Abstract
The subgenomic (sg) mRNAs of arteriviruses (order Nidovirales) form a 5′- and 3′-coterminal nested set with the viral genome. Their 5′ common leader sequence is derived from the genomic 5′-proximal region. Fusion of sg RNA leader and “body” segments involves a discontinuous transcription step. Presumably during minus-strand synthesis, the nascent RNA strand is transferred from one site in the genomic template to another, a process guided by conserved transcription-regulating sequences (TRSs) at these template sites. Subgenomic RNA species are produced in different but constant molar ratios, with the smallest RNAs usually being most abundant. Factors thought to influence sg RNA synthesis are size differences between sg RNA species, differences in sequence context between body TRSs, and the mutual influence (or competition) between strand transfer reactions occurring at different body TRSs. Using an Equine arteritis virus infectious cDNA clone, we investigated how body TRS activity affected sg RNA synthesis from neighboring body TRSs. Flanking sequences were standardized by head-to-tail insertion of several copies of an RNA7 body TRS cassette. A perfect gradient of sg RNA abundance, progressively favoring smaller RNA species, was observed. Disruption of body TRS function by mutagenesis did not have a significant effect on the activity of other TRSs. However, deletion of body TRS-containing regions enhanced synthesis of sg RNAs from upstream TRSs but not of those produced from downstream TRSs. The results of this study provide considerable support for the proposed discontinuous extension of minus-strand RNA synthesis as a crucial step in sg RNA synthesis.
The synthesis of subgenomic (sg) mRNAs is a common mechanism that positive-strand RNA viruses have evolved to express structural and auxiliary proteins that do not have to be translated directly from the genomic mRNA (reviewed in reference 29). Subgenomic RNAs are produced either by internal initiation of transcription on a minus-strand template (28), by premature termination of minus-strand synthesis (46, 60), or by an RNA recombination-like process of discontinuous RNA synthesis, as it is found in coronaviruses and arteriviruses, members of the order Nidovirales (5, 24, 48). The nidovirus genome and sg mRNAs form a 3′-coterminal nested set of 3 to 10 RNA species, depending on the particular virus species. As reported recently, in some nidovirus genera, including okaviruses (8) and toroviuses (47, 59), the viral sg transcripts have different 5′-terminal sequences. However, coronavirus and arterivirus sg mRNAs and torovirus sg mRNA2 contain a common 5′ leader sequence, which is derived from the 5′-proximal region of the genome. The 3′-proximal segment of such a coronavirus or arterivirus sg mRNA species (which is referred to as the mRNA body) is unique, albeit partially overlapping the bodies of other sg mRNAs (Fig. 1A). The leader is thought to be joined to an sg RNA body in a recombination-like RNA strand transfer process that is governed by short conserved transcription-regulating sequences (TRSs). These are present at the 3′ end of the leader (leader TRS) and at the 5′ end of each of the body regions in the 3′-proximal region of the genome (body TRSs) (Fig. 1A). Base pairing between the arterivirus leader TRS (in the plus strand) and the complement of the body TRSs (in the minus strand) was shown to be essential for sg mRNA synthesis (34, 35, 56).
FIG. 1.
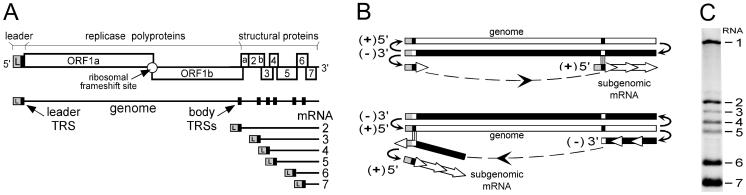
(A) Schematic diagram of the genome organization and expression of EAV, the arterivirus prototype. The regions of the genome specifying the leader (L) sequence, the replicase gene (ORFs 1a and 1b), and the structural protein genes are indicated. The nested set of EAV mRNAs (genome and sg mRNAs 2 to 7) is depicted below. The black boxes in the genomic RNA indicate the positions of leader and major body TRSs. (B) Alternative models for nidovirus discontinuous sg RNA synthesis. The discontinuous step may occur during either plus-strand or minus-strand RNA synthesis. In the latter case, sg mRNAs would be synthesized from an sg minus-strand template. For details, see the text. (C) Northern hybridization analysis of intracellular EAV RNA resolved by denaturing agarose gel electrophoresis. As a probe, 32P-labeled oligonucleotide E154 was used, which is complementary to the 3′ end of all viral plus-strand RNA molecules (see Materials and Methods).
Two opposing models have been proposed to explain the discontinuous step in arterivirus and coronavirus sg RNA synthesis (Fig. 1B). According to the leader-primed transcription model, sg RNA synthesis would be primed by free leader molecules that attach, by means of TRS-TRS base pairing, to the body TRS complements in the genomic minus strand, after which the leader primer would be elongated to produce an sg mRNA (3, 23, 49). This model was partly based on the fact that sg minus-strand RNAs were initially not detected in cells infected with the coronavirus Mouse hepatitis virus (MHV) (25). However, after the subsequent discovery of such molecules in coronavirus-infected cells (38, 41), an alternative transcription model was proposed (39). According to this model, the discontinuous step in transcription does not occur during plus-strand but during minus-strand RNA synthesis. The body TRSs in the plus-strand template would attenuate minus-strand synthesis, after which the nascent minus strand, having a TRS complement at its 3′ end, would be transferred to base pair with the leader TRS, which may be presented by a stem-loop structure (6, 52, 56). The sg minus strands would then be completed by addition of the leader complement and be used as templates for the synthesis of sg plus strands (mRNAs). In recent years, this model of Sawicki and Sawicki has gained considerable experimental support from both biochemical and genetic studies (4, 34, 37), and consequently, it will be used as our working model in this paper.
The smallest sg RNA species tend to be the most abundant ones in all viruses that produce a nested set of multiple sg RNAs (29). In general (see below), nidovirus sg RNAs are no exception to this rule: both sg plus and minus strands are produced in different and constant molar amounts in the infected cells, and the shorter species are usually more abundant then the larger ones (7, 11, 24, 27) (Fig. 1C). For the coronavirus MHV, it has been proposed that relative sg RNA abundance is exclusively determined by the size of the leader TRS-body TRS duplex (45). However, other studies of coronaviruses (1, 13, 15, 21) and arteriviruses (10) have shown that relative sg RNA abundance does not correlate with the size of the leader TRS-body TRS duplex, and moreover, this hypothesis was experimentally refuted for MHV by van der Most et al. (54).
For the arterivirus Equine arteritis virus (EAV), we recently established that the stability of the leader TRS-body TRS duplex is an important determinant of sg RNA abundance, although it is clear that other factors are also involved (34, 35, 56). The fact that the relative molar ratios of coronavirus sg minus strands are similar to those of the plus strands (4, 14, 37, 38, 41) indicates, in the frame of the discontinuous minus-strand synthesis model, that sg RNA abundance may be regulated at the level of (discontinuous) minus-strand synthesis. If body TRSs indeed are attenuators of minus-strand synthesis, then more strand transfer events are likely to occur at 3′-proximal body TRSs than at 3′-distal body TRSs, which are further away from the site at which minus-strand synthesis is initiated. Consequently, strand transfer reactions at 3′-proximal TRSs would suppress the transcriptional activity of more-upstream (in the plus sense), 3′-distal TRSs, but not vice versa.
If all body TRSs had identical flanking sequences and, consequently, identical strand transfer potential, then a gradient of sg RNA abundance, progressively favoring the smallest sg RNAs, would be expected. However, every TRS in a nidovirus genome is situated in a unique primary and higher-order sequence context that influences its activity in sg RNA synthesis (1, 2, 15, 17, 32, 33). This may partially explain why the sg RNA gradient in nidovirus-infected cells is not perfect. For example, in some coronaviruses, the smallest sg RNA is less abundant then the next larger sg RNA (9, 41). Also, sg mRNA2 of the arterivirus EAV is considerably more abundant than mRNAs 3 to 5 (11). Alternatively, if the production of sg mRNAs were regulated primarily at the level of plus-strand synthesis, then their abundance would depend mainly on their relative sizes (because smaller sg minus-strand templates would be copied faster) (40) and not on their order in the nested set. An alternative explanation is that the primary and/or higher-order sequence contexts of body TRSs solely determine their activity and regulate sg mRNA abundance. In the latter two scenarios, body TRS activity will not influence the activity of other body TRSs.
To investigate these issues in the arterivirus system, we have now produced derivatives of an EAV infectious cDNA clone in which body TRS-flanking sequences were standardized by engineering one, two, three, or four direct repeats of a 417-nucleotide (nt) RNA7 body TRS cassette in the 3′-proximal part of the genome. To study the influence of body TRS activity on the activity of up- and downstream body TRSs in these replicons, all possible combinations of wild-type and mutant TRSs were tested in the context of the three-repeat construct. Furthermore, we explored the influence of body TRS position and sg RNA size by analyzing constructs with different deletions in the 3′-proximal region of the genome.
MATERIALS AND METHODS
Construction of plasmids.
The constructs with repeated RNA7 body TRS cassettes were assembled as follows (Fig. 2A). First, the three-repeat construct (mABC) was made. To obtain cassettes A and B, a 417-nt fragment (nt 12229 to 12645) from EAV full-length cDNA clone pEAV030HNB, a derivative of clone pEAV030H (55) that contained an engineered BspEI restriction site at nt 12228 to 12233 (56), was PCR amplified with two combinations of primers that contained additional restriction sites. The amplified fragments contained the RNA7 body TRS, its up- and downstream flanking sequences, and the complete open reading frame (ORF) of the EAV nucleocapsid (N) protein gene. Cassette A was obtained by amplification of the fragment with oligonucleotides E251 (5′-AGATGGCCATGGCCGGACCTGTTCCC-3′), containing an NcoI restriction site, as the forward primer and E252 (5′-CTATTCGAATTCTTACGGCCCTGCTG-3′), containing an EcoRI restriction site, as the reverse primer. Cassette B was obtained by amplification of the fragment with oligonucleotides E253 (5′-AGATGGGAATTCCCGGACCTGTTCCC-3′), containing an EcoRI restriction site, as the forward primer and E254 (5′-CTATTCTCCGGATTACGGCCCTGCTG-3′), containing a BspEI restriction site, as the reverse primer.
FIG. 2.
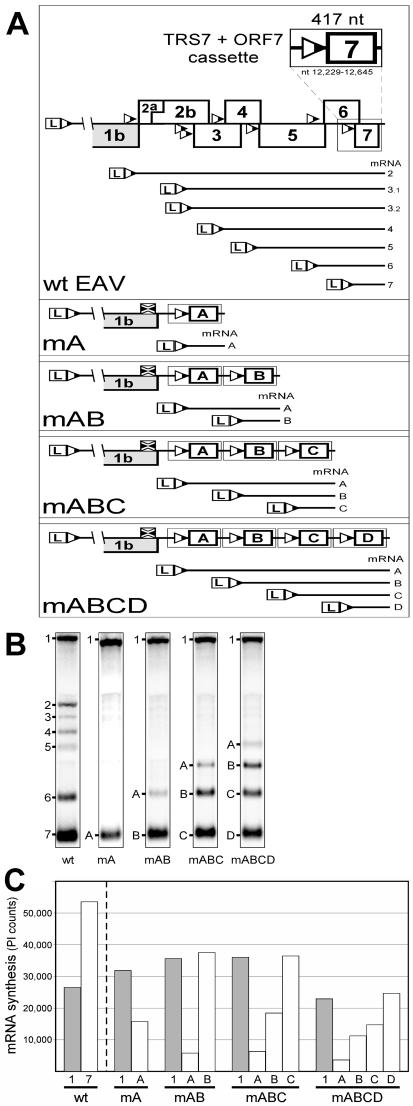
(A) Scheme of constructs with repeated RNA7 body TRS cassettes. The upper panel shows a close-up view of the 3′-proximal quarter of the EAV genome, where the structural gene ORFs (2a to 7) and body TRSs are located. The TRSs are indicated with triangles. The nested set of sg mRNAs, including the two major subspecies of sg mRNA3 (33), is shown below. The contents of the TRS7 cassette (see Materials and Methods) are depicted above. The lower panel shows the composition of the constructs with one (mA) to four (mABCD) repeats of the TRS7 cassette. For each construct, the corresponding sg RNAs are shown. In all constructs, the RNA2 TRS was knocked out by mutation (depicted by crossed squares). (B) Northern analysis of EAV-specific RNA isolated from cells transfected with RNA transcribed either from the wild-type (wt) EAV infectious cDNA clone or from the constructs with repeated TRS7 cassettes. (C) Amounts of genomic and sg mRNAs produced by the wild-type EAV construct and by the TRS7 repeat constructs. The average (of two experiments) amounts of RNAs are shown.
All PCRs were performed with Pfu DNA polymerase (Stratagene), and the sequences of PCR-generated products were confirmed by sequence analysis with a GeneAmp PCR system 2400 (Perkin-Elmer), ABI Prism kit (Perkin-Elmer), and ABI Prism 310 genetic analyzer (Perkin-Elmer). Cassettes A and B were cloned into pBluescript-based shuttle vector pM92128m2NB that contained the pEAV030H-derived sequence from nt 9149 to nt 12845 of the EAV genome, including two engineered restriction sites (NcoI at nt 9822 to 9827 and BspEI at nt 12228 to 12233) and an RNA2 body TRS knockout mutation (nt 9711 to 9716) (33). Cassettes A and B were inserted directly upstream of the BspEI restriction site. The EAV ORF7 region downstream of this site (nt 12229 to 12645) was identical to cassettes A and B and was termed cassette C. Cassettes A and B were cloned between the NcoI and EcoRI and and EcoRI and BspEI restriction sites, respectively, to obtain pM92128ABC. In this manner, three identical cassettes were cloned directly head to tail, replacing the complete EAV ORF2a-ORF6 region, which also contains the body TRSs for sg mRNAs 3 to 6 (the RNA2 body TRS inside the replicase gene was inactivated by mutagenesis; see above). The unique restriction sites flanking each of the cassettes allowed easy modification of the construct.
Subsequently, the insert from pM92128ABC containing the three cassettes was transferred to the EAV full-length cDNA clone (construct mABC). Derivatives of mABC with one or multiple mutated body TRSs (Fig. 3A) were obtained by replacing cassettes with analogous 417-nt fragments amplified from constructs containing RNA7 body TRS mutations. The mAB construct was obtained by deleting the EcoRI-BspEI fragment from mABC. The mABCD construct was engineered by insertion of the 417-nt fragment amplified with primers E252 and E253 (both with additional EcoRI sites) into the EcoRI site of mABC. The mA construct resulted from deletion of the NcoI-BspEI fragment from pM92128ABC and transfer of this deletion to the full-length cDNA clone.
FIG. 3.
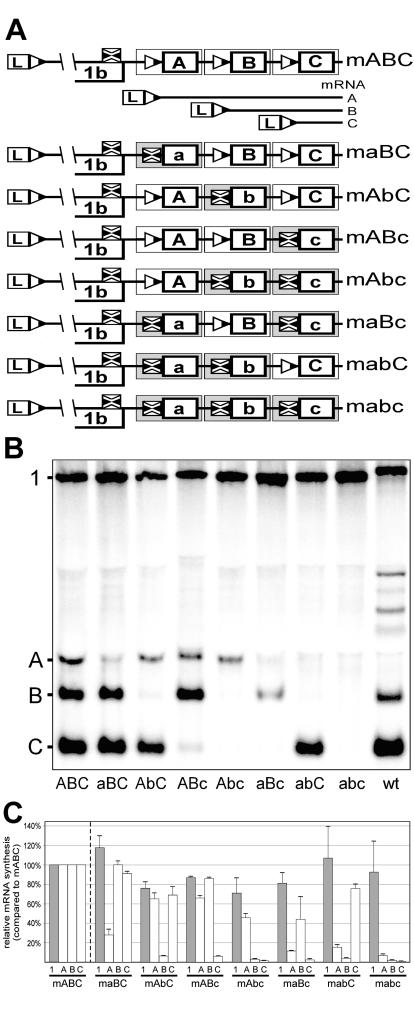
(A) Scheme of seven mutant three-repeat constructs, representing all possible combinations of a wild-type (wt) body TRS and a mutant body TRS. The wild-type TRSs are indicated by triangles and capital letters, and the mutant TRSs are indicated by crossed squares and lowercase letters. (B) Northern analysis of EAV-specific RNA isolated from cells transfected with RNA transcribed either from the wild-type EAV infectious cDNA clone or from the wild-type and seven mutant three-repeat constructs of the first series (TRS mutations 5′-UCAACU-3′ to 5′-UgAAgU-3′). (C) Relative amounts of genomic and sg mRNAs produced by the mutant three-repeat constructs. The amounts of each RNA species (genomic and subgenomic) produced by the mutant constructs were independently related to the amounts of the corresponding RNA species produced by the wild-type three-repeat construct (mABC), which were set at 100%. The average (of three experiments) amounts of RNAs are shown.
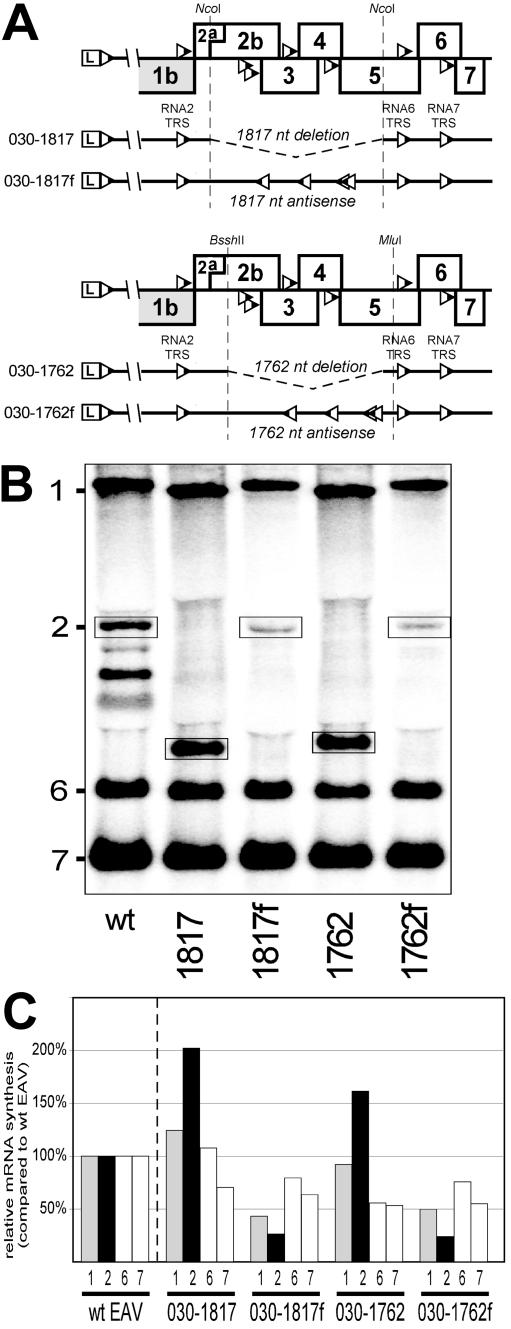
Deletion constructs 030-1615, 030-1717, 030-1927, 030-2207, 030-2282, and 030-2319 (see Fig. 6) were described by Molenkamp et al. (30). The constructs in which deletions were filled with (presumed) “TRS-free” sequences (see Fig. 7A) were generated by cloning a 1,817-nt NcoI fragment (nt 9822 to 11639) and a 1,762-nt BssHII-MluI fragment (nt 9976 to 11738) from the EAV cDNA sequence back into clone pEAV030HNB in the reverse orientation. The resulting constructs were named 030-1817f and 030-1762f, respectively, with the corresponding deletion constructs being termed 030-1817 and 030-1762, respectively.
FIG. 6.
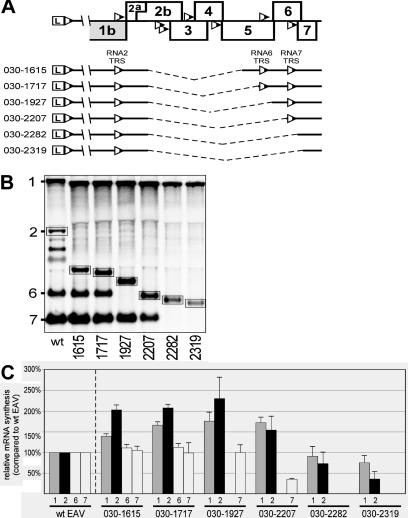
(A) Scheme of the deletion constructs (30). See the text for details. (B) Northern analysis of EAV-specific RNA isolated from cells transfected with RNA transcribed either from the wild-type EAV infectious cDNA clone or from the deletion constructs. The RNA2 bands are boxed. (C) Relative amounts of genomic and sg mRNAs produced by the deletion constructs. Amounts of each RNA species (genomic and subgenomic) produced by the deletion constructs were independently related to the amounts of the corresponding RNA species produced by the wild-type construct, which were set at 100%. The average (of three experiments) amounts of RNAs are shown.
FIG. 7.
(A) Scheme of the constructs bearing TRS-free antisense EAV sequences. The antisense sequences were inserted into either the NcoI restriction site (030-1817f) or between the BssHII and MluI restriction sites (030-1762f). Note the reverse orientations of the body TRSs 3.1, 3.2, 4, and 5 in these constructs. The corresponding deletion constructs (030-1817 and 030-1762) are also shown. (B) Northern analysis of EAV-specific RNA isolated from cells transfected with RNA transcribed either from the wild-type EAV infectious cDNA clone or from the TRS-free insertion and corresponding deletion constructs. The RNA2 bands are boxed. (C) Relative amounts of genomic and sg mRNAs produced by the TRS-free insertion and corresponding deletion constructs. See the legend to Fig. 6 for the calculation of RNA levels. The average (of two experiments) amounts of RNAs are shown.
RNA transfection and immunofluorescence assays.
Following in vitro transcription from full-length cDNA clones, EAV RNA was introduced into BHK-21 cells by electroporation as described by van Dinten et al. (55). To determine transfection efficiencies, transfected cells were seeded on coverslips and fixed with 3% paraformaldehyde in phosphate-buffered saline at 14 to 16 h posttransfection. Immunofluorescence assays were carried out as described previously (53). To visualize the nuclei for cell counting, nuclear DNA was stained with 5 μg of Hoechst B2883 (Sigma) per ml. Cells were counted with the Scion Image software (Scion corporation), and the percentage of transfected cells was calculated on the basis of the number of cells positive for the EAV replicase component nsp3 (36).
RNA isolation and analysis.
For RNA analysis, cells were lysed at 14 h posttransfection. Isolation of intracellular RNA was performed with the acidic phenol method as described by Pasternak et al. (33). Total cytoplasmic RNA was resolved in denaturing agarose-formaldehyde gels. Hybridization of dried gels with the radioactively labeled oligonucleotide probe E154 (5′-TTGGTTCCTGGGTGGCTAATAACTACTT-3′), which is complementary to the 3′ end of the EAV genome and recognizes all viral mRNA molecules (genomic and subgenomic), and phosphoimager quantitation of individual bands were performed as described by Pasternak et al. (33).
RESULTS
Activity of the EAV RNA7 body TRS is strongly influenced by its position in an EAV replicon.
In the wild-type situation, each EAV body TRS is located in a unique primary and higher-order sequence context, which is assumed to modulate its transcriptional activity. Thus, the wild-type genome, with its array of nonidentical body TRSs, is not an ideal system to study the effects that a body TRS can exert on up- and downstream body TRSs.
In order to standardize body TRS context, a cassette that contained the sg RNA7 body TRS (5′-UCAACU-3′), 21 nt of its natural upstream flanking sequence, and 388 nt of its downstream flanking sequence, including the complete EAV N protein gene which is normally expressed from sg mRNA7, was designed (Fig. 2A). The 21-nt upstream flanking sequence sufficed to obtain abundant transcription of sg mRNA7, albeit at a somewhat reduced level compared to that of the wild-type genome (see below). One to four copies of this cassette were cloned head to tail directly downstream of the EAV replicase gene, the 3′-proximal cassette being the “natural” ORF7 and the others replacing the part of the genome that contains all envelope protein genes and body TRSs (Fig. 2A). Because the sg mRNA2 body TRS is located within the replicase gene, it could not be inactivated by deletion; instead, all constructs contained the previously described sg RNA2 body TRS knockout mutation (33), which is translationally silent with respect to the replicase gene (5′-UCAACU-3′ to 5′-UuAAuU-3′). The body TRSs inserted in the constructs with the one to four direct repeats were designated TRS A, TRS B, TRS C, and TRS D (in the 5′-3′ direction), and the replicon constructs were named mA, mAB, mABC, and mABCD (Fig. 2A). It should be noted that TRSs B, C, and D are located in identical sequence contexts over a distance of more than 400 nt up- and downstream. TRS A, on the other hand, shares only 21 nt of upstream flanking sequence (but still 400 nt of the downstream sequence) with the other TRSs. The synthesis of one to four sg mRNA species (sg mRNAs A to D) from the body TRSs in these constructs was expected. The complete N protein gene was preserved at the 5′ end of sg mRNAs A to D in order to avoid the instability that may result from the fact that an mRNA lacks a 5′ ORF.
BHK-21 cells were transfected with RNA that had been in vitro transcribed from the replicon constructs with one to four repeats of the RNA7 body TRS cassette. As a control, we used construct WTM−, a derivative of the EAV cDNA clone bearing a frameshift mutation in the membrane (M) protein gene (30). This mutation prevented virus particle formation and, consequently, the spread of virus to neighboring cells (30), which was important because the replicon constructs used in this study were all unable to produce infectious progeny. Construct WTM− produced wild-type levels of all sg RNAs (Fig. 2B and data not shown) and was used as a control for EAV sg mRNA synthesis throughout this study. At the end of the first cycle of virus replication, intracellular RNA was isolated as described above, and transfection efficiencies (usually 20 to 40%) were determined by immunofluorescence assay. The amounts of viral RNA (genomic and subgenomic) were calculated by phosphoimager scanning of denaturing formaldehyde-agarose gels hybridized with oligonucleotide probe E154, which recognizes all virus-specific plus-strand RNA molecules. The data were normalized for transfection efficiency, and each of the constructs was tested in duplicate.
Figures 2B and 2C show that, regardless of the number of TRSs in a construct, a perfect gradient of sg mRNA abundance was obtained, progressively favoring the smallest transcripts. In other words, identical body TRSs that were located in identical 400-nt up- and downstream sequence contexts (TRSs B, C, and D) but differed in their relative order (and, therefore, in the sizes of the corresponding sg mRNAs) displayed different transcriptional activities. TRS A, which has a unique (upstream) sequence context among the four TRSs (see above and Fig. 2A), produced approximately three times more sg RNA A in mA than in the other three constructs. This clearly confirmed that sg RNA abundance is determined by factors other than stability of the leader TRS-body TRS duplex and the primary sequence and/or structural context of the body TRS. Even after standardization of body TRS sequence contexts, sg RNA levels were different and correlated with either the relative order of the TRSs in the genome or sg RNA size or both. To distinguish between these possibilities, an additional series of mutants were produced, which will be discussed in the next section.
At the same time, our analysis showed that overall body TRS context, not necessarily involving immediate flanking sequences, also plays a role in the regulation of sg RNA synthesis. Compared to sg mRNA7 produced by the wild-type control, sg mRNA A of mA was 3.5 times less abundant and sg mRNAs B of mAB and C of mABC were 1.5 times less abundant (Fig. 2C). All these sg RNAs were of the same size and each was produced from the most 3′-proximal body TRS in the construct. However, except for the 21 nt immediately 5′ of the TRS, these TRSs had different upstream flanking sequences, which apparently influenced sg mRNA abundance. In contrast, TRS B in mAB and TRS C in mABC, which were located in identical up- and downstream sequence contexts, directed the synthesis of almost equal amounts of sg mRNAs.
Remarkably, in the mABCD construct, the amounts of genomic and sg RNAs were somewhat decreased compared to those of equivalent RNA species (in terms of size and relative position of the body TRS with respect to the 3′ end) from the other constructs (Fig. 2C). This may imply that the level of EAV-specific RNA molecules was limited by the availability of a (virus- or host-encoded) factor which is involved in both genome replication and sg RNA synthesis.
Effect of different body TRS mutations in the context of the three-repeat construct.
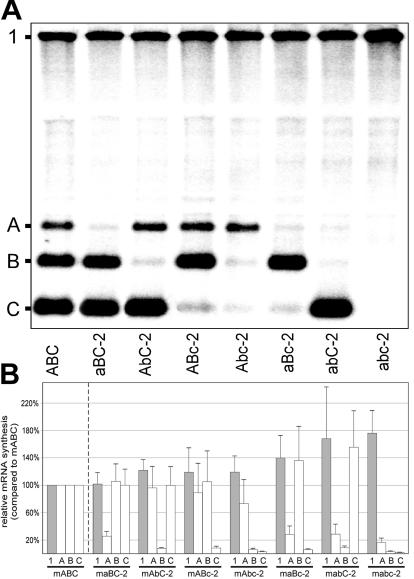
The directionality of the discontinuous minus-strand synthesis model predicts that disruption of the attenuating function of a body TRS would enhance the activity of upstream but not of downstream body TRSs. Due to the reduced attenuation of minus-strand synthesis by the mutant TRS, more minus-strand-synthesizing complexes would reach upstream TRSs. We previously identified body TRS nucleotide substitutions that strongly reduced body TRS activity (33, 34, 56). We now analyzed whether these substitutions can modulate the activity of flanking body TRSs by generating two panels of mutant three-repeat constructs (Fig. 3A), representing all possible combinations of wild-type and mutant body TRSs.
In the first panel, mutant body TRSs carried a double C-to-G substitution (C2 to G and C5 to G; 5′-UCAACU-3′ to 5′-UgAAgU-3′) which, in the context of the EAV full-length clone, blocked sg mRNA7 synthesis to a level that could not be detected by reverse transcription-PCR methods (56; G. van Marle, W. J. M. Spaan, and E. J. Snijder, unpublished data). The constructs were named according to the particular combination of wild-type and mutant TRSs, which was indicated with capital and lowercase letters, respectively (for example, construct mAbC contained wild-type TRSs A and C and a mutant TRS B; Fig. 3A). The amounts of sg mRNAs and genome produced by these mutant constructs were compared to those of construct mABC, which contained three wild-type body TRS7 cassettes. The experiments were repeated three times, and average relative sg RNA amounts were calculated.
Figures 3B and 3C show that, as predicted, sg mRNA synthesis from mutant body TRSs was efficiently blocked in all constructs. The low levels of residual sg RNA synthesis from mutant TRSs in some of the constructs can be explained by RNA recombination and will be discussed in the next section. The “downregulation” of sg mRNA synthesis from some of the wild-type TRSs in constructs containing mutant TRSs (e.g., sg mRNA A in mAbc or B in maBc compared to mABC) can probably also be explained by RNA recombination. Remarkably, though, the introduction of mutant body TRSs did not dramatically increase transcription from either up- or downstream TRSs or genome replication, suggesting that body TRSs (or their inactivation by mutagenesis) did not influence the activity of their neighbors.
The absence of any effect of the C2-to-G and C5-to-G body TRS substitutions on the activity of up- and downstream body TRSs can be explained by the fact that these nucleotides participate in the base pairing interaction that forms the leader TRS-body TRS duplex (34). This conclusion was based on the observation that the reduction of mRNA7 synthesis that was caused by the replacement of either cytosine in the RNA7 body TRS could be efficiently complemented by introduction of the same mutation into the leader TRS, thus restoring the possibilities for duplex formation. This demonstrated that, despite the replacement of these nucleotides in the body TRS, attenuation of minus-strand RNA synthesis at the body TRS could still occur. In the absence of a matching leader TRS, e.g., in our mutant three-repeat constructs, all of which contained a wild-type leader TRS, attenuation of minus-strand synthesis at a mutant body TRSs should yield a nascent minus stand with a 3′ end that is unable to base pair with the leader TRS. Although this would reduce the synthesis of the corresponding sg RNA species, this scenario predicts that the number of minus-strand-synthesizing complexes reaching flanking body TRSs would not change, and consequently, their transcriptional activity would not be affected.
Based on the hypothesis outlined above, we designed a second panel of mutant three-repeat constructs, now with a combination of four body TRS mutations (5′-UCAACU-3′ to 5′-aguACa-3′) (33) that also abolished sg mRNA7 synthesis to a level that could not be detected by reverse transcription-PCR (A. O. Pasternak and E. J. Snijder, unpublished data). A body TRS-specific defect was previously attributed to two of these mutations, U1 to A and A3 to U. Introduction of the same mutation in the leader TRS did not restore sg RNA synthesis, suggesting that nascent minus strands carrying these mutations in their 3′-terminal TRS complement were either not produced at all or were somehow unable to engage in the base pairing interaction with the leader TRS. In any case, in the context of the full-length genome, the phenotype of these mutations was fundamentally different from that of the C2 and C5 replacements used in the first panel of mutants (34).
The nomenclature of the mutants in the second series was analogous to that in the first series (Fig. 3A), resulting in maBC-2, mAbC-2, and so on. We anticipated that the attenuating potential of the mutant body TRS would now be affected, leading to enhanced transcriptional activity of upstream body TRSs. Remarkably, however, such polar effects of mutant downstream body TRSs on their upstream neighbors were again not observed (Fig. 4), and the results were strikingly similar to those obtained with the first panel of body TRS mutants (Fig. 3), suggesting that the second mutant TRS had also retained its attenuating potential for minus-strand synthesis. Consequently, we postulate that sg RNA synthesis from the 5′-aguACa-3′ mutant body TRS is blocked at an intermediate stage, i.e., between attenuation of minus-strand synthesis and base pairing with the leader TRS, for example, because an interaction with body TRS-specific protein factors is affected by the nucleotide substitutions. In agreement with this, and again suggesting the involvement of common factors in genome replication and sg mRNA synthesis, in the second series of mutant constructs (lanes maBc-2, mabC-2, and mabc-2 in Fig. 4B), there seemed to be a general tendency towards enhanced genomic and sg RNA synthesis compared to mABC. This enhancement seemed to occur in an unpolar fashion and apparently at the expense of sg mRNA synthesis from the mutant TRSs. However, a direct comparison in a single experiment between the genomic RNA levels of the mabc and mabc-2 constructs (for which the largest difference was expected, based on the comparison of Fig. 3C and 4B) did not reveal statistically significant differences (data not shown).
FIG. 4.
(A) Northern analysis of EAV-specific RNA isolated from cells transfected with RNA transcribed either from the wild-type (wt) EAV infectious cDNA clone or from the wild-type and seven mutant three-repeat constructs of the second series (TRS mutations 5′-UCAACU-3′ to 5′-aguACa-3′). (B) Relative amounts of genomic and sg mRNAs produced by the mutant three-repeat constructs of the second series. See the legend to Fig. 3 for the calculation of RNA levels. The average (of three experiments) amounts of RNAs are shown.
Homologous recombination in constructs with multiple RNA7 body TRS cassettes.
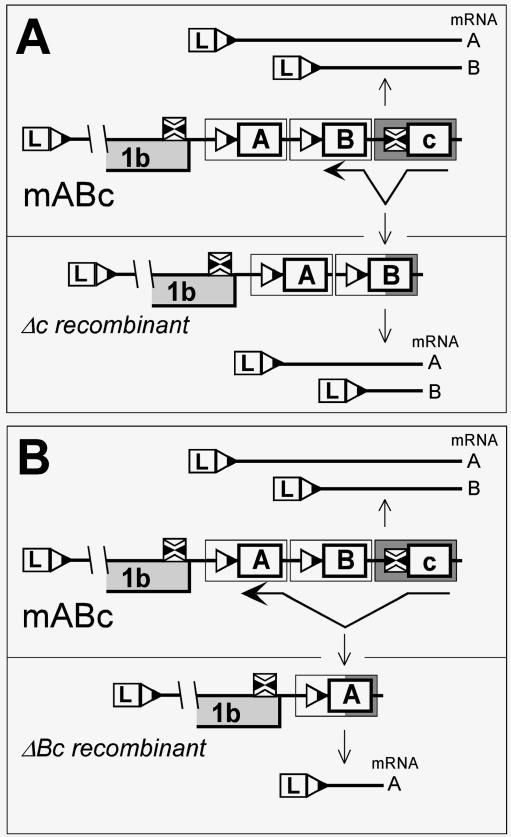
The mutations introduced into the body TRSs of both series of three-repeat constructs efficiently knocked out sg mRNA7 synthesis when tested in the context of the wild-type viral genome (33, 56). However, small amounts of sg mRNAs seemed to be produced from the same mutant TRSs in a number of three-repeat constructs (Fig. 3B and 4A). We interpreted this intriguing phenomenon as follows. The sequence similarity in the three direct repeats of the 417-nt RNA7 body TRS cassette in mABC and its mutant derivatives will promote RNA recombination, based on RNA-dependent RNA polymerase (RdRp) template switches that may be either intra- or intermolecular and that may occur during either genome replication or sg RNA synthesis. During genome replication (see Fig. 5 for an example), replication-competent recombinant genomes that have precisely one or two cassettes deleted or inserted may arise. If the deleted segment contains a mutant TRS and at least one upstream wild-type body TRS is present, then the synthesis of the sg mRNA that was abolished due to the TRS mutations will appear to be partially restored. The sizes of the sg transcripts produced from the wild-type TRSs in such recombinant genomes will be reduced by one (Fig. 5A) or two (Fig. 5B) cassettes and consequently will match the size of the sg mRNAs whose transcription was initially inactivated by the TRS mutations. Likewise, if a wild-type TRS-containing cassette is inserted by an RNA recombination event downstream of a mutated TRS, sg mRNA molecules from downstream TRSs will become one or two cassettes larger, and the synthesis of the larger sg mRNA will appear to be partially restored. Interestingly, the observed restoration of the synthesis of larger sg mRNA species due to the presence of active downstream TRSs (see, e.g., lane abC in Fig. 4A) suggests the insertion of one or two cassettes by either a “backward jump” of the RdRp complex or an intermolecular template switch. Similar homologous RNA recombination events may also occur during sg RNA synthesis.
FIG. 5.
Schematic representation of the proposed homologous recombination in the TRS7 repeat constructs. For clarity, mABc is shown as a parental construct. Both upper panels show its composition as well as the sg mRNA molecules (A and B) produced. Synthesis of the smallest sg mRNA molecule (RNA C) is knocked out by TRS mutation in the parental construct. Homologous recombination may result in deletion of either one (A) or two (B) TRS cassettes, rendering the sg mRNAs one (Δc recombinant) or two (ΔBc recombinant) cassettes smaller, respectively, as depicted in the lower panels. The sizes of sg mRNAs B (Δc recombinant) and A (ΔBc recombinant) would match the size of sg mRNA C, which would appear to be partially restored. Analogous homologous recombination may result in insertion of one or two cassettes.
If the above explanation of our observations is correct, then it is expected that no residual sg RNA synthesis would be observed for constructs mabc and mabc-2, because all their body TRSs are mutant and their “repair” by recombination events involving wild-type body TRS cassettes is impossible. The results in Fig. 3B and 4A indeed showed that the three-repeat construct required at least one functional TRS to activate the synthesis of specific sg mRNAs, suggesting that constructs with repeated TRS cassettes indeed underwent RNA recombination. Although this phenomenon clearly affected our quantitative analysis of sg mRNA synthesis by these constructs, the Northern blot analyses shown in Fig. 3B and 4A make it clear that this influence must have been very moderate. Thus, we are confident that the main conclusions of this work are not affected by the presence of a small population of recombinant genomes (and their transcripts) at the time of RNA isolation.
Effects of deletion of body TRS-containing sequences on sg mRNA abundance.
The data obtained with the RNA7 body TRS-repeat constructs suggested that, although body TRSs are probably part of the attenuation signals for minus-strand synthesis, the six-nucleotide body TRS itself is not sufficient to attenuate nascent minus-strand synthesis. Also, the effect of TRS point mutations or combinations of TRS point mutations is apparently not strong enough to disrupt attenuation and thereby enhance the activity of upstream TRSs. Thus, we concluded that a larger genome segment, containing the TRS and its flanking sequences, might be required for attenuation. In order to remove such an attenuation signal and enhance the activities of upstream body TRSs, it would be necessary to delete a relatively large region containing one or more body TRSs.
To test this hypothesis, we used a previously engineered series of deletion constructs (30) that lacked 1.6 to 2.3 kb of genome sequence from the structural protein-coding region of the EAV full-length cDNA clone (Fig. 6A). The common 5′ border of these deletions was a BalI restriction site located 0.3 kb downstream of the RNA2 body TRS (which should leave enough of its 3′-flanking sequence to ensure optimal sg mRNA2 synthesis). In all these constructs, the RNA2 TRS was the only body TRS upstream of the deletion, whereas the variable 3′ border of the deletion was located 3′ of the RNA5, RNA6, or RNA7 body TRS. Thus, the RNA3.1 to RNA5 body TRSs were deleted in all constructs and the RNA6 and RNA7 body TRSs in some constructs. Consequently, these constructs were expected to produce sg mRNAs 2, 6, and 7 (030-1615 and 030-1717), sg mRNAs 2 and 7 (030-1927 and 030-2207), or sg mRNA2 only (030-2282 and 030-2319), with the size of mRNA2 being variable due to the fact that this transcript contained the engineered deletions. According to the discontinuous minus-strand synthesis model, we expected upregulation of RNA2 body TRS activity and unchanged sg mRNA synthesis from the RNAs 6 and 7 body TRSs upon deletion of an interjacent part of the genome that contained a number of other body TRSs. The six deletion constructs were tested in three independent experiments along with a wild-type control (Fig. 6B), and the average relative amounts of genome RNA and the sg mRNAs derived from body TRSs 2, 6, and 7 were determined (Fig. 6C).
Four of six deletion constructs (030-1615, 030-1717, 030-1927, and 030-2207) showed upregulation of sg RNA synthesis from the RNA2 body TRS (1.5- to 2.3-fold compared to the wild-type control; Fig. 6B and 6C). In the same four constructs, genome replication was also enhanced 1.4- to 1.75-fold. At the same time, production of sg mRNAs 6 and 7 remained at wild-type levels, with the exception of the 030-2207 construct, which synthesized three times less sg mRNA7, apparently because, as in the RNA7 TRS three-repeat constructs discussed above, only 21 nt of the natural upstream flanking sequence of the RNA7 body TRS had been retained.
The data obtained with the deletion constructs were consistent with the discontinuous minus-strand synthesis model: the transcription of the sg mRNA species produced from the body TRS upstream of the deletion was enhanced, whereas the production of sg mRNA species from the body TRSs downstream of the deletion remained the same. Furthermore, the data again suggest the coordinated regulation of genome replication and sg mRNA synthesis: the enhanced genome replication can be explained by the fact that, due to the deletion of several body TRSs, more RdRp complexes were available to synthesize a full-length minus strand. Because the full-length RNA is also the template for sg RNA2 synthesis, one could argue that the variations in RNA2 synthesis in these constructs were solely due to the variation in genome replication. However, it seems that the modulations of these two processes in the deletion constructs were at least partially independent, because the RNA2/RNA1 ratios varied quite substantially, from 0.17 ± 0.05 in the 030-2319 construct to 0.53 ± 0.08 in the 030-1615 construct. A comparison of lanes 030-1927 and 030-2207 in Fig. 6C also argues against that view; while the amount of genome was comparable, sg mRNA2 synthesis surprisingly dropped 1.5-fold in construct 030-2207. Furthermore, if the variations in sg mRNA synthesis were due solely to the variations in genome replication, one would expect upregulation of sg mRNA 6 and 7 production in the 030-1615, 030-1717, 030-1927, and 030-2207 constructs, which was not observed.
In the remaining two constructs, 030-2282 and 030-2319, which contained the largest deletions, both genome replication and sg mRNA2 synthesis were downregulated. Reduced genome replication can be explained by the fact that the region of the genome downstream of the 3′ border of the 030-2207 deletion (nt 12228) contained sequences required for optimal replication. Note that deletion of an additional 200 nt downstream resulted in a construct with severely impaired genome replication (30). Unexpectedly, in constructs 030-2282 and (especially) 030-2319, sg mRNA2 synthesis was reduced to a larger extent than genome replication. It is possible that in the 3′ end of the genome, the cis-acting signals required for sg mRNA synthesis are partially distinct from those for genome replication, resembling the situation in the coronavirus MHV (26).
Although the data obtained with the deletion constructs can be explained by the discontinuous minus-strand synthesis model, the possibility remains that, in addition to the relative order of body TRSs (or, more precisely, the number of downstream body TRSs), the size of specific EAV RNA molecules also influences or determines their relative abundance. Indeed, only the production of those RNA species that had become smaller (RNA1 and sg mRNA2) was enhanced, and no effects were seen on the production of the RNA species (mRNA6 and mRNA7) that had retained their original size (Fig. 6). Based on the deletion constructs tested so far, we could not exclude this possibility, although it was clear that RNA size, if it does play a role, is certainly not the sole factor governing RNA synthesis. For example, in the constructs with the smallest RNA1 and RNA2 sizes (030-2282 and 030-2319), their abundance was decreased instead of upregulated. Genome levels in the other four deletion constructs were enhanced 1.4- to 1.75-fold, whereas genome size was reduced by a maximum of 17%. Also, the upregulation of sg and genome RNA abundance in the second series of mutant three-repeat constructs (see above) cannot be explained by changes in RNA size. Nevertheless, we wanted to assess the importance of RNA size and therefore produced a final set of constructs (Fig. 7) in which the deleted sequences were replaced with (putative) “TRS-free” inserts of identical size.
Insertion of TRS-free sequences between adjacent body TRSs.
To test whether it is the number of downstream body TRSs or the size of the sg transcript that plays a major role in the regulation of sg mRNA abundance, we decided not only to delete a body TRS-containing fragment from the genome but to also replace it with a putatively TRS-free (i.e., devoid of 5′-UCAACU-3′ sequences) sequence of the same size. In this manner, the size of the genome and the sg mRNAs produced from body TRSs upstream of the replacement would remain the same. If the number of downstream body TRSs determines sg mRNA abundance, then the production of sg mRNAs from body TRSs upstream of the heterologous replacement would be enhanced, as in the deletion constructs discussed above. If, on the other hand, sg mRNA size is more important, then sg RNA levels would remain the same.
To investigate this issue, four constructs were produced (Fig. 7A). Two of them, 030-1817 and 030-1762, contained deletions of 1,817 and 1,762 nt, respectively. Like some of the deletion mutants described in the previous paragraph, both constructs lacked the sequences containing the body TRSs for mRNAs 3.1, 3.2, 4, and 5, so that the RNA2 body TRS was the only body TRS upstream of the deletion, making sg mRNA2 synthesis the readout of the experiment. In the other two constructs, 030-1817f and 030-1762f, the antisense copy of the deleted fragment was inserted at the site of the deletion, as described in Materials and Methods. These antisense EAV sequences were devoid of 5′-UCAACU-3′ sequences and were therefore, at least in theory, considered TRS free. Their insertion exactly restored the sizes of sg mRNA2 and genomic RNA to the wild-type values. These constructs were tested in two independent experiments along with the wild-type construct.
The amounts of sg RNAs in the new deletion constructs (030-1817 and 030-1762) did not differ much from those of the deletion constructs described in the previous chapter (Fig. 7B and 7C); mRNA2 amounts were upregulated 1.5 to 2-fold, whereas amounts of sg mRNAs 6 and 7 remained on the wild-type level or were even slightly downregulated (030-1762). In the 030-1817 construct, genome replication was enhanced by 40%, whereas in the 030-1762 construct, it was not increased at all. In contrast to what was expected, sg mRNA2 synthesis by the 030-1817f and 030-1762f constructs was not enhanced at all, but instead was decreased threefold compared to the wild-type value. Also, genome replication was reduced more than twofold. The reasons for this unexpected reduction in sg and genomic RNA synthesis are puzzling. It is possible that the insertion of minus-sense sequences in plus-sense viral RNA (or vice versa) caused instability of the viral RNA molecules. Alternatively, because the inserted sequence is an antisense copy of a segment containing several body TRSs, it is possible that, in these constructs, the synthesis of plus instead of minus strands was now attenuated at the body TRS complements.
DISCUSSION
Interplay between nidovirus body TRSs.
The present study is the first to specifically address the relative abundance of arterivirus sg mRNAs. Previously, several studies have been conducted to investigate this issue for coronaviruses. However, most of these studies relied on sg RNA synthesis from helper virus-dependent replicons, which were based on defective interfering RNA genomes. Obviously, this is a less straightforward experimental system than the full-length cDNA clone used in this study for EAV. Joo and Makino (19) inserted two identical body TRSs in an MHV replicon and observed that the downstream TRS inhibited sg RNA synthesis from the upstream TRS. When the downstream TRS was mutagenized, resulting in underproduction of the corresponding sg RNA, its polar attenuating effect on the upstream one was less pronounced. Removal of the downstream TRS restored sg RNA synthesis from the upstream body TRS. Because these effects were observed only when body TRSs were inserted close to one another, not when they were separated by more than 100 nt, the authors proposed that their findings resulted from steric hindrance between (putative) scanning transcription factors that bind to body TRSs. Similar results were obtained in the bovine coronavirus replicon system (22), where tandem placement of two or three RNA7 body TRSs into a bovine coronavirus replicon resulted in the almost exclusive synthesis of sg RNA from the downstream TRS. These findings argue against the exclusive role of size in the regulation of sg RNA abundance, because prominent effects were observed despite negligible sg RNA size differences.
Van Marle et al. (57) inserted equivalent oligonucleotide cassettes representing either wild-type or mutant RNA3 body TRSs at three well-separated positions in an MHV replicon (in contrast to the above studies, where TRSs were placed close to one another). In these constructs, the middle body TRS always produced the most abundant sg RNA species, irrespective of the functionality of the other TRSs. The most upstream TRS was always the least active one, and the amount of sg RNA produced by the downstream TRS was intermediate. These findings argued once more against an exclusive regulatory role of sg RNA size. At the same time, they underscored the important role of body TRS-flanking sequences in transcriptional regulation, a notion that is also supported by more recent data obtained for transmissible gastroenteritis virus by Alonso et al. (1). Similar effects were observed by Hsue and Masters (15) when they inserted an extra MHV RNA7 body TRS in the 3′ untranslated region of the MHV genome by targeted homologous recombination. When placed ectopically, this RNA7 TRS was eight times less active than in its natural position, despite the fact that it was the most 3′-proximal body TRS in this mutant, thus yielding the smallest sg RNA species. The above results might be partially explained by TRS positional effects. However, using an MHV replicon system, Jeong et al. (17) have shown that this effect was determined by differences in body TRS sequence context and not directly by the position of the TRS in the genome itself.
In this study, using the EAV full-length genome, we standardized the sequence contexts of body TRSs to eliminate any potential effects of their immediate flanking sequences on sg mRNA transcription. As expected, this resulted in a perfect gradient of sg mRNA abundance (Fig. 2), progressively favoring RNA species produced from more 3′-proximal TRSs, in accordance with the proposed role of the body TRS as an attenuator of minus-strand synthesis. However, similar results were obtained for brome mosaic virus, in which sg mRNA synthesis is initiated internally on a genomic minus-strand template (28) and attenuation of minus-strand synthesis does not play a role. Insertion of multiple promoter elements into brome mosaic virus RNA3 resulted in an sg RNA gradient progressively favoring the smaller transcripts (12). This result may be explained by differences in the time required to synthesize sg mRNAs of different sizes. For coronaviruses, it has been proposed that sg mRNA abundance reflects both the relative abundance of the corresponding sg minus-strand templates and the relative size of an sg mRNA (4).
RNA and protein determinants of body TRS activity.
It was unclear which factor was the primary determinant of the sg mRNA gradient that we obtained for EAV (Fig. 2). To clarify this issue, we engineered two series of EAV three-repeat constructs (Fig. 3 and 4), similar to the approach followed previously for MHV by van Marle et al. (57). In that study, downstream functional TRSs suppressed sg RNA synthesis from upstream TRSs, whereas upstream TRSs had little or no effect on downstream TRSs. Since these TRSs were separated by considerable distances (361 to 761 nt), it was unlikely that steric hindrance occurred. Likewise, the novel MHV body TRS inserted by Hsue and Masters (15) exerted polar attenuating effects on upstream TRSs over a distance of at least 2,076 nt, reducing sg mRNA 7 and 6 transcription by one-half and one-third, respectively. Such polar attenuating effects of 3′-proximal sg RNAs on the synthesis of 3′-distal ones are again not specific for nidoviruses, as they were also observed in the luteovirus barley yellow dwarf virus (20), when the sg RNA1 promoter was duplicated and placed ectopically in the genome. Although the mechanism of barley yellow dwarf virus sg RNA synthesis is obviously different from that of nidoviruses, it has been argued that it might involve premature termination of negative-strand synthesis (20), which would explain the observed polar effects.
In contrast to the studies discussed above, we did not observe any polar effects of mutations in 3′-proximal body TRSs on 3′-distal ones in the EAV replicon system. In the context of the discontinuous minus-strand synthesis model, we previously speculated (34) that body TRS-specific transcription defects, which could not be compensated for by restoration of the base pairing possibilities with the leader TRS, might derive from impaired attenuation of minus-strand synthesis at the body TRS. Body TRS substitutions inducing such defects were included in the mutant TRS in our second series of three-repeat constructs (Fig. 4), but polar effects of mutant downstream TRSs on their upstream neighbors were not observed. This argues against the above hypothesis and suggests that a larger RNA domain (TRS plus flanking sequences) may determine attenuation. This contradicts the findings of van Marle et al. (57), where a single point mutation in the MHV RNA3 body TRS was apparently sufficient to impair its attenuating function. Possibly, that mutation specifically disrupted a higher-order RNA structure necessary for attenuation, although we could not achieve this effect for EAV even by mutating four of six TRS nucleotides. Alternatively, the sequence or structural requirements for attenuation in the MHV defective interfering RNA system and the EAV genome may be different.
Although our TRS mutations apparently could not impair attenuation of minus-strand synthesis, we believe that body TRS-specific transcription defects (34) could derive from impaired binding of a regulatory factor that normally interacts either with the body TRS in the plus-strand template or with its complement in the nascent minus strand. Consequently, minus-strand synthesis would still be attenuated in such body TRS mutants, but a subsequent step in the mechanism, mediated by this regulatory factor, would be blocked. Introduction of an sg RNA promoter was found to impair genome replication in the context of both a full-length barley yellow dwarf virus genome (20) and an MHV defective-interfering-RNA-based replicon (16). In that study, sg RNA synthesis impaired genome replication in cis but not in trans, contradicting the hypothesis of Sethna et al. (41) that genome replication and sg mRNA synthesis compete for a limited amount of trans-acting factors. On the other hand, van Marle et al. (57) did not observe any effects of sg RNA synthesis on the accumulation of similar MHV replicons. It is possible that the single point mutation that van Marle et al. used to knock out the TRS activities did not interfere with the binding of a putative regulatory factor.
The components of the viral RdRp complex are obvious candidates to play a role in the regulation of nidovirus sg mRNA synthesis. One or several components of this complex may interact with body TRSs and commit the RdRp complex to sg mRNA synthesis in a fashion similar to the mechanism of initiation of eukaryotic DNA-dependent RNA polymerase II transcription. The affinity of such regulatory components for body TRSs, together with the relative order of body TRSs in the genome, may be the major factors that determine sg mRNA levels, whereas RdRp complexes not committed to sg mRNA synthesis would be available to replicate the genomic RNA. When all RdRp complexes are engaged in genomic and sg mRNA synthesis, a further increase in total viral RNA synthesis would be impossible. This would explain the downregulation of the synthesis of all RNA species of construct mABCD. On the other hand, in the second series of mutant mABC derivatives (Fig. 4), the removal of a putative TRS-linked recognition signal for the RdRp complex may have resulted in a (modest) increase in the capacity to synthesize other RNA species.
Interestingly, genome replication of the replicase mutant EAV030F (55, 58), which carries a single mutation (Ser-2429 to Pro) in the nsp10 helicase (42), was recently found to be enhanced compared to that of wild-type EAV (A. O.Pasternak and E. J. Snijder, unpublished data). EAV030F is almost completely deficient in sg RNA synthesis, producing several hundred times fewer sg positive and negative strands than wild-type virus (58; A. O.Pasternak and E. J. Snijder, unpublished data). In this mutant, genome replication appears to have been enhanced following the loss of sg mRNA synthesis. Remarkably, specific point mutations in the predicted zinc finger domain of another EAV replicase subunit, nsp1, which is necessary for sg mRNA synthesis but fully dispensable for genome replication (51), were also recently found to shift the balance between genome replication and sg mRNA synthesis (M. A. Tijms, J. C. Dobbe, C. C. Posthuma, A. E. Gorbalenya, and E. J. Snijder, unpublished data). Taken together, these data suggest that specific replicase components, possibly including nsp10 or nsp1, may regulate sg mRNA synthesis by interacting (directly or indirectly) with body TRSs. Interestingly, the nsP2 replicase subunit of an alphavirus, Semliki Forest virus, was proposed to target the replicase to the sg promoter (50). In addition, host factors may also interact with TRSs and influence sg mRNA synthesis (43, 44, 61, 62).
Technical limitations of our current assays.
A number of studies on coronavirus and arterivirus sg mRNA synthesis (1, 2, 15, 17, 18, 31-35, 54, 56) argued that the stability of both the leader TRS-body TRS duplex and the body TRS sequence context must synergistically define its activity. In the present study we tried to standardize both of these factors and still observed dramatic differences in transcriptional activity. It was our aim to elucidate which factors caused these differences, but unfortunately throughout this study we were unable to separate the influence of two tightly linked factors, the relative order of the body TRSs and the size differences between the sg mRNAs that they specify. It is quite likely that these factors, alone or (more likely) in combination (4), are important, but with two different approaches, the mutant three-repeat constructs (Fig. 3 and 4) and the constructs in which TRS-free sequences were inserted (Fig. 7), we still could not separate their roles.
Clearly, we are approaching the limits of what reverse genetics systems and the biochemical analysis of transfected cells can reveal about the complex replication and transcription mechanism that is used by nidoviruses. One particular shortcoming of our current system is that we routinely measure the amount of sg mRNAs, which represents the end stage of a multistep transcription process that is governed by a number of factors. To dissect the influence of these factors, it will be necessary to develop readout systems for the individual steps. Thus, it should become possible to analyze and measure reaction intermediates, like the nascent sg minus strand that is thought to be transferred from the body TRS to the leader TRS (Fig. 1B). This would clarify which specific step of the process is affected by specific mutations. So far, detection of minus strands (genomic and subgenomic) has been hampered by the fact that they are only present in minor quantities in arterivirus-infected cells (10, 58). This prevents their reliable detection and quantitation, a problem that is enhanced by the nested-set structure of nidovirus plus- and minus-strand RNAs, which makes it hard, e.g., to design a one-step assay that would exclusively detect a molecule like the nascent minus strand prior to antileader addition. It is obvious that, in the long run, in vitro reconstitution systems for nidovirus sg RNA synthesis will be required to progress towards the next level of functional dissection and mechanistic understanding.
Acknowledgments
We acknowledge Richard Molenkamp and Babette Rozier for kindly providing the deletion constructs and Jessika Dobbe for technical assistance. We are grateful to Marieke Tijms, Richard Molenkamp, Erwin van den Born, and Volker Thiel for helpful discussions.
A.O.P. was supported by grant 700-31-020 from the Council for Chemical Sciences of The Netherlands Organization for Scientific Research.
REFERENCES
- 1.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, S., and S. Makino. 1998. Characterizations of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology 243:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., S. A. Stohlman, and M. M. C. Lai. 1983. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J. Virol. 48:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian, D. A., and W. J. M. Spaan. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, R. Y., R. Krishnan, and D. A. Brian. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 70:2720-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., L. Kuo, R. R. R. Rowland, C. Even, K. S. Faaberg, and P. G. W. Plagemann. 1993. Sequences of 3′ end of genome and of 5′ end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′ leader and bodies of seven subgenomic mRNAs. J. Gen. Virol. 74:643-659. [DOI] [PubMed] [Google Scholar]
- 8.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:927-935. [DOI] [PubMed] [Google Scholar]
- 9.de Groot, R. J., R. J. ter Haar, M. C. Horzinek, and B. A. van der Zeijst. 1987. Intracellular RNAs of the feline infectious peritonitis coronavirus strain 79-1146. J. Gen. Virol. 68:995-1002. [DOI] [PubMed] [Google Scholar]
- 10.den Boon, J. A., M. F. Kleijnen, W. J. M. Spaan, and E. J. Snijder. 1996. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J. Virol. 70:4291-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deVries, A. A. F., E. D. Chirnside, P. J. Bredenbeek, L. A. Gravestein, M. C. Horzinek, and W. J. M. Spaan. 1990. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 18:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann, M. A., R. Y. Chang, S. Ku, and D. A. Brian. 1993. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology 196:163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, M. A., P. B. Sethna, and D. A. Brian. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 64:4108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsue, B., and P. S. Masters. 1999. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J. Virol. 73:6128-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong, Y. S., and S. Makino. 1992. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J. Virol. 66:3339-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong, Y. S., J. F. Repass, Y. N. Kim, S. M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joo, M., and S. Makino. 1992. Mutagenic analysis of the coronavirus intergenic consensus sequence. J. Virol. 66:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koev, G., B. R. Mohan, and W. A. Miller. 1999. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 73:2876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konings, D. A., P. J. Bredenbeek, J. F. Noten, P. Hogeweg, and W. J. M. Spaan. 1988. Differential premature termination of transcription as a proposed mechanism for the regulation of coronavirus gene expression. Nucleic Acids Res. 16:10849-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, M. M. C., R. S. Baric, P. R. Brayton, and S. A. Stohlman. 1984. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl. Acad. Sci. USA 81:3626-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, M. M. C., C. D. Patton, and S. A. Stohlman. 1982. Replication of mouse hepatitis virus: negative-stranded RNA and replicative form RNA are of genome length. J. Virol. 44:487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, Y. J., X. Zhang, R. C. Wu, and M. M. C. Lai. 1996. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J. Virol. 70:7236-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meulenberg, J. J. M., E. J. de Meijer, and R. J. M. Moormann. 1993. Subgenomic RNAs of Lelystad virus contain a conserved leader- body junction sequence. J. Gen. Virol. 74:1697-1701. [DOI] [PubMed] [Google Scholar]
- 28.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 29.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 31.Nelsen, C. J., M. P. Murtaugh, and K. S. Faaberg. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 73:270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasternak, A. O., A. P. Gultyaev, W. J. M. Spaan, and E. J. Snijder. 2000. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 74:11642-11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription- regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawicki, D., T. Wang, and S. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 38.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Biol. Med. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 40.Schaad, M. C., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169-8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seybert, A., L. C. van Dinten, E. J. Snijder, and J. Ziebuhr. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 74:9586-9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen, X., and P. S. Masters. 2001. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc. Natl. Acad. Sci. USA 98:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, S. T., P. Huang, H. P. Li, and M. M. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 19:4701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shieh, C. K., L. H. Soe, S. Makino, M. F. Chang, S. A. Stohlman, and M. M. Lai. 1987. The 5′-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology 156:321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 47.Snijder, E. J., M. C. Horzinek, and W. J. M. Spaan. 1990. A 3′-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J. Virol. 64:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snijder, E. J., and Meulenberg, J. J. M. 2001. Arteriviruses, p. 1205-1220. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 49.Spaan, W. J. M., H. Delius, M. Skinner, J. Armstrong, P. J. M. Rottier, S. Smeekens, B. A. M. van der Zeijst, and S. G. Siddell. 1983. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 2:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suopanki, J., S. G. Sawicki, D. L. Sawicki, and L. Kaariainen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Virol. 79:309-319. [DOI] [PubMed] [Google Scholar]
- 51.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. USA 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van den Born, E., A. P. Gultyaev, and E. J. Snijder. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10:424-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Most, R. G., R. J. de Groot, and W. J. M. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dinten, L. C., J. A. den Boon, A. L. M. Wassenaar, W. J. M. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation which abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base- pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Marle, G., W. Luytjes, R. G. van der Most, T. van der Straaten, and W. J. M. Spaan. 1995. Regulation of coronavirus mRNA transcription. J. Virol. 69:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Marle, G., L. C. van Dinten, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 73:5274-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, X., and M. M. Lai. 1995. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J. Virol. 69:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, X., H. P. Li, W. Xue, and M. M. Lai. 1999. Formation of a ribonucleoprotein complex of mouse hepatitis virus involving heterogeneous nuclear ribonucleoprotein A1 and transcription-regulatory elements of viral RNA. Virology 264:115-124. [DOI] [PubMed] [Google Scholar]