Abstract
Multidomain proapoptotic BAX and BAK, once activated, permeabilize mitochondria to trigger apoptosis, whereas antiapoptotic BCL-2 members preserve mitochondrial integrity. The BH3-only molecules (BH3s) promote apoptosis by either activating BAX-BAK or inactivating antiapoptotic members. Here, we present biochemical and genetic evidence that NOXA is a bona fide activator BH3. Using combinatorial gain-of-function and loss-of-function approaches in Bid−/−Bim−/−Puma−/−Noxa−/− and Bax−/−Bak−/− cells, we have constructed an interconnected hierarchical model that accommodates and explains how the intricate interplays between the BCL-2 members dictate cellular survival versus death. BID, BIM, PUMA and NOXA directly induce stepwise, bimodal activation of BAX-BAK. BCL-2, BCL-XL and MCL-1 inhibit both modes of BAX-BAK activation by sequestering activator BH3s and “BH3-exposed” monomers of BAX-BAK, respectively. Furthermore, autoactivation of BAX and BAK can occur independently of activator BH3s through downregulation of BCL-2, BCL-XL and MCL-1. Our studies lay a foundation on targeting the BCL-2 family for treating diseases with dysregulated apoptosis.
Central players of the mitochondrion-dependent apoptotic program are the BCL-2 family proteins1–3, consisting of (1) multidomain antiapoptotic BCL-2, BCL-XL and MCL-1, (2) multidomain proapoptotic BAX and BAK, and (3) proapoptotic BH3-only molecules (BH3s). Multidomain members contain all four BCL-2 homology domains (BH1-4) whereas BH3s only share sequence homology within the BH3 domain. Multiple apoptotic stimuli culminate in mitochondrial outer membrane permeabilization (MOMP), resulting in the release of cytochrome c into the cytosol to activate caspases4, 5. The decision of a given cell in undergoing MOMP is determined by the interplays among these three BCL-2 subfamilies1–3, 6, 7. BAX and BAK are the essential effectors responsible for MOMP whereas BCL-2, BCL-XL and MCL-1 preserve mitochondrial integrity8, 9. BH3s are death sentinels that relay upstream apoptotic signals to initiate apoptosis by either activating BAX-BAK directly (“activator” BH3s) or inactivating BCL-2, BCL-XL and MCL-1 (“inactivator” or “sensitizer” BH3s)6, 9–15. BH3s execute their function through binding of their BH3 domains into the hydrophobic binding groove of multidomain proapoptotic or antiapoptotic members16–22.
BID, BIM and PUMA are “activator BH3s” that directly interact with BAX-BAK to induce the stepwise structural reorganization and oligomerization of BAX-BAK6, 8, 9, 14, 15. One important intermediate step of BAX-BAK activation driven by activator BH3s is to expose the BH3 domain of BAX-BAK such that the “BH3-exposed” BAX-BAK monomer can bind to the hydrophobic dimerization pocket of another BAX-BAK molecule, initiating homo-dimerization and subsequent homo-oligomerization23, 24. Antiapoptotic BCL-2 members (BCL-2s) inhibit apoptosis principally through sequestering tBID-BIM-PUMA from activating BAX-BAK, providing a frontline protection against apoptotic insults6, 9. Additionally, antiapoptotic BCL-2s can also sequester partially activated, BH3-exposed, monomeric BAX-BAK to prevent their homo-oligomerization2, 3, 7, serving as a fail-safe mechanism. The ability of antiapoptotic BCL-2s to sequester tBID-BIM-PUMA is further modulated by “inactivator” BH3s (BAD and NOXA) through high-affinity, competitive binding. Specifically, BAD and NOXA displace sequestered tBID-BIM-PUMA from BCL-2-BCL-XL and MCL-1, respectively, to activate BAX-BAK indirectly6, 10–13.
Two non-mutually exclusive models have been proposed concerning how BH3s activate BAX-BAK2, 3. The direct activation model states that the “activator” subgroup of BH3s can directly induce the conformational changes of BAX-BAK6, 8, 9, 14, 25, 26. The indirect model proposes that BAX and BAK are kept in check by the antiapoptotic BCL-2 proteins and activation of BAX-BAK occurs by default as long as all the antiapoptotic BCL-2s are neutralized by BH3s27. The generation of Bid−/−Bim−/−Puma−/− triple knockout (TKO) mice provides in vivo evidence supporting the direct activation model15. However, the observation that double deficiency of Bax and Bak incurs more severe embryonic lethality than triple deficiency of Bid, Bim and Puma15, 28 suggest that either additional activators of BAX-BAK exist in other cell types, or BAX-BAK can be activated through an alternative mechanism that is entirely independent of BH3s. Alternatively, non-apoptotic functions of BAX-BAK, such as regulation of mitochondrial fission/fusion or endoplasmic reticulum (ER) calcium homeostasis29, 30, may account for the more severe embryonic lethality of Bax−/−Bak−/− mice.
Here, we demonstrate that NOXA is a bona fide activator of BAX and BAK that directly interacts with BAX-BAK to drive the homo-oligomerization of BAX-BAK even in the absence of BID, BIM and PUMA. Due to the unique high expression of NOXA in mouse embryonic fibroblasts (MEFs), Bid−/−Bim−/−Puma−/− MEFs succumbed to diverse intrinsic apoptotic signals whereas Bid−/−Bim−/−Puma−/−Noxa−/− MEFs were completely resistant to growth factor deprivation or ER stress. Surprisingly, BAX and BAK could be “autoactivated” by DNA damage in transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs through downregulation of BCL-2, BCL-XL and MCL-1. Liberation of the small fraction, “BH3-exposed” BAK/BAX monomers from the antiapoptotic BCL-2s is sufficient to induce a “feed-forward” amplification loop for the initiation of mitochondrial apoptosis. Interestingly, BCL-XL is superior to BCL-2 and MCL-1 in preventing DNA damage-induced apoptosis due to its dual inhibition of BAX and BAK as well as higher protein stability. Altogether, we propose an interconnected hierarchical model that accommodates and explains how the intricate interplays between BH3s, multidomain antiapoptotic and multidomain proapoptotic BCL-2 members dictate cellular decision of survival versus death.
RESULTS
NOXA directly activates BAX and BAK independently of BID, BIM and PUMA
Genetic loss-of-function studies indicate that BID, BIM and PUMA are required for the activation of BAX and BAK in neurons and T-cells15. However, it remains unclear whether BID, BIM and PUMA represent the full repertoire of activator BH3s in all cell types, and whether BAX and BAK can be activated to induce apoptosis in the absence of activator BH3s. To address these questions, we first examined fibroblasts generated from Bid−/−Bim−/−Puma−/− TKO mouse embryos. Although TKO MEFs were significantly more resistant to apoptosis than wild-type MEFs, they eventually succumbed to various apoptotic stimuli (Supplementary Fig. 1). These data suggest the existence of yet unidentified activator(s) of BAX and BAK in fibroblasts. During the process of searching for additional activator(s) of BAX-BAK, we found that recombinant NOXA protein generated using wheat germ extract (WGE) but not reticulocyte lysates was capable of inducing cytochrome c release from mitochondria isolated from wild-type but not Bax−/−Bak−/− cells in a dose-dependent manner (Fig. 1a, b). The cytochrome c releasing activity of NOXA was abrogated by BH3 mutations (Fig. 1a), supporting the importance of BH3 domain in activating BAX-BAK. We have previously demonstrated that in vitro transcribed, translated (IVTT) mouse NOXA protein using reticulocyte lysates failed to induce cytochrome c efflux from mitochondria6. Because we encountered difficulty in generating IVTT human NOXA protein using reticulocyte lysates, we exploited WGE. Both human and mouse NOXA generated using WGE exhibited comparable cytochrome c releasing activity (Fig. 1c). Notably, BAD protein could not induce cytochrome c release irrespective of the IVTT system employed (Fig. 1c). These data suggest that either WGE activates or reticulocyte lysates inhibit the cytochrome c releasing activity of NOXA. NOXA protein generated using reticulocyte lysates could not induce cytochrome c release even in the presence of WGE (Supplementary Fig. 2a). In contrast, the cytochrome c releasing activity of NOXA protein generated using WGE was abrogated by reticulocyte lysates (Supplementary Fig. 2a), supporting that an inhibitory factor or activity is present in reticulocyte lysates. Interestingly, the inhibitory effect of reticulocyte lysates appears to be specific for NOXA (Supplementary Fig. 2a, b).
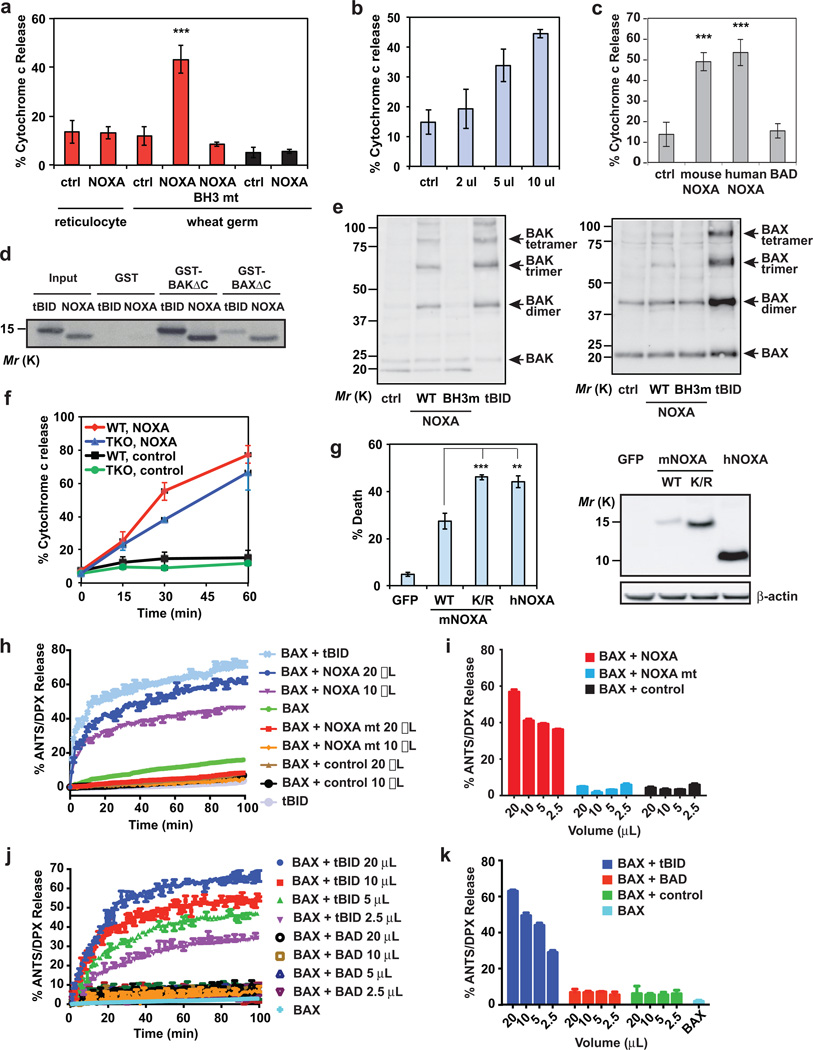
Figure 1. NOXA directly activates BAX and BAK independently of BID, BIM and PUMA.
(a) Mitochondria isolated from wild-type or Bax−/−Bak−/− MEFs were incubated with the indicated IVTT proteins generated using reticulocyte lysates or wheat germ extract (WGE) at 30°C for 30 min, after which the release of cytochrome c was quantified by ELISA assays (mean ± s.d., n = 3 independent experiments). (b) Isolated wild-type mitochondria were incubated with the indicated amounts of IVTT mouse NOXA protein (WGE) and the release of cytochrome c was quantified (mean ± s.d., n = 3 independent experiments). (c) Isolated wild-type mitochondria were incubated with the indicated IVTT proteins (WGE) and the release of cytochrome c was quantified (mean ± s.d., n = 3 independent experiments). (d) Radiolabeled IVTT NOXA or tBID protein was incubated with GST, GST-BAXΔC or GST-BAKΔC protein immobilized on glutathione beads. The precipitates and input were analyzed by Nu-PAGE and autoradiography. (e) Isolated wild-type mitochondria were incubated with the indicated IVTT proteins for 30 min then treated with BMH crosslinker. The BAX and BAK homo-oligomers were detected by anti-BAX and anti-BAK immunoblots, respectively. (f) Mitochondria isolated from wild-type or Bid−/−Bim−/−Puma−/− MEFs were incubated with IVTT NOXA (WGE) at 30°C for the indicated times and release of cytochrome c was quantified (mean ± s.d., n = 3 independent experiments). (g) Bid−/−Bim−/−Puma−/− MEFs were infected with the indicated retrovirus. Cell death was quantified by annexin-V staining at 30 h (mean ± s.d., n = 3 independent experiments). The expression of the indicated proteins was detected by an anti-HA immunoblot. (h and i) ANTS/DPX (fluorophore/quencher)-encapsulated liposomes were incubated with recombinant BAX protein plus the indicated IVTT proteins generated using WGE. The release of entrapped fluorophore monitored with time is shown in (h) (mean ± s.d., n = 3 independent experiments). The release of entrapped fluorophore at 60 min is shown in (i) (mean ± s.d., n = 3 independent experiments). (j and k) ANTS/DPX-encapsulated liposomes were incubated with recombinant BAX protein plus the indicated IVTT proteins (WGE). The release of entrapped fluorophore monitored with time is shown in (j) (mean ± s.d., n = 3 independent experiments). The release of entrapped fluorophore at 60 min is shown in (k) (mean ± s.d., n =3 independent experiments). **, P < 0.01; ***, P < 0.001 (Student’s t-test). Uncropped images of blots are shown in Supplementary Fig. S7.
We next examined whether NOXA can interact and activate BAX-BAK directly. Indeed, direct interaction between NOXA and BAX or BAK was detected by GST-pull-down assays (Fig. 1d). Furthermore, wild-type NOXA but not BH3 mutant NOXA induced the formation of BAX or BAK homo-oligomers (Fig. 1e). More importantly, NOXA induced cytochrome c efflux in Bid−/−Bim−/−Puma−/− TKO mitochondria (Fig. 1f). Overexpression of mouse NOXA protein did not induce robust apoptosis likely due to its labile nature. Supporting this notion, overexpression of mutant NOXA with substitutions of all lysine residues with arginine that stabilized NOXA protein significantly enhanced the death-inducing activity of mouse NOXA (Fig. 1g). Interestingly, human NOXA protein was more stable than mouse NOXA protein and consequently induced more apoptosis (Fig. 1g). To further validate that NOXA is a bona fide activator BH3, liposome permeabilization assays were performed (Fig. 1h, i). As previously reported31, 32, recombinant BAX protein was unable to induce liposomal release of fluorophore unless it was activated by tBID (Fig. 1h). By analogy, wild-type NOXA but not BH3 mutant NOXA could induce BAX-dependent liposome permeabilization (Fig. 1h, i). In contrast, BAD failed to activate BAX to permeabilize liposome (Fig. 1j, k). These data strongly support that NOXA can directly activate BAX to induce MOMP.
Noxa deficiency further protects Bid−/−Bim−/−Puma−/− MEFs and small intestine from apoptosis
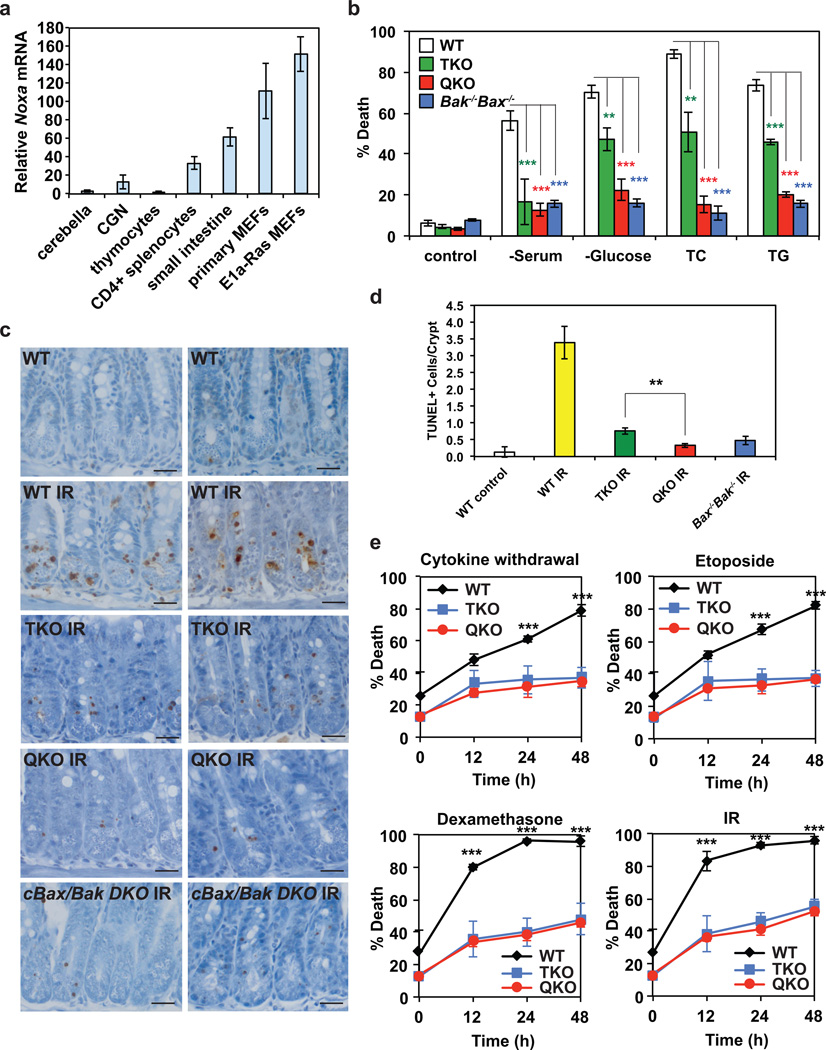
We reasoned that differential expressions of NOXA in various cell types might contribute to the different apoptotic phenotypes of Bid−/−Bim−/−Puma−/− neurons, T cells, and MEFs. Quantitative RT-PCR demonstrated that NOXA was highly expressed in MEFs compared to neurons and lymphocytes (Fig. 2a), which is consistent with previous observations that deficiency of Noxa protects transformed MEFs but not lymphocytes from DNA damage-induced apoptosis33–35. To determine whether NOXA is the missing activator of BAX and BAK in Bid−/−Bim−/−Puma−/− MEFs, we generated Bid−/−Bim−/−Puma−/−Noxa−/− quadruple knockout (QKO) MEFs. Primary Bid−/−Bim−/−Puma−/−Noxa−/− MEFs were more resistant than Bid−/−Bim−/−Puma−/− MEFs to apoptosis triggered by serum or glucose deprivation, and ER stress (tunicamycin or thapsigargin) (Fig. 2b). Remarkably, the protection conferred by quadruple deficiency of Bid, Bim, Puma and Noxa was comparable to that of double deficiency of Bax and Bak (Fig. 2b). The reason why deletion of Noxa further protects Bid−/−Bim−/−Puma−/− MEFs is not due to compensatory induction of Noxa (Supplementary Fig. 2c). Altogether, these data indicate that NOXA, along with BID, BIM and PUMA, are activators of BAX-BAK in MEFs. Because intermediate expression of NOXA was detected in small intestine (Fig. 2a) where deficiency of Noxa was reported to confer protection against ionizing radiation34, we next examined whether deficiency of Noxa further protects Bid−/−Bim−/−Puma−/− small intestine from irradiation. Similar to Bid−/−Bim−/−Puma−/− mice, Bid−/−Bim−/−Puma−/−Noxa−/− mice were born at a less-than-expected Mendelian ratio and exhibit the same developmental defects including persistent interdigital webs and imperforate vagina (Supplementary Fig. 2d–e and Supplementary Table 1). Although triple deletion of Bid, Bim and Puma provided profound inhibition of irradiation-induced apoptosis in small intestine, only quadruple deletion of Bid, Bim, Puma and Noxa or double deletion of Bax and Bak completely blocked apoptosis (Fig. 2c, d). Consistent with the lower NOXA expression in lymphocytes (Fig. 2a) and the absence of BAX-BAK activation detected in Bid−/−Bim−/−Puma−/− T-cells as reported15, Bid−/−Bim−/−Puma−/− T-cells were as resistant as Bid−/−Bim−/−Puma−/−Noxa−/− T-cells to various apoptotic signals including cytokine withdrawal, glucocorticoid, and genotoxic stress (Fig. 2e). Of note, Noxa expression is lower both at the basal level and in response to genotoxic stress in T-cells than MEFs (Supplementary Fig. 2f). Total body irradiation-induced apoptosis in spleen was greatly reduced in both Bid−/−Bim−/−Puma−/− and Bid−/−Bim−/−Puma−/−Noxa−/− mice (Supplementary Fig. 3a). Triple deletion of Bid, Bim and Puma or quadruple deletion of Bid, Bim, Puma, and Noxa almost completely rescued splenic CD4+ T-cells from irradiation (Supplementary Fig. 3b). Collectively, these data suggest that although NOXA can directly activate BAX and BAK, Noxa deficiency only confers resistance to apoptosis in tissues or cell types that express sufficient NOXA for the activation of BAX- and BAK-dependent death program.
Figure 2. Noxa deficiency further protects Bid−/−Bim−/−Puma−/− mouse embryonic fibroblasts or small intestine from apoptosis.
(a) The mRNA levels of Noxa in the indicated tissues or cells were assessed by qRT-PCR. Data are normalized against 18S rRNA (mean ± s.d., n = 3 independent experiments). (b) Primary MEFs generated from E13.5 wild-type, Bid−/−Bim−/−Puma−/− TKO, Bid−/−Bim−/−Puma−/−Noxa−/− QKO, or Bax−/−Bak−/− DKO mouse embryos were untreated, or cultured in the absence of serum or glucose for 3 days, or in the presence of tunicamycin (TC) or thapsigargin (TG) for 2 days. Cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). (c and d) Apoptosis in the small intestinal crypts of wild-type (n = 3), Bid−/−Bim−/−Puma−/− TKO (n = 3), Bid−/−Bim−/−Puma−/−Noxa−/− QKO (n = 2), or conditional Bax and Bak DKO (n = 2) mice at 8 to 17 weeks of age at 4 h after 18 Gy whole body irradiation was assessed by TUNEL staining (brown, magnification 400×). 300 small intestinal crypts from each mouse were analyzed. Representative light microscopy images are shown in (c). Scale bars, 50 µm. The number of TUNEL positive cells in the crypts was quantified and summarized in (d) (mean ± s.d.). (e) CD4+ T cells purified from the spleens of wild-type (n = 3), Bid−/−Bim−/−Puma−/− TKO (n = 3), or Bid−/−Bim−/−Puma−/−Noxa−/− QKO mice (n = 3) at 8 to 10 weeks of age were cultured in the absence of cytokine, in the presence of etoposide, in the presence of dexamethasone, or after exposure to 2.5 Gy γ-irradiation. Cell death was quantified by annexin-V staining at the indicated times (mean ± s.d.). **, P < 0.01; ***, P < 0.001 (Student’s t-test).
Transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs still succumb to DNA damage-induce apoptosis
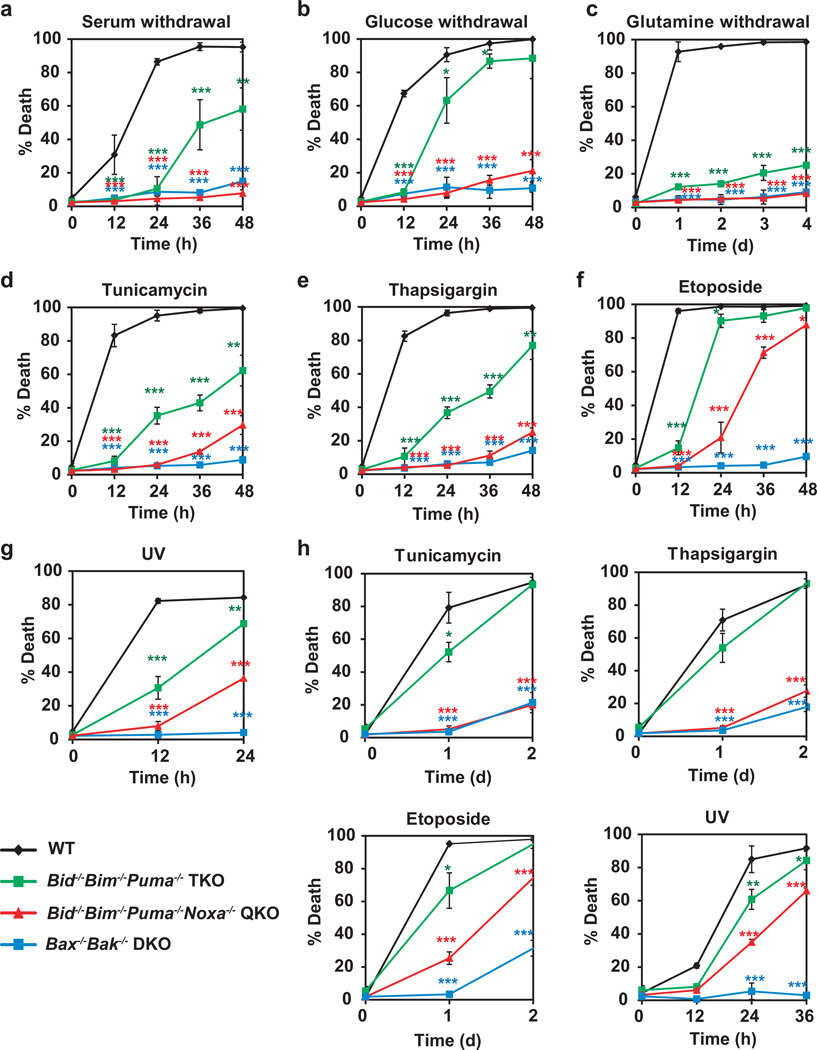
To study DNA damage-induced apoptosis, primary MEFs were transformed with E1A and Ras because primary MEFs undergo cell cycle arrest instead of apoptosis upon DNA damage36. Similar to primary MEFs, E1A/Ras-transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs were nearly as resistant as Bax−/−Bak−/− MEFs to growth factor deprivation and ER stress (Fig. 3a–e). Consistent with the notion that transformed cells are more dependent on glutamine for survival, transformation of primary MEFs with E1A/Ras sensitizes these cells to glutamine deprivation-induced apoptosis (Fig. 3c). Deletion of Bid, Bim and Puma greatly reduced whereas deletion of Bid, Bim, Puma and Noxa completely blocked glutamine deprivation-induced apoptosis in E1A/Ras-transformed MEFs (Fig. 3c). Surprisingly, quadruple deficiency of Bid, Bim, Puma and Noxa failed to provide similar protection against etoposide or UV as double deficiency of Bax and Bak (Fig. 3f, g). To exclude the possibility that regulation of BAX-BAK activation is unique in E1A/Ras-transformed cells, primary MEFs were transformed with SV40. Again, SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs were sensitive to etoposide and UV while they were as resistant as SV40-transformed Bax−/−Bak−/− MEFs to ER stress (Fig. 3h). Together, these data suggest that either an additional activator of BAX-BAK is induced or an alternative mechanism is involved in DNA damage-induced activation of BAX-BAK in transformed MEFs.
Figure 3. Quadruple deficiency of Bid, Bim, Puma and Noxa abrogates apoptosis in transformed mouse embryonic fibroblasts triggered by growth factor deprivation and ER stress but not genotoxic stress.
(a–g) E1A/Ras-transformed wild-type, Bid−/−Bim−/−Puma−/− TKO, Bid−/−Bim−/−Puma−/−Noxa−/− QKO, or Bax−/−Bak−/− DKO MEFs were untreated, or cultured in the absence of serum (a), glucose (b) or glutamine (c), or in the presence of tunicamycin (d), thapsigargin (e) or etoposide (f), or irradiated with UV-C (g). Cell death was quantified by annexin-V staining at the indicated times (mean ± s.d., n = 3 independent experiments). (h) SV40-transformed wild-type, Bid−/−Bim−/−Puma−/− TKO, Bid−/−Bim−/−Puma−/−Noxa−/− QKO, or Bax−/−Bak−/− DKO MEFs were untreated, or cultured in in the presence of tunicamycin, thapsigargin or etoposide, or irradiated with UV-C. Cell death was quantified by annexin-V staining at the indicated times (mean ± s.d., n = 3 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t-test).
DNA damage activates BAX and BAK-dependent mitochondrial apoptosis in transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs
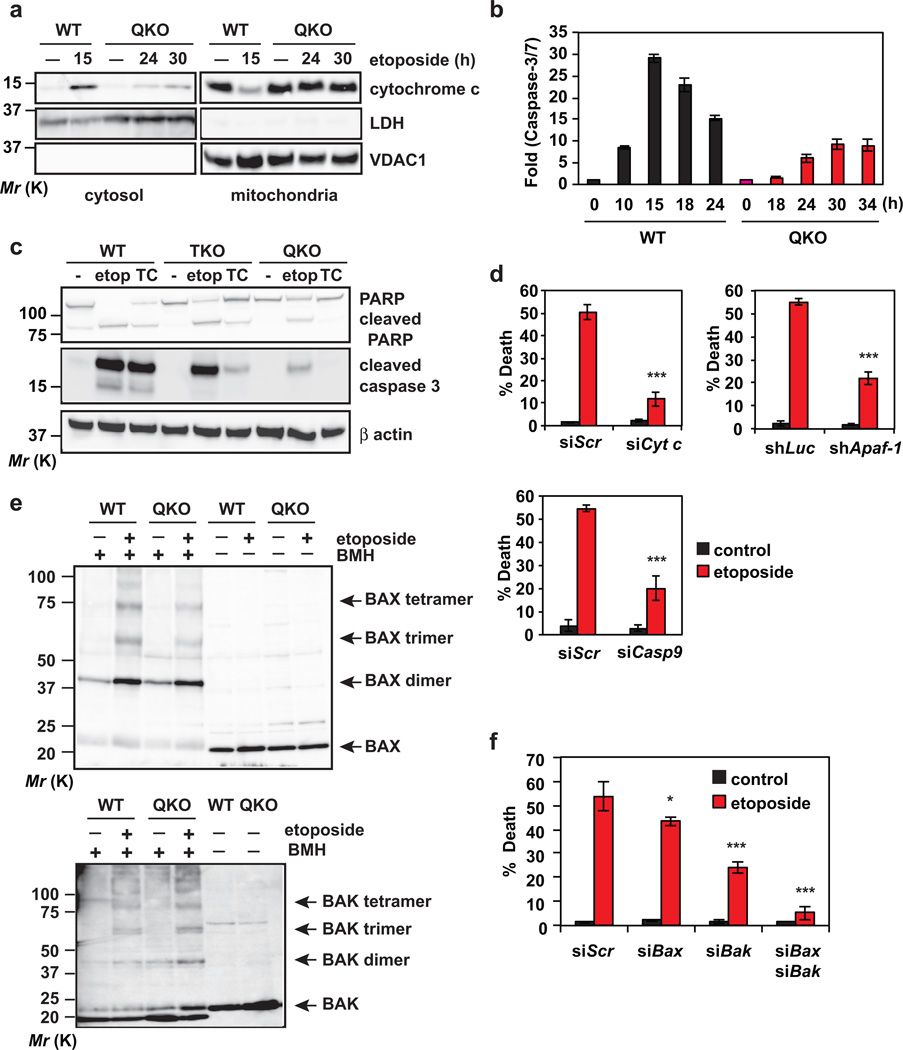
To characterize DNA damage-induced cell death in transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs, we first examined cytochrome c-initiated caspase activation. In SV40-transformed wild-type MEFs, etoposide induced cytochrome c efflux from the mitochondria to the cytosol within 15 hours (Fig. 4a), which coincides with the peak of caspase activation (Fig. 4b). In contrast, cytochrome c translocation was not detected in Bid−/−Bim−/−Puma−/−Noxa−/− cells until 24 hours and was to a much lesser extent than wild-type cells (Fig. 4a). Accordingly, lower caspase activity was detected in Bid−/−Bim−/−Puma−/−Noxa−/− cells than wild-type cells (Fig. 4b). Cleavage of PARP and Caspase-3 was detected in QKO cells upon etoposide but not ER stress (Fig. 4c). In support of the involvement of cytochrome c-APAF-1-Caspase-9 axis in DNA damage-induced apoptosis in QKO cells, knockdown of cytochrome c, Apaf-1 or Caspase-9 protected QKO cells from etoposide-induced apoptosis (Fig. 4d and Supplementary Fig. 4a). We next determined whether BAX and BAK are activated in QKO cells upon DNA damage. Indeed, both BAX and BAK homo-oligomers were detected in QKO cells upon etoposide at a later time point (Fig. 4e). Consequently, knockdown of either Bax or Bak partially inhibited whereas knockdown of both Bax and Bak near completely blocked etoposide-induced apoptosis in QKO cells (Fig. 4f and Supplementary Fig. 4a). Together, these data indicate that DNA damage can activate BAX-BAK in transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs. The low amplitude of cytochrome c efflux and caspase activation suggest that this mode of BAX and BAK activation is less efficient than that involves BID, BIM, PUMA and NOXA.
Figure 4. DNA damage activates BAX and BAK-dependent mitochondrial apoptosis in transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO mouse embryonic fibroblasts.
(a) SV40-transformed wild-type or Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs, untreated or treated with etoposide for the indicated times, were subjected to subcellular fractionation. Cytosolic and mitochondrial fractions were analyzed by anti-cytochrome c, anti-LDH and anti-VDAC1 immunoblots. LDH and VDAC1 serve as cytosolic and mitochondrial controls, respectively. (b) SV40-transformed wild-type or Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs, untreated or treated with etoposide for the indicated times, were analyzed for Caspase-3/7 activities (mean ± s.d., n = 3 independent experiments). (c) SV40-transformed wild-type, Bid−/−Bim−/−Puma−/− TKO, or Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs, untreated or treated with etoposide (etop) or tunicamycin (TC), were analyzed by anti-PARP, anti-cleaved Caspase-3, and anti-actin immunoblots. (d) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs were infected with retrovirus expressing shRNA against luciferase or Apaf-1, or transfected with scrambled siRNA (siScr) or siRNA against Cytochrome c or Caspase-9. After 48 h, cells were untreated or treated with etoposide for 36 h. Cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). (e) Mitochondria isolated from SV40-transformed wild-type or Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs untreated or treated with etoposide for 15 h (WT) or 36 h (QKO) were subjected to BMH crosslinking. The BAX and BAK homo-oligomers were detected by anti-BAX and anti-BAK immunoblots, respectively. (f) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs were transfected with scrambled siRNA (siScr) or siRNA against Bax and/or Bak. After 48 h, cells were untreated or treated with etoposide for 36 h. Cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). *, P < 0.05; ***, P < 0.001 (Student’s t-test). Uncropped images of blots are shown in Supplementary Fig. S7.
BAX and BAK can be autoactivated in the absence of activator BH3s through downregulation of BCL-2, BCL-XL and MCL-1
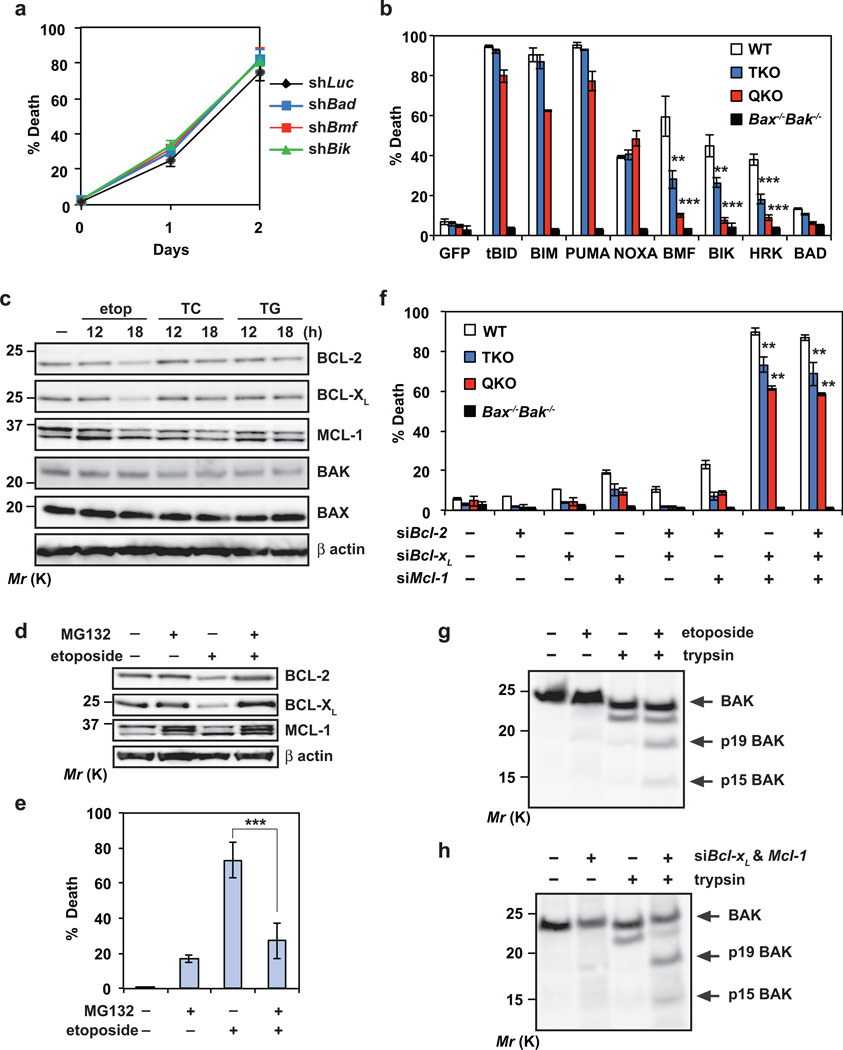
To elucidate how DNA damage activates BAX-BAK in QKO cells, we first investigated whether other known BH3s are involved. Knockdown of Bad, Bmf, Bik or Hrk failed to protect QKO cells from etoposide-induced apoptosis (Fig. 5a and Supplementary Fig. 4b–d). Moreover, overexpression of BAD, BMF, BIK or HRK induced minimal apoptosis in QKO cells (Fig. 5b), supporting the proposed hierarchy of BH3s where activator BH3s function downstream of inactivator BH3s6. Consistent with the notion that tBID, BIM, PUMA and NOXA are activator BH3s, overexpression of these BH3s effectively induced spontaneous apoptosis in QKO cells (Fig. 5b). The inability of BH3s other than BID, BIM, PUMA and NOXA in killing QKO cells raises a possibility that an alternative, BH3-indpendent mechanism may be involved in the activation of BAX-BAK in QKO cells upon DNA damage.
Figure 5. BAX and BAK can be autoactivated by DNA damage independently of activators BID, BIM, PUMA and NOXA through downregulation of BCL-2, BCL-XL and MCL-1.
(a) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs were infected with retrovirus expressing shRNA against luciferase, Bad, Bmf or Bik. After 48 h, cells were untreated or treated with etoposide for 36 h. Cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). (b) SV40-transformed wild-type, Bid−/−Bim−/−Puma−/− TKO, Bid−/−Bim−/−Puma−/−Noxa−/− QKO, or Bax−/−Bak−/− MEFs were infected with retrovirus expressing GFP or the indicated BH3-only proteins to induce spontaneous apoptosis. NOXA denotes human NOXA. Cell death was quantified by annexin-V staining at 30 h (mean ± s.d., n = 3 independent experiments). (c) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs, untreated or treated with etoposide, tunicamycin (TC) or thapsigargin (TG), were subjected to immunoblot analysis using the indicated antibodies. (d) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs were untreated or treated with etoposide and/or MG132 for 18h, and subjected to immunoblot analysis using the indicated antibodies. (e) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs were untreated or treated with etoposide and/or MG132 for 36h. Cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). (f) SV40-transformed wild-type, Bid−/−Bim−/−Puma−/− TKO, Bid−/−Bim−/−Puma−/−Noxa−/− QKO, or Bax−/−Bak−/− MEFs were transfected with scrambled siRNA (siScr) or siRNA against Bcl-2, Bcl-xL and/or Mcl-1 to induce spontaneous apoptosis. After 2 days, cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). (g) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs were untreated or treated with etoposide in the presence of the pancaspase inhibitor Q-VD-OPh to preserve cell integrity upon apoptosis induction. After 24 h, cells were permeabilized with digitonin and subjected to limited trypsin proteolysis. The BAK cleavage products were detected by an anti-BAK (G23) immunoblot. (h) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs transfected with scrambled siRNA or siRNA against Bcl-xL and Mcl-1 were subjected to limited trypsin proteolysis. The BAK cleavage products were detected by an anti-BAK (G23) immunoblot. **, P < 0.01; ***, P < 0.001 (Student’s t-test). Uncropped images of blots are shown in Supplementary Fig. S7.
Although most BAX and BAK do not interact with antiapoptotic BCL-2s in viable cells6, 7, those complexed with antiapoptotic BCL-2s likely have already exposed their BH3 domain that bind to the hydrophobic dimerization groove of antiapoptotic BCL-2s3, 7. Foreseeably, if antiapoptotic BCL-2s are downregulated by DNA damage in QKO MEFs, the “BH3-exposed” monomers of BAX-BAK will probably be liberated to activate other native molecules of BAX-BAK, initiating “autoactivation”. Indeed, BCL-2, BCL-XL and MCL-1 were decreased in QKO MEFs upon etoposide treatment (Fig. 5c). In contrast, no concurrent downregulation of antiapoptotic BCL-2s was detected in QKO MEFs upon ER stress (Fig. 5c). Significantly, proteasome inhibition mitigated DNA damage-induced decrease of antiapoptotic BCL-2s, and consequently protected QKO cells from etoposide-induced apoptosis (Fig. 5d, e). Of note, etoposide-induced downregulation of BCL-2 and BCL-XL was also observed in wild-type MEFs but not in T-cells (Supplementary Fig. 5, a, b). Interestingly, UV but not ionizing radiation could induce downregulation of antiapoptotic BCL-2s in QKO cells (Supplementary Fig. 5c). Consequently, QKO cells were almost as resistant as Bax−/−Bak−/− cells to ionizing radiation but succumbed to UV (Supplementary Fig. 5d and Fig. 3h).
To further confirm that loss of antiapoptotic BCL-2s can lead to autoactivation of BAX-BAK and apoptosis, siRNA-mediated knockdown of antiapoptotic BCL-2s was performed. Concurrent knockdown of Bcl-2, Bcl-xL and Mcl-1 induced significant apoptosis after 2 days in wild-type, TKO and QKO cells but not in Bax−/−Bak−/− cells in the absence of any death stimulus (Fig. 5f and Supplementary Fig. 4a). Interestingly, knockdown of both Bcl-xL and Mcl-1 was sufficient to induce spontaneous apoptosis. Of note, knockdown of Bcl-2, Bcl-xL or Mcl-1 did not sensitize QKO cells to overexpression of BAD, BMF, BIK or HRK (Supplementary Fig. 5e). Given that the basal levels of activator BH3s are low in healthy cells and most of the activator BH3s are induced by intrinsic death signals, deficiency of activator BH3s appears to provide more protection against apoptosis triggered by intrinsic death signals than knockdown of antiapoptotic BCL-2s. It is noteworthy that overexpression of NOXA induces more apoptosis than knockdown of Mcl-1 in MEFs (Fig. 5b, f), supporting that NOXA does not simply induce apoptosis by inactivating MCL-1.
Liberation of the “BH3-exposed” BAK monomers from antiapoptotics initiates a feed-forward amplification loop for the activation of mitochondrial apoptosis
It was reported that the BH3 domain of BAK is inaccessible in viable cells and cleavage of BAK at the BH3 domain by limited proteolysis can reflect the exposure of the BH3 domain in BAK23, 37. Cleavage of BAK at the BH3 domain by trypsin generates a ~15 kDa C-terminal fragment (p15) that is tethered to the mitochondria through the C-terminal transmembrane anchor. Indeed, cleaved BAK p15 was detected in QKO cells upon etoposide treatment (Fig. 5g), suggesting that the “BH3-exposed” BAK is liberated from the antiapoptotic BCL-2s in QKO cells upon DNA damage. Furthermore, BAK cleavage at the BH3 domain was observed in QKO cells upon knockdown of both Bcl-xL and Mcl-1 (Fig. 5h). Of note, only low levels of BAK p15 could be detected because the “BH3-exposed” BAK likely binds to the hydrophobic groove of another BAK molecule immediately23. Limited proteolysis of BAK also revealed another feature of BAK activation, i.e. exposure of the α1 helix or BH4 domain, leading to the generation of a 19 kDa mitochondrially targeted C-terminal fragment p19 (Fig. 5g, h). These data suggest that liberation of the “BH3-exposed” BAK from antiapoptotic BCL-2s may induce a “feed-forward” mechanism to activate other native molecules of BAX-BAK. Accordingly, even a small fraction of “BH3-exposed” BAX-BAK monomers that are set free from the antiapoptotic BCL-2s is sufficient to initiate the mitochondrial apoptotic program. Collectively, these data support a model in which BAX and BAK can undergo autoactivation upon DNA damage through downregulation of antiapoptotic BCL-2s in transformed Bid−/−Bim−/−Puma−/−Noxa−/− MEFs.
BCL-XL is superior to BCL-2 and MCL-1 in preventing DNA damage-induced apoptosis
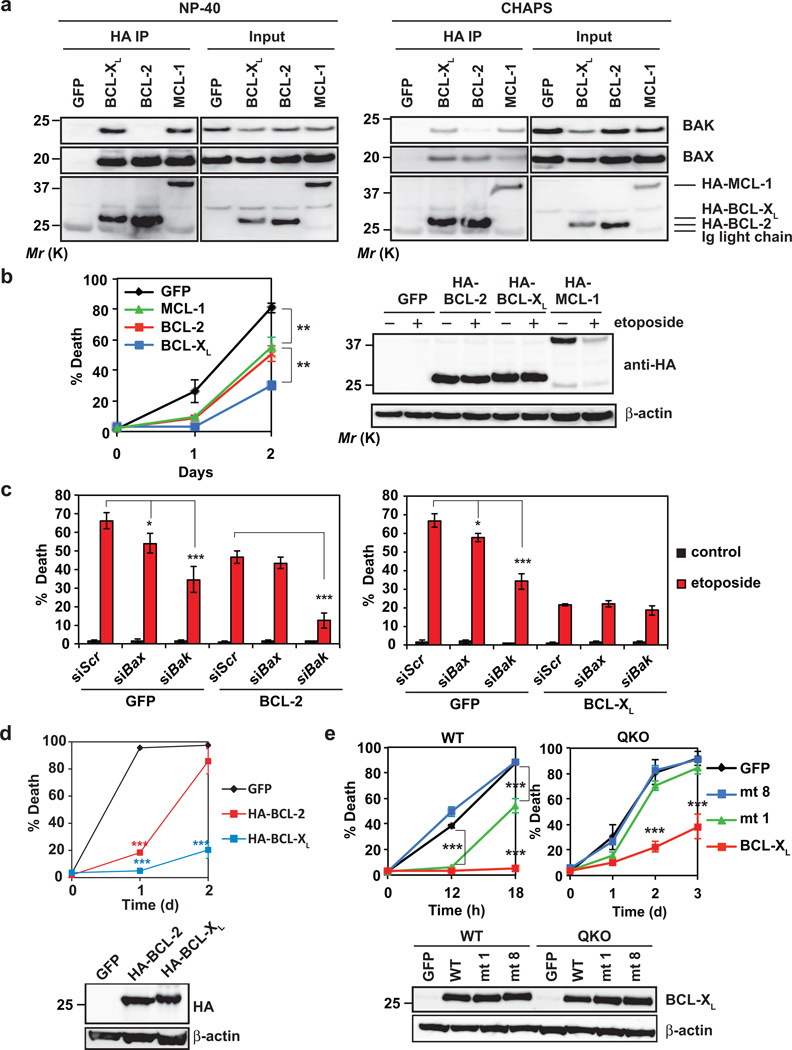
The loss-of-function studies of antiapoptotic BCL-2s suggest that BCL-2 may be less important than BCL-XL and MCL-1 in gauging cellular survival (Fig. 5f). This could be due to the fact that BCL-2 only interacts with BAX but not BAK37, 38. In QKO cells, co-immunoprecipitation assays demonstrated that BAX interacted with BCL-2, BCL-XL and MCL-1 whereas BAK only interacted with BCL-XL and MCL-1 but not BCL-2 (Fig. 6a). The same interaction profiles were also observed in WT and TKO MEFs (Supplementary Fig. 5f), which is consistent with previously published results27, 38. Notably, the same results were obtained using either CHAPS or NP-40 buffer except that more heterodimers were detected in the presence of NP-40 than CHAPS (Fig. 6a). Consistent with the notion that most endogenous BAX-BAK do not interact with antiapoptotic BCL-2s in viable cells6, 7, only a portion of BAX-BAK was co-precipitated with antiapoptotic BCL-2s (Fig. 6a).
Figure 6. BCL-XL is superior to BCL-2 and MCL-1 in preventing DNA damage-induced apoptosis due to its dual inhibition of BAX and BAK as well as higher protein stability.
(a) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs stably expressing GFP, HA-BCL-2, HA-BCL-XL or HA-MCL-1 were subjected to anti-HA immunoprecipitation in 0.2% NP-40 or 1% CHAPS lysis buffer. The input (5%) and immunoprecipitates were analyzed by anti-BAX, anti-BAK, and anti-HA immunoblots. (b) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs stably expressing GFP, HA-BCL-2, HA-BCL-XL or HA-MCL-1 were untreated or treated with etoposide. Cell death was quantified by annexin-V staining at the indicated times (mean ± s.d., n = 3 independent experiments). The expression of HA-BCL-2, HA-BCL-XL or HA-MCL-1 was detected by an anti-HA immunoblot with or without etoposide treatment for 12 h. (c) SV40-transformed Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs stably expressing GFP, BCL-2 or BCL-XL were transfected with scrambled siRNA (siScr) or siRNA against Bax or Bak. After 48 h, cells were untreated or treated with etoposide for 36 h. Cell death was quantified by annexin-V staining (mean ± s.d., n = 3 independent experiments). (d) SV40-transformed wild-type MEFs stably expressing HA-tagged BCL-2 or BCL-XL were untreated or treated with etoposide. Cell death was quantified by annexin-V staining at the indicated times (mean ± s.d., n = 3 independent experiments). The expression of HA-tagged BCL-2 and BCL-XL was detected by an anti-HA immunoblot. (e) SV40-transformed wild-type or Bid−/−Bim−/−Puma−/−Noxa−/− QKO MEFs stably expressing GFP, wild-type BCL-XL, BCL-XL mutant 1 (F131V/D133A) or BCL-XL mutant 8 (G138E/R139L/I140N) were untreated or treated with etoposide. Cell death was quantified by annexin-V staining at the indicated times (mean ± s.d., n = 3 independent experiments). The expression of BCL-XL was analyzed by an anti-BCL-XL immunoblot. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t-test). Uncropped images of blots are shown in Supplementary Fig. S7.
We next investigated whether antiapoptotic BCL-2s might display differential activity against DNA damage in QKO cells. Indeed, overexpression of BCL-2 provided less protection to QKO cells against etoposide-induced apoptosis than BCL-XL (Fig. 6b). Surprisingly, MCL-1 also provided less protection than BCL-XL even though both interact with BAX and BAK. The lesser protection conferred by MCL-1 is likely due to its labile nature, which is supported by the observation that overexpressed MCL-1 was rapidly degraded by etoposide within 12 hours (Fig. 6b). To further assess whether the differential activity of BCL-2 versus BCL-XL in regulating apoptosis is due to the inability of BCL-2 to inhibit BAK, we silenced Bax or Bak in QKO cells overexpressing BCL-2 or BCL-XL. Knockdown of Bak but not Bax further protected BCL-2-overexpressing QKO cells from etoposide-induced apoptosis (Fig. 6c), suggesting that etoposide is capable of activating BAK in BCL-2-overexpressing cells. In contrast, knockdown of either Bax or Bak had no effect on BCL-XL in protecting QKO cells against etoposide (Fig. 6c). Even in wild-type cells, overexpression of BCL-2 only delays whereas overexpression of BCL-XL completely blocks etoposide-induced apoptosis (Fig. 6d). The delayed death kinetics observed in BCL-2 overexpressed cells further supports the lower efficiency of BAK autoactivation in triggering mitochondrial apoptosis. It is noteworthy that transformed Bax−/−Bak−/− MEFs still succumb to DNA damage-induced programmed necrotic death between 2–3 days that cannot be inhibited by overexpression of BCL-XL39. In aggregate, BCL-XL is superior to BCL-2 and MCL-1 in preventing DNA damage-induced apoptosis due to its dual inhibition of BAX and BAK as well as higher protein stability.
Binding to BAX and BAK is required for BCL-XL to inhibit apoptosis in QKO cells
To further illustrate that BCL-XL inhibits etoposide-induced apoptosis in QKO cells through sequestering BAX-BAK but not BH3s, we employed reported, distinct BCL-XL mutants that differentially interact with BH3s or BAX-BAK9, 40. The BCL-XL mutant 1 (F131V/D133A) known to interact with BH3s but not BAX-BAK protected wild-type but not QKO cells from etoposide-induced apoptosis (Fig. 6e). Conversely, the BCL-XL mutant 8 (G138E/R139L/I140N) known to interact with neither BH3s nor BAX-BAK failed to inhibit apoptosis in both wild-type and QKO cells (Fig. 6e). Together, these data suggest that BCL-XL protects wild-type cells through sequestration of either activator BH3s or multidomain BAX-BAK. In the absence of activator BH3s, BCL-XL binds to BAX-BAK to inhibit apoptosis. In summary, our data support a model in which antiapoptotic BCL-2s keep both activator BH3s and the “BH3-exposed” BAX-BAK monomers in check in viable cells. In response to intrinsic death signals, BID, BIM, PUMA and NOXA are activated through transcriptional regulation and/or posttranslational modifications, triggering the homo-oligomerization of BAX-BAK to initiate apoptosis. In addition, etoposide and UV decrease antiapoptotic BCL-2s in transformed MEFs, resulting in autoactivation of BAX-BAK even in the absence of BID, BIM, PUMA and NOXA (Supplementary Fig. 6).
DISCUSSION
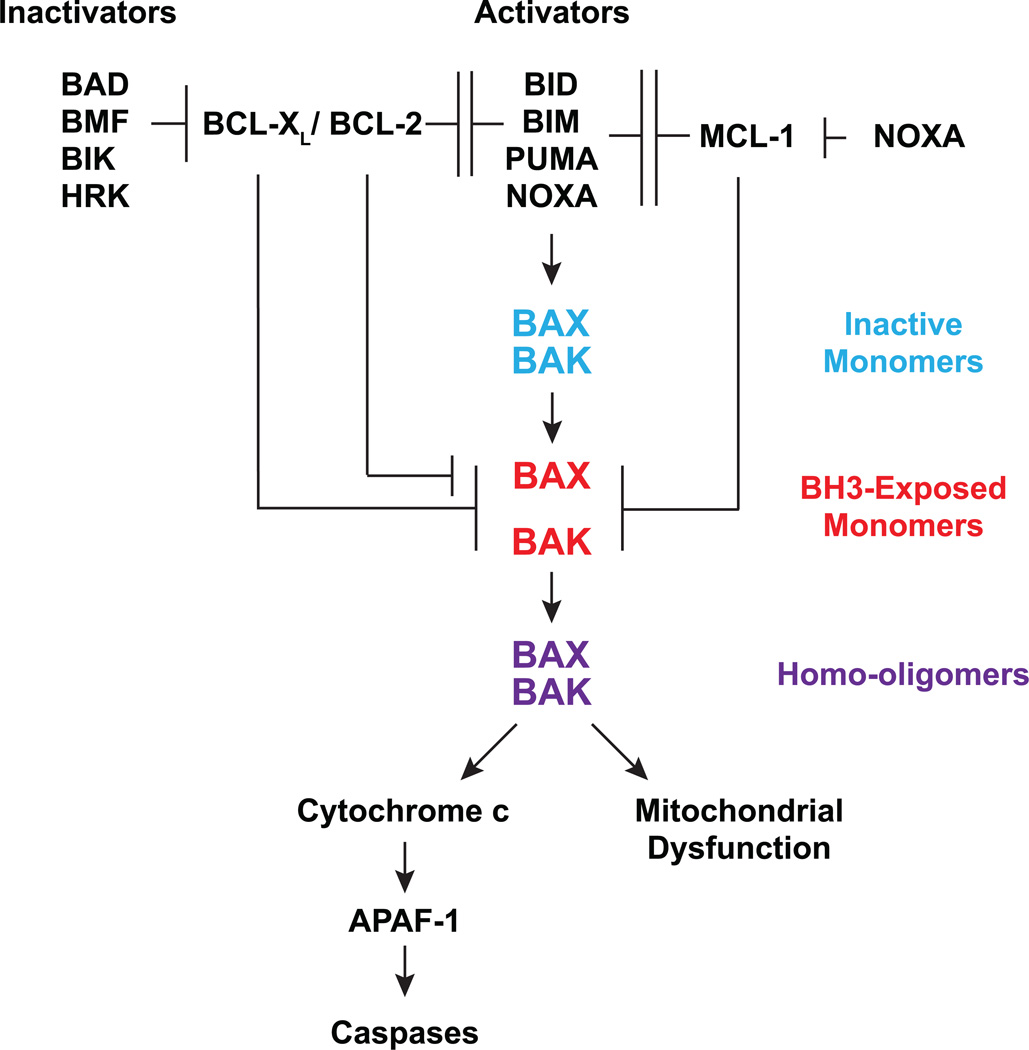
Our results support an interconnected hierarchical model in which the BCL-2 family proteins regulate mitochondrial apoptosis (Fig. 7). BID, BIM, PUMA and NOXA activate BAX-BAK directly to induce stepwise, bimodal activation of BAX-BAK. Activator BH3s convert inactive BAX-BAK monomers to “BH3-exposed” BAX-BAK monomers, initiating the activation of BAX-BAK. Subsequently, activator BH3s prevent antiapoptotic BCL-2s from sequestering “BH3-exposed” monomers of BAX-BAK, leading to homo-oligomerization of BAX-BAK. The antiapoptotic BCL-2s inhibit both modes of BAX-BAK activation. First, they sequester activator BH3s to prevent the initiation of BAX-BAK activation, providing frontline protection6, 9. Second, the antiapoptotic BCL-2s can also sequester “BH3-exposed” BAX-BAK monomers to prevent the homo-oligomerization of BAX-BAK, serving as a fail-safe mechanism or the second line of defense. The interaction between activator BH3s and antiapoptotic BCL-2s confers mutual inhibition because it not only prevents activator BH3s from activating BAX-BAK but also refrains the antiapoptotics from sequestering the “BH3-exposed” BAX-BAK monomers. Conceivably, BID, BIM and PUMA can prevent BCL-2, BCL-XL and MCL-1 from sequestering BAX-BAK whereas NOXA can only inhibit MCL-1. This difference may contribute to the lower death-inducing activity of NOXA in comparison with BID, BIM and PUMA (Fig. 5b). Consequently, Noxa deficiency only confers resistance to apoptosis in tissues or cell types that express sufficient NOXA for the activation of BAX-BAK. The remaining BH3s, including BAD, BMF, BIK and HRK, cannot activate BAX-BAK directly and appear to promote apoptosis by preventing BCL-2 and BCL-XL from sequestering activator BH3s. NOXA is unique among all BH3s in the sense that it can prevent MCL-1 from sequestering BID, BIM and PUMA due to its high binding affinity to MCL-1. The observation that deficiency of BID, BIM, PUMA and NOXA abrogates apoptosis triggered by overexpression of BAD, BMF, BIK or HRK (DP5) supports the BH3 hierarchy entailing upstream “inactivator” BH3s and downstream “activator” BH3s6.
Figure 7.
A schematic depicts the interconnected hierarchical model in which the BCL-2 family proteins regulate mitochondrion-dependent cell death.
The interaction between activator BH3s and multidomain BAX-BAK has been debated for decades due to low binding affinity27. Recent biophysical demonstration of BID, BIM or PUMA bound to BAX or BAK have helped revolve this controversy18–22. It is now recognized that the binding of activator BH3s to BAX-BAK has to be transient and dynamic in order to induce stepwise, bimodal activation of BAX-BAK14, 18, 19, 22, 24, 41. The interaction between activator BH3s and BAX-BAK should be “hit-and-run” because the same binding pocket of BAX-BAK is utilized for homo-oligomerization. Hence, the low binding affinity between activator BH3s and BAX-BAK renders a simple solution. In contrast, BH3s bind tightly to the antiapoptotic BCL-2s. By analogy, the antiapoptotic BCL-2s function like “decoy” death receptors that form inert stable complexes with BH3s.
Because BID, BIM and PUMA also bind and inactivate all the antiapoptotic BCL-2s, it was proposed that BAX-BAK can be autoactivated once they are released from the antiapoptotic BCL-2s by BID, BIM and PUMA (indirect activation model)27. The identification of NOXA as an activator of BAX-BAK argues against the indirect activation model because NOXA exhibits high binding affinity only to MCL-16, 11–13. Nonetheless, an unequivocal evidence of indirect activation of BAX-BAK is not revealed until our generation of Bid−/−Bim−/−Puma−/−Noxa−/− QKO cells. We demonstrated that autoactivation of BAX-BAK can occur in transformed QKO cells when antiapoptotic BCL-2s are simultaneously decreased upon DNA damage. However, autoactivation of BAX-BAK appears less efficient in part due to the fact that only a small fraction of BAX-BAK is readily bound to antiapoptotic BCL-2s whereby BH3 exposure is required for BAX-BAK to interact with antiapoptotic BCL-2s. Therefore, activator BH3s probably function as catalysts for BAX-BAK activation by inducing the BH3 exposure of BAX-BAK, and at the same time restrain antiapoptotic BCL-2s. The presence of heterodimers between antiapoptotic BCL-2s and BAX-BAK in QKO cells suggests that exposure of the BH3 domain in BAX-BAK can be generated independently of activator BH3s. Potential mechanisms include heat, changes in intracellular pH, oxidative stress, protein misfolding, or post-translational modifications42. It is possible that these heterodimers have important non-apoptotic functions in the maintenance of cellular homeostasis. Retrotranslocation of BAX from the mitochondria to the cytosol mediated by BCL-XL appears to offer a means to reduce the BAX and BCL-XL heterodimers43, which may not only gauge apoptosis but also fine-tune non-apoptotic activities.
Many studies have employed peptides derived from the BH3 domains of BH3s10–13, 41, 44. In principal, most BH3 peptides mimic full-length proteins in inhibiting antiapoptotic BCL-2s. However, not all of the BH3 peptides can recapitulate full-length proteins in activating BAX-BAK. For example, while full-length PUMA protein has potent cytochrome c releasing activity6, conflicting results have been reported with regard to the PUMA BH3 peptide12, 13, 21, 45, 46. Here, we show that full-length NOXA protein is capable of permeabilizing wild-type mitochondria and liposome in the presence of BAX. Similar to PUMA, inconsistent results have been obtained from NOXA BH3 peptide. NOXA BH3 peptide was shown to bind to BAK and induce BAK-dependent liposome permeabilization in one report but not the other19, 47. One plausible explanation is that different peptide preparations result in different alpha helicity. Alternatively, regions other than the BH3 domain may enhance the interaction between PUMA-NOXA and BAX-BAK directly or indirectly.
The identification of NOXA as a missing activator of BAX-BAK and the creation of Bid−/−Bim−/−Puma−/−Noxa−/− QKO cells provide an unprecedented opportunity in solving the puzzle of BCL-2 family signaling network. Our studies comparing cells deficient for upstream apoptotic initiators (activator BH3s) to those deficient for downstream apoptotic effectors (BAX and BAK) have helped resolve previously elusive, even contradictory, observations concerning the BCL-2 family proteins. Targeting the BCL-2 family to promote apoptosis holds great promise for cancer therapy48–53. With recent advances in the development of small molecule inhibitors of antiapoptotic BCL-2s and establishment of a comprehensive functional roadmap of BCL-2 family proteins, this goal is within reach.
METHODS
Generation of knockout mice
Bid−/−, Bim−/−, Puma−/−, Noxa−/−, Bax−/−, Bak−/−, and BaxF/F mice were described previously15, 25, 34, 54. All of the mice were on mixed 129Sv and C57Bl/6 background. To generate Bid−/−Bim−/−Puma−/− or Bid−/−Bim−/−Puma−/−Noxa−/− mice, Bid−/−Bim+/−Puma−/−Noxa+/− mice were crossed with Bid−/−Bim+/−Puma−/−Noxa+/− or Bid−/−Bim+/−Puma−/−Noxa−/− mice. In addition, intercrosses of Bid−/−Bim+/−Puma−/− mice or Bid−/−Bim+/−Puma−/−Noxa−/− mice were performed. To generate conditional Bax and Bak DKO mice, 4-hydroxytamoxifen (50 mg/kg) was administered by intraperitoneal injection into BaxF/−Bak−/−Rosa26CreERT2+/+ mice every other day for a total of 5 times. Deletion of Bax was confirmed by genotyping and an anti-BAX immunoblot. Animal experiments were performed in accordance to the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center.
Plasmid construction, retrovirus production, siRNA, and shRNA
The indicated Bcl-2 members were cloned into pSG5 (Stratagene) or pCDNA3 (Life Technologies) for in vitro transcription and translation reactions and the retroviral expression vector MSCV-IRES-GFP (pMIG) or MSCV-Puro (Clontech). BCL-XL mutants were described previously40. The NOXA mutants were generated by PCR-based site-directed mutagenesis. The BH3 mutant of NOXA contains L27E & L78E mutations. The K/R mutant of NOXA contains K5R, K29R, K33R, K53R, K56R, K72R, K84R, K90R and K97R mutations. All the constructs were confirmed by DNA sequencing. The production of retroviruses was described previously6. The following siRNA oligos were purchased from Ambion Silencer Select oligos (Applied Biosystems): Bax, 5’-GGAUGAUUGCUGACGUGGAtt-3’; Bak, 5’- AUAUCAUACUGCAUCAUUAtt-3’; cytochrome c, 5’- UGAUCUUCGCUGGAAUUAAtt-3’; Bcl-2, 5’- CAUUAUAAGCUGUCACAGAtt-3’; Bcl-xL, 5’- UCAGUUUAGUGAUGUCGAAtt-3’; Mcl-1, 5’-CCAAGAAAGCUUCAUCGAAtt-3’; and Hrk 5’-GGAUGAGAGGUUUCUAUUUtt-3’ and 5’-GGAGGAAGCUGGUUCCUGUtt-3’. The siRNA oligos against Caspase-9 were purchased from Dharmacon: 5’- UGGAUGCUGUGUCAAGUUU-3’. The scramble siRNA oligos were purchased from Applied Biosystems (Part number 4390844) and Dharmacon (D-001206-13-20). siRNA oligos were reverse-transfected using lipofectamine™ RNAiMAX (Life Technologies) to a final concentration of 10 nM. Retrovirus-mediated knockdown constructs were generated using pSuper-Retro-Puro or pSuperior-Retro-Puro according to the manufacture’s instruction (Oligoengine). The shRNA target sequence for Bad is GTCCTGGTGGGATCGAAAC. The shRNA target sequence for Bmf is GTCCTGGTGGGATCGAAAC. The shRNA target sequence for Bik is GATTCGAAGCCTCACCAAC. The shRNA target sequence for Apaf-1 is GAACGGTGAAGGTGTGGAA.
Cell culture, viability assay, Caspase 3/7 activity assay, and TUNEL assay
Mouse embryonic fibroblasts (MEFs) were generated from E13.5 embryos and cultured in IMEM supplemented with 20% fetal bovine serum (Life Technologies). All the MEFs utilized in the experiments were within the first 3 passages. Apoptosis was induced in primary MEFs by 1 µg/ml tunicamycin, 2 µM thapsigargin, or cultured in the absence of serum or glucose. To generate E1A/Ras-transformed MEFs, primary MEFs were infected with retrovirus expressing E1A-IRES-Ras as described previously55, followed by selection under 2 µg/ml of puromycin. Pools of puromycin-resistant cells were utilized for cell death assays. SV40 transformation was described previously9. Apoptosis was induced in E1A/Ras- or SV40-transformed MEFs by 10 µg/ml etoposide, 1 µg/ml tunicamycin, 2 µM thapsigargin, UV-C irradiation (250 or 500 J/m2), 20 Gy γ-irradiation, or cultured in the absence of serum, glucose, or glutamine. Cell death was quantified by annexin-V (BioVision) staining, followed by flow cytometric analyses. Flow cytometry was performed using either a FACSCalibur (BD Biosciences) or a LSRFortessa (BD Biosciences). Data were analyzed using CellQuest Pro (BD Biosciences) or FACSDiva (BD Biosciences). P values for statistical analyses were obtained using Student’s t test. The activity of Caspase 3/7 in cells was assessed using the Caspase-Glo 3/7 Assay System (Promega) according to the manufacturer’s instructions.
CD4+ T cells were isolated using anti-CD4 conjugated magnetic beads (Miltenyi). CD4+ T cells purified from the spleens of wild-type (two female and one male), Bid−/−Bim−/−Puma−/− TKO (two female and one male), or Bid−/−Bim−/−Puma−/−Noxa−/− QKO (three female) mice at 8 to 10 weeks of age were cultured in the absence of cytokine, in the presence of etoposide, in the presence of dexamethasone, or after exposure to 2.5 Gy γ-irradiation. Cell death was quantified by annexin-V staining at the indicated times. Wild-type (two female and one male), Bid−/−Bim−/−Puma−/− TKO (one female and two male), or Bid−/−Bim−/−Puma−/−Noxa−/− QKO (two female and one male) mice at 8 to 10 weeks of age were irradiated with 5 Gy total body irradiation. 3 days later, total numbers of splenocytes and CD4+ splenocytes were assessed. The percentage of survival was defined as the numbers of viable cells from irradiated mice divided by those from unirradiated ones with the same genotypes
Apoptosis in the small intestinal crypts of wild-type (three female), Bid−/−Bim−/−Puma−/− TKO (one female and two male), Bid−/−Bim−/−Puma−/−Noxa−/− QKO (one female and one male), or conditional Bax and Bak DKO (one female and one male) mice at 8 to 17 weeks of age at 4 h after 18 Gy whole body irradiation was assessed by TUNEL assays. The immunohistochemical TUNEL assay was performed at Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center. Slides were manually deparaffinized in xylene, re-hydrated in series of alcohol dilutions (100%, 95% and 70%) and tap water, placed in Discovery XT autostainer (Ventana Medical Systems), treated with Proteinase K (20 µg/ml in PBS) for 12 minutes and then incubated with TdT-biotin-dUTP labeling mix (Roche) for 1 hour. The detection was performed with DAB detection kit (Ventana Medical Systems) according to manufacturer instruction. Slides were counterstained with hematoxylin and coverslipped with Permount (Fisher Scientific). TUNEL positive cells were quantified in 300 small intestinal crypts in each mouse under light microscopy.
In vitro transcription and translation, cytochrome c release assay, protein crosslinking, and GST pull-down assay
In vitro transcription and translation (IVTT) reactions were performed with TNT T7 Quick Coupled Transcription/Translation Reticulocyte Lysate or Wheat Germ Extract Systems (L1170 and L4140, Promega) according to the manufacture’s instructions. For the Wheat Germ Extract System, SG5- and pCDNA3-based constructs were linearized with XbaI and NdeI, respectively. All the constructs generated full-length BCL-2 family proteins with expected molecular weights validated by autoradiography. Mitochondria purification and cytochrome c release assays were performed as described previously6. Quantification of cytochrome c release was performed using colorimetric ELISA assays (MCTC0, R&D Systems). To obtain the percentage of cytochrome c release, the amount of cytochrome c present in mitochondrial supernatant was divided by total cytochrome c present in both mitochondrial supernatant and pellet. Crosslinking of BAX or BAK was performed as described previously6. Briefly, 5 or 10 mM BMH (Pierce) in DMSO was added to mitochondria for 30min at room temperature. Approximately 2 µg of GST, GST-BAKΔC (aa 1–183), or GST-BAXΔC (aa 1–171) proteins were immobilized on glutathione-agarose beads (Sigma), followed by incubation with IVTT [35S]-methionine-labeled BH3-only proteins in buffer (0.2 % Triton X-100, 140 mM NaCl, 5 mM MgCl2, 50 mM Tris, pH 8.0). The bound complexes were analyzed by 10 % Nu-PAGE (Life Technologies) and autoradiography.
Antibodies, immunoblot analysis, immunoprecipitation, and limited trypsin proteolysis
Cells were lysed in RIPA buffer. Protein concentration was determined by BCA kit (Pierce). 25–50 µg of proteins were resolved by 10% NuPAGE (Life Technologies), transferred onto PVDF membrane (Immobilon-P, Millipore). Antibody detection was accomplished using enhanced chemiluminescence method (Western Lightning, PerkinElmer) and LAS-3000 Imaging system (FUJIFILM). Antibodies used for immunoblot analysis are listed as followed: anti-BAK (NT, Upstate), anti-BAK (G23, Santa Cruz Biotechnology), anti-BAX (N-20, Santa Cruz Biotechnology), anti-HA (12CA5), anti-BCL-XL (#2762, Cell Signaling Technology), anti-BCL-2 (3F11), anti-MCL-1 (#600-401-394, Rockland), anti-cytochrome c (#556433, Pharmingen), anti-LDH (#100-1173, Rockland), anti-VDAC1 (ab16814, Abcam), anti-cleaved Caspase-3 (#9661, Cell Signaling Technology), anti-PARP (#9542, Cell Signaling Technology), anti-BAD (C-20, Santa Cruz), anti-APAF-1 (AB16941, Upstate Biotechnology), anti-Caspase-9 (#9504, Cell Signaling Technology), and anti-actin (Chemicon). The dilution for all of the primary antibodies for immunoblot analysis was 1/1,000. For co-immunoprecipitation, cells were lysed in 0.2% NP40 isotonic buffer (0.2% NP40, 142.5 mM KCl, 5 mM MgCl2, 1 mM EGTA, 20 mM HEPES at pH 7.5) or 1 % CHAPS buffer (1 % CHAPS, 142.5 mM KCl, 2 mM CaCl2, 20 mM Tris-Cl, pH 7.4) supplemented with complete protease inhibitors (Roche). Cell lysates were immunoprecipitated with anti-HA antibody (12CA5) and analyzed by 10 % NuPAGE (Life Technologies) and immunoblots using the indicated antibodies. To assess BAK cleavage by limited trypsin proteolysis, cells (5×105) were resuspended in 100 µl of PBS containing 0.02% digitonin and 30 µg/ml trypsin on ice for 30 min. Following quenching of protease activity with 100 µg/ml soybean trypsin inhibitor (Sigma), samples were analyzed by anti-BAK (G23) immunoblots. All of the immunoblots shown are representative images from at least 2 independent experiments.
Reverse transcription and quantitative PCR
Total RNA was extracted from cells using TRIZOL (Life Technologies) according to the manufacturer’s instructions. Reverse transcription was performed with oligo-dT plus random decamer primers (Ambion) using Superscript II (Life Technologies). Quantitative PCR was performed with SYBR green master mix (Applied Biosystems) in duplicates using the indicated gene specific primers. Quantitative PCR was performed on a ViiA™ 7 Real-Time PCR System (Applied Biosystems). Data were analyzed as described previously by normalization against GAPDH or 18S rRNA55. The primers for quantitative PCR are listed below: Noxa, 5’- CCAGATTGGGGACCTTAGTCTCC-3’ and 5’-AGTTGAGCACACTCGTCCTTCAAG-3’; Bmf, 5’-CTTGTGGGGTGACAGAGGAACC-3’ and 5’-GGTCTCGGTTCTGCTGGTGT-3’; Bik, 5’-GAGAGACGTGGACCTCATGGAG-3’ and 5’-TGAGGCTTCGAATCAAGCTCCTG-3’; and Hrk, 5’-GTGCTCACAGGGCTTAGGAG-3’ and 5’-GTACTGCTGCAAGGAGAGGAG-3’.
Liposomal Release Assay
Liposomes were prepared, and release assays were performed as previously described32, 56. Liposomal composition reflects a mixture of the following molar percentages of lipids (Avanti Polar Lipids): phosphatidylcholine, 48%; phosphatidylethanolamine, 28%; phosphatidylinositol, 10%; dioleoyl phosphatidylserine, 10%; and tetraoleoyl cardiolipin, 4%. Aliquots of mixed lipids (1 mg total) are stored in glass at −20°C under nitrogen, and before use, resuspended in liposome assay buffer (10 mM HEPES, 200 mM KCl, and 1 mM MgCl2 [pH 7]) containing 12.5 mM of the fluorescent dye ANTS (8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt) and 45 mM of the quencher DPX (p-xylene-bis-pyridinium bromide). The resulting slurry is vortexed for 10 min and freeze-thawed five times in liquid nitrogen and a 40°C water bath, respectively. The solution is then passed through an Avanti Mini-Extruder Set (#610000) equipped with a 100 nm filter, followed by passage through a Sepharose column (GE Healthcare) to remove residual ANTS/DPX. The liposomes are brought up to a volume of 3 ml to produce a final liposome stock. For the liposomal release assay, recombinant BAX (400 nM) is combined with IVTT tBID, wild-type NOXA or BH3 mutant NOXA at the indicated protein: ligand ratios in a 96 well black flat-bottom plate (Costar) and then 10 µl of liposomes are added to a final volume of 100 µl in liposome assay buffer. Liposomal release is quantified based on the increase in fluorescence that occurs when the ANTS fluorophore is separated from the DPX quencher upon release from the liposomes into the supernatant. Fluorescence intensity (λex = 355 nm and λem = 520 nM) is measured over time at 32°C using a Tecan Infinite M1000 plate reader. Fluorescence measurements are recorded each minute until the release measurements plateau, at which point the liposomes are quenched with 0.2% Triton X-100 (100% release). Maximal ligand-induced BAX-mediated release was determined by treating liposomes with a mixture of tBID and BAX (400 nM). The percentage release of ANTS/DPX was calculated according to the equation ([F − F0] / [F100 − F0]) × 100, where F0 and F100 are baseline and maximal fluorescence, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We apologize to all the investigators whose research could not be appropriately cited owing to space limitation. We thank Hsiu-Fang Chen and Song Han for technical assistance. This work was supported by grants to E. Cheng from the NIH (R01CA125562) and the American Cancer Society (118518-RSG-10-030-01-CCG), and to E. Gavathiotis from the NIH (R01CA178394). This work was also supported by the NIH P30CA008748.
Footnotes
AUTHOR CONTRIBUTIONS
H.C.C. designed and conducted experiments, and analyzed data. E.H.C. designed research, analyzed data, and supervised the project. H.C.T., M.K, Y.H., H.K., A.Y., Y.T.G. and DER conducted experiments. J.J.H and E.G. supervised some experiments. H.C.T., D.R., P.C., S.T., and C.P.L. generated essential reagents.
REFERENCES
- 1.Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 2.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 3.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 4.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 5.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 6.Kim H, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 7.Westphal D, Kluck RM, Dewson G. Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 2014;21:196–205. doi: 10.1038/cdd.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei MC, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng EH, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 10.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 12.Kuwana T, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Certo M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren D, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 17.Czabotar PE, et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci U S A. 2007;104:6217–6222. doi: 10.1073/pnas.0701297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czabotar PE, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Moldoveanu T, et al. BID-induced structural changes in BAK promote apoptosis. Nature structural & molecular biology. 2013;20:589–597. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci U S A. 2013;110:E986–E995. doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards AL, et al. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chemistry & biology. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brouwer JM, et al. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol Cell. 2014;55:938–946. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Dewson G, et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell. 2008;30:369–380. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desagher S, et al. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis SN, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 28.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol. Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Developmental cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scorrano L, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 31.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 32.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Villunger A, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 34.Shibue T, et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 2003;17:2233–2238. doi: 10.1101/gad.1103603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naik E, Michalak EM, Villunger A, Adams JM, Strasser A. Ultraviolet radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J Cell Biol. 2007;176:415–424. doi: 10.1083/jcb.200608070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 37.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 38.Willis SN, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu HC, et al. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci U S A. 2009;106:1093–1098. doi: 10.1073/pnas.0808173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng EH, Levine B, Boise LH, Thompson CB, Hardwick JM. Bax-independent inhibition of apoptosis by Bcl-XL. Nature. 1996;379:554–556. doi: 10.1038/379554a0. [DOI] [PubMed] [Google Scholar]
- 41.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochimica et biophysica acta. 2011;1813:521–531. doi: 10.1016/j.bbamcr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 43.Edlich F, et al. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu NY, Sukumaran SK, Kerk SY, Yu VC. Baxbeta: a constitutively active human Bax isoform that is under tight regulatory control by the proteasomal degradation mechanism. Mol Cell. 2009;33:15–29. doi: 10.1016/j.molcel.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 46.Du H, et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 2011;286:491–501. doi: 10.1074/jbc.M110.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai H, et al. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol. 2011;194:39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 49.Tse C, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 50.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature medicine. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 51.Davids MS, Letai A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3127–3135. doi: 10.1200/JCO.2011.37.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walensky LD. From mitochondrial biology to magic bullet: navitoclax disarms BCL-2 in chronic lymphocytic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:554–557. doi: 10.1200/JCO.2011.37.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson MA, Huang D, Roberts A. Targeting BCL2 for the treatment of lymphoid malignancies. Seminars in hematology. 2014;51:219–227. doi: 10.1053/j.seminhematol.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Takeuchi O, et al. Essential role of BAX, BAK in B cell homeostasis and prevention of autoimmune disease. Proc Natl Acad Sci U S A. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda S, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–2409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.