Abstract
Transposable elements (TEs) are major components of vertebrate genomes, with major roles in genome architecture and evolution. In order to characterize both common patterns and lineage-specific differences in TE content and TE evolution, we have compared the mobilomes of 23 vertebrate genomes, including 10 actinopterygian fish, 11 sarcopterygians, and 2 nonbony vertebrates. We found important variations in TE content (from 6% in the pufferfish tetraodon to 55% in zebrafish), with a more important relative contribution of TEs to genome size in fish than in mammals. Some TE superfamilies were found to be widespread in vertebrates, but most elements showed a more patchy distribution, indicative of multiple events of loss or gain. Interestingly, loss of major TE families was observed during the evolution of the sarcopterygian lineage, with a particularly strong reduction in TE diversity in birds and mammals. Phylogenetic trends in TE composition and activity were detected: Teleost fish genomes are dominated by DNA transposons and contain few ancient TE copies, while mammalian genomes have been predominantly shaped by nonlong terminal repeat retrotransposons, along with the persistence of older sequences. Differences were also found within lineages: The medaka fish genome underwent more recent TE amplification than the related platyfish, as observed for LINE retrotransposons in the mouse compared with the human genome. This study allows the identification of putative cases of horizontal transfer of TEs, and to tentatively infer the composition of the ancestral vertebrate mobilome. Taken together, the results obtained highlight the importance of TEs in the structure and evolution of vertebrate genomes, and demonstrate their major impact on genome diversity both between and within lineages.
Keywords: genome size, transposon, mobile DNA, retrotransposon
Introduction
The genome sequences of mammals and other vertebrates have been shown to be significantly repetitive, with a strong contribution of transposable elements (TEs) to genome size and architecture (Deininger et al. 2003; Kazazian 2004; Feschotte and Pritham 2007; Böhne et al. 2008; Kordis 2009). TEs can disrupt host sequences and serve as substrates for homologous recombination, generating DNA rearrangements such as deletions, duplications, inversions, and translocations (Burns and Boeke 2012). Such rearrangements can be deleterious for the host through the alteration of gene-coding potential and regulation, or through the modification of other important genomic sequences (Kazazian 2004). TEs are therefore sources of mutations and genetic diseases in human and other organisms (Vorechovsky 2010; Hancks and Kazazian 2012).
On the other hand, there is convincing evidence that TEs are important for the function and evolution of genes, gene networks, and genomes (Böhne et al. 2008; Feschotte 2008; Ellison and Bachtrog 2013; Xie et al. 2013). In particular, many exons and regulatory sequences of host genes, and even entire new RNA and protein-coding genes are derived from TEs, a phenomenon called molecular domestication (Volff 2006; Rebollo et al. 2012; Jacques et al. 2013; Kapusta et al. 2013). One prominent example of a TE-derived gene with important function in vertebrates is the RAG1 protein, which together with RAG2 catalyzes the V(D)J somatic recombination responsible for the diversity of antigen-binding regions in immunoglobulins and T-cell receptors (Agrawal et al. 1998; Hiom et al. 1998; Kapitonov and Jurka 2005).
The propensity of TEs to transpose and increase their copy number is attenuated by host genome defense mechanisms such as DNA methylation and Piwi-interacting small RNAs (piRNAs) (Levin and Moran 2011). TE expansion phases can alternate with reduced activity (Le Rouzic and Capy 2005; Goodier and Kazazian 2008; Brookfield 2011). TEs can multiply in genomes either after introduction through horizontal transfer or mutational activation of resident copies, until the host becomes able to regulate their activity, for example, through the production of specific piRNAs (Le Rouzic and Capy 2009; Evgen’ev 2013). Prolonged reduced TE activity might lead to the elimination of the element.
On the basis of their mechanisms of transposition, TEs are ordered in two main classes, which are themselves split into orders, superfamilies, families, and subfamilies (Finnegan 1989; Wicker et al. 2007). Class I (retrotransposons) is composed of five orders (Malik et al. 1999; Eickbush and Jamburuthugoda 2008). Two orders harbor long DNA repeats: Long terminal repeat (LTR) retrotransposons (including endogenous retroviruses [ERVs]; Gifford and Tristem 2003), which show classical flanking LTRs in direct orientation, and Dictyostelium intermediate repeat sequence (DIRS) elements, with more complex long repeats that can be inverted and internally repeated. The three remaining orders, LINEs, SINEs (long/short interspersed nuclear elements), and Penelope (PLE)-like elements, are non-LTR retrotransposons. Autonomous retrotransposons encode a reverse transcriptase, while noncoding nonautonomous elements like the SINE sequences are mobilized in trans by proteins from autonomous elements. Prominent retrotransposon superfamilies in vertebrates include Gypsy, BEL/Pao (De la Chaux and Wagner 2011), and Copia (LTR retrotransposons), as well as DIRS1/Ngaro (DIRS retrotransposons; Poulter and Goodwin 2005). Vertebrate LINE superfamilies are LINE1, CR1 like (including the CR1, L2, and Rex1-Babar families; Chalopin et al. 2013; Kojima and Jurka 2013a, 2013b), RetroTransposable Element (RTE), Jockey, Dong/Rex6 (Volff et al. 2001; Novick et al. 2009), and R2 (Kapitonov and Jurka 2009; Kojima and Jurka 2013a, 2013b; Luchetti and Mantovani 2013).
Class II transposons (DNA transposons) are divided into two subclasses depending on the number of DNA strands that are cut during transposition (Wicker et al. 2007). Subclass I, in which both DNA strands are cleaved, contains TIR (terminal inverted repeat) transposons and Crypton elements, the former building the most abundant and diverse order. Autonomous TIR elements encode a transposase and move through a “cut and paste” mechanism. Crypton elements use a tyrosine recombinase in a transposition mechanism probably involving recombination between a circular intermediate and the DNA target. Subclass II elements include Helitrons (which replicate via a rolling-circle mechanism; Kapitonov and Jurka 2001) and Maverick/Polinton transposons (self-synthesizing transposons; Feschotte and Pritham 2005; Kapitonov and Jurka 2006). Nonautonomous elements such as miniature inverted transposable elements use the enzymatic machinery of autonomous DNA transposons to transpose.
With half of known extant vertebrate species, teleost fishes represent a very diverse group of animals at the morphological, ecological, and also genomic levels (Volff 2005; Nelson 2006; Ravi and Venkatesh 2008; Sarropoulou and Fernandes 2011). For example, teleost fishes show a wide range of genome sizes (from 0.32 to 133 billion base pairs; Gregory 2001). Different teleost models have been developed to study vertebrate development (medaka and zebrafish; Wittbrodt et al. 2002), cancer (platyfish; Schartl et al. 2013), speciation and behavior (cichlids and stickleback; Jones et al. 2012), and genome structure and evolution (fugu and tetraodon; Jaillon et al. 2004). Several of the studied teleost species, such as the Atlantic salmon, rainbow trout, or Nile Tilapia, are also of economic interest. Some studies have suggested that major differences in TE content exist between vertebrate sublineages, and that teleost fish genomes present a higher diversity of TEs than other vertebrate genomes (Volff et al. 2003; Duvernell et al. 2004; Furano et al. 2004; Basta et al. 2007; Böhne et al. 2008; Novick et al. 2009; Kojima and Jurka 2011). However, information on TE diversity and evolution in fish and other vertebrates is still incomplete. We therefore took advantage of the growing amount of available genomic data to perform a systematic comparative analysis of TE content and activity in fish species and other vertebrate sublineages. Our study uncovered common TE patterns in vertebrates, but also major differences in TE activity and evolution that very likely contributed to lineage-specific genomic and organismal diversity in vertebrates.
Materials and Methods
Genomic Data Sets
To build TE libraries, we collected genome sequences of amphioxus (Branchiostoma_floridae_v2.0.assembly.fasta, http://genome.jgi-psf.org/Brafl1/Brafl1.download.ftp.html, last accessed January 30, 2015), Oikopleura (Odioica_reference_v3.fa, http://www.genoscope.cns.fr/externe/GenomeBrowser/Oikopleura/, last accessed January 30, 2015), lamprey (Petromyzon_marinus.Pmarinus_7.0.70.dna.toplevel.fa from Ensembl, http://www.ensembl.org/index.html, last accessed January 30, 2015), elephant shark (EsharkAssembly, http://esharkgenome.imcb.a-star.edu.sg, last accessed January 30, 2015), fugu (Takifugu_rubripes.FUGU4.66.dna.toplevel.fa, Ensembl), tetraodon (Tetraodon_nigroviridis.TETRAODON8.73.dna.toplevel.fa, Ensembl), stickleback (Gasterosteus_aculeatus.BROADS1.68.dna.toplevel.fa, Ensembl), tilapia (Oreochromis_niloticus.Orenil1.0.68.dna.toplevel.fa, Ensembl), platyfish (Xiphophorus_maculatus.Xipmac4.4.2.69.dna.nonchromosomal.fa, Ensembl), medaka (Oryzias_latipes.MEDAKA1.73.dna.toplevel.fa, Ensembl), Atlantic cod (Gadus_morhua.gadMor1.73.dna.toplevel.fa, Ensembl), zebrafish (Danio_rerio.Zv9.66.dna.toplevel.fa, Ensembl), European eel (draft genome version 1, www.zfgenomics.org/sub/eel, last accessed January 30, 2015), spotted gar assembly accession update (http://www.ncbi.nlm.nih.gov/assembly/GCF_000242695.1/, last accessed January 30, 2015, Genbank Assembly), African coelacanth (Latimeria_chalumnae.LatCha1.72.dna_toplevel.fa, Ensembl), and Chinese soft-shell turtle (Pelodiscus_sinensis.PelSin_1.0.73.dna.toplevel.fa, Ensembl).
For tetrapods (except for turtle, see above) and Ciona, we directly used premasked genomes and RepeatMasker outfiles (“.out” and “.align”) from RepeatMasker Genomic Datasets (http://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html, last accessed January 30, 2015): Ciona (ci2), frog (xenTro2), American alligator (allMis0), green anole (anoCar2), zebra finch (taeGut1), chicken (galGal3), platypus (ornAna1), opossum (monDom5), mouse (mm9), and human (hg19). For premasked genomes, genome sizes correspond to the golden path available on the Ensembl server. For others, genome sizes were calculated during the masking process.
Construction of Species-Specific TE Libraries
We established species-specific TE libraries by combining automatic and manual annotations for the following species: amphioxus, lamprey, elephant shark, fugu, tetraodon, stickleback, tilapia, platyfish, zebrafish (manual and Repbase sequences; Jurka 2000), and spotted gar. Manual annotation involved searching the downloaded genomes, using TBLASTN (Altschul et al. 1990) using TE proteins from different superfamilies as queries. In this process, reverse transcriptases were used to find retrotransposons, and transposases to detect DNA transposons. The longest sequences derived from BLAST hits containing TE-specific features such as TIRs or LTRs as well as characteristic open reading frames were kept for further analyses, including molecular phylogeny-based positioning of the elements in the TE classification. Censor (Jurka et al. 1996) was also used to identify the sequences. Automatic annotation was performed using the RepeatModeler software (Smit, AFA, Hubley, R. RepeatModeler software weblink: http://www.repeatmasker.org, last accessed January 30, 2015) with default parameters. For the coelacanth, we used and reannotated the library from Amemiya et al. (2013).
Genome Masking
Amphioxus, Oikopleura, lamprey, elephant shark, fugu, tetraodon, stickleback, tilapia, platyfish, medaka, cod, zebrafish, spotted gar, coelacanth, and soft-shell turtle genomes were locally masked using RepeatMasker version 3.3.0 (Smit, AFA, Hubley, R, and Green, P. RepeatMasker Open-3.0. 1996–2010; http://www.repeatmasker.org, last accessed January 30, 2015) with “-a” and “-lib” default parameters.
Copy Number and Genome Coverage Estimation
Copy number and genome coverage were calculated on RepeatMasker outfiles (.out). Copy number corresponds to the listed number of insertions in the masked genomes. Total copy number and coverage for each superfamily were calculated using a custom script. Additionally, a second calculation was performed, including only sequence insertions longer than 80 nucleotides and sharing more than 80% of identity with the reference sequence from the species-specific library (supplementary information, Supplementary Material online). This eliminated very short and divergent sequences. After such filtering, the estimated total genome coverage by TEs was reduced, particularly in elephant shark, platyfish, European eel, spotted gar, turtle, alligator, and all mammals. This suggests that these genomes contain a significant number of very short and/or degenerated elements.
Sequence Alignments and Phylogenetic Reconstructions
Consensus TE nucleotide sequences were retrieved from TE libraries, translated into proteins using Augustus (human and chicken models; Stanke et al. 2004) and FGENESH (fish and zebrafish models, http://www.softberry.com/, last accessed January 30, 2015), and aligned using Clustal omega (Sievers et al. 2011). Phylogenetic trees were reconstructed using maximum likelihood with optimized parameters and default aLRT (nonparametric branch support) using the Seaview interface (Gouy et al. 2010).
Kimura Distance-Based Distribution Analysis of TE Copies in Genomes
Kimura distances between genome copies and TE consensus from the library were determined using RepeatLandscape (https://github.com/caballero/RepeatLandscape, last accessed January 30, 2015) on alignments included in .align files after genome masking with RepeatMasker. The rates of transitions and transversions were calculated on alignments and transformed to Kimura distance (Kimura 1980) by using the following equation: K = −1/2 ln(1 − 2p − q) – 1/4 ln(1 − 2q), where q is the proportion of sites with transversions and p the proportion of sites with transitions. For tetrapods (except for turtle) and Ciona, TE landscapes are available on http://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html (last accessed January 30, 2015).
Estimation of Relationship between Genome Size and Percentage of TEs
We performed a linear regression model to test the relationship between TE content and genome size. We tested for an interactive and an additive effect of the taxa (included in the model as a two-class factor: Sarcopterygians and actinopterygians) to determine if this relationship is different in sarcopterygians and actinopterygians. We used t-tests to determine if the intercepts and slopes were statistically different from zero, and Pearson’s correlation coefficient to estimate the strength of the correlation. A set of 13 species (extracted from Amemiya et al. 2013) was used to test for a potential phylogenetic effect on the correlation between genome size and percentage of TEs using a Brownian correlation matrix (Felsenstein 1985; Díaz-Uriarte and Garland 1998).
Results
Diversity of Global TE Content in Vertebrate Genomes
We analyzed 23 vertebrate genomes including the genomes of 11 sarcopterygians (4 mammals: Human, mouse, opossum, and platypus; 2 birds: chicken and zebra finch; 3 reptiles: Mississipi alligator, green anole, and Chinese soft-shell turtle; 1 amphibian: xenopus, aka western clawed frog; 1 coelacanth), 10 actinopterygians (spotted gar, European eel, zebrafish, cod, medaka, platyfish, tilapia, stickleback, tetraodon and fugu), 1 chondrichthyan (elephant shark), and the jawless sea lamprey. Two urochordates (Ciona and Oikopleura) and one cephalochordate (amphioxus) were used as nonvertebrate chordate outgroups (figs. 1–4).
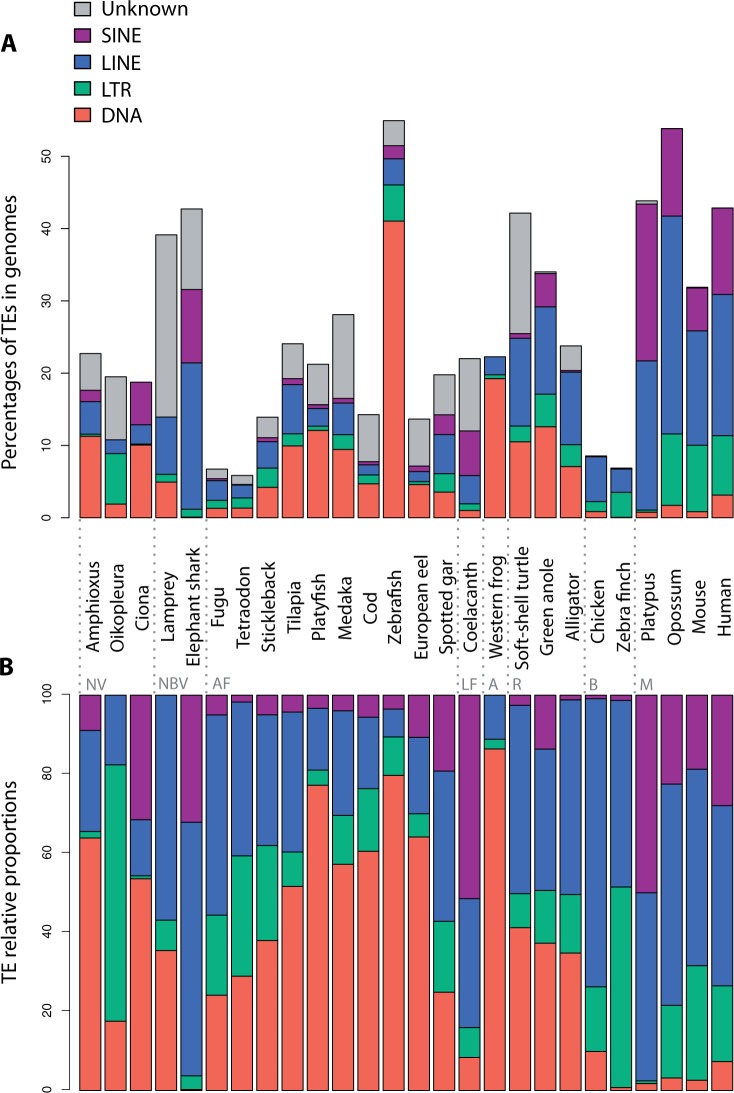
Fig. 1.—
Total amount and relative proportions of DNA transposons, LTR, LINE, and SINE retrotransposons in vertebrate genomes. The amount of DNA transposons, LTR, LINE, and SINE retrotransposons, and unclassified elements (Unknown), as well as their respective proportions were estimated based on RepeatMasker outfiles. (A) Percentages of TEs in the different vertebrate and nonvertebrate genomes studied. (B) Relative TE proportions in genomes (unclassified elements were not included). For more clarity, PLE elements have been included into LINE elements and DIRS sequences into LTR retrotransposons, and subclass I and subclass II DNA transposons have been grouped. NV, nonvertebrates; NBV, nonbony vertebrates; AF, actinopterygian fish; LF, lobe-finned fish; A, amphibians; R, nonbird reptiles; B, birds; M, mammals.
Fig. 2.—
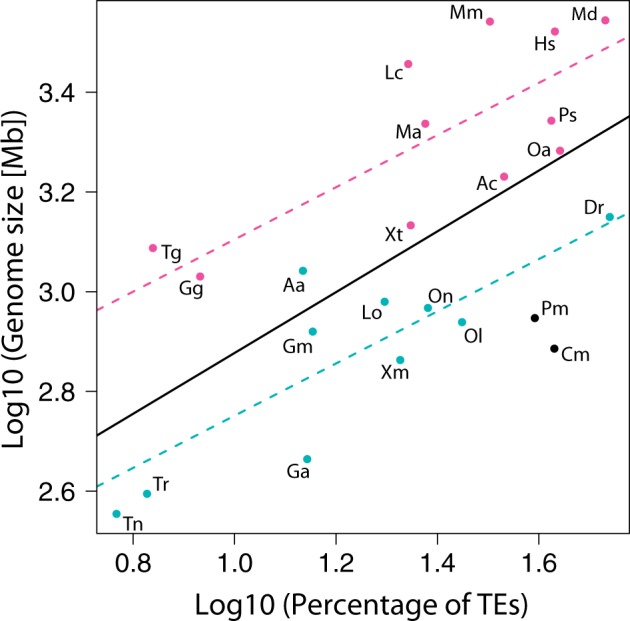
Correlation between genome size and transposable element content in vertebrates. Actinopterygian fish are represented by blue dots, sarcopterygians by pink dots, nonbony vertebrates by black dots. Pearson’s correlations were estimated taking into account all vertebrates (black line), only actinopterygian fish (blue dashed line), and only sarcopterygians (pink dashed line). Aa, Anguilla anguilla (European eel); Ac, Anolis carolinensis (green anole); Am, Alligator mississippiensis (American alligator); Cm, Callorhinchus milii (elephant shark); Dr, Danio rerio (zebrafish); Ga, Gasterosteus aculeatus (stickleback); Gg, Gallus gallus (chicken); Gm, Gadus morhua (Atlantic cod); Hs, Homo sapiens (human); Lc, Latimeria chalumnae (African coelacanth); Lo, Lepisosteus oculatus (spotted gar); Md, Monodelphis domestica (opossum); Mm, Mus musculus (mouse); Oa, Ornithorhynchus anatinus (platypus); Ol, Oryzias latipes (medaka); On, Oreochromis niloticus (tilapia); Pm, Petromyzon marinus (lamprey); Ps, Pelodiscus sinensis (Chinese soft-shell turtle); Tg, Taeniopygia guttata (zebra finch); Tr, Takifugu rubripes (fugu); Tn, Tetraodon nigroviridis (Tetraodon); Xm, Xiphophorus maculatus (platyfish); Xt, Xenopus tropicalis (western clawed frog).
Fig. 3.—
Diversity and abundance of TE superfamilies in vertebrates. Presence/absence of TE superfamilies was determined using automatic annotation, manual verification, and literature information. Presence of superfamilies is shown by full squares with gray gradient. Absence is represented by dashed squares. The premasked genome of the western clawed frog does not provide details within the CR1-like superfamily. Superfamilies having been domesticated at least once in the bony vertebrate lineage are shown by asterisks. Divergence times between species were estimated using the TimeTree public database (Hedges et al. 2006) and literature data based on fossil records.
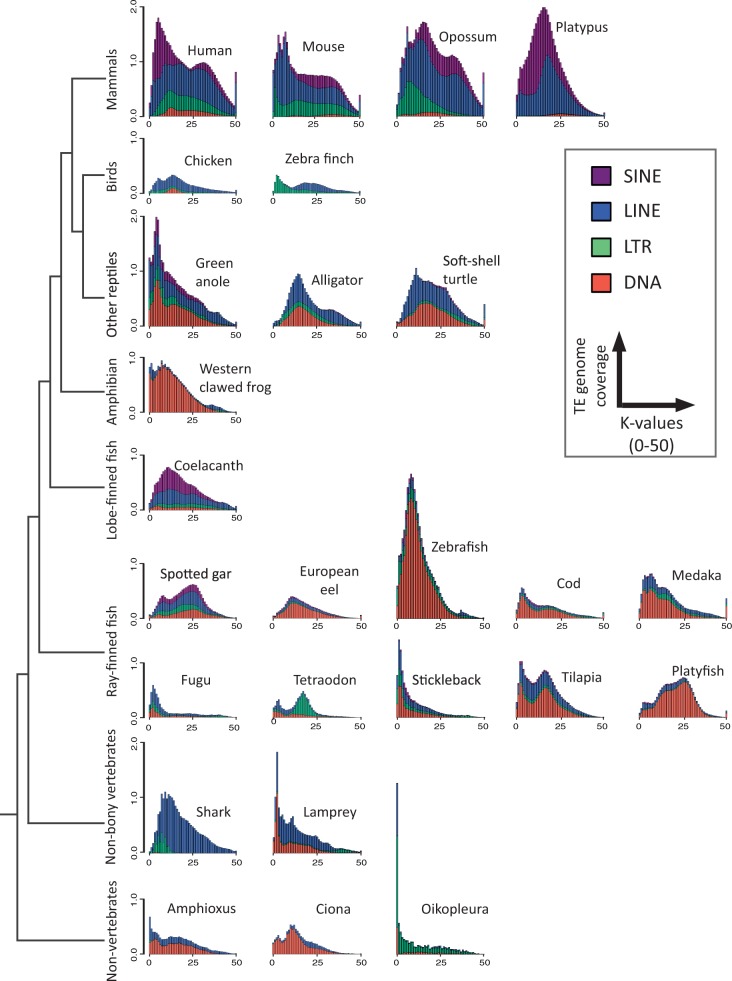
Fig. 4.—
Kimura distance-based copy divergence analysis of transposable elements in vertebrate and nonvertebrate genomes. The graphs represent genome coverage (y axis) for each type of TEs (DNA transposons, SINE, LINE, and LTR retrotransposons) in the different genomes analyzed, clustered according to Kimura distances to their corresponding consensus sequence (x axis, K-value from 0 to 50). Copies clustering on the left of the graph do not diverge very much from the consensus sequence of the element and potentially correspond to recent copies, while sequences on the right might correspond to ancient/degenerated copies. For more clarity, PLE elements have been included into LINE elements and DIRS sequences into LTR retrotransposons, and subclass I and subclass II DNA transposons have been grouped. For tetrapods (except for turtle) and Ciona, TE landscapes are also available on http://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html, last accessed January 30, 2015).
The global contribution of TEs to vertebrate genomes was analyzed. The results presented in fig. 1A confirmed that TE content is variable in the species studied. The genomes of nonbony vertebrates (lamprey and elephant shark), some fish species, coelacanth, xenopus, nonbird reptiles, and mammals contain a high fraction of TEs (>20% of the genome). In contrast, the compact genomes of pufferfishes (fugu and tetraodon) and birds are poor in TEs. TE content is very variable between sequenced fish genomes, with around a 10-fold difference between compact pufferfish genomes and the TE-rich genome of zebrafish.
Contribution of TEs to Vertebrate Genome Size
We tested the relationship between TE content and genome size in vertebrates. We analyzed our set of 23 vertebrate species and observed a positive correlation between TE content and genome size (fig. 2 and supplementary information, Supplementary Material online), statistically supported by a t-test (t = 0.523, P < 0.0001). We detected an additive effect of the taxa (t = 6.488, P < 0.001), but with similar slopes. This indicated that for a same genome size, relative TE contribution was more important in actinopterygian fish than in sarcopterygians (fig. 2), or that for a similar TE content, sarcopterygians have larger genomes than actinopterygian fish. A shift between actinopterygian and sarcopterygian regression lines was also observed when other types of repeats were included in the study (data not shown). These results suggest that low copy number and/or nonrepeated sequences contribute more significantly to genome size in sarcopterygians than in actinopterygian fish. We tested a possible effect of the phylogeny on the correlation (species were weighted depending on phylogenetic tree) using a Brownian correlation matrix (Felsenstein 1985; Díaz-Uriarte and Garland 1998). This was performed on a smaller data set of 13 vertebrate species (human, mouse, opossum, platypus, zebra finch, chicken, green anole, western clawed frog, African coelacanth, tilapia, fugu, zebrafish, and elephant shark) based on the phylogeny published by Amemiya et al. (2013). We did not detect any phylogenetic signal and again found a significant positive correlation between genome size and percentage of TEs.
Relative Contribution of Different Types of TEs to Vertebrate Genomes
The relative contribution of major types of TEs, that is, LTR, LINE, and SINE retrotransposons as well as DNA transposons, was estimated in the genomes analyzed (fig. 1B). According to their TE composition, genomes were classified into four main categories: 1) genomes with predominance of DNA transposons: Amphioxus, Ciona, most teleost fish (tilapia, platyfish, medaka, cod, zebrafish, and European eel), and xenopus; 2) genomes with predominance of LINEs and SINEs: Nonbony vertebrates (lamprey, elephant shark), some actinopterygian fish (fugu and spotted gar), coelacanth, chicken, and all mammals; 3) genomes with predominance of LTR retrotransposons: Oikopleura; and 4) genomes with no predominance of any particular type of TEs, including some teleost fish (tetraodon and stickleback), nonbird reptiles, and zebra finch. Some genomes were particularly poor in DNA transposons, with a mobilome almost exclusively constituted by retroelements (elephant shark, coelacanth, birds, and mammals).
Distribution of TE Superfamilies in Vertebrates
Some TE superfamilies, including vertebrate ERVs, Penelope-like, LINE1, and CR1-like retrotransposons, as well as Tc-Mariner and hobo, Ac and Tam3 (hAT), DNA transposons, were found to be present in all vertebrate lineages (fig. 3). ERVs, which are remnants of retroviral past infections, are very abundant in amniotes but have lower copy numbers in other lineages. Penelope-like elements were detected in all species studied, but with low copy numbers in mammals. The LINE1 superfamily presented contrasting genome coverage between lineages, constituting over 5% of the genome of marsupials and placental mammals but with low copy numbers in monotremes and birds. Although the CR1-like superfamily is globally ubiquitous in vertebrates, its constituting families CR1, LINE2, and Rex1/Babar show a more patchy distribution. Finally, Tc-Mariner and hAT are widespread DNA transposons superfamilies.
Several other TE superfamilies were detected in the majority of species analyzed but with punctual absence considered as putative lineage-specific loss events: Gypsy retrotransposons have been lost in birds, RetroTransposable Element (RTE), retrotransposons in chicken and western clawed frog, PiggyBac transposons in platypus (and with very low copy numbers in tetrapods), and Helitron transposons in birds. Interestingly, many TE superfamilies are present in fish but absent from some tetrapod sublineages. This is the case for Copia retrotransposons and for Maverick and Harbinger DNA transposons, which are not present in mammals and birds. DIRS retrotransposons are absent from alligator, birds, and mammals. Jockey retrotransposons were only detected in alligator and anole among tetrapods. R2 retrotransposons are absent from the vast majority of terrestrial tetrapods. The EnSpm DNA transposon superfamily, already described in zebrafish (Bao and Jurka 2008), was detected as short noncoding sequences in coelacanth but not in most tetrapods.
Finally, several superfamilies showed more patchy distributions, revealing multiple events of loss (or gain) of TEs. This is the case for BEL/Pao retrotransposons, which are absent from mammals, birds/alligator, turtle, European eel, and elephant shark. Although the R4-like Rex6/Dong elements were detected in most fish genomes and are strongly represented in the green anole, they were not found in birds/alligator, turtle, frog, or eel. Many DNA transposon superfamilies also have a patchy distribution in vertebrates (fig. 3).
Teleost Genomes Contain the Highest Diversity of TE Superfamilies in Vertebrates
With an average of 24 superfamilies present in each species studied, the actinopterygian lineage, including teleost fishes (9 species in this study), is the lineage showing the highest TE diversity in vertebrates (fig. 3). All superfamilies found in vertebrates are represented in at least one actinopterygian species, and most of them are present in all teleost genomes (Gypsy, BEL/Pao, ERV, DIRS, Penelope, Rex6/Dong, R2, LINE1, RTE, LINE2, Rex1/Babar, Jockey, Helitron, Maverick, Zisupton, Tc-Mariner, hAT, Harbinger, PiggyBac, and EnSpm). A strong genomic contribution of LINE2 retrotransposons as well as Tc-Mariner and hAT transposons was observed. Teleost fishes are the only vertebrates that contain Zisupton transposons (Böhne et al. 2012). Differences between teleost species were visible. With 27 superfamilies, zebrafish and cod presented the highest TE diversity. Absence of many DNA transposons was observed in some species, particularly in fugu, tetraodon, stickleback, tilapia, and platyfish.
Loss of TE Superfamilies in the Sarcopterygian Lineage
Within sarcopterygians, major lineage-specific differences in TE superfamily content were observed. With 26 TE superfamilies in its genome, the coelacanth presented the highest TE richness, with a diversity similar to that observed in actinopterygian fish (fig. 3). In contrast, tetrapods showed, on average, only 14 superfamilies, with 21 superfamilies in xenopus (amphibian), 15–18 in nonbird reptiles, 7–9 in birds, and 11–14 in mammals. This suggests elimination of ancestral TE families during tetrapod evolution. Reduction of TE diversity in tetrapods is particularly associated with, but not restricted to, loss of DNA transposon superfamilies.
In amphibians, the TE landscape in western clawed frog is essentially composed of five DNA transposon superfamilies (Tc-Mariner, hAT, Harbinger, PiggyBac, and T2/Kolobok) and CR1-like retrotransposons. Nonbird reptile genomes show a particularly high copy number of CR1/LINE3, Gypsy/Ty3, Penelope, Tc-Mariner, and hAT elements. Many types of TEs are absent in birds: Only 7–9 TE superfamilies have been maintained in the two species studied, with predominance of ERVs and CR1 retrotransposons. Finally, in mammals, the same TE superfamilies were found in the three sublineages (monotremes, marsupials, and placentals). However, LINE2 elements are predominant in the platypus (monotreme), while LINE1 is the most reiterated non-LTR retrotransposon in opossum and placental mammals (therians). In addition, some low copy number DNA transposons (PiggyBac, MuDr, and Merlin), possibly acquired by horizontal transfer, were detected in human but not in the mouse.
Lineage-Specific TE Activity during Vertebrate Evolution
For the genomes studied, Kimura distances (K-values; Kimura 1980) were calculated for all TE copies of each element in order to estimate the “age” and transposition history of TEs (fig. 4). Copy divergence is correlated with the age of activity: Very similar copies (low K-values) are indicative of rather recent activity (on the left part of the graph), while divergent copies (high K-values) have been generated by more ancient transposition events (on the right part of the graph). Results were grouped for the four different types of TEs (DNA transposons, LTR, LINE, and SINE retrotransposons) (fig. 4).
Mammalian profiles are characterized by a strong predominance of retroelements compared with DNA transposons. In the human genome, one major ancient transposition burst, mainly involving LINEs, was detected, as well as a more recent important expansion of SINEs that was not associated with any concomitant increase in LINE copy number. This contrasts with the situation in the mouse genome, where evidence for more recent LINE (and LTR) amplification was observed, but without strong increase in SINE copy number. Two major concomitant LINE/SINE amplifications were detected in opossum (in addition to a more recent LTR burst) and one in the platypus. In contrast to the situation in human and opossum, the identification of LINE elements with very low K-values in mouse and platypus genomes suggests the presence of recent, possibly active copies. Ancient divergent LINE elements with high K-values were found in therians but not in the platypus (monotreme). Including publicly available mammalian Kimura profiles in the analysis confirmed that mammalian genomes are mostly shaped by non-LTR retrotransposons and a regular activity of LTR retrotransposons over time (supplementary information, Supplementary Material online).
Both bird species studied showed two major transposition bursts: In chicken, an initial burst involving LINE and DNA transposons was followed by a second burst of only LINEs; in the zebra finch, the oldest burst was LINE elements and the youngest burst was due to LTR retroelements. Ancient TE copies but few recent elements were detected in the two bird species.
In contrast to the situation observed in birds and mammals, DNA transposons have been very active during the evolution of the three nonbird reptile species analyzed. LINEs have also strongly contributed to the genomes of these species. Profiles were relatively similar in alligator and turtle, with ancient and “middle-aged” bursts of transposition and few recent copies. In contrast, the genome of the green anole has undergone a younger general burst of transposition and contains recent copies from all four types of TEs.
The genome of xenopus, the only representative of the amphibian lineage included in this study, has been predominantly shaped by DNA transposons, with a lot of recent copies present indicating a lot of recent activity. In addition, a young and small amplification of LINEs has occurred in xenopus.
The coelacanth genome is dominated by LINEs and SINEs, with at least one major middle-aged transposition burst and some recent LINE copies.
In actinopterygian fish, the spotted gar, which is a nonteleost species, has a genome that has been shaped by all four major types of TEs, with two major bursts of activity and few recent copies. Within teleosts, significant interspecific differences in profiles were observed, with generally one or two general bursts of transposition. Some genomes are dominated by rather recent copies (cod, medaka, stickleback, fugu), some by ancient copies (platyfish), and some by both (eel, tilapia, tetraodon), with no clear phylogenetic signal: Both pufferfishes show clearly different patterns, as it is also the case for the related species medaka and platyfish. Teleost genomes generally contain fewer ancient copies (K-values >25) than mammalian genomes, suggesting differences in the dynamics of TE elimination. In contrast to gar, most teleost genomes studied have been strongly shaped by DNA transposons. This is particularly the case for zebrafish, which shows the higher amplification of DNA transposons among the vertebrates studied here. LINEs significantly contributed to the genome of several species including medaka, tilapia, and fugu, while a significant middle-aged burst of LTR elements (around K-value 19) was detected in tetraodon. High number of recent TE copies, suggesting activity, was particularly identified in zebrafish, tilapia, stickleback, and pufferfishes.
The elephant shark Kimura profile is made up of LINE retrotransposons and a smaller contribution from a recent burst of LTR retrotransposons. The genome of the lamprey is dominated by DNA transposons and LINE retrotransposons, with many young DNA transposon copies. An ancient burst of LTR retrotransposons was also detected.
In nonvertebrate species included in this work, Ciona and amphioxus genomes mostly contain DNA transposons and LINE retrotransposons, while Oikopleura is mainly composed of LTR retrotransposons. Active copies are probably present in these species, with a very recent strong burst of DNA transposons and LTR retrotransposons in Oikopleura.
Discussion
Using species-specific TE libraries, we have analyzed retrotransposons and DNA transposons in sequenced genomes from species covering major branches of the vertebrate lineage. This study uncovered an important inter- and intralineage diversity concerning the nature, genomic contribution, activity, and evolution of TEs in vertebrate genomes.
Diversity of TE Contribution to Genome Size in Vertebrates
TEs and other repeats make up an important part of most vertebrate genomes. However, the global contribution of TEs is variable between lineages: for example, the genome of mammals contains many more TEs than the genomes of birds. Variability in TE content is also observed within lineages: In teleost fish, the genome coverage of TEs is 10 × higher in zebrafish (55% of the genome) than in the pufferfish tetraodon (6%). Short TE-related sequences strongly contribute to some vertebrate genomes including those of mammals (Hancock 2002). It is of course important to note that we focused on already sequenced genomes; other particularly TE-rich (or TE-poor) vertebrate genomes are still to be sequenced, for example, the large genomes of salamanders and lungfish (Dufresne and Jeffery 2011; Metcalfe and Casane 2013). In addition, our evaluation of TE content is certainly an underestimation: We worked on assembled genome drafts, which often do not include TE-rich regions of the genomes like centromeres or other heterochromatic regions. Our methods of analysis were very conservative, and may have missed other types of TEs, or very old and divergent elements. Using alternative methods, it has been estimated that TE content in the human genome might be as high as 66–69% (De Koning et al. 2011).
Factors influencing genome size and DNA content variation between species are multiple, including whole genome duplications, segmental duplications, deletions, and DNA repeat proliferation (Parfrey et al. 2008). It has been established that TEs and other DNA repeats play an important role in genome size diversity (Petrov 2001; Kidwell 2002; Ågren and Wright 2011). In insects, both satellite sequences and TEs have been implicated in genome size variation (Vieira et al. 1999, 2002; Kidwell 2002; Bosco et al. 2007). Accordingly, we confirmed a correlation between TE content and genome size in vertebrates, indicating that larger genomes tend to have more TEs than smaller genomes. Such a correlation was also observed after testing separately for actinopterygian fish and sarcopterygians. However, the results obtained suggest that TEs (and other types of repeats) contribute more significantly to genome size in actinopterygian fish than in sarcopterygians. Sarcopterygian genomes might contain a more important fraction of low copy number or nonrepeated elements, or of very divergent repeat sequences that were identified neither as reiterated nor as TEs in our study.
Inter- or intralineage differences in TE contribution to genomes might be explained by variability in TE activity, which can be influenced by transposition rates of TEs present in the genomes, competition between TEs, and variations in host-mediated defense mechanisms against mobile elements (Le Rouzic and Capy 2006). The elimination rate of TEs is also an important parameter, species with a slow rate of DNA loss tending to increase their genome size (Petrov 2001; Sun et al. 2012). Indeed, the Kimura distance-based comparative analysis performed in this work suggested strong variability in TE activity between vertebrates, with important differences in the number of recent potentially active elements. For example, the number of recent copies is low in human and opossum but higher in the mouse. The genome of the green anole continues to sustain strong transposition activity while there are almost no active copies in the alligator anymore. In addition, lineage-specific differences in TE elimination rates might be also involved. For instance, large mammalian genomes contain more ancient divergent and fractionated TE copies than fish genomes, suggesting differences in the dynamics of DNA elimination. This has been already proposed by Blass et al. (2012) based on the analysis of non-LTR retrotransposons in the genome of the three-spine stickleback. Within actinopterygians, differences were even observed between related fish species: The genome of the platyfish contains many more old TE copies than that of the related medaka. Differences in TE elimination between fish species might be supported by the genome architecture of the pufferfish: In this compact and TE-poor genome, all types of repeats are excluded from euchromatic gene-rich regions and accumulate in particular heterochromatic compartments, a structure generally not observed in other fish species (Dasilva et al. 2002; Fischer et al. 2005).
Population-level processes might have played a role in differences in TE content and genome size observed in fish and other vertebrates (Lynch and Conery 2003). Accordingly, freshwater fish species, which have smaller effective population sizes than marine fish species, have generally larger genomes (Yi and Streelman 2005). In the green anole lizard Anolis carolinensis, full-length L1 retrotransposon inserts are more likely to be fixed in populations of small effective size (Tollis and Boissinot 2013).
TE Landscape Diversity in Extant Vertebrate Species
Mobilome diversity in vertebrates is manifested not only by global variations in TE content between lineages and species but also by differences in the types and superfamilies of TEs present in genomes, and by their differential colonization success. In this study, we showed that major differences exist between and even within lineages regarding the different types of TEs present in the mobilome. For example, the genomes of mammals, birds, coelacanth, and elephant shark have been almost exclusively shaped by retroelements and contain few DNA transposons, while DNA transposons are the most prominent type of TEs in teleost fishes and xenopus genomes (but LTR retrotransposons are amplified in the giant genomes of plethodontid salamanders; Sun et al. 2012). Within mammals, LTR elements constitute a significant part of mobilome in therians (human, mouse, and opossum) but not in platypus (monotreme), and sublineage-specific transposition bursts have been observed (SINEs in primates, DNA transposons in hyrax, and also in bats; Pritham and Feschotte 2007). In fishes, TE composition of zebrafish and tetraodon is extremely different, and each fish species possesses its own Kimura distance-based TE profile in terms of relative contribution of each type of TE. Divergent repeat landscapes have been also observed in snake genomes (Castoe et al. 2011).
This diversity is also observed considering the number of TE superfamilies. Many examples of lineage-specific loss (or gain) of TE superfamilies have been identified. Some vertebrate lineages contain many TE superfamilies, including teleost fishes (24 TE superfamilies on average per genome), the coelacanth (26 TE superfamilies), and xenopus (amphibian, 21 TE superfamilies). In contrast, a strong reduction of TE diversity was observed in mammals (11–14 TE superfamilies) and the two bird species studied (7–9 superfamilies). Low TE content and diversity have been also reported in turkey (Dalloul et al. 2010), flycatchers (Ellegren et al. 2012), duck (Huang et al. 2013), and falcons (Zhan et al. 2013). The three nonbird reptile species analyzed showed an intermediate TE richness, with 15–18 superfamilies. These results suggest a reduction of TE diversity through elimination of TE superfamilies in the sarcopterygian lineages having led to mammals and birds. In birds, the small genome size and low TE content suggest that loss of certain TE families might be a consequence of general constraints acting toward a reduction of noncoding DNA content in the genome. In contrast, the situation in the large genomes of mammals might be explained in terms of competition between TEs (Abrusán and Krambeck 2006; Le Rouzic and Capy 2006). The loss of certain TE superfamilies might be associated with the extreme success of specific families of LINE and SINE non-LTR retrotransposons, for example, LINE1 and Alu sequences in primate genomes. As a result of competition for genomic resources, resident successful families might have supplanted and eliminated other types of TEs. Alternatively, extinction of some TE families might have been driven by mutational inactivation or through the development of a new specific defense systems by the host, allowing the massive opportunistic expansion of the remaining active TE families.
Even when the same TE superfamily or family is present in different genomes, differences in copy number might contribute to lineage divergence. This is the case even for TE superfamilies present in all vertebrate lineages, including ERVs, Penelope-like, LINE1, and CR1-like retrotransposons, and Tc-Mariner and hAT DNA transposons. For example, ERVs are very abundant in amniotes but present lower copy numbers in other vertebrates. Penelope-like elements display very modest copy numbers in mammals compared with other lineages. The LINE1 superfamily is a major component of the genome of marsupials and placental mammals, but is poorly represented in monotremes and birds.
Toward an Inference of the Ancestral Vertebrate Mobilome?
This analysis provides a framework for a first attempt to infer the ancestral vertebrate mobilome, that is, TE composition in terms of diversity in the last common ancestor (LCA) of the vertebrate species studied. This is a very difficult task, particularly because TEs can also be introduced through horizontal transfer into lineages.
If we assume a major mode of vertical transmission, we can infer that many superfamilies of autonomous TEs were present in the genome of the vertebrate LCA. TE superfamilies found in almost or all species studied were probably represented in the ancestral vertebrate mobilome, including Gypsy LTR retrotransposons, Penelope-like retrotransposons, LINE1, RTE, and CR1-like non-LTR retrotransposons, as well as Helitron, Tc-Mariner, hAT, and PiggyBac DNA transposons. Some SINE elements are also widespread, like the V-SINE elements (Ogiwara et al. 2002; Piskurek and Jackson 2011), but many have been formed in specific vertebrate sublineages. The LCA mobilome probably also included superfamilies present in jawless vertebrates, chondrichtyans, actinopterygians, amphibians, and reptiles but lost in birds and mammals, including Copia and DIRS LTR retrotransposons as well as Maverick and Harbinger DNA transposons.
However, the possible acquisition of TEs by horizontal gene transfer (HGT) must also be considered particularly for DNA transposons, even if HGT events have been thought to be rather rare in vertebrates (Syvanen 2012; Wallau et al. 2012). HGT has been widely described in insects (Sormacheva et al. 2012). In vertebrates, the Space Invaders (SPIN), DNA transposon, which presents a patchy distribution in vertebrates, has been transmitted horizontally several times in mammals and other tetrapods (Pace et al. 2008; Gilbert et al. 2012). HGT of the non-LTR retrotransposon BovB has been reported between reptiles and within mammals (Kordis and Gubensek 1999; Walsh et al. 2013). Horizontal transmission of Tc-Mariner elements possibly also occurred between teleosts and lampreys (Kuraku et al. 2012). More putative cases have been also reported (Novick et al. 2010; Thomas et al. 2011; Oliveira et al. 2012; Gilbert et al. 2013). Retroviruses, which infect vertebrates, have been proposed to serve as vectors for HGT (Yohn et al. 2005; Piskurek and Okada 2007). According to their patchy distribution, several TE superfamilies are candidates for HGT: MuDr in human; Merlin in stickleback, zebrafish, western clawed frog, and human (Feschotte 2004); Chapaev in the green anole, fugu, and platyfish; and P in platyfish and coelacanth. Alternatively, these TEs might have been lost repeatedly during the evolution of the vertebrate lineage. Much work is required to determine the modes of acquisition and loss of these elements in vertebrates.
Conclusions
In this work, we present an overview of content, diversity, activity, and evolution of TEs in the vertebrate lineage. The results obtained highlight inter- and intralineage diversity, showing that differential TE activity and evolution have strongly contributed to genome divergence in vertebrates. TEs can also mediate diversity through lineage-specific events of molecular domestication, leading to new gene regulations and functions (Böhne et al. 2008). The functional consequences of lineage-specific TE expansion on genome architecture and regulation remain to be investigated. Further work on individual TE families and subfamilies, which are more relevant to assess true events of loss and gain during evolution, will uncover new aspects of TE dynamics in vertebrate and allow the discovery of new cases of horizontal transfer.
Supplementary Material
Supplementary tables S1 and S2, figures S1 and S2, and information are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org, last accessed January 2015).
Acknowledgments
This work was supported by PhD grants from the French Ministry for Higher Education and Research and from the “Ligue Nationale contre le Cancer” (to D.C.). We thank the CBP and PSMN centers at the ENS Lyon, especially Emmanuel Quemener and Thomas Bellembois, for technical IT supports; Emmanuelle Lerat and Clément Goubert for help and advices; Marie Sémon for helpful comments; and Ian Warren for careful reading of the manuscript.
Literature Cited
- Abrusán G, Krambeck HJ. Competition may determine the diversity of transposable elements. Theor Popul Biol. 2006;70:364–375. doi: 10.1016/j.tpb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Ågren JA, Wright SI. Co-evolution between transposable elements and their hosts: a major factor in genome size evolution? Chromosome Res. 2011;19:777–786. doi: 10.1007/s10577-011-9229-0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Jurka J. EnSpm DNA transposons in Zebrafish. Repbase Reports. 2008;8:823. [Google Scholar]
- Basta HA, Buzak AJ, McClure MA. Identification of novel retroid agents in Danio rerio, Oryzias latipes, Gasterosteus aculeatus and Tetraodon nigroviridis. Evol Bioinform Online. 2007;3:179–195. [PMC free article] [PubMed] [Google Scholar]
- Blass E, Bell M, Boissinot S. Accumulation and rapid decay of non-LTR retrotransposons in the genome of the three-spine stickleback. Genome Biol Evol. 2012;4:687–702. doi: 10.1093/gbe/evs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- Böhne A, et al. Zisupton—a novel superfamily of DNA transposable elements recently active in fish. Mol Biol Evol. 2012;29:631–645. doi: 10.1093/molbev/msr208. [DOI] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto JT, Markov TA. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics. 2007;177:1277–1290. doi: 10.1534/genetics.107.075069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield JFY. Host-parasite relationships in the genome. BMC Biol. 2011;9:67. doi: 10.1186/1741-7007-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol Evol. 2011;3:641–653. doi: 10.1093/gbe/evr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin D, et al. Evolutionary active transposable elements in the genome of the coelacanth. J Exp Zool B Mol Dev Evol. 2013;322:322–333. doi: 10.1002/jez.b.22521. [DOI] [PubMed] [Google Scholar]
- Dalloul RA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasilva C, et al. Remarkable compartmentalization of transposable elements and pseudogenes in the heterochromatin of the Tetraodon nigroviridis genome. Proc Natl Acad Sci U S A. 2002;99:13636–13641. doi: 10.1073/pnas.202284199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Chaux N, Wagner A. BEL/Pao retrotransposons in metazoan genomes. BMC Evol Biol. 2011;11:154. doi: 10.1186/1471-2148-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Díaz-Uriarte R, Garland T., Jr Effects of branch length errors on the performance of phylogenetically independent contrasts. Syst Biol. 1998;47:654–672. doi: 10.1080/106351598260653. [DOI] [PubMed] [Google Scholar]
- Dufresne F, Jeffery N. A guided tour of large genome size in animals: what we know and where we are heading. Chromosome Res. 2011;19:925–938. doi: 10.1007/s10577-011-9248-x. [DOI] [PubMed] [Google Scholar]
- Duvernell DD, Pryor SR, Adams SM. Teleost fish genomes contain a diverse array of L1 retrotransposon lineages that exhibit a low copy number and high rate of turnover. J Mol Evol. 2004;59:298–308. doi: 10.1007/s00239-004-2625-8. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- Ellison CE, Bachtrog D. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science. 2013;342:846–850. doi: 10.1126/science.1239552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen’ev MB. What happens when Penelope comes?: an unusual retroelement invades a host species genome exploring different strategies. Mob Genet Elements. 2013;3:e24542. doi: 10.4161/mge.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol Biol Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. Non-mammalian c-integrases are encoded by giant transposable elements. Trends Genet. 2005;21:551–552. doi: 10.1016/j.tig.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Fischer C, et al. Diversity and clustered distribution of retrotransposable elements in the compact genome of the pufferfish Tetraodon nigroviridis. Cytogenet Genome Res. 2005;110:522–536. doi: 10.1159/000084985. [DOI] [PubMed] [Google Scholar]
- Furano AV, Duvernell D, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Hernandez SS, Flores-Benabib J, Smith EN, Feschotte C. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol Biol Evol. 2012;29:503–515. doi: 10.1093/molbev/msr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Waters P, Feschotte C, Schaack S. Horizontal transfer of OC1 transposons in the Tasmanian devil. BMC Genomics. 2013;14:134. doi: 10.1186/1471-2164-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Gregory TR. The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells Mol Dis. 2001;27:830–843. doi: 10.1006/bcmd.2001.0457. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JM. Genome size and the accumulation of simple repeats: implications of new data from genome sequencing projects. Genetica. 2002;115:93–103. doi: 10.1023/a:1016028332006. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Huang Y, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat Genet. 2013;45:776–783. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques PÉ, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P. CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. R2 non-LTR retrotransposons in the bird genome. Repbase Rep. 2009;9:1329. [Google Scholar]
- Kapusta A, et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Jurka J. Crypton transposons: identification of new diverse families and ancient domestication events. Mob DNA. 2011;2:12. doi: 10.1186/1759-8753-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima KK, Jurka J. Non-LTR retrotransposons from green anole. Repbase Rep. 2013a;4:1615. [Google Scholar]
- Kojima KK, Jurka J. Non-LTR retrotransposons from the western painted turtle. Repbase Rep. 2013b;9:2217. [Google Scholar]
- Kordis D. Transposable elements in reptilian and avian (sauropsida) genomes. Cytogenet Genome Res. 2009;127:94–111. doi: 10.1159/000294999. [DOI] [PubMed] [Google Scholar]
- Kordis D, Gubensek F. Horizontal transfer of non-LTR retrotransposons in vertebrates. Genetica. 1999;107:121–128. doi: 10.1023/a:1004082906518. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Qiu H, Meyer A. Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol Evol. 2012;4:929–936. doi: 10.1093/gbe/evs069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Capy P. The first step of transposable elements invasion: parasitic strategy vs. genetic drift. Genetics. 2005;169:1033–1043. doi: 10.1534/genetics.104.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Capy P. Population genetics models of competition between transposable element subfamilies. Genetics. 2006;174:785–793. doi: 10.1534/genetics.105.052241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Capy P. Theorical approaches to the dynamics of transposable elements in genomes, populations, and species. Genome Dyn Stab. 2009;4:1–19. [Google Scholar]
- Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchetti A, Mantovani B. Non-LTR R2 element evolutionary patterns: phylogenetic incongruences, rapid radiation and the maintenance of multiple lineages. PLoS One. 2013;8:e57076. doi: 10.1371/journal.pone.0057076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Metcalfe CJ, Casane D. Accommodating the load: the transposable element content of very large genomes. Mob Genet Elements. 2013;3:e24775. doi: 10.4161/mge.24775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the world. New York: John Wiley and Sons; 2006. [Google Scholar]
- Novick P, Smith J, Ray D, Boissinot S. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene. 2010;449:85–94. doi: 10.1016/j.gene.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Novick PA, Basta H, Floumanhaft M, McClure MA, Boissinot S. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol Biol Evol. 2009;26:1811–1822. doi: 10.1093/molbev/msp090. [DOI] [PubMed] [Google Scholar]
- Ogiwara I, Miya M, Ohshima K, Okada N. V-SINEs: a new superfamily of vertebrate SINEs that are widespread in vertebrate genomes and retain a strongly conserved segment within each repetitive unit. Genome Res. 2002;12:316–324. doi: 10.1101/gr.212302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira SG, Bao W, Martins C, Jurka J. Horizontal transfers of Mariner transposons between mammals and insects. Mob DNA. 2012;3:14. doi: 10.1186/1759-8753-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposons in mammals and other tetrapods. Proc Natl Acad Sci U S A. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Katz LA. The dynamic nature of eukaryotic genomes. Mol Biol Evol. 2008;25:787–794. doi: 10.1093/molbev/msn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. Evolution of genome size: new approaches to an old problem. Trends Genet. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- Piskurek O, Jackson DJ. Tracking the ancestry of a deeply conserved eumetazoan SINE domain. Mol Biol Evol. 2011;28:2727–2730. doi: 10.1093/molbev/msr115. [DOI] [PubMed] [Google Scholar]
- Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci U S A. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter RT, Goodwin TJ. DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet Genome Res. 2005;110:575–588. doi: 10.1159/000084991. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V, Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr Opin Genet Dev. 2008;18:544–550. doi: 10.1016/j.gde.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Sarropoulou E, Fernandes JM. Comparative genomics in teleost species: knowledge transfer by linking the genomes of model and non-model fish species. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6:92–102. doi: 10.1016/j.cbd.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Schartl M, et al. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45:567–572. doi: 10.1038/ng.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormacheva I, et al. Vertical evolution and horizontal transfer of CR1 non-LTR retrotransposons and Tc1/mariner DNA transposons in Lepidoptera species. Mol Biol Evol. 2012;29:3685–3702. doi: 10.1093/molbev/mss181. [DOI] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, et al. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol. 2012;4:168–183. doi: 10.1093/gbe/evr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen M. Evolutionary implications of horizontal gene transfer. Annu Rev Genet. 2012;46:341–358. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
- Thomas J, Sorourian M, Ray D, Baker RJ, Pritham EJ. The limited distribution of Helitrons to vesper bats supports horizontal transfer. Gene. 2011;474:52–58. doi: 10.1016/j.gene.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Tollis M, Boissinot S. Lizards and LINEs: selection and demography affect the fate of L1 retrotransposons in the genome of the green anole (Anolis carolinensis) Genome Biol Evol. 2013;5:1754–1768. doi: 10.1093/gbe/evt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira C, Lepetit D, Dumont S, Biémont C. Wake up of transposable elements following Drosophila simulans worldwide colonization. Mol Biol Evol. 1999;16:1251–1255. doi: 10.1093/oxfordjournals.molbev.a026215. [DOI] [PubMed] [Google Scholar]
- Vieira C, Nardon C, Arpin C, Lepetit D, Biémont C. Evolution of genome size in Drosophila. Is the invader’s genome being invaded by transposable elements? Mol Biol Evol. 2002;19:1154–1161. doi: 10.1093/oxfordjournals.molbev.a004173. [DOI] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Volff JN, Bouneau L, Ozouf-costaz C, Fischer C. Diversity of retrotransposable elements in compact pufferfish genomes. Trends Genet. 2003;19:674–678. doi: 10.1016/j.tig.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Volff JN, Körting C, Froschauer A, Sweeney K, Schartl M. Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates. J Mol Evol. 2001;52:351–360. doi: 10.1007/s002390010165. [DOI] [PubMed] [Google Scholar]
- Vorechovsky I. Transposable elements in disease-associated cryptic exons. Hum Genet. 2010;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- Wallau GL, Ortiz MF, Loreto EL. Horizontal transposons transfer in Eukarya: detection, bias and perspectives. Genome Biol Evol. 2012;4:801–811. doi: 10.1093/gbe/evs055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A. 2013;110:1012–1016. doi: 10.1073/pnas.1205856110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Shima A, Schartl M. Medaka—a model organism from the far East. Nat Rev Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- Xie M, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013;45:836–841. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Streelman JT. Genome size is negatively correlated with effective population size in ray-finned fish. Trends Genet. 2005;21:643–646. doi: 10.1016/j.tig.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Yohn CT, et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, et al. Peregrine and saker falcon genome sequences provide insights into evolution of a predatory lifestyle. Nat Genet. 2013;45:563–566. doi: 10.1038/ng.2588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.