 7E, during the Z4 event, constitutive heterochromatin shows a burst of transcription. Generally, gene activation is accompanied by ‘open’ chromatin with enrichment of active marks and loss of repressive marks on their regulatory regions.42 In fact, we saw increased levels of activating histone marks, H3K27ac, reduced levels of DNA methylation, along with clustering around nucleoli (Fig. 7E). At the same time, repressive marks, H3K9me2, and DNA methylation were still abundant in the acetylated active heterochromatin. These results suggest that the transcriptional burst is induced from ‘open’ heterochromatin, but the heterochromatin immediately returns to its silent conformation.
In addition to the constitutive heterochromatin, we also observed ‘zHC’ that comprise a few hundred ‘ZAGs’, including Zscan4, which show a burst of transcription during Z4 event. Although zHC is not located in the constitutive heterochromatin region and forms scattered clusters throughout the genome (Fig. 3E), we show that in regular mESCs zHC is transcriptionally silenced and shares features of heterochromatin such as high levels of DNA methylation and the lack of activating histone marks. However, as summarized in Fig. 7E, during the Z4 event, zHC also shows a burst of transcription, with increased levels of activating histone marks, H3K27ac, and reduced levels of DNA methylation (Fig. 7E). Because H3K27ac marks distal enhancers associated with active genes34 and is important for recruiting Pol II to regulatory regions to initiate transcription,38 it is likely that the burst of transcription of ZAGs from the zHC is controlled by H3K27ac.
The rapid and profound changes in heterochromatin during Z4 event are rather unusual, even considering the already unusual chromatin features of regular mESCs,43 such as a bivalent structure,44,45 dynamic plasticity,46 and non-CpG methylation.47 These chromatin changes specific to Z4 event suggest that mESCs undergo rapid derepression followed by the immediate rerepression of both constitutive heterochromatin and zHC during the Z4 event (Fig. 7E). This notion is further supported by our finding that both activating chromatin remodelling complexes (HATs, SWI/SNF) and repressing chromatin remodelling complexes (HDAC1, HDAC2, LSD1/KDM1A, NuRD) gather on heterochromatin during Z4 event. Simultaneous modifications of both constitutive heterochromatin and zHC are also intriguing, as it has been shown that different enzymes are involved in the histone methylation of constitutive heterochromatin and facultative heterochromatin (including zHC): Suv39h1/h2 for constitutive heterochromatin and G9a for facultative heterochromatin.48 It remains to be clarified whether these enzymes are involved in Z4 events.
4.2. Biological significance of Z4 event
It has been shown that Z4 event is required for the maintenance of telomere length and genome stability.17 Also, increasing the frequency of Z4 event by the forced expression of Zscan4 in mESCs can improve their quality (i.e. the extent of chimerism when injected into blastocysts).18 Our finding that Z4 event is accompanied by the activation of both constitutive heterochromatin and zHC was therefore not anticipated, because it is generally thought that the relaxation of heterochromatin leads to genome instability and deterioration of cells.49–51 For example, the transcription of retrotransposons, which are usually repressed, is known to produce insertional mutations and genome instability.52 Why then would mESCs undergo Z4 event? One speculative possibility is that going through a usually forbidden transition to heterochromatin activation permits the repair of DNA damage in the heterochromatin. It is known that when DNA in tightly packed heterochromatin is damaged, the heterochromatin must be opened so that the DNA repair machinery can access the DNA.53 The otherwise harmful effects of transcriptional burst from heterochromatin could be countered by blocking the translation of the RNAs: we have shown previously that Z4 event is accompanied by the block of global protein synthesis through the activation of two-cell-specific Eif1a-like genes.19
Dynamic changes of heterochromatin during Z4 event are also consistent with the significant biological ‘corrections’ such as the extension of telomeres17 and the alleviation of karyotype abnormality.18 For example, transcription of telomeres is known to be positively correlated with telomere length.22,54 Also, activation and clustering of pericentromeric heterochromatin would be consistent with involvement of the Z4 event in chromosome segregation, because this is indeed the chromosome region where the assembly of kinetochore and other transactions critical for chromosome segregation occur.55
Of course, the unusual properties of Z4 event must have a function apart from the maintenance of artificially produced mESCs. The Z4 event can be considered a normal part of mammalian development and tissue homeostasis. For example, the burst of Zscan4 expression can be observed in late two-cell embryos during mouse preimplantation development;16 in four to eight cell embryos during human preimplantation development;56 and in rare adult cells that most likely include tissue stem cells.57 The burst of Zscan4 is required for preimplantation embryo development, as its knockdown causes developmental defects.16 In the mouse preimplantation embryos, Z4 event seems to occur during the late two-cell embryo—a stage when genomes derived from sperm and oocyte initiate massive transcription.58 During that short period, major satellites and retrotransposons located in constitutive heterochromatin,32,33 and many ZAGs located in zHC13,18 also show a burst of transcription. Clustering of pericentromeric heterochromatin has also been observed in two-cell embryos, although they appear to surround nucleoli more uniformly.59,60
4.3. Z4 event versus 2C state
Recent studies have revealed that in standard culture conditions mESCs fluctuate among distinct transient states, resulting in their heterogenous mixture.10,11 However, these states do not seem to be intrinsically transient, because mESCs can be maintained stably in a specific state depending on the culture condition, for example in a naive pluripotent state or a primed pluripotent state.7–9 Considering its short duration and its role in genome stability, we call a short burst of Zscan4 transcription ‘Zscan4 event’ or ‘Z4 event’ in this paper, so that it can be operationally distinguished from various pluripotency states. The need for such distinction seems to be further supported by the data presented in this manuscript. However, our view seems to contradict with the recent view expressed by some other authors who inferred that Zscan4+ cells, in which two-cell-specific Zscan4 is expressed, are essentially the same as cells in the 2C state.13,61,62 The ‘two-cell (2C) state’ is one of the transient mESC states identified recently as a totipotent state using the expression of a MuERV-L retrotransposon as a marker.13 However, there are some differences between them. For example, unlike mESCs in the totipotent 2C state,13 mESCs undergoing a Z4 event (i.e. Zscan4+ mESCs) contribute inefficiently to chimeric mice when injected into blastocysts and thus seem to temporarily lack pluripotency.18
4.4. Molecular functions of Zscan4
Previously we have shown that the ectopic expression of Zscan4 per se can trigger biological events that occur specifically during Z4 events in mESCs. For example, the length of telomeres can be extended within a few days after ectopic expression of Zscan4,17 and the efficiency of contribution to chimeric mice can be dramatically improved by the forced expression of Zscan4.18 We have also shown that the forced expression of Zscan4 induces the expression of Tmem92, Tcstv1, Tcstv3, and other ZAGs.18 However, it has not been clear how Zscan4 functions and is molecularly involved in these biological events.
Here we have shown that at least some of the heterochromatin changes associated with Z4 events can be triggered by ectopic expression of Zscan4. It is interesting to note that telomeric heterochromatin shows an increase of transcription and H3K27ac as well as a decrease of DNA methylation during a Z4 event. Because it is known that such epigenetic changes of telomeric heterochromatin cause the extension of telomeres by recombination,22,54 these data suggest the molecular mechanism for rapid extension of telomeres by the forced expression of Zscan4.17
With the identification of Zscan4-associated protein complexes by Immunoprecipitation–Mass spectrometry analyses, we have revealed that Zscan4 forms complexes with many distinct proteins, including both activating and repressing chromatin remodellers. The association of Trim28 (Kap1) also indicates the localization of at least some of the Zscan4 protein complexes on heterochromatin. The notion that Zscan4 functions through specific protein–protein interaction is consistent with the known molecular function of the SCAN domain as a mediator of protein–protein interaction.63
4.5. Conclusion
Here we have shown that mESCs undergo dramatic epigenetic changes to regulate transient activation and repression of specific heterochromatin regions, a process that is likely an intrinsic part of a special mechanism for the maintenance of genome stability. In contrast to S. Pombe where constitutive heterochromatin is transcribed in S phase of every cell cycle,25 Z4 events occur infrequently in mESCs cultured in the standard condition. Thus, the Z4 event can be seen as a non-periodic checkpoint, a ‘reset’ mechanism that promotes proper chromosome dynamics in stem cells.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjounals.org.
Funding
This work was in part supported by the Keio University Medical Science Fund—The Mitsunada Sakaguchi Laboratory, the Takeda Science Foundation, and the Intramural Research Program of the National Institutes of Health (AG000656, AG000702), National Institute on Aging. Funding to pay the Open Access publication charges for this article was provided by the Keio University Medical Science Fund–The Mitsunada Sakaguchi Laboratory.
Supplementary Material
Acknowledgements
We thank lab members and David Schlessinger for discussion and critical reading of the manuscript.
References
- 1.Evans M.J., Kaufman M.H.. 1981, Establishment in culture of pluripotential cells from mouse embryos, Nature, 292, 154–6. [DOI] [PubMed] [Google Scholar]
- 2.Martin G.R. 1981, Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells, Proc. Natl Acad. Sci. USA, 78, 7634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley A., Evans M., Kaufman M.H., Robertson E.. 1984, Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines, Nature, 309, 255–6. [DOI] [PubMed] [Google Scholar]
- 4.Suda Y., Suzuki M., Ikawa Y., Aizawa S.. 1987, Mouse embryonic stem cells exhibit indefinite proliferative potential, J. Cell Physiol., 133, 197–201. [DOI] [PubMed] [Google Scholar]
- 5.Festuccia N., Osorno R., Wilson V., Chambers I.. 2013, The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states, Curr. Opin. Genet. Dev., 23, 504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martello G., Smith A.. 2014, The nature of embryonic stem cells, Annu. Rev. Cell Dev. Biol., 30, 647–75. [DOI] [PubMed] [Google Scholar]
- 7.Kalkan T., Smith A.. 2014, Mapping the route from naive pluripotency to lineage specification, Philos. Trans. R Soc. Lond. B Biol. Sci., 369, 20130540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying Q.L., Wray J., Nichols J. et al. 2008, The ground state of embryonic stem cell self-renewal, Nature, 453, 519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nichols J., Smith A.. 2009, Naive and primed pluripotent states, Cell Stem Cell, 4, 487–92. [DOI] [PubMed] [Google Scholar]
- 10.Canham M.A., Sharov A.A., Ko M.S., Brickman J.M.. 2010, Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript, PLoS Biol., 8, e1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers I., Silva J., Colby D. et al. 2007, Nanog safeguards pluripotency and mediates germline development, Nature, 450, 1230–4. [DOI] [PubMed] [Google Scholar]
- 12.Morgani S.M., Canham M.A., Nichols J. et al. 2013, Totipotent embryonic stem cells arise in ground-state culture conditions, Cell Rep., 3, 1945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macfarlan T.S., Gifford W.D., Driscoll S. et al. 2012, Embryonic stem cell potency fluctuates with endogenous retrovirus activity, Nature, 487, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J.C.. 1993, Derivation of completely cell culture-derived mice from early-passage embryonic stem cells, Proc. Natl Acad. Sci. USA, 90, 8424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longo L., Bygrave A., Grosveld F.G., Pandolfi P.P.. 1997, The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism, Transgenic Res., 6, 321–8. [DOI] [PubMed] [Google Scholar]
- 16.Falco G., Lee S.L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S.. 2007, Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells, Dev. Biol., 307, 539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalzman M., Falco G., Sharova L.V. et al. 2010, Zscan4 regulates telomere elongation and genomic stability in ES cells, Nature, 464, 858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amano T., Hirata T., Falco G. et al. 2013, Zscan4 restores the developmental potency of embryonic stem cells, Nat. Commun., 4, 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung S.S., Wong R.C., Sharov A.A., Nakatake Y., Yu H., Ko M.S.. 2013, Repression of global protein synthesis by eif1a-like genes that are expressed specifically in the two-cell embryos and the transient zscan4-positive state of embryonic stem cells, DNA Res., 20, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter M.G., Stagg C.A., Falco G. et al. 2008, An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells, Gene Expr. Patterns, 8, 181–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storm M.P., Kumpfmueller B., Bone H.K. et al. 2014, Zscan4 is regulated by PI3-kinase and DNA-damaging agents and directly interacts with the transcriptional repressors LSD1 and CtBP2 in mouse embryonic stem cells, PLoS One, 9, e89821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blasco M.A. 2007, The epigenetic regulation of mammalian telomeres, Nat. Rev. Genet., 8, 299–309. [DOI] [PubMed] [Google Scholar]
- 23.Richards E.J., Elgin S.C.. 2002, Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects, Cell, 108, 489–500. [DOI] [PubMed] [Google Scholar]
- 24.Saksouk N., Simboeck E., Dejardin J.. 2015, Constitutive heterochromatin formation and transcription in mammals, Epigenet. Chromatin, 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloc A., Martienssen R.. 2008, RNAi, heterochromatin and the cell cycle, Trends Genet., 24, 511–7. [DOI] [PubMed] [Google Scholar]
- 26.Trojer P., Reinberg D.. 2007, Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell, 28, 1–13. [DOI] [PubMed] [Google Scholar]
- 27.Arney K.L., Fisher A.G.. 2004, Epigenetic aspects of differentiation, J. Cell Sci., 117, 4355–63. [DOI] [PubMed] [Google Scholar]
- 28.Gendrel A.V., Heard E.. 2014, Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation, Annu. Rev. Cell Dev. Biol., 30, 561–80. [DOI] [PubMed] [Google Scholar]
- 29.Olson L.E., Bedja D., Alvey S.J., Cardounel A.J., Gabrielson K.L., Reeves R.H.. 2003, Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1, Cancer Res., 63, 6602–6. [PubMed] [Google Scholar]
- 30.Nishiyama A., Xin L., Sharov A.A. et al. 2009, Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors, Cell Stem Cell, 5, 420–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki M., Jing Q., Lia D., Pascual M., McLellan A., Greally J.M.. 2010, Optimized design and data analysis of tag-based cytosine methylation assays, Genome Biol., 11, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kigami D., Minami N., Takayama H., Imai H.. 2003, MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos, Biol. Reprod., 68, 651–4. [DOI] [PubMed] [Google Scholar]
- 33.Probst A.V., Okamoto I., Casanova M., El Marjou F., Le Baccon P., Almouzni G.. 2010, A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development, Dev. Cell, 19, 625–38. [DOI] [PubMed] [Google Scholar]
- 34.Creyghton M.P., Cheng A.W., Welstead G.G. et al. 2010, Histone H3K27ac separates active from poised enhancers and predicts developmental state, Proc. Natl Acad. Sci. USA, 107, 21931–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadler M.B., Murr R., Burger L. et al. 2011, DNA-binding factors shape the mouse methylome at distal regulatory regions, Nature, 480, 490–5. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y., Sharov A.A., McDole K. et al. 2011, Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells, Science, 334, 1706–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lienert F., Mohn F., Tiwari V.K. et al. 2011, Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells, PLoS Genet., 7, e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin Q., Yu L.R., Wang L. et al. 2011, Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation, EMBO J., 30, 249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai A.Y., Wade P.A.. 2011, Cancer biology and NuRD: a multifaceted chromatin remodelling complex, Nat. Rev. Cancer, 11, 588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer R., Brero A., von Hase J., Schroeder T., Cremer T., Dietzel S.. 2005, Common themes and cell type specific variations of higher order chromatin arrangements in the mouse, BMC Cell Biol., 6, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guenatri M., Bailly D., Maison C., Almouzni G.. 2004, Mouse centric and pericentric satellite repeats form distinct functional heterochromatin, J. Cell Biol., 166, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlic R., Chung H.R., Lasserre J., Vlahovicek K., Vingron M.. 2010, Histone modification levels are predictive for gene expression, Proc. Natl Acad. Sci. USA, 107, 2926–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stancheva I. 2011, Revisiting heterochromatin in embryonic stem cells, PLoS Genet., 7, e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein B.E., Mikkelsen T.S., Xie X. et al. 2006, A bivalent chromatin structure marks key developmental genes in embryonic stem cells, Cell, 125, 315–26. [DOI] [PubMed] [Google Scholar]
- 45.Mikkelsen T.S., Ku M., Jaffe D.B. et al. 2007, Genome-wide maps of chromatin state in pluripotent and lineage-committed cells, Nature, 448, 553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T.. 2006, Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells, Dev. Cell, 10, 105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lister R., Pelizzola M., Dowen R.H. et al. 2009, Human DNA methylomes at base resolution show widespread epigenomic differences, Nature, 462, 315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinkai Y., Tachibana M.. 2011, H3K9 methyltransferase G9a and the related molecule GLP, Genes Dev., 25, 781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ting D.T., Lipson D., Paul S. et al. 2011, Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers, Science, 331, 593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carone D.M., Lawrence J.B.. 2013, Heterochromatin instability in cancer: from the Barr body to satellites and the nuclear periphery, Semin. Cancer Biol., 23, 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egger G., Liang G., Aparicio A., Jones P.A.. 2004, Epigenetics in human disease and prospects for epigenetic therapy, Nature, 429, 457–63. [DOI] [PubMed] [Google Scholar]
- 52.Kazazian H.H. Jr., Goodier J.L.. 2002, LINE drive. retrotransposition and genome instability, Cell, 110, 277–80. [DOI] [PubMed] [Google Scholar]
- 53.Cann K.L., Dellaire G.. 2011, Heterochromatin and the DNA damage response: the need to relax, Biochem. Cell Biol., 89, 45–60. [DOI] [PubMed] [Google Scholar]
- 54.Schoeftner S., Blasco M.A.. 2008, Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II, Nat. Cell Biol., 10, 228–36. [DOI] [PubMed] [Google Scholar]
- 55.Obeso D., Pezza R.J., Dawson D.. 2014, Couples, pairs, and clusters: mechanisms and implications of centromere associations in meiosis, Chromosoma, 123, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vassena R., Boue S., Gonzalez-Roca E. et al. 2011, Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development, Development, 138, 3699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ko S.B., Azuma S., Yokoyama Y. et al. 2013, Inflammation increases cells expressing ZSCAN4 and progenitor cell markers in the adult pancreas, Am. J. Physiol. Gastrointest. Liver Physiol., 304, G1103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamatani T., Carter M.G., Sharov A.A., Ko M.S.. 2004, Dynamics of global gene expression changes during mouse preimplantation development, Dev. Cell, 6, 117–31. [DOI] [PubMed] [Google Scholar]
- 59.Probst A.V., Almouzni G.. 2008, Pericentric heterochromatin: dynamic organization during early development in mammals, Differentiation, 76, 15–23. [DOI] [PubMed] [Google Scholar]
- 60.Akiyama T., Suzuki O., Matsuda J., Aoki F.. 2011, Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos, PLoS Genet., 7, e1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torres-Padilla M.E., Chambers I.. 2014, Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage, Development, 141, 2173–81. [DOI] [PubMed] [Google Scholar]
- 62.Morgani S.M., Brickman J.M.. 2014, The molecular underpinnings of totipotency, Philos. Trans R Soc. Lond. B Biol. Sci., 369, 20130549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edelstein L.C., Collins T.. 2005, The SCAN domain family of zinc finger transcription factors, Gene, 359, 1–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.