SUMMARY
Everolimus, an inhibitor of the mammalian target of rapamycin (mTOR), is effective in treating tumors harboring alterations in the mTOR pathway. Mechanisms of resistance to everolimus remain undefined. Resistance developed in a patient with metastatic anaplastic thyroid carcinoma after an extraordinary 18-month response. Whole-exome sequencing of pretreatment and drug-resistant tumors revealed a nonsense mutation in TSC2, a negative regulator of mTOR, suggesting a mechanism for exquisite sensitivity to everolimus. The resistant tumor also harbored a mutation in MTOR that confers resistance to allosteric mTOR inhibition. The mutation remains sensitive to mTOR kinase inhibitors.
A better understanding of the mechanisms of sensitivity and resistance to anticancer therapies may improve patient selection and allow the development of rational treatment designs. One approach involves studying paired biopsy samples of pretreatment and drug-resistant tumors obtained from patients with exquisite sensitivity or unusually durable responses to therapy.
Everolimus is a Food and Drug Administration–approved oral allosteric inhibitor of mTOR. Tumors that exhibit a dependency on the mTOR pathway might have enhanced sensitivity to mTOR inhibition. Inactivating mutations in the tumor-suppressor genes TSC1, TSC2, and STK11 result in mTOR-pathway activation and are targetable by TOR inhibitors in hamartoma syndromes1–3 and in malignant perivascular epithelioid-cell tumors.4 In a phase 2 study of everolimus in urothelial carcinoma, whole-genome sequencing in a patient who had a durable complete remission revealed a somatic TSC1 mutation.5 We recently identified an additional mechanism of exquisite sensitivity to everolimus in a patient with metastatic urothelial carcinoma: activating mutations in mTOR itself.6 Although mechanisms of sensitivity to everolimus are beginning to be identified, mechanisms of clinically acquired resistance to everolimus remain unknown.
We identified a patient with metastatic anaplastic thyroid cancer, an aggressive neoplasm associated with a median survival of 5 months, who had exquisite sensitivity to everolimus. The patient, who was enrolled in a phase 2 study of everolimus for thyroid cancer (ClinicalTrials.gov number, NCT00936858), had a near-complete response that lasted for 18 months, followed by progressive disease. Of seven patients with anaplastic thyroid cancer treated with everolimus in this trial to date, this patient was the only one who had a response. To identify potential genomic mechanisms of exquisite sensitivity and acquired resistance to everolimus, we performed whole-exome sequencing on the pretreatment and drug-resistant tumors.
CASE REPORT
The patient is a 57-year-old woman who had noted a rapidly enlarging mass on the left side of her neck in 2010. She underwent a total thyroidectomy and central neck dissection, which revealed a 3.8-cm anaplastic thyroid cancer arising in a background of an oncocytic variant of poorly differentiated thyroid cancer (Fig. 1A, and Fig. S1 in Supplementary Appendix 1, available with the full text of this article at NEJM.org). Resection margins were positive, and 3 of 12 lymph nodes were involved. At 3 weeks after surgery, the serum thyroglobulin level was 17.2 ng per milliliter, with undetectable thyroglobulin antibodies.
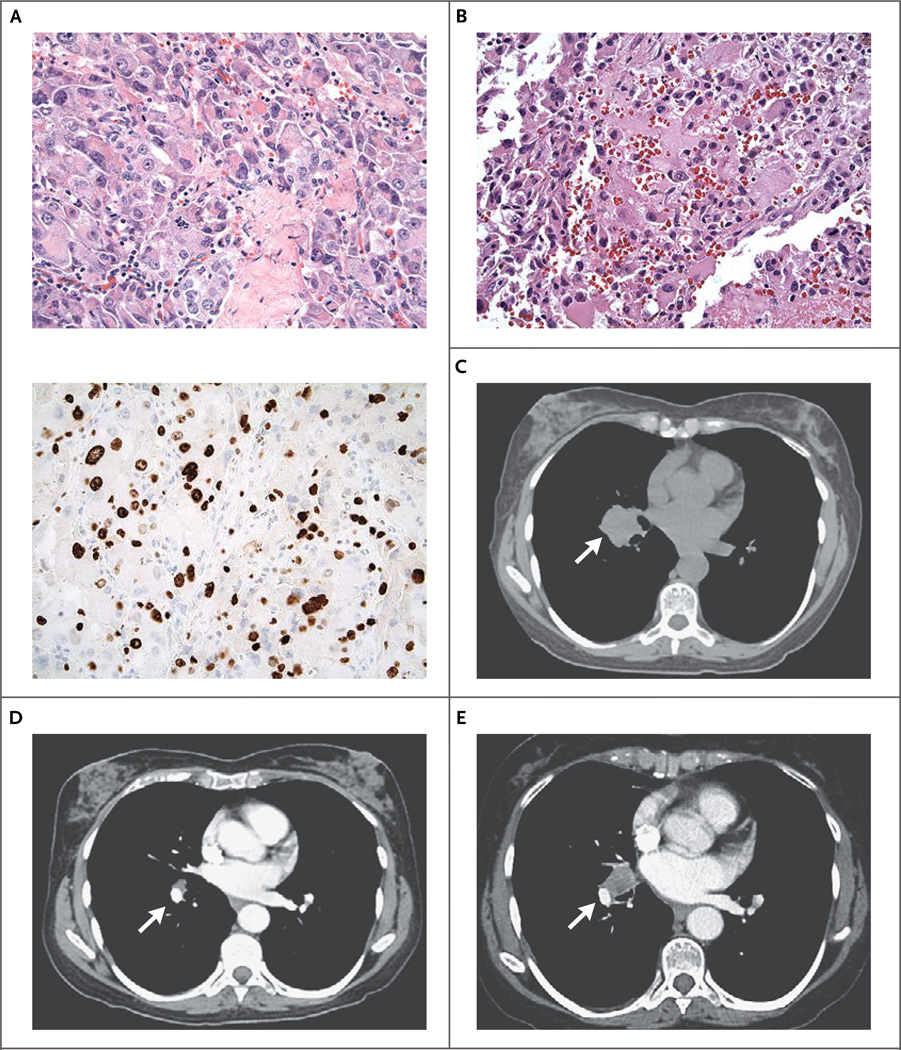
Figure 1. Histologic Findings and Computed Tomographic (CT) Scans in a Patient with Metastatic Anaplastic Thyroid Carcinoma.
Hematoxylin and eosin staining of a total-thyroidectomy specimen (Panel A, top) shows anaplastic thyroid carcinoma; MIB-1 staining (Panel A, bottom) reveals the high proliferative rate of the tumor. A histologic section (hematoxylin and eosin) of an enlarged mediastinal lymph node, obtained after 18 months of a response to everolimus monotherapy, shows recurrent thyroid carcinoma (Panel B). Axial CT scans of the chest show a right-sided hilar mass (arrow) before treatment with everolimus (Panel C), 6 months after treatment initiation (Panel D), and at the time of everolimus resistance, 18 months after treatment initiation (Panel E).
The patient received concurrent radiation therapy and weekly carboplatin and paclitaxel chemotherapy. The serum thyroglobulin level at 4 weeks after the completion of chemotherapy and radiation therapy was 12.0 ng per milliliter. Restaging scans obtained 3 months later revealed a new, right-sided hilar mass (Fig. 1C), and the patient enrolled in a phase 2 clinical trial of everolimus, which was administered at a dose of 10 mg daily. Within 6 months, follow-up scans showed that the lesion had greatly diminished in size (from 3.0 by 2.6 cm to 1.1 by 0.8 cm) (Fig. 1D).
After 18 months of a sustained response to everolimus, scans revealed progressive disease (Fig. 1E). The patient underwent a mediastinoscopy with removal of an enlarged lymph node, which contained metastatic anaplastic thyroid cancer (Fig. 1B, and Fig. S1 in Supplementary Appendix 1). Whole-exome sequencing was performed on biopsy samples of the pretreatment and resistant tumors as well as on a blood sample.
METHODS
OVERSIGHT
The study was approved by the institutional review board of the Dana–Farber/Harvard Cancer Center. The patient provided written informed consent for sequencing.
HISTOLOGIC STUDIES
Tumor sections were deparaffinized and stained with antibodies against pS6 (phosphorylated ribosomal protein S6) and TSC2 with the use of standard protocols. For details, see the Methods section in Supplementary Appendix 1.
WHOLE-EXOME SEQUENCING
Whole-exome sequencing was performed on the pretreatment tumor, the resistant tumor, and blood. The mean depth of coverage for the pretreatment and resistant tumors was 300× and 375×, respectively. Somatic point mutations, small insertions or deletions (indels), and copy-number alterations, detected in the tumor DNA but not the germline DNA, were identified (Table S1 in Supplementary Appendix 2 [described in Supplementary Appendix 1]). Evaluation for rearrangements was not performed.
LABORATORY STUDIES
Expression plasmids containing mTOR complementary DNA (cDNA) were generated, and site-directed mutagenesis, stable transfections, analysis of cell-growth inhibition, and immunoblot studies were performed. For details, see the Methods section in Supplementary Appendix 1.
RESULTS
MUTATIONS IN TSC2 AND MTOR
The pretreatment tumor contained a somatic nonsense mutation in the tumor-suppressor gene TSC2 that inactivates the gene, allowing for activation of the mTOR pathway7,8 and resulting in sensitivity to mTOR inhibition in some cancers1,2,4,9 (Fig. 2A). This truncating mutation (Q1178⋆) is known to inactivate TSC2 by eliminating the guanosine triphosphatase–activating protein domain near the C-terminal, a domain essential for inhibiting mTOR complex 1.8,10 Somatic mutations in TSC2 have been identified in many tumor types, including cancers of the kidney and bladder as well as hamartomas and malignant perivascular epithelioid-cell tumors in patients with tuberous sclerosis complex. TSC2 mutations have not previously been reported in any type of thyroid cancer. TSC2 Q1178⋆ has been identified twice in the germline of patients with tuberous sclerosis complex,11 but somatic mutations at this locus have not been reported.
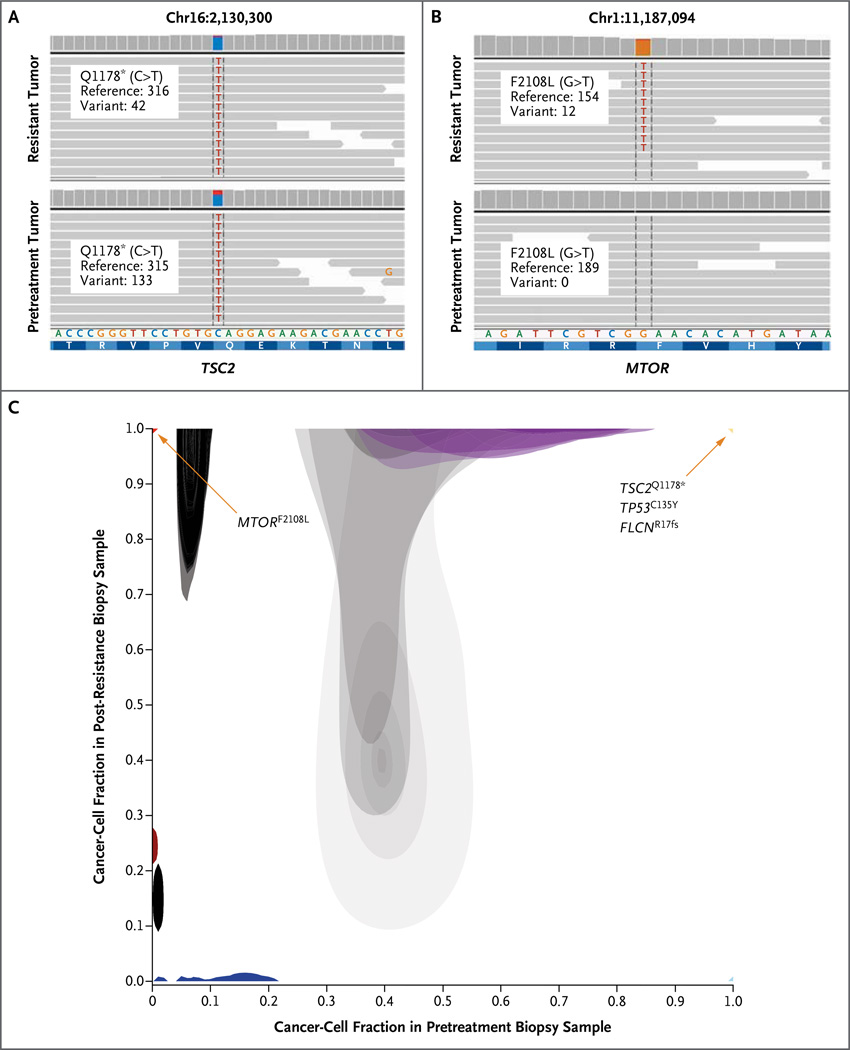
Figure 2. Mutations in TSC2 and MTOR in Anaplastic Thyroid Carcinoma Revealed by Whole-Exome Sequencing.
Whole-exome sequencing of the tumor tissue before treatment and after the development of resistance to everolimus revealed a TSC2 nonsense mutation in both tumors (Panel A) and an F2108L mutation in MTOR in the resistant tumor that was undetectable in the pretreatment tumor (Panel B). Representative genome images from the Integrative Genomics Viewer (Broad Institute), along with the number of reads for the reference allele and the variant allele, are shown for each alteration. A comparison of the proportion of cancer cells harboring specific mutations in the pretreatment biopsy sample (Panel C, x axis) versus those in the post-resistance biopsy sample (y axis) is shown. Mutations in TSC2, TP53, and FLCN were present in all cancer cells from both biopsy samples (yellow triangle at upper right), whereas the MTOR mutation was detected in all cancer cells in the post-resistance biopsy sample but was not detected in the pretreatment biopsy sample (red triangle at upper left). Additional mutations were detected in all cancer cells in the pretreatment biopsy sample but in no cancer cells in the post-resistance biopsy sample (light blue triangle at lower right). Other mutations were predicted to occur in a subgroup of the cancer cells in the pretreatment biopsy but in none of the cancer cells in the post-resistance biopsy sample (dark blue) or in a subgroup of the cancer cells in the post-resistance biopsy sample but in none of the cancer cells in the pretreatment biopsy sample (dark red). Shaded areas denote Bayesian posterior probability distributions over cancer-cell fraction values for each mutation. Gray shading indicates mutations with cancer-cell fraction distributions having considerable uncertainty, with lighter shading indicating greater uncertainty.
In addition to the TSC2 mutation, 317 somatic coding single-nucleotide variants and 44 coding indels were identified (Table S1 in Supplementary Appendix 2). These included a common mutation in TP53 (C135Y) and an N-terminal frame shift in FLCN (R17fs), a tumor-suppressor gene that encodes folliculin. Germline mutations in folliculin result in the Birt–Hogg–Dubé syndrome, an autosomal dominant disorder characterized by fibrofolliculomas, renal and pulmonary cysts, and renal cancer. Folliculin is involved in TSC2 and mTOR signaling12–15; in some studies, inactivation of FLCN has been shown to result in increased mTOR activity,14 raising the possibility that this mutation, along with the TSC2 mutation, may contribute to sensitivity to everolimus.
Genomewide copy-number analysis showed near-haploidization of the cancer genome with retention of chromosome 7, a feature that appears to be pathognomonic of oncocytic follicular thyroid carcinoma,16 This finding is consistent with the putative origin of this anaplastic thyroid cancer (Fig. S2 in Supplementary Appendix 1).
The TSC2 nonsense mutation persisted in the resistant tumor (Fig. 2A). Immunohistochemical analysis of tissue from the resistant tumor revealed the absence of TSC2 and the presence of pS6, a downstream target of mTOR. These findings are indicative of persistent mTOR-pathway activity (Fig. S3 in Supplementary Appendix 1). Immunohistochemical analysis of the pretreatment tissue could not be completed for technical reasons. Mutations in TP53 and FLCN also persisted in the resistant tumor.
The resistant tumor also had a somatic mutation in MTOR (MTORF2108L). This mutation was not detected in the pretreatment tumor despite robust sequence coverage of this locus (189×) (Fig. 2B). To identify the relative change in the frequency of each genomic alteration from the pretreatment tumor to the resistant tumor, the fraction of tumor cells harboring a given alteration in each pair of samples was estimated (Fig. 2C, and Table S1 in Supplementary Appendix 2). Although the TSC2, TP53, and FLCN alterations were estimated to be present in 98 to 100% of the cancer cells in both the pretreatment and resistant tumors, the estimated proportion of cancer cells with MTORF2108L was 0% in the pretreatment tumor as compared with 96% in the resistant tumor (Fig. 2C). To our knowledge, MTOR mutations have not previously been identified in patients with acquired resistance to everolimus, and this particular mutation has not been described in patients.17
RESISTANCE TO ALLOSTERIC mTOR INHIBITORS RESULTING FROM mTORF2108L
Although MTORF2108L has not been described previously, the homologous mutation was characterized nearly 20 years ago in fission yeast.18 In a mutagenesis screen to identify mutations in tor2 (the fission yeast homologue of MTOR) that conferred resistance to rapamycin, tor2F2049L was one of five rapamycin-resistant mutants identified. All five tor2 residues identified have been conserved in TOR proteins across species, including mTOR.18 In a yeast two-hybrid assay, tor2F2049L did not bind FKBP (FK506 binding protein)–rapamycin, suggesting a mechanism for rapamycin resistance in fission yeast.18
On the basis of these findings, we hypothesized that mTORF2108L causes resistance to allosteric mTOR inhibition by preventing the binding of the drug to the protein. Indeed, structural studies confirm that mTORF2108L occurs in the FKBP–rapamycin binding (FRB) domain, the region of the protein that is required to bind to rapamycin and its analogues. As shown in Figures 3A and 3B, the substitution of a leucine for a phenylalanine is predicted to prevent binding of the drug to the protein by means of steric hindrance.
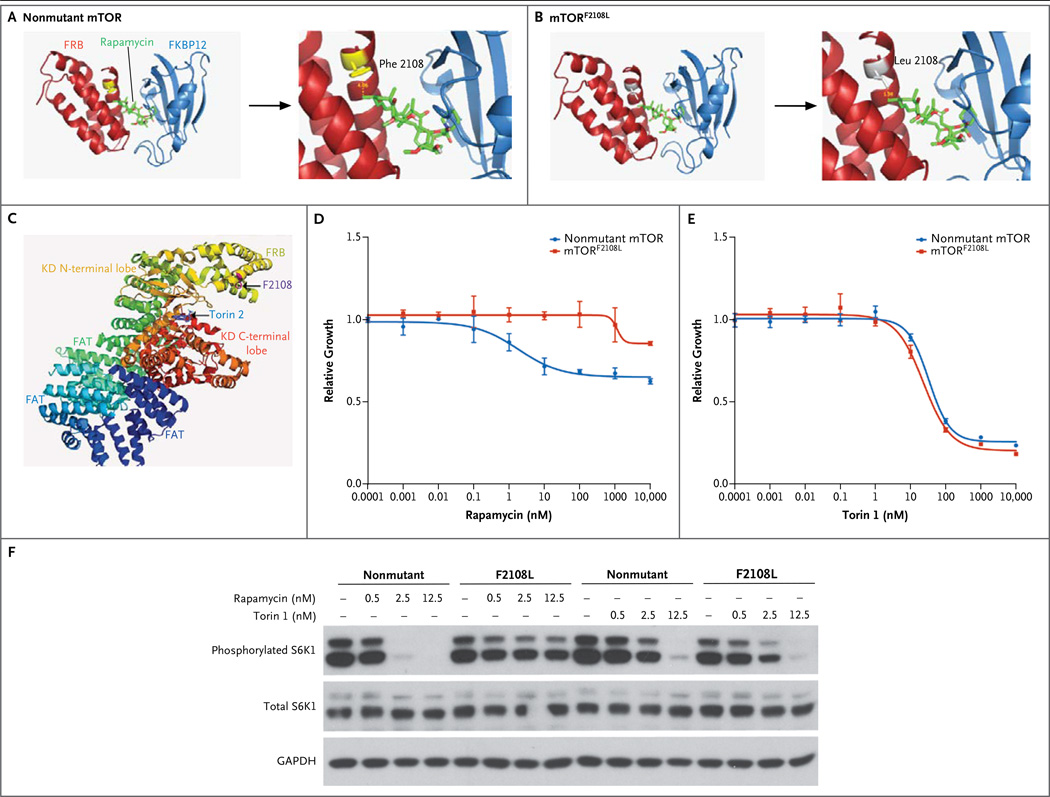
Figure 3. Effects of Nonmutant mTOR and mTORF2108L.
Ribbon rendition of mTOR crystal structure (PDB 1FAP) for nonmutant mTOR (Panel A) and mTORF2108L (Panel B) shows the FKBP (FK506 binding protein)–rapamycin binding (FRB) domain, FKBP12, and rapamycin. Enlarged views show that the substitution of leucine for phenylalanine sterically hinders rapamycin binding (Panels A and B, right side). Ribbon rendition of mTOR crystal structure (PDB 4JSX) (Panel C) highlights the kinase domain (KD) C-terminal lobe, KD N-terminal lobe, FRB domain, and FAT domain. As shown, residue F2108 is not expected to be involved with binding of torin 2. Growth-inhibition curves are shown for rapamycin (Panel D) and torin 1 (Panel E) in human embryonic kidney (HEK) 293T cells stably expressing nonmutant mTOR or MTORF2108L. Constructs expressing mTORF2108L or nonmutant mTOR were expressed in HEK 293T cells (Panel F). The levels of phosphorylated and total S6 kinase 1 (S6K1), the downstream target of mTOR, are shown for the HEK 293T cells after treatment with 0 nM, 0.5 nM, 2.5 nM, or 12.5 nM of rapamycin or torin 1, as indicated. GAPDH denotes glyceraldehyde 3-phosphate dehydrogenase.
To confirm that mTORF2108L confers resistance to allosteric mTOR inhibition, the F2108L mutation was introduced into nonmutant mTOR, and the mutant cDNA was stably expressed in human embryonic kidney (HEK) 293T cells. Cells expressing mTORF2108L were significantly more resistant to inhibition with rapamycin than were cells expressing nonmutant mTOR (Fig. 3D). We next examined the effect of the mutant on the phosphorylation of endogenous S6 kinase 1 (S6K1), a downstream target of mTOR. At baseline, cells expressing mTORF2108L and those expressing nonmutant mTOR had similar levels of phosphorylated S6K1. Treatment with rapamycin, however, completely inhibited phosphorylation of S6K1 in cells expressing nonmutant mTOR but had virtually no effect in cells expressing mTORF2108L (Fig. 3F).
SENSITIVITY OF mTORF2108L TO DIRECT TOR KINASE INHIBITION
Because mTORF2108L occurs in the FRB domain rather than the active site, we hypothesized that this mutant protein should remain sensitive to inhibition by direct ATP-competitive TOR kinase inhibitors. As shown in the structural model in Figure 3C, there is no predicted effect of the F2108L mutation on binding of torin 1, a direct TOR inhibitor. In contrast to cells treated with rapamycin, cells expressing mTORF2108L and those expressing nonmutant mTOR were equally sensitive to treatment with torin 1 (Fig. 3E). Similarly, inhibition of S6K1 phosphorylation by torin 1 was equivalent in cells expressing mTORF2108L and those expressing nonmutant mTOR (Fig. 3F). Taken together, these results indicate that mTORF2108L remains sensitive to direct kinase inhibition.
DISCUSSION
This report of a nonsense mutation in TSC2 in a tumor from a patient with anaplastic thyroid cancer who had a response to everolimus supports the notion that cancers of diverse types with mTOR-pathway–activating mutations are sensitive to mTOR inhibitors.1–6 Once resistance emerged in this patient, the mechanism of acquired resistance to everolimus was identified as a mutation in mTOR that prevented everolimus from binding to mTOR. The mutant mTOR remained sensitive to direct mTOR kinase inhibition.
Anaplastic thyroid cancer is a highly aggressive and rapidly fatal disease, for which there is no adequate treatment. To date, limited molecular profiling studies in this disease have identified a few targetable alterations. Oncogenic BRAF mutations have been described, and a response to the RAF inhibitor vemurafenib was observed in a patient with BRAF-mutant anaplastic thyroid cancer.19 Additional targetable alterations include mutations in PIK3CA, PTEN, KRAS, NRAS, and ALK.20,21 Genomic profiling of anaplastic thyroid cancer to screen for mTOR-pathway alterations may identify subgroups of patients who are potential candidates for enrollment in clinical trials of mTOR-directed therapies. Indeed, several activating alterations in the mTOR pathway — including alterations in MTOR, TSC1 or TSC2, STK11, and RHEB — have been observed recurrently in multiple cancer types,5,6,17 providing the rationale for the development of so-called basket trials of mTOR inhibitors in patients with diverse tumor types who have somatic mTOR-pathway alterations.
Although cancers driven by a dominant oncogene frequently have dramatic responses to targeted kinase inhibitors, the tumors invariably become resistant to these agents. The majority of known mechanisms of clinical resistance involve secondary mutations in the target kinase. Such mutations has been described for ABL, KIT, EGFR, ALK, BRAF, MEK, PDGFRA, FLT3, and ROS1.22–24 Indeed, in the case of every kinase inhibitor for which a resistance mechanism has been described, there is clinical evidence that resistance can occur through a secondary alteration in the target kinase. Our results indicate that acquired resistance to mTOR inhibition can occur through the same mechanism. It is likely that additional secondary mutations in the mTOR FRB domain will be identified in patients, as suggested by studies in fission yeast18 and experience with other tumor types.23,24 We speculate that these patients would be ideal candidates for clinical trials of mTOR kinase inhibitors.
Serial biopsies to profile tumors that develop resistance to targeted therapies have the potential not only to uncover mechanisms of therapeutic resistance but also to suggest effective follow-up treatment. Ultimately, a comprehensive knowledge of mechanisms of acquired resistance, coupled with the ability to diagnose the relevant mechanisms in situ, may lead to the development of therapeutic strategies, including targeted combinations, that are capable of producing longterm responses in many cancers.
Supplementary Material
Acknowledgments
Supported by grants from the Next Generation Fund at the Broad Institute of Massachusetts Institute of Technology and Harvard (to Dr. Wagle), Novartis Pharmaceuticals (to Dr. Lorch), the Starr Cancer Consortium (to Dr. Garraway), and the National Cancer Institute (to Drs. Kwiatkowski and Garraway).
We thank Tyler Haddad for help in gathering clinical data, Vicky Vergara for assistance with tumor specimens, and Sheila Fisher and Stacey Gabriel from the Broad Institute Genomics Platform for sequencing studies.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Bissler JJ, McCormack FX, Young LR, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies DM, de Vries PJ, Johnson SR, et al. Sirolimus therapy for angiomyolipoma in tuberous sclerosis and sporadic lymphangioleiomyomatosis: a phase 2 trial. Clin Cancer Res. 2011;17:4071–4081. doi: 10.1158/1078-0432.CCR-11-0445. [DOI] [PubMed] [Google Scholar]
- 3.Klümpen H-J, Queiroz KCS, Spek CA, et al. mTOR inhibitor treatment of pancreatic cancer in a patient with Peutz-Jeghers syndrome. J Clin Oncol. 2011;29(6):e150–e153. doi: 10.1200/JCO.2010.32.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner AJ, Malinowska-Kolodziej I, Morgan JA, et al. Clinical activity of mTOR inhibition with sirolimus in malignant perivascular epithelioid cell tumors: targeting the pathogenic activation of mTORC1 in tumors. J Clin Oncol. 2010;28:835–840. doi: 10.1200/JCO.2009.25.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagle N, Grabiner BC, Van Allen EM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–1356. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 9.Goncharova EA, Goncharov DA, Li H, et al. mTORC2 is required for proliferation and survival of TSC2-null cells. Mol Cell Biol. 2011;31:2484–2498. doi: 10.1128/MCB.01061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoogeveen-Westerveld M, Ekong R, Povey S, et al. Functional assessment of TSC2 variants identified in individuals with tuberous sclerosis complex. Hum Mutat. 2013;34:167–175. doi: 10.1002/humu.22202. [DOI] [PubMed] [Google Scholar]
- 11.Niida Y, Lawrence-Smith N, Banwell A, et al. Analysis of both TSC1 and TSC2 for germline mutations in 126 unrelated patients with tuberous sclerosis. Hum Mutat. 1999;14:412–422. doi: 10.1002/(SICI)1098-1004(199911)14:5<412::AID-HUMU7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Hartman TR, Nicolas E, Klein-Szanto A, et al. The role of the Birt-Hogg-Dubé protein in mTOR activation and renal tumorigenesis. Oncogene. 2009;28:1594–1604. doi: 10.1038/onc.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsun ZY, Bar-Peled L, Chantranupong L, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasumi Y, Baba M, Ajima R, et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci U S A. 2009;106:18722–18727. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba M, Hong SB, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corver WE, Ruano D, Weijers K, et al. Genome haploidisation with chromosome 7 retention in oncocytic follicular thyroid carcinoma. PLoS One. 2012;7(6):e38287. doi: 10.1371/journal.pone.0038287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabiner BC, Nardi V, Birsoy K, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz MC, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 19.Rosove MH, Peddi PF, Glaspy JA. BRAF V600E inhibition in anaplastic thyroid cancer. N Engl J Med. 2013;368:684–685. doi: 10.1056/NEJMc1215697. [DOI] [PubMed] [Google Scholar]
- 20.Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE. Targeted nextgeneration sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1852–E1860. doi: 10.1210/jc.2013-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murugan AK, Xing M. Anaplastic thyroid cancers harbor novel oncogenic mutations of the ALK gene. Cancer Res. 2011;71:4403–4411. doi: 10.1158/0008-5472.CAN-10-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Awad MM, Engelman JA, Shaw AT. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med. 2013;369:1173. doi: 10.1056/NEJMc1309091. [DOI] [PubMed] [Google Scholar]
- 23.Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 24.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.