Abstract
In response to genotoxic attacks, cells activate sophisticated DNA repair pathways such as nucleotide excision repair (NER), which consists of damage removal via dual incision and DNA resynthesis. Using permanganate footprinting as well as highly purified factors, we show that NER is a dynamic process that takes place in a number of successive steps during which the DNA is remodeled around the lesion in response to the various NER factors. XPC/HR23B first recognizes the damaged structure and initiates the opening of the helix from position −3 to +6. TFIIH is then recruited and, in the presence of ATP, extends the opening from position −6 to +6; it also displaces XPC downstream from the lesion, thereby providing the topological structure for recruiting XPA and RPA, which will enlarge the opening. Once targeted by XPG, the damaged DNA is further melted from position −19 to +8. XPG and XPF/ERCC1 endo-nucleases then cut the damaged DNA at the limit of the opened structure that was previously “labeled” by the positioning of XPC/HR23B and TFIIH.
To counteract the detrimental effect of genotoxic attacks, cells activate sophisticated and specific DNA repair pathways. Damage induced by UV radiation, environmental agents, and anticancer drugs are removed by two distinct nucleotide excision repair (NER)1 subpathways, namely global genome repair (GGR), which eliminates lesions from the entire genome, and transcription-coupled repair (TCR), a specialized pathway that repairs damages on a transcribed strand of active genes (1–3). Human NER involves the ordered action of factors in dual incision and DNA repair resynthesis steps (4). Any mutation that affects either the enzymatic activity or the ordered assembly of the dual incision complex leads to genetic disorders such as xeroderma pigmentosum, trichothiodystrophy, or Cockayne syndrome (5, 6).
In global genome repair, the dual incision is a multistep process that results from the coordinated action of XPC/HR23B, TFIIH, XPA, RPA, XPG, and XPF/ERCC1, resulting in the removal of the damaged oligonucleotide (4, 7, 8). After being recognized by the XPC/HR23B complex, the damaged DNA structure is targeted by TFIIH, which recruits the other factors upon the addition of ATP (9–11). The unwound DNA is then incised by the two endonucleases XPG and XPF/ERCC1 on the 3′ and 5′ side of the lesion, respectively (12–15), leaving a gap structure that is filled up by the DNA polymerase ε or δ and the accompanying factors PCNA, RF-C, RPA, and DNA ligase I (16). Whether or not the NER reaction occurs by sequential arrival of the various factors or by a pre-assembled complex referred to as the repairosome or the holoenzyme is still under debate (17–19). Although the hypothesis of the sequential assembly, which has gained a lot of support from recent biological studies, seems to be more accepted, the order of assembly of the NER factors on the damaged DNA and their contribution to the DNA remodeling to allow the repair are not fully understood (10, 20, 21). As an example, to further learn about the role of TFIIH and its XPB and XPD helicases in NER, it is necessary to determine how it associates with the damaged DNA and recruits the additional factors such as XPA and RPA to promote the formation of an open intermediate essential for the dual incision by XPG and XPF endonucleases. In the present study, we have focused our attention on the damaged DNA itself, trying to understand how the various NER factors target it and remodel it to finally allow its opening and dual incision. Thus, we have set up a permanganate footprinting assay for analyzing the formation of single-stranded DNA regions induced by the NER factors and, furthermore, a site-specific protein-DNA photo cross-linking assay for defining the location of some NER factors around the lesion.
Using purified factors, we were able to precisely determine the role of each factor in changing the DNA conformation at the various stages of the NER reaction, including damaged DNA recognition, ATP-dependent DNA unwinding, stabilization of the open structure, and oligonucleotide incision.
EXPERIMENTAL PROCEDURES
Cloning and Vector Constructions
The XPA, ERCC1, XPF, HR23B, and XPC cDNAs were amplified from a human cDNA library and inserted in the pVL1392 vector (Pharmingen). A His6 tag was added N-terminally to XPA and XPF. The resulting vectors were recombined with baculovirus DNA (BaculoGold DNA, Pharmingen) in Sf9 cells, and viral stocks were prepared by a three-step growth amplification procedure. The human RPA and XPG cDNAs were cloned into expressing vectors (22, 23).
Purification of the Repair Factors
XPF/ERCC1 complex purification was performed as described previously (14). Briefly, Sf9 cells were infected with ERCC1 and XPF-His-tagged baculoviruses. Cells extracts were prepared in buffer A (20 mM Tris-HCl, pH 7.9, 0.15 M NaCl, 20% glycerol, 0.1% Nonidet P-40, 5 mM MgCl2, 5 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, and 1× protease mixture inhibitor) and centrifuged (15,000 rpm for 40 min at 4 °C). The supernatant was dialyzed for 4 h against buffer B (same as buffer A except for 5% glycerol, 0.02% Nonidet P-40, and 0.5 mM dithiothreitol) and applied to a phosphocellulose column (P11; Whatman), and the flow-through was then applied to a heparin Ultrogel column (Sepracor, Villeneuve la Garenne, France). Following extensive washes, the protein complexes were eluted at 0.6 M NaCl and further incubated for 4 h with 500 μl of the metal affinity resin (Talon; Clontech) in the presence of imidazole (1 mM). After sequential washing with buffer B containing 0.6 M NaCl and 1 mM imidazole, 5 mM imidazole, and, finally, 0.1 M NaCl and 20 mM imidazole, the complex was eluted in buffer B containing 0.1 M NaCl and 0.2 M imidazole. Mutants XPF-D676A and XPF-D720A were purified according to published procedures (15). The XPC/HR23B complex or its individual components (XPC and HR23B) were expressed and purified from insect cells (24). The recombinant heterotrimeric RPA complex was produced in Escherichia coli and purified (22). Recombinant XPG was produced in insect cells and purified as described (23). All of the NER factors were dialyzed against buffer C (50 mM Tris-HCl, pH 7.8, 10% glycerol, 0.1 mM EDTA, 0.5 mM dithiothreitol, and 50 mM KCl).
XPA insect cell extract was applied on a heparin Ultrogel resin (Sepracor) pre-equilibrated in buffer A and washed with buffer A containing 0.3 M NaCl. XPA was then eluted in buffer A containing 0.5 M NaCl, dialyzed for 3 h against buffer D (50 mM Tris-HCl pH 7.8, 10% glycerol, and 0.3 M KCl) and incubated for 2 h at 4 °C with 200 μl of metal affinity resin (Talon; Clontech). After packing the column, the resin was subsequently washed with buffer D containing 20 mM imidazole and 50 mM imidazole. XPA was eluted in buffer D containing 0.1 M imidazole and next with buffer D containing 0.1 M EDTA. The fractions were pooled and dialyzed against buffer C. TFIIH was purified from HeLa cells (25).
Dual Incision Assay
The single cisplatin lesion (Pt-GTG) plasmid, called 105.TS (26), was used for dual incision (7) in a 15-μl reaction mixture containing XPC/HR23B (10 ng), XPA (25 ng), RPA (50 ng), XPG (7.5 ng), XPF/ERCC1 (6.25 ng), and TFIIH (20 ng) in the presence of 2 mM ATP. Following 10 min of pre-incubation at 30 °C, 30 ng of the Pt-GTG plasmid was added, and the reactions were further incubated for 90 min. The excised fragment was detected on 14% urea-PAGE after annealing with the complementary oligonucleotide and the addition of four radiolabeled [α-32 P]dCMP residues by Sequenase V2.1 (USB) (26).
KMnO4 Footprinting Assays
The damaged strand probe was obtained upon AgeI digestion of the 105.TS plasmid (26) and radiolabeling at the 3′end in a Klenow reaction, the Pt adduct being located at 156 bp from the labeled end. For the non-damaged strand, 105.TS was digested by BssHII, labeled, and then digested by HindIII. The resulting 165-bp fragment in which the lesion is located 92 bp from the labeled 3′ end was purified by the “crush and soak “method after migration in a 5% nondenaturating PAGE (27).
Reactions (75 μl) were carried out in 20 mM Hepes/KOH, pH 7.6, 60 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 0.3 mM EGTA, 0.4% polyvinyl alcohol, and 0.4% polyethylene glycol 10000 buffer containing the labeled cisplatinated probe (40 fmol) and, when indicated, 5 mM ATP, XPC/HR23B (40 ng), XPA (80 ng), RPA (60 ng), XPG (10 ng), XPF/ERCC1 (6.25 ng), and TFIIH (40 ng). After incubation at 30 °C for 15 min, 3 μl of 120 mM KMnO4 was added, and oxidation was allowed to proceed for 3 min at room temperature before reduction by adding 6 μl of 14.6 M β-mercaptoethanol for 5 min in ice. After organic extraction and ethanol precipitation, dried pellets were resuspended in 100 μl of a solution containing 1 M piperidine, 1 mM EDTA, and 1 mM EGTA and incubated at 90 °C for 25 min. Samples were next ethanol precipitated, and final pellets were recovered in 10 μl of loading buffer and analyzed in 8% urea PAGE (damaged strand probe) or in 12% urea PAGE (undamaged strand probe). In parallel, dideoxy sequencing reactions were performed.
Protein-DNA Photo Cross-linking
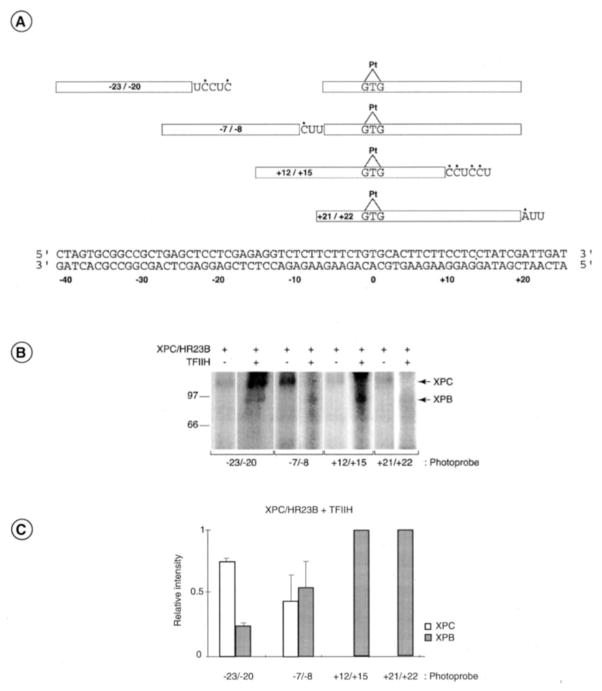
The synthesis of the photoreactive nucleotide AB-dUTP, complex assembly, and analysis of the photo cross-linked polypeptides were done as described previously (28, 29). For the synthesis of the photoprobes that place two AB-dUMPs on the 3′ side of the cisplatin (photoprobes +12/+15 and +21/+22), we used cisplatinated oligonucleotides of various lengths designed to allow incorporation at the appropriate positions (details are provided the legend to Fig. 6A). For the synthesis of the photoprobes that place two AB-dUMPs on the 5′ side of the cisplatin (photoprobes −23/−20 and −8/−7), we used both a cisplatinated oligonucleotide and an additional 5′ oligonucleotide for directing the incorporation of AB-dUMP. Fill-in with DNA polymerase and ligation were done as described previously (28, 29). After gel purification of the photoprobes, complexes were assembled with either XPC/HR23B alone or XPC/HR23B and TFIIH in the presence of ATP (1 mM).
Fig. 6. Photo Cross-linking of XPC and XPB to specific positions along the damaged DNA.
A, four cisplatinated photoprobes (−23/−20, −8/−7, +12/+15, and +21/+22) that place two photoreactive nucleotides AB-dUMP (U) in juxtaposition to radiolabeled nucleotides (*) at specific locations along the damaged DNA (Pt, position 0) were synthesized and used in the photo cross-linking experiments. B, XPC/HR23B alone or XPC/HR23B and TFIIH in the presence of 1 mM ATP were incubated with each photoprobe, and the complexes were UV irradiated and processed to determine the polypeptides that cross-link to the various photoprobes. The position of XPC, XPB, and the molecular weight markers are indicated. C, relative intensities of the XPC and XPB cross-linking signals in three independent experiments using our four photoprobes when both XPC/HR23B and TFIIH were present in the reactions. S.D. is indicated.
RESULTS
In Vitro Reconstitution of DNA Opening in the NER Reaction
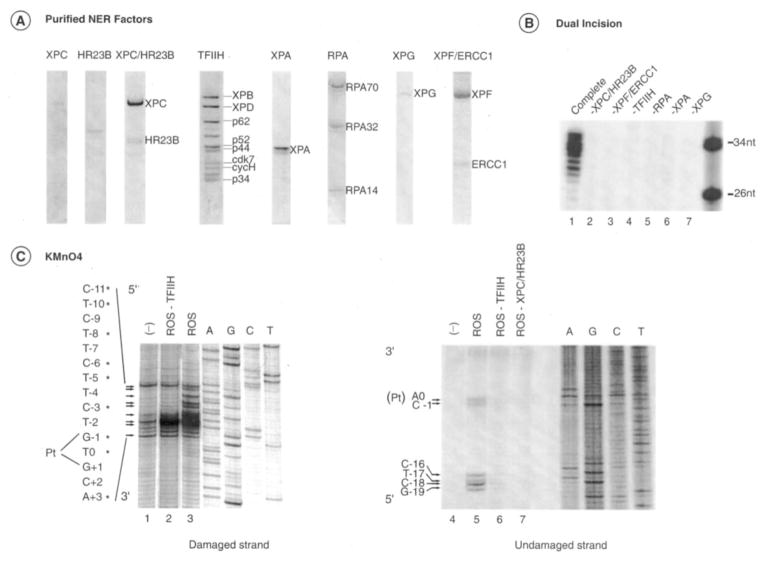
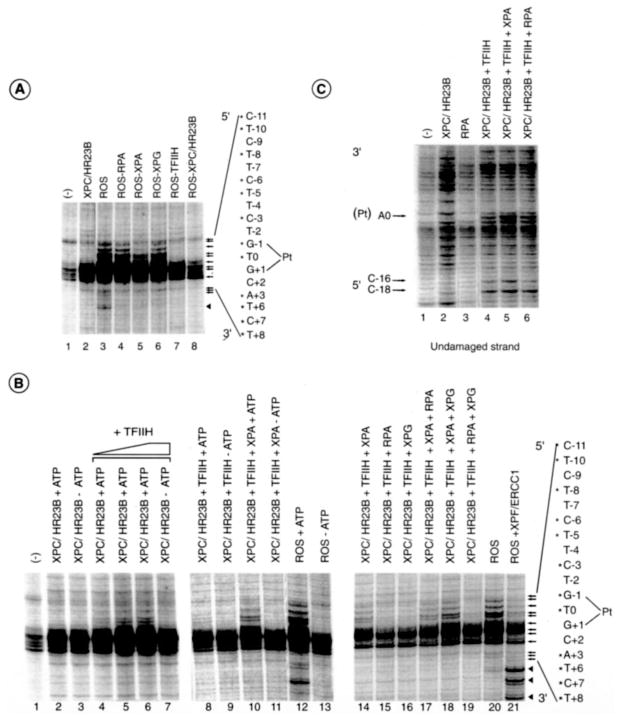
To investigate the DNA opening step of the NER reaction, we set up a permanganate sensitivity assay that allows the detection of single-stranded DNA around the cisplatinated DNA by generating breaks in phosphate backbone upon piperidine treatment. In such an assay, KMnO4 oxidized the pyrimidine residues in single-stranded DNA preferentially over those in double-stranded DNA, and the oxidized products could be converted to strand breaks by piperidine treatment (30). In this type of assay, only changes in DNA structure upon protein binding are detected. We thus applied this assay to damaged DNA carrying a single GTG 1,3-intrastrand cisplatin adduct and tested the changes in footprinting patterns upon incubation together with the highly purified factors XPC/HR23B, XPA, RPA, TFIIH, XPG, and XPF/ERCC1 (Fig. 1A) (7, 26). This set of factors is both necessary and sufficient to remove the damaged region, and the omission of any one of the factors prevents the dual incision reaction around the cisplatinated adduct (Fig. 1B). Together, they can open the DNA in the absence of XPF/ERCC1 and form the reconstituted opening system (ROS) (Fig. 1C). We thus observed a modification of the pattern of the single-stranded damaged DNA with an increase of the sensitivity around the cisplatin lesion at positions C−3, G−1, and T0 and the appearance of distinctively TFIIH-promoted bands at positions T−5, C−6, T−8, and T−10, (Fig. 1C, left panel, and as summarized in Fig 7A). In the absence of any NER factors, permanganate-sensitive sites were found at positions C−11, C−3, G−1, T0, G+1, C+2, and A+3, arising from the distortion induced by the damage itself (lane 1). We also noticed the appearance of breaks induced by the permanganate treatment in the undamaged strand upon the addition of the ROS factors at positions A0, C−1, C−16, T−17, C−18, and G−19 (lane 5). In the absence of TFIIH and XPC/HR23B, KMnO4 sensitive sites were not detected (lanes 6 and 7).
Fig. 1. NER factors used in dual incision and damaged DNA opening.
A, the purified NER factors XPC, HR23B, XPC/HR23B, TFIIH, RPA, XPG, and XPF/ERCC1 were stained with Coomassie Blue. B, dual incision with the purified factors. Repair factors were individually omitted as shown in lanes 2–7. In lane 1, the repair reaction was performed in the presence of all NER factors. nt, nucleotide. C, KMnO4 modifications. The nucleotides marked with asterisks match up sequentially with the arrows that point to bands of interest. The damaged strand (left panel, lanes 1–3) and undamaged strand probes (right panel, lanes 4–7) were incubated with ROS containing XPC/HR23B, TFIIH, XPA, RPA, and XPG. When indicated, ROS lacks either TFIIH (lanes 2 and 6) or XPC/HR23B (lane 7). Lane 1, bovine serum albumin (100 μg). An autoradiography of the gel is shown on the far right with dideoxy sequencing reaction A, G, C, and T ladders as markers. Pt, position 0.
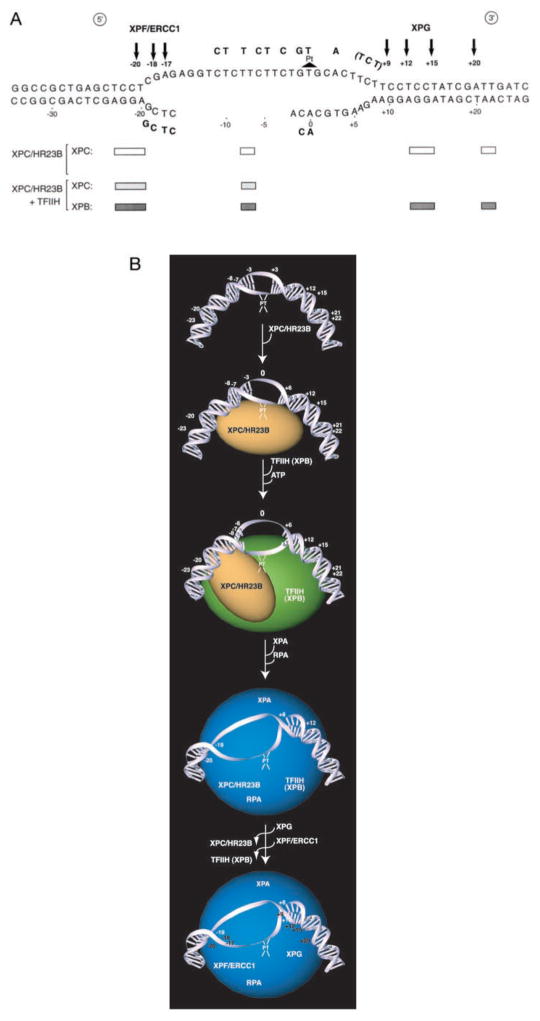
Fig. 7. Opening of the DNA around the Pt-GTG crosslink by the NER factors.
A, opening around the Pt-GTG during NER, as evidenced by reactivity to KMnO4, is depicted by placement of these residues above or below the axis of the duplex sequence. The position of the incision sites by XPG and XPF/ERCC1 endo-nucleases are indicated by arrows. The bottom part of the figure summarizes the photo cross-linking data with XPC/HR23B, either alone (open box) or in the presence of TFIIH and ATP (grey box for XPC, black box for XPB). B, a model representing the relocalization of XPC in the presence of TFIIH and the various steps leading to the removal of the damaged oligonucleotide. Each step is characterized by an increase in the remodeling of damaged DNA that is progressively opened by the repair machinery. In the absence of TFIIH, XPC approaches the damaged DNA from nucleotides −20 to +20, the contact at positions −8/−7 being the strongest. In the presence of TFIIH, XPC cross-links strongly to positions −23/−20 and weakly to positions −8/−7, but does not cross-link to positions on the 3′ end of the cisplatin adduct. The fully open complex (−19 to +8) is used as a template for incision by XPF/ERCC1 and XPG as indicated by arrows.
XPC Facilitates DNA Opening and Recruits the NER Factors
We next investigated the ability of each factor to separately target the damaged DNA. XPC/HR23B induces local changes in the footprinting pattern at positions G-1 and T-0 (Fig. 2A, lane 7 and C, lane 2). Upon longer exposure, minor changes are also detected over an extensive DNA region from about −20 to +20 (data not shown), further suggesting some “breathing” of the double helix in the region where other factors associate with the DNA (see below). The addition of XPF/ERCC1 to the damaged DNA generates a strong band at position C−3, as well as at positions G−14/G−16 and T+18/G+20 on each side of the lesion, respectively (lane 6), the specificity of which will be discussed below. We noticed, however, that the intensity of the G−1 band is slightly lower, whereas the C−11 band is slightly higher in the absence of NER factors (compare lanes 1–5); when considering weaker bands from A+3 to T+15 for example, they have the same intensity in both conditions.
Fig. 2. Contribution of the NER factors to the DNA opening.
A, XPC/HR23B and XPF/ERCC1 target the damaged DNA. Each NER factor was added separately to the damaged strand probe and tested for KMnO4 modification as indicated by arrows at the right of the panel. Bands corresponding to the 3′ incision are indicated by an arrowhead. B, combinations of the NER factors (except XPC/HR23B) as indicated at the top of the panel. C, XPC (40 ng) and HR23B (40 ng), either alone or in combination (lanes 5 and 9), or the purified XPC/HR23B complex from the co-infected extract (lanes 2 and 6) were tested for KMnO4 modifications (left panel) in the absence (lanes 2–5) or presence of XPA, RPA, TFIIH, and XPG (lanes 6–9). Dual incision in the presence of HR23B (10 – 40 ng) and XPC (10, 40 and 160 ng) either alone or in combination (XPC, 10 ng; XPC together with 50, 100, and 150 ng of HR23B). Pt, position 0; nt, nucleotide.
We addressed the question of whether any combination of the TFIIH, XPA, RPA, and XPG factors was able to contribute to helix opening in the absence of XPC/XR23B. No significant change in the footprinting pattern was observed using the combination of these factors under standard conditions (Fig. 2B). Only upon longer exposure did we observe a minor change at positions T−5 and C−6 in the presence of XPA and TFIIH (data not shown); the XPA-TFIIH complex was found to exhibit some affinity for damaged DNA (31). Altogether, the above results are fully consistent with an early role for the XPC/HR23B complex in recognition of the damage and recruitment of the other NER factors suggested by other studies (9, 10).
We also analyzed the contribution of each component of the XPC/HR23B complex (24, 32). The addition of either XPC or HR23B to the damaged DNA provides similar permanganate footprinting patterns, as indicated by the opening at positions G−1 and T+6 (Fig. 2C, lanes 2–5) when the preformed XPC/HR23B complex is used. Upon the addition of XPC/HR23B, the G-1 band is increased 5.5-fold, whereas with either XPC or HR23B G−1 is increased only 2-fold (Fig. 2C; compare lane 2 with lanes 3 and 4). Although either XPC or HR23B similarly target the damaged DNA (Fig. 2C, left panel, lanes 3 and 4), the addition of XPA, RPA, XPG, and TFIIH to HR23B alone does not promote the opening (lane 8), suggesting that HR23B did not allow the recruitment of the unwinding factors and/or their proper positioning. However when XPA, RPA, XPG, and TFIIH are added together with XPC, either alone or in combination with HR23B, permanganate-induced bands are detected at positions T−10, T−8, C−6, T−5, C−3, G−1, T0, and T+6 (Fig. 2C, lanes 7 and 9, and also Fig. 7A), emphasizing the essential role of XPC in recruiting other NER factors. In agreement with the footprint data, the addition of HR23B alone to XPA, RPA, XPG, TFIIH, or XPF/ERCC1 does not allow the removal of the damaged oligonucleotide unless XPC is added (Fig. 2C, right panel).
TFIIH Is Recruited to the XPC/HR23B-DNA Complex and Further Advances DNA Opening
Having shown that XPC has an early role in open complex formation, we next addressed the contribution of RPA, XPA, XPG, and TFIIH to DNA opening by incubating the damaged DNA with the ROS system in which one NER factor was systematically omitted. The absence of TFIIH resulted in the loss of the extension of the footprinting pattern induced by XPC/HR23B (Fig. 3A, compare lane 7 with lanes 2 and 3), whereas the omission of RPA or XPG did not significantly affect the overall footprinting pattern (Fig. 3A, lanes 4, 6, and 3, respectively). We especially noticed that, in the absence of XPA, the footprinting intensity at positions T−10, T−8, C−6, and T−5 as well as at positions near the damage site is 2-fold weaker (Fig. 3A, lane 5). Once XPC/HR23B and TFIIH are bound to the damaged DNA, additional NER factors are no longer required to extend the footprinting pattern on the 5′ side. Only on the 3′ side are all NER factors imperative (Fig. 3A, lanes 2–6).
Fig. 3. Ordered remodeling of the damaged DNA upon NER factor recruitment.
A, KMnO4 modification in which NER factors were individually omitted. B, ATP induces DNA unwinding by TFIIH (lanes 1–13). Recruitment of XPA, RPA, and XPG to the XPC/HR23B-TFIIH-damaged DNA complex is depicted (lanes 14–19); the addition of XPF/ERCC1 to ROS is shown (lanes 20–21). C, RPA does not modify the KMnO4 footprinting of the undamaged strand already targeted by XPC/HR23B and TFIIH. The nucleotides marked with asterisks match up sequentially with the arrows that point to bands of interest. Bands corresponding to the 3′ incision are indicated by an arrowhead. Pt, position 0.
We next addressed the role of ATP, which is required for the activity of the XPB and XPD helicases of TFIIH (33–35) in modulating the footprinting pattern of the damaged DNA around the lesion (36). We first observed that ATP does not modify the XPC/HR23B footprinting pattern of the damaged DNA (Fig. 3B, lanes 2 and 3). The addition of TFIIH in the absence of ATP did not allow modification of the XPC/HR23B-induced DNA footprint (Fig. 3B, lanes 3 and 7). In the presence of ATP, TFIIH opens the DNA according to the typical footprinting pattern in which permanganate breaks are found at both positions T−5 and C−6 and near the damage (Fig. 3B, lanes 4 – 6). The footprinting pattern induced by XPC/HR23B and TFIIH is even increased in the presence of XPA at positions T0, G−1, C−3, T−5, and C−6, and additional bands appeared at positions T−8 and T−10 (Fig. 3B, lanes 8–11). The addition of either RPA or XPG in the absence of XPA only slightly modified the opening profile around the damage and at positions T−5 and C−6 (lanes 15–16). Even if all of the factors are present, the addition of ATP is indispensable for dual incision (e.g. complete ROS) (Fig. 3B, lanes 12 and 13). The addition of RPA to a reaction containing XPC, TFIIH, and XPA did not modify the footprinting pattern of the damaged strand (lanes 14 and 17), whereas the addition of XPG increased the KMnO4 sensitivity pattern detected in the presence of XPA. Indeed, scanned T−5 and C−6 bands are increased 2.5–3-fold (Fig. 3B, lanes 14 and 18). When the double combination RPA+XPG is tested, we observed a slight increase at positions T−5 and C−6 (Fig. 3B, lanes 15, 16, and 19). Although RPA was shown to crosslink damaged DNA (37, 38), under our experimental conditions it does not modify the single-stranded structure induced by XPC and TFIIH (Fig. 3C, lanes 4 and 6). Indeed RPA, either alone or in combination, does not modify the permanganate footprinting pattern of both the damaged and the undamaged DNA strand (Fig. 3, B and C).
The XPG-XPF/ERCC1 Connection for Incision
Upon the addition of all five NER core factors, including XPC-HR23B, TFIIH, XPA, RPA, and XPG, we detected an optimal (both quantitatively and qualitatively) footprinting pattern (Fig. 3B, lane 20). In such a case, T−5 and C−6 are 3-fold higher (compare Fig. 3B, lanes 18 and 20). In the presence of XPF/ERCC1 we observed a 2-fold decrease of the bands at positions T−5, C−6, T−8, T−10, and near the lesion (mainly at positions G−1 and T0), as well as the appearance of other bands at positions T+12, T+15, and A+20 (Fig. 3B, lane 20 and 21; see also below and Fig. 7A).
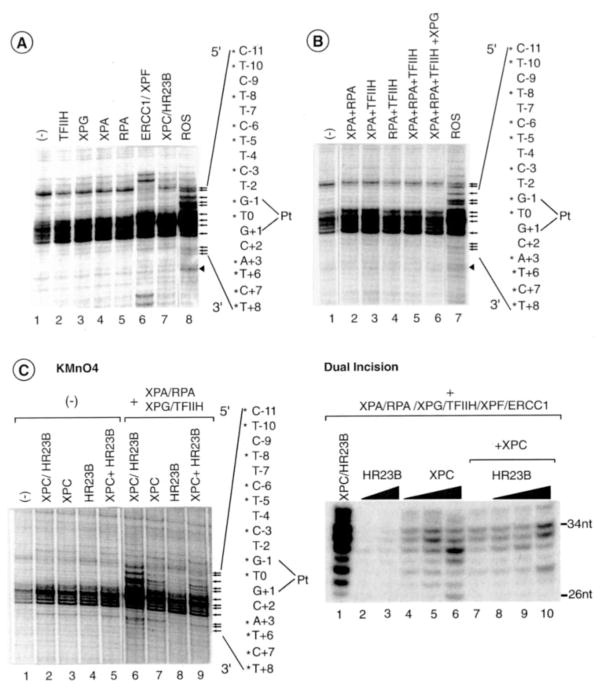
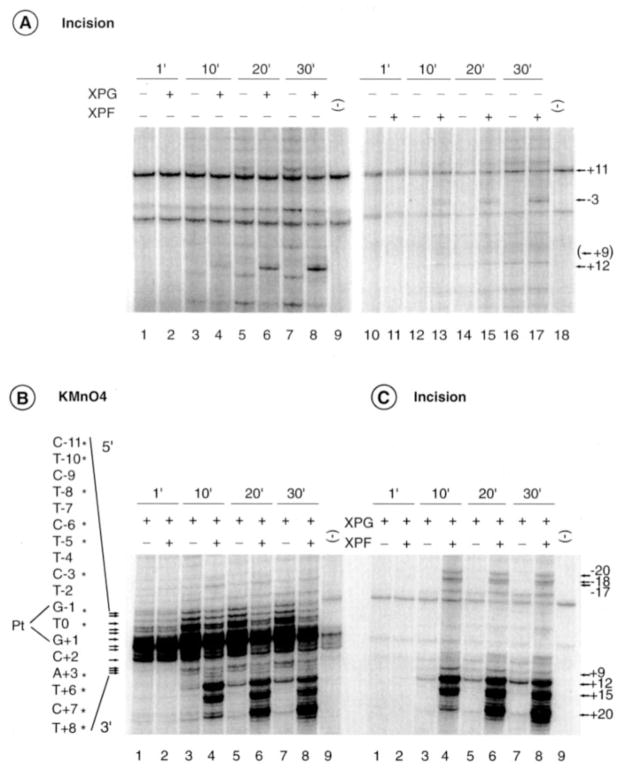
At this stage of our investigation we were wondering whether the additional bands induced by XPG and XPF/ERCC1 endonucleases, either alone or in combination, were the result of either an extension of the DNA opening or incisions in the damaged DNA. Therefore, the ability of either XPG or XPF/ERCC1 to incise the DNA on each side of the cisplatin lesion was tested. Both experiments were conducted in parallel under the same experimental conditions, except that after the incision reaction only one set of samples was KMnO4-treated. When XPG is added to the intermediate complex containing XPC/HR23B, TFIIH, XPA, and RPA, we observe some cut at position +12 (at t = 10 min) (Fig. 4A, lanes 4, 6, and 8). When added to the same intermediate complex, XPF/ERCC1 endonuclease did not significantly hydrolyze the damaged DNA in the presence of the four NER factors (Fig. 4A, lanes 10–18). We did, however, notice some incision at position −3 that probably results from an inappropriate positioning of XPF/ERCC1 on the intermediate complex in the absence of XPG (Fig. 4A, lanes 16–17; see also Fig. 2A, lane 6). We should mention that, in such a case, both XPG and XPF endonuclease activities are rather weak (compare, for example, Fig. 4, A and B, the relative intensity of C−11 and T+12 at t = 10 min).
Fig. 4. XPG and XPF/ERCC1 extend the DNA opening and excise the damaged oligonucleotide.
A, time course of DNA incision in which XPC/HR23B, TFIIH, XPA, RPA and damaged strand probe were incubated in the presence or absence of either XPG (lanes 1–9) or XPF/ERCC1 (lanes 10–18). Kinetics of KMnO4 modification (B) and DNA incision (C) upon the addition of XPF/ERCC1 to the five NER factors, including XPG. The 3′ and 5′ incisions are indicated on each side of the panel. The nucleotides marked with asterisks match up sequentially with the arrows that point to bands of interest. Pt, position 0.
Importantly, the addition of XPF/ERCC1, together with XPG, modified the KMnO4 footprinting pattern (Fig. 4B). An increase of the 3′ incision cuts were obtained at positions +12, +15, and +20 with a progressive decrease of the KMnO4-induced cuts on the 5′ side of the lesion and the appearance of new bands at positions −20, −18, and −17. To discriminate between bands produced by KMnO4-induced hydrolysis and bands resulting from the XPG and/or XPF/ERCC1 nucleases activities, DNA products were analyzed on a sequencing gel after 1′, 10′, 20′, and 30′ of incision reaction in the absence of permanganate treatment. Upon the addition of XPF/ERCC1, we observed the formation of incision-specific bands on both sides of the opened DNA bubble at positions −20, −18, −17, +12, +15, and +20 (Fig. 4C, lanes 4, 6 and 8, and Fig. 7A). Our data show that both XPG and XPF work in concert and that the hydrolysis occurs at the limit of the opened DNA structure.
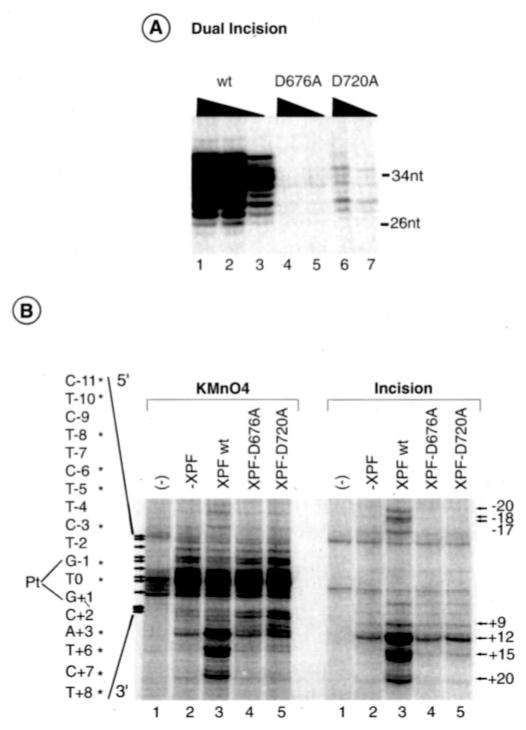
We next investigated whether the XPG endonuclease activity was regulated by either the physical presence of XPF or the enzymatic activity of XPF/ERCC1. We thus analyzed whether XPF-D676A and XPF-D720A, two XPF mutants impaired in nuclease activity and the dual incision reaction (Fig. 5A) but not DNA binding, can affect the activity of XPG to cut damaged DNA (39). When KMnO4 assays were carried out in the presence of XPF-D676A and XPF-D720A/ERCC1, we detected a 5′ footprinting profile very similar to the one observed in the absence of XPF/ERCC1 (Fig. 5B, left panel). However, we should notice that the presence of the XPF mutants significantly increases the bands at positions C+7, T+8, C+10, C+11, and T+12, with the latter one being an XPG-induced sensitive site as shown in the incision assay (Fig. 5B, right panel). Except for this one site, it is interesting to note that the other XPG-sensitive sites (at positions +15 and +20) require the presence of an active XPF, showing once more the synergy between both nucleases in the elimination of the damaged oligonucleotide.
Fig. 5. Mutant XPF/ERCC1 enhances DNA opening.
A, dual incision assays using either XPF/ERCC1 wild type (wt) (12, 6, or 3 ng), XPF-D676A and XPF-D720A/ERCC1 (50 and 18 ng), in addition to the five NER factors. nt, nucleotide. B, KMnO4 modifications (left panel) and DNA incisions (right panel) in the presence of XPF-D676A, -D720A, and -wt/ERCC1 as indicated at the top of the panel. KMnO4-induced cuts as well as incisions are indicated on each side of the panel. The nucleotides marked with asterisks match up sequentially with the arrows that point to bands of interest. Pt, position 0.
Photo Cross-linking of XPC and XPB around the Damaged DNA
We have used site-specific protein-DNA photo cross-linking to determine the location of XPC/HR23B and TFIIH subunits, the two factors that initiate the formation of the pre-incision complex between nucleotides −30 and +30 around the cisplatin damage. Four photoprobes placing the photoreactive nucleotide AB-dUMP at specific locations around the damage were designed (Fig. 6A). The DNA repair pre-incision complex containing either XPC/HR23B alone or XPC/HR23B and TFIIH was incubated with the various photoprobes in the presence of ATP and further UV-irradiated. Under those conditions, the photoreactive nitrene of AB-dUMP can be covalently crosslinked to the polypeptides located at a distance of ~10 A from the DNA backbone (40, 41, 59). The samples were then digested with exonucleases to remove non-crosslinked DNA tails, and the crosslinked polypeptides were analyzed by SDS-PAGE. To ensure the specificity of the crosslinked bands, all of the assays were carried out in parallel with photoprobes lacking the cisplatin adduct, which did not result in the formation of covalent protein-DNA complexes (data not shown). A crosslinking signal was considered specific when its intensity was significantly higher in reactions containing the damaged photoprobe as compared with the undamaged photoprobe.
The interaction of the DNA repair factors XPC/HR23B and TFIIH with the damaged DNA is much weaker that the interaction of the basal transcription factors (including TFIIH) and RNA polymerase II with promoter DNA (28), which explains the low intensity of the cross-linking signals in Fig. 6B. When XPC/HR23B alone was used in the reactions, we repeatedly obtained cross-linking of XPC at all the positions tested (Fig. 6B). When TFIIH and ATP were added to reactions containing XPC/HR23B, we obtained cross-linking of XPB at all three positions (cross-link at position +21/+22 is always weak) as well as cross-linking of XPC at the two positions located upstream of the damage (at position −23/−20 and −8/−7). Fig. 6C shows the relative intensity of the XPC and XPB cross-linking signals obtained in three independent experiments using our four photoprobes in the presence of both XPC/HR23B and TFIIH. Whereas the cross-linking of XPB is weaker than that of XPC at positions −23/−20, both factors cross-link with similar efficiencies at positions −8/−7. Notably, only XPB cross-linked to both photoprobes +12/+15 and +21/+22. XPC did not crosslink to photoprobes +12/+15 and +21/+22 in the presence of TFIIH. Together, these results indicate that XPC makes extensive DNA contacts around the lesion in the absence of TFIIH but is displaced from the 3′ side of the lesion when TFIIH associates with the XPC/HR23B-DNA complex, suggesting that the entry of TFIIH induces a relocalization of XPC in the complex. According to SDS-PAGE, our results also indicate that XPB is the TFIIH subunit that comes in closest contact with the open DNA.
DISCUSSION
Our results indicate that NER is a dynamic process that takes place in a number of successive steps starting from DNA damage recognition to its elimination (10, 36, 42, 43), during which process the repair factors change the conformation of the damaged DNA (Fig. 7B). Previous studies have shown that a cisplatinated DNA fragment is intrinsically bent by ~25–30 degrees (44, 45, 46). Our permanganate footprinting experiments further indicate that the DNA is melted from positions −3 and +3 around the cisplatin adduct (13, 42). This peculiar DNA conformation is recognized and bound by the XPC/HR23B complex. Our photo cross-linking experiments indicate that XPC approaches the damaged DNA over a region from about −20 to +20, extending the DNA opening from −3 to +6 and changing the DNA conformation according to the permanganate-sensitive positions upstream (positions −21 to −14) and downstream (positions +9 to +20) of the lesion. Binding of XPC/HR23B then allows the recruitment of TFIIH in an ATP-independent manner (the present study and Ref. 11). In the presence of ATP, TFIIH further opens the damaged DNA from −6 to +6. It is noteworthy that the XPB helicase of TFIIH is the subunit that more closely approaches the damaged DNA. As shown by our crosslinking results, the association of TFIIH with the XPC/HR23B-DNA complex displaces XPC, which does not contact the DNA downstream of the lesion in the presence of TFIIH. Presumably, this is the consequence of ATP addition that allows both XPB and XPD helicases to unwind the damaged DNA, resulting in an intermediate complex structurally ready to recruit the other NER factors.
Although both XPC and HR23B separately yield a footprint when bound to the damaged DNA, HR23B alone does not accurately remodel the damaged DNA to recruit the other NER factors, underlining its secondary role in the in vitro dual incision (24, 47, 48). We especially noticed that the addition of TFIIH to reactions that already contain HR23B, even in the presence of ATP, did not support unwinding of the damaged DNA. By contrast, XPC recruits TFIIH even in the absence of HR23B (49).
The “opening” of the DNA around the damage between positions −6 and +6 is next stabilized by XPA, which increases the KMnO4 sensitivity of the pyrimidines already exposed by XPC and TFIIH binding. The present study thus reveals a contribution of XPA in further enlarging the DNA opening initiated by XPC and TFIIH.
The influence of RPA on the DNA unwinding activity of TFIIH and the final opening of the damaged DNA remains unclear. Contrary to certain previous proposals, (21, 50–53), under our experimental conditions neither XPA nor RPA nor the preformed XPA-RPA complex (data not shown) was able to initiate the opening of the damaged DNA and the formation of the dual incision complex. Although RPA was shown to be positioned on the 5′ side of the damaged DNA (37) to protect single-stranded DNA from nonspecific hydrolysis, the present study failed to demonstrate the commitment of RPA toward changing the damaged DNA conformation and/or intermediate complex stabilization. It remains, however, that the presence of RPA, together with XPC, TFIIH, and XPA, is crucial for the dual incision and DNA resynthesis steps (7, 54, 55) (11). Whereas RPA might position both XPG and XPF endonucleases, it is also involved in the recruitment of factors such as RF-C and PCNA to take over the DNA resynthesis step (56) (57).
The nucleoprotein complex that results from the ordered addition of XPC/HR23B, TFIIH, XPA, and RPA is then structurally ready to be targeted by XPG and XPF/ERCC1 endo-nucleases that will remove the damaged oligonucleotide. XPG first joins and stabilizes the complex. Indeed, although other investigators observed XPG cuts whenever the substrate contains a 5′ single-stranded, 3′ double-stranded structure (Refs. 12, 13, and 58, and our results) indicate that XPG alone is unable to bind and hydrolyze the damaged DNA. It appears that although it is associated with the four NER factors and thus provides a DNA substrate with a branch point located at +9 and a 5′ single-stranded structure, XPG will cut the DNA at position +12. Then, the addition of XPF/ERCC1 most likely promotes XPG incision to additional sites at positions +15 and +20. Such a gain in specificity probably results from the repositioning of certain factors and the release of others (11).
XPF/ERCC1, either alone or in combination with the four NER factors of the intermediate complex, may hydrolyze non-specifically the damaged DNA as well as structures containing single-strand tracks (Refs. 14 and 39, and the present study). The specificity of XPF in hydrolyzing on the 5′ side is provided only after XPG has joined the complex. Both XPF and XPG work in concert to remove the damaged oligonucleotide. Whether or not the specificity of both nucleases would require additional (and not yet identified) factors to allow a single and specific cut remains unanswered. The recruitment of XPG and XPF/ERCC1 has been shown previously to induce the release of both XPC/HR23B and TFIIH, respectively (11). Here, we show that both XPG and XPF endonucleases target and hydrolyze the damaged DNA at positions already protected by XPC and TFIIH.
It is interesting to note how the various factors deal with the damaged DNA; it is recognized by XPC, unwound by XPB and XPD (part of TFIIH), its open structure is then stabilized by XPA, and it is hydrolyzed by XPG and XPF. Mutations in the genes encoding these factors cause xeroderma pigmentosum, which is associated in some cases with CS and TTD. Strikingly, mutations in the HR23B, RPA, and ERCC1 genes have not yet been associated with NER defects; the function of these factors in DNA opening/remodeling is necessary but subordinate. How mutations in any one of the six genes responsible for XP disturb the dual incision in NER has to be further examined, not only to establish a genotype/phenotype relationship but also to elucidate the role of each of them in DNA repair.
Acknowledgments
We thank T. Riedl and S. Dubaele for fruitful discussions and critical reading of the manuscript and R. Wood, J. H. J. Hoeijmakers, and S. Clarkson for providing some recombinant baculoviruses and antibodies. We are grateful to M. Chipoulet and A. Fery for high technical expertise, I. Kolb-Cheynel and J. L. Weickert for the production of recombinant baculoviruses, and B. Boulay and S. Metz for their high expertise in designing and producing the figures.
Footnotes
This work was supported by European Economic Community Grants QLG1–1999 and QLRT-1999-02002) and grants from l’Association de la Recherche sur le Cancer and the Commissariat à l’Énergie Atomique (to J. M. E.) and the Canadian Institutes of Health Research (to B. C.).
The abbreviations used are: NER, nucleotide excision repair; ROS, reconstituted opening system; RPA, replication protein A; XPA/C/F/G, xeroderma pigmentosum group A, C, F, or G.
References
- 1.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Spivak G, Hanawalt PC. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 4.De Laat WL, Jaspers NG, Hoeijmakers JH. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 5.Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JHJ. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler KW, editors. McGraw-Hill Inc; New York: 1998. pp. 245–274. [Google Scholar]
- 6.Lehmann AR. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 7.Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 8.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 9.Sugasawa K, Ng J, Masutani C, Iwai S, van der Spek P, Eker A, Hanaoka F, Bootsma D, Hoeijmakers J. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 10.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 11.Riedl T, Hanaoka F, Egly JM. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnovan A, Davies AA, Moggs JG, West SC, Wood RD. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 13.Evans E, Fellows J, Coffer A, Wood RD. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Laat WL, Appeldoorn E, Jaspers NGJ, Hoeijmakers JHJ. J Biol Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 15.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, Rademakers S, de Rooij J, Jaspers NG, Hoeijmakers JH, Wood RD. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 16.Shivji MKK, Podust VN, Hubsher U, Wood RD. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 17.Svejstrup JQ, Wang Z, Feaver WJ, Wu X, Bushnell DA, Donahue TF, Friedberg EC, Kornberg RD. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 18.Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W. Science. 1999;284:958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- 19.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson CW, Linn S, Reinberg D. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 20.Araujo SJ, Nigg EA, Wood RD. Mol Cell Biol. 2001;21:2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reardon JT, Sancar A. Mol Cell Biol. 2002;22:5938–5945. doi: 10.1128/MCB.22.16.5938-5945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henricksen LA, Umbricht CB, Wold MS. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 23.O’Donovan A, Scherly D, Clarkson SG, Wood RD. J Biol Chem. 1994;269:15965–15968. [PubMed] [Google Scholar]
- 24.Sugasawa K, Masutani C, Uchida A, Maekawa T, van der Spek PJ, Bootsma D, Hoeijmakers JH, Hanaoka F. Mol Cell Biol. 1996;16:4852–4861. doi: 10.1128/mcb.16.9.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerard M, Fischer L, Moncollin V, Chipoulet JM, Chambon P, Egly JM. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 26.Frit P, Kwon K, Coin F, Auriol J, Dubaele S, Salles B, Egly JM. Mol Cell. 2002;10:1391–1401. doi: 10.1016/s1097-2765(02)00732-3. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. pp. 6.46–6.49. [Google Scholar]
- 28.Douziech M, Coin F, Chipoulet JM, Arai Y, Ohkuma Y, Egly JM, Coulombe B. Mol Cell Biol. 2000;20:8168–8177. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert F, Coulombe B. Methods Mol Biol. 2001;148:383–393. doi: 10.1385/1-59259-208-2:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Gralla JD. J Biol Chem. 1995;270:1277–1281. doi: 10.1074/jbc.270.3.1277. [DOI] [PubMed] [Google Scholar]
- 31.Nocentini S, Coin F, Saijo M, Tanaka K, Egly JM. J Biol Sci. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 32.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, Hoeijmakers JHJ, Hanaoka F. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JHJ, Chambon P, Egly JM. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 34.Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 35.Coin F, Bergmann E, Tremeau-Bravard A, Egly JM. EMBO. 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweizer U, Hey T, Lipps G, Krauss G. Nucleic Acids Res. 1999;27:3183–3189. doi: 10.1093/nar/27.15.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermanson-Miller IL, Turchi JJ. Biochemistry. 2002;41:2402–2408. doi: 10.1021/bi0112863. [DOI] [PubMed] [Google Scholar]
- 39.Enzlin JH, Scharer OD. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persinger J, Bartholomew B. Methods Mol Biol. 2001;148:363–381. doi: 10.1385/1-59259-208-2:363. [DOI] [PubMed] [Google Scholar]
- 41.Bartholomew B, Kassavetis GA, Braun BR, Geiduschek EP. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Constantinou A, Gunz D, Evans E, Lalle P, Bates PA, Wood RD, Clarkson SG. J Biol Chem. 1999;274:5637–5648. doi: 10.1074/jbc.274.9.5637. [DOI] [PubMed] [Google Scholar]
- 43.Mu D, Wakasugi M, Hsu DS, Sancar A. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 44.Anin MF, Leng M. Nucleic Acids Res. 1990;18:4395–4400. doi: 10.1093/nar/18.15.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellon SF, Coleman JH, Lippard SJ. Biochemistry. 1991;40:8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 46.van Garderen CJ, van Houte LP. Eur J Biochem. 1994;225:1169–1179. [PubMed] [Google Scholar]
- 47.Sugasawa K, Ng JM, Masutani C, Maekawa T, Uchida A, van der Spek PJ, Eker AP, Rademakers S, Visser C, Aboussekhra A, Wood RD, Hanaoka F, Bootsma D, Hoeijmakers JH. Mol Cell Biol. 1997;17:6924–6931. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batty D, Rapic’-Otrin V, Levine AS, Wood RD. J Mol Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 49.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. J Biol Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- 50.Jones CJ, Wood RD. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 51.Wakasugi M, Sancar A. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 52.Patrick SM, Turchi JJ. Biochemistry. 1998;37:8808–8815. doi: 10.1021/bi9730590. [DOI] [PubMed] [Google Scholar]
- 53.Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aboussekhra A, Biggerstaff M, Shivji MKK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hübscher U, Egly JM, Wood RD. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 55.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 56.de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG, Hoeijmakers JH. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuzhakov A, Kelman Z, Hurwitz J, O’Donnell M. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hohl M, Thorel F, Clarkson SG, Scharer OD. J Biol Chem. 2003;278:19500–19508. doi: 10.1074/jbc.M213155200. [DOI] [PubMed] [Google Scholar]
- 59.Forget D, Langelier MF, Thérien C, Trinh V, Coulombe B. Mol Cell Biol. 2004;24:1122–1131. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]