Significance
Population heterogeneity can make the treatment of tumors more challenging. Whereas a therapeutic agent may be effective against one fraction of a population, it may be less effective against another fraction. Although heterogeneity can be genetic and attributed to mutations, there can also be nongenetic heterogeneity, where a clonal population can harbor distinct subpopulations. Here, we identified a single gene, p21, that was responsible for population heterogeneity in cell cycle activity and explain that this heterogeneity can arise from regulatory relationships of p21 with Cyclin-dependent kinase 2 (CDK2) and E3 ubiquitin ligases. We suggest that, instead of using CDK inhibitors (CKIs) in cancer therapy, CKIs themselves should be targeted. Given concurrently with chemotherapy agents, CKI inhibitors would reduce tumor heterogeneity and thus increase chemotherapy efficacy.
Keywords: tumor heterogeneity, cell dormancy, synthetic uORF, nongenetic cell heterogeneity, positive feedback loop
Abstract
Phenotypic heterogeneity within a population of genetically identical cells is emerging as a common theme in multiple biological systems, including human cell biology and cancer. Using live-cell imaging, flow cytometry, and kinetic modeling, we showed that two states—quiescence and cell cycling—can coexist within an isogenic population of human cells and resulted from low basal expression levels of p21, a Cyclin-dependent kinase (CDK) inhibitor (CKI). We attribute the p21-dependent heterogeneity in cell cycle activity to double-negative feedback regulation involving CDK2, p21, and E3 ubiquitin ligases. In support of this mechanism, analysis of cells at a point before cell cycle entry (i.e., before the G1/S transition) revealed a p21–CDK2 axis that determines quiescent and cycling cell states. Our findings suggest a mechanistic role for p21 in generating heterogeneity in both normal tissues and tumors.
A population of genetically identical cells can exhibit phenotypic heterogeneity (1, 2). That is, even when cells with the same DNA sequence and epigenetic markings experience the same environment, there can be cell to cell variability. Researchers have reported that cells within a clonal population can vary by size and morphology (3), cell cycle activity (4, 5), lifespan (6), and receptor sensitivity to cytokines (7, 8). Furthermore, population heterogeneity can determine biological outcome: instead of a homogenous stem cell population where cells renew equally, a subpopulation can remain dormant until needed (5), a bacterial colony can harbor a subpopulation resistant to antibiotics (3), and a clonal tumor can harbor a subpopulation that survives radiation or chemotherapy (9). Although many instances of nongenetic population heterogeneity have been documented, fewer underlying regulatory mechanisms governing this heterogeneity have been reported.
When cells exhibit heterogeneity in cell cycle activity, one subpopulation actively cycles, whereas another subpopulation remains quiescent. The main activators of the cell cycle are Cyclin-dependent kinases (CDKs) bound to Cyclin proteins, and a major class of cell cycle inhibitors is CDK inhibitors (CKIs). One CKI, p21, was first discovered in the mid-1990s as an inhibitor of CDKs in G1 phase (10) and a factor that was transcriptionally activated by p53 (11). Since its discovery, much of the research involving p21 has focused on its role in arresting the cell cycle in response to activation by p53 after DNA damage. However, cells without exogenous DNA damage still express p21. For example, single-cell analysis of p21 and CDK2 activity led Spencer et al. (12) to propose that p21 is involved in the cellular decision to enter the cell cycle under normal conditions.
Because p21 is a potent inhibitor of Cyclin-CDK complexes, the cell must actively regulate p21 levels under normal conditions to promote the transition from G1 to S phase. At this stage of the cell cycle, CDK2 activity begins to increase and causes inactivation of the anaphase promoting complex (APC). The inactivation of the APC allows Skp2 levels to increase, because Skp2 is a target of the APC-Cdh1 complex. The E3 ubiquitin ligase complex of Skp1/Cullin/F box (SCF) and Skp2 recognizes p21 that has bound Cyclin E-CDK2 complexes and been phosphorylated by CDK2. SCF/Skp2 then ubiquitinates p21, targeting it for proteasomal degradation (13). Thus, p21 both regulates and, through the action of E3 ubiquitin ligase complexes that target p21, is regulated by active CDK2 bound to Cyclin E.
The p21–CDK2 control scheme is an example of a double-negative feedback loop. When stochastic gene expression leads to fluctuations in factors involved in positive or double-negative feedback regulation, distinct cellular states within a population can arise (1, 14–18). Because of the role of p21 in the double-negative feedback regulation of cell cycle activity, we hypothesized that p21 controlled population heterogeneity in quiescent and cycling cell states. In mammalian systems, there are few examples where inactivation of a single gene leads to loss of heterogeneity in cell cycle activity. Here, in comparing wild type (WT), deficient, and ectopically restored p21 genetic backgrounds, we reveal a direct role for low basal levels of p21 in controlling population heterogeneity in cell cycle activity.
Results
Live-Cell Imaging Revealed p21-Dependent Population Heterogeneity.
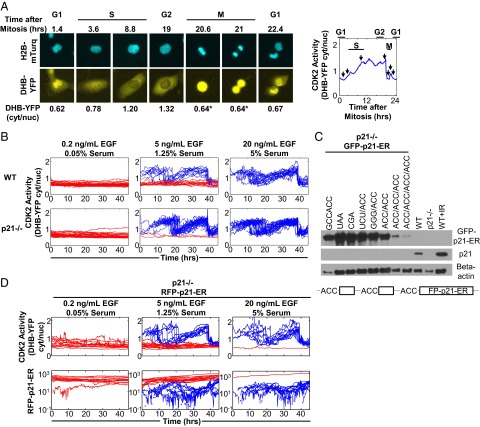
To gauge cell cycle activity, we tracked the activity of CDK2 in single cells by using a fluorescent reporter consisting of the C-terminal CDK2 phosphorylation domain of DNA helicase B (DHB) fused to YFP (19). In G0 or G1 phase cells, the unphosphorylated reporter is located primarily in the nucleus (Fig. 1A and Fig. S1A). During the G1/S transition, S, and G2 phases, CDK2 phosphorylates the reporter, causing it to translocate from the nucleus to the cytoplasm (Movie S1). As a result, the cytoplasmic-to-nuclear ratio of DHB-YFP can be used to monitor CDK2 activity and cell cycle progression (12).
Fig. 1.
p21 causes population heterogeneity in cell cycle activity. (A) Single-cell tracking of cell cycle progression using a DHB-YFP reporter of CDK2 activity. (Left) CDK2-dependent translocation of DHB-YFP from the nucleus to the cytoplasm occurs during G1/S, S, and G2 phases. DHB-YFP returns to the nucleus after mitosis (M) and remains during early G1. H2B-mTurquoise (H2B-mTurq) is the nuclear marker. *During mitosis, after nuclear membrane collapse, the cytoplasmic-to-nuclear ratio (cyt/nuc) is not well-defined. (Right) CDK2 activity (cyt/nuc DHB-YFP) vs. time for cell in Left; arrows indicate depicted time points. (B) Single-cell traces of CDK2 activity in MCF10A WT and p21-deficient (−/−) cycling (blue) and quiescent (red) cells. Traces for cycling cells were aligned to the second mitosis. B and D show results for populations grown under varying growth factor concentrations (EGF and serum). (C) Immunoblot of p21. The right three immunoblot lanes show WT and p21−/− cells without ionizing irradiation (IR) and WT with 10 Gy IR. The left eight immunoblot lanes show expression of GFP-p21-ER in p21−/− cells. Expression levels were tuned by varying translation initiation site bases preceding the GFP-p21-ER gene and/or synthetic uORFs. Slashes separate specified bases preceding each of one to three uORFs and bases preceding GFP-p21-ER. Bases without a slash precede GFP-p21-ER without any uORF. Expression of RFP-p21-ER in D was tuned with two uORFs (ACC/ACC/ACC and schematic) to approximate basal expression. (D) Traces for CDK2 activity and RFP-p21-ER expressed in p21−/− cells.
Using live-cell imaging to monitor the reporter in MCF10A cells, we could study growth factor-dependent (here synonymous with “mitogen-dependent”) heterogeneity in cell cycle activity. For typical experiments, researchers often desire maximal proliferation and therefore provide 20 ng/mL EGF and 5% (vol/vol) serum. However, this culture-optimized level of mitogenic stimulation is likely supraphysiological, and in comparison, human serum has been reported to contain 1–4 ng/mL EGF (20). In our experiments, when we supplemented cells with 20 ng/mL EGF and 5% (vol/vol) serum, almost all cells were actively cycling (Fig. 1B, Upper Right and Movie S1). In contrast, as expected, at a low level of stimulation (0.2 ng/mL EGF, 0.05% serum), all cells were quiescent (Fig. 1B, Upper Left) (when describing quiescence, here we do not distinguish G0 phase cells from those in prolonged G1). However, at an intermediate level of stimulation [5 ng/mL EGF, 1.25% (vol/vol) serum], we observed heterogeneity in cell cycle activity with 43% of cells in a cycling state and 57% of cells in a quiescent state (Fig. 1B, Upper Center and Movie S2).
In contrast, when we tracked the cell cycle activity of MCF10A cells deficient in p21, this heterogeneity was largely abrogated (Fig. 1B, Lower Center and Movie S3); 92% of cells were cycling. p21 is commonly known as a cell cycle inhibitor that is induced by the transcription factor p53 in response to DNA damage (Fig. 1C, WT+IR). However, even under normal, undamaged conditions, cells express a background or basal level of p21 (Fig. 1C, WT). We hypothesized that, in undamaged WT cells, the basal level of p21 was responsible for heterogeneity in cell cycle activity. This notion would be supported if expression of p21 independent of p53 restored population heterogeneity in p21-deficient cells. To test this hypothesis, we expressed p21 in cells with a p21-deficient genetic background using vectors where expression did not require p53 (i.e., constitutive expression). The ectopic p21 was expressed as a fusion with red fluorescent protein (RFP) to enable live single-cell fluorescence detection. The p21 was also fused to an estrogen receptor (ER) domain (RFP-p21-ER), and therefore, nuclear localization could be induced by the addition of 4-hydroxytamoxifen (4-OHT). Because the activity of p21 depends on its interactions with factors in the nucleus, we were thus able to induce the activity of p21 by addition of 4-OHT at the beginning of experiments. To achieve physiologically relevant levels of p21 expression, we used synthetic upstream open reading frames (uORFs) and different translation initiation site sequences (21, 22). We generated vectors that expressed the p21 fusion proteins over a broad range of levels (Fig. 1C and Fig. S1B). To approximate the relatively low basal level in undamaged WT cells, we chose a vector using two uORFs (Fig. 1C and Fig. S1 B–D, vector ACC/ACC/ACC). After ectopically expressing this low level of p21 in p21-deficient cells, we observed that, in the quiescent populations, p21 was sustained at high levels (Fig. 1D, Lower, red). In cycling populations, p21 was expressed at lower levels and exhibited oscillatory dynamics out of phase with CDK2 activity (Fig. 1D, Lower, blue and Fig. S1E). The single-cell dynamics, which indicated that p21 decreases at the start of S phase and increases at G2, were in line with previous studies that used synchronized populations to show the cell cycle-dependent regulation of p21 (23–25). Finally, at intermediate growth factor stimulation [5 ng/mL EGF, 1.25% (vol/vol) serum], ectopic p21 restored the heterogeneity in cell cycle activity exhibited by WT cells (Fig. 1D, Upper Center and Movie S4). Under these conditions, 52% of cells were actively cycling, whereas 48% remained quiescent.
Cellular Incorporation of BrdU Revealed a Range of Growth Factor Stimulation That Supported p21-Dependent Cell Cycle Heterogeneity.
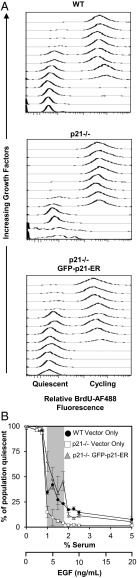
Having established p21-dependent heterogeneity at an intermediate growth factor concentration using live-cell imaging, we next investigated the growth factor concentration range where quiescent and cycling states could coexist. We administered a 48-h pulse of BrdU to cells incubated in 12 different growth factor conditions and used flow cytometry to evaluate the fraction of each population that remained quiescent vs. actively cycling. Cells that did not incorporate any BrdU over a 48-h period were considered quiescent (Fig. S2A).
At low growth factor stimulation (between 0 ng/mL EGF, 0% serum and 3 ng/mL EGF, 0.75% serum), both WT and p21-deficient cells were largely quiescent (Fig. 2A, Top and Middle), and at high growth factor stimulation [between 8 ng/mL EGF, 2% (vol/vol) serum and 20 ng/mL EGF, 5% (vol/vol) serum], both WT and p21-deficient cells were mostly cycling (Fig. 2A, Top and Middle). However, over an intermediate range of stimulation [between 4 ng/mL EGF, 1% serum and 7 ng/mL EGF, 1.75% (vol/vol) serum], the WT population distributions were bimodal in BrdU incorporation, indicating that both quiescent and cycling states coexisted (Fig. 2A, Top). When the percentage of each population that remained quiescent was plotted against growth factor concentration, it could be seen that the response manifested as a graded change in the heterogeneity of the population (Fig. 2B). In contrast, p21-deficient populations exhibited significantly less heterogeneity at all growth factor levels. They responded to changes in growth factor stimulation with more switch-like behavior (Fig. 2 A, Middle and B) (represented by the steeper drop in the quiescent subpopulation)—when stimulation surpassed a threshold level (4 ng/mL EGF, 1% serum), the populations switched from being predominantly quiescent to predominantly cycling.
Fig. 2.
p21 enables coexistence of cycling and quiescent cells over a range of growth factor stimulation. (A) Histograms for cell populations (y axes show numbers of cells); the cycling subpopulation is distinguished from the quiescent subpopulation by BrdU incorporation. (B) Percentage of quiescent cells from A. The shaded region indicates conditions at which 25–75% of WT cells remained quiescent. Data are represented as means ± SDs of triplicate samples.
In line with results from the live-cell imaging experiments, in BrdU incorporation experiments, ectopic expression of p21 fused to GFP and the ER (GFP-p21-ER) also restored heterogeneity to p21-deficient populations (Fig. 2 A, Bottom and B). At the same intermediate growth factor stimulation that supported heterogeneity in WT cells [between 4 ng/mL EGF, 1% serum and 7 ng/mL EGF, 1.75% (vol/vol) serum], populations expressing ectopic p21 exhibited bimodality in BrdU incorporation. Ectopic p21 expression caused the sharp, switch-like response to growth factors observed in p21-deficient cells to become more graded, closely resembling the response of the WT population (Fig. 2B). This result further supported the notion that p21 bestows population heterogeneity in cycling and quiescent states.
Population Distributions in p21 Expression and Cell Cycle Phase Reflected Reversibility and Growth Factor-Dependent Heterogeneity in Cell States.
We next sought to determine whether the growth factor-dependent cycling and quiescent states were reversible. Reversibility would suggest that the heterogeneity in cell cycle activity was not caused by mutations or heritable epigenetic markings. We used flow cytometry to analyze population distributions in GFP-p21-ER and GFP-ER expression and cell cycle phase (Fig. S2 B–E). When flow cytometry analysis could not detect GFP-p21-ER expression in cells, we identified those populations as GFP-low. In the live-cell imaging and BrdU incorporation experiments, we expressed GFP-p21-ER using the ACC/ACC/ACC uORF-containing RNA leader sequence (Fig. 1C and Fig. S1B) to reproduce low basal expression levels. Although we could distinguish GFP-high and -low subpopulations, we could not confidently quantify their fractions in the total population because the GFP-p21-ER expression level was low. Thus, for analysis of GFP-p21-ER expression by flow cytometry, we used a leader that generated a moderate expression level (Fig. 1C, UCU/ACC and Fig. S1 B–D). We obtained similar results in experiments using the ACC/ACC/ACC and the UCU/ACC RNA leaders, but here, we have chosen to plot the results for the UCU/ACC leader.
To evaluate reversibility, we changed growth factor stimulation from maximal levels [20 ng/mL EGF, 5% (vol/vol) serum] to submaximal levels (0 ng/mL EGF, 0% serum and 4 ng/mL EGF, 1% serum) before returning to maximal stimulation. In line with our previous results, decreasing the growth factor stimulation levels shifted population distributions toward subpopulations in G1 with high p21 (Fig. S2B). When maximal stimulation was restored, the population distributions returned to ones dominated by cycling cells expressing low p21 (Fig. S2C), indicating reversibility in cycling and quiescent states.
We also sought to determine whether GFP-p21-ER expression levels were associated with distinct cell cycle-phase distributions. By staining DNA with propidium iodide (PI), we analyzed the cell cycle phase of each GFP-high and -low subpopulation. At all stimulation levels, the cells that highly expressed GFP-p21-ER were predominantly in G1 (2 N DNA content, and here, we do not distinguish G1 from G0) and, to a lesser extent, G2/M (4 N DNA content) (Fig. S2 B and C). No cells expressing high levels of GFP-p21-ER were found in S phase (0% 2 N < DNA < 4 N). In contrast, with intermediate and maximal growth factor stimulation, cells that were expressing low or undetectable levels of GFP-p21-ER were actively cycling, which was evidenced by the sizable population in S phase (∼30% 2 N < DNA < 4 N).
However, did the population distributions in GFP-p21-ER and cell cycle phase reflect population heterogeneity in cycling and quiescent states? Keep in mind that, even if all or nearly all cells in an unsynchronized population were cycling [e.g., the population under conditions of 20 ng/mL EGF and 5% (vol/vol) serum] (Fig. S2B), the cells in G1 and, to a lesser extent, G2/M would still express a high level of GFP-p21-ER. This high GFP-p21-ER expression occurs because, regardless of whether it is cycling, p21 is stabilized at these points during the cell cycle. However, a heterogeneous population consisting of a fraction of cells that is cycling and a fraction that is quiescent would exhibit an even greater percentage of cells with stabilized p21 (i.e., the GFP-high subpopulation). The existence of a quiescent G1-arrested population would further increase the percentage of GFP-high cells in the total population, because p21 would be stabilized in all of the quiescent cells as well as a fraction of the cycling cells. Thus, at an intermediate stimulation condition that supported heterogeneity in cell cycle activity (Fig. 1), the increased fraction of cells expressing high levels of GFP-p21-ER (4 ng/mL EGF, 1% serum; 41% GFP-high) over that of the maximal stimulation condition [20 ng/mL EGF, 5% (vol/vol) serum; 33% GFP-high] can be attributed to the quiescent subpopulation (Fig. S2B). We consider this increased fraction of GFP-high cells in the heterogeneous population to be telling and that, in general, flow cytometry analysis of p21 and cell cycle population distributions supported the results from live-cell tracking and BrdU incorporation experiments. As a control, we also expressed GFP-ER in p21−/− cells. Under all conditions tested, GFP-ER expression was unimodal (Fig. S2 D and E). Thus, the growth factor-dependent regulation of GFP-p21-ER was likely dependent on the p21 domain.
Kinetic Model of Double-Negative Feedback Regulation Simulated Bistability in Cell Cycle States.
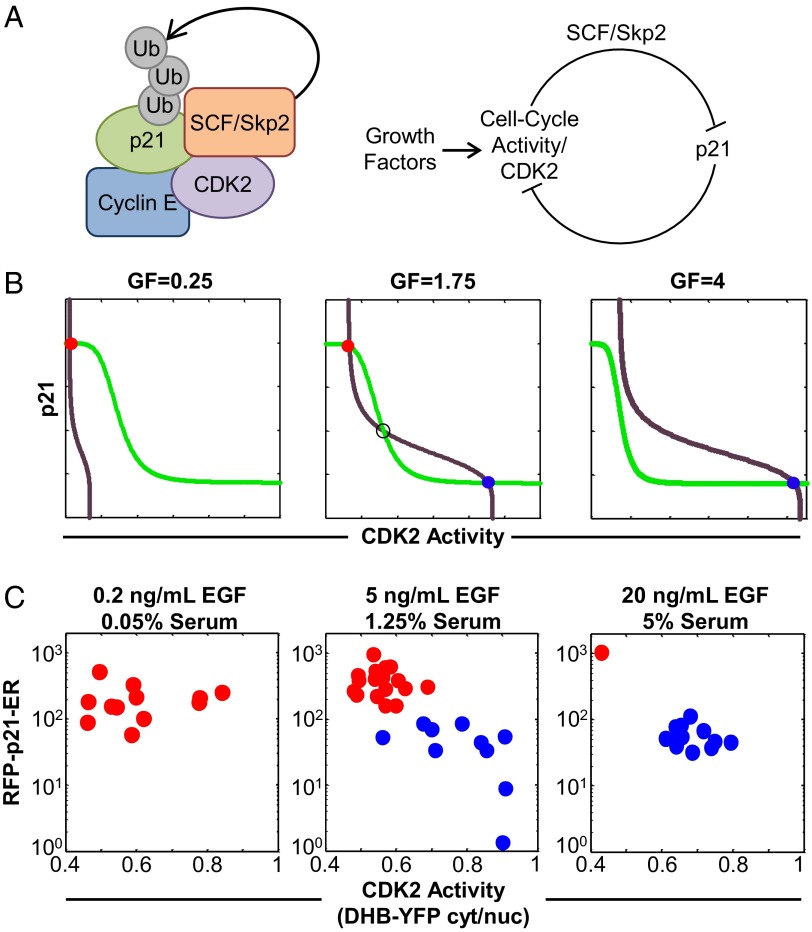
We postulated that the bistability in reversible cell states arose from double-negative feedback regulation involving CDK2 and p21 (Fig. 3 A and B). We created a kinetic model to show the potential bistability in levels of p21 and CDK2 activity at a single point in the cell cycle—namely, a period in G1 before the G1/S transition, when it is typically believed that cells have not committed to the next cell cycle (i.e., the restriction point).
Fig. 3.
Double-negative feedback regulation explains p21-dependent heterogeneity in cell cycle activity. (A) Schematic of (Left) interacting factors and (Right) double-negative feedback regulation. Ub, ubiquitin. (B) Steady-state balance plots generated by the model: steady-state p21 (green) and CDK2 (purple) activity levels, where intersections indicate stable quiescent (red) and cycling (blue) states at three growth factor (GF) concentrations. Open circles indicate an unstable steady state. At intermediate stimulation (GF = 1.75 relative units), two states (high p21/low CDK2 activity and low p21/high CDK2 activity) can stably coexist in a single population. (C) Live-cell imaging: RFP-p21-ER vs. CDK2 activity for cycling (blue) and quiescent (red) cells. Values for cycling cells were determined at a point in G1 3 h after mitosis and before cell cycle entry.
The level of active CDK2 complexed with Cyclin E or Cyclin A (represented by CDK2a in our equations) is affected by (i) growth factor stimulation, (ii) inhibition by p21, and (iii) positive feedback [a positive feedback loop was previously modeled by Yao et al. (17) and involved pRb, E2F, and Cyclin E]. The concentration of p21 is affected by its interaction with the Cyclin E-CDK2 complex through the action of SCF/Skp2 and other E3 ubiquitin ligase complexes (Fig. 3A and SI Text). The parameters used in the model are defined in Table S1.
We generated steady-state balance plots by solving for CDK2 activity over a range of p21 concentrations (Fig. 3B, purple lines) and then solving for p21 concentration over a range of CDK2 activity values (Fig. 3B, green lines) at three different growth factor concentrations. Each intersection of the CDK2 and p21 lines represented a steady state of the system. The system was monostable at both high and low growth factor concentrations (Fig. 3B, Left and Right). At intermediate growth factor concentration, three intersection points existed (Fig. 3B, Center), indicating that the system was bistable. The points corresponding to high p21/low CDK2 activity (Fig. 3B, Center, red circle) and low p21/high CDK2 activity (Fig. 3B, Center, blue circle) represented quiescent and cycling stable steady states, respectively.
Because our live-cell imaging experiments tracked p21, CDK2 activity, and cell cycle phase over multiple cell generations, we could examine levels of p21 and CDK2 activity within the G1 window described by our model. Under all stimulation conditions, we found that high p21/low CDK2 activity was associated with G1-arrested quiescent cells (Fig. 3C), whereas low p21/high CDK2 activity was associated with cycling cells in early G1, 3 h after mitosis and before the G1/S transition. Most importantly, under the intermediate stimulation condition [5 ng/mL EGF, 1.25% (vol/vol) serum], we found that coexisting quiescent and cycling states were also characterized by high p21/low CDK2 activity and low p21/high CDK2 activity, respectively. Therefore, together, modeling and experiments indicated that cell states were reflected by each cell’s position along a p21–CDK2 axis. Additionally, although there can be numerous other factors that distinguish G1 proliferating cells from G1 quiescent cells, the convergence between modeling and experimental results suggests that p21-dependent population heterogeneity can be explained, at least in part, by double-negative feedback regulation.
Although modeling p21 as the sole CKI was sufficient to describe the coexistence of two positions along a p21–CDK2 axis, there are two other kinase inhibitor proteins (KIPs), p27 and p57, regulating CDK2 that needed to be taken into account to appropriately model the CDK2 activity in our experiments (SI Text and Fig. S3). In particular, by including the contributions from p27 and p57, we were able to simulate the behavior of p21-deficient cells. Like p21, KIPs are regulated by CDK2 through the SCF/Skp2 E3 ubiquitin ligase, and they also inhibit CDK2 by binding to the CDK2-Cyclin E complex (13). In our model, the kinetic constants governing CDK2 and KIP regulation were the same as those involving p21.
Using this expanded model, we generated stimulus–response curves to evaluate bistability over a range of growth factor concentrations for three p21 generation rates. For each growth factor concentration, we plotted the corresponding steady-state CDK2 activity (Fig. S3 B and C) or CKI concentration (Fig. S3D). As with our previous experimental (Figs. 1B and 3C) and modeling results (Fig. 3B), when p21 was present in the system (kgen,p21 = 1 or 2), the system was bistable at intermediate growth factor concentrations (Fig. S3 B, Center and Right, C, Center and Right, and D, Center and Right). In the p21-deficient case (kgen,p21 = 0), the system remained monostable at all growth factor concentrations (Fig. S3 B, Left, C, Left, and D, Left).
Without the positive feedback on CDK2 (k4 = 0), the change in CDK2 activity in p21-deficient cells was more graded (Fig. S3B, Left). However, when the model included the positive feedback on CDK2 (k4 = 0.25), there was a sharp increase in CDK2 activity at a particular intermediate growth factor concentration in the p21-deficient case (Fig. S3C, Left). This increase also reflected the switch-like transition between quiescent and cycling states that we previously observed in p21-deficient cells at conditions between 3 ng/mL EGF and 0.75% serum and 4 ng/mL EGF and 1% serum (the sharp decrease in the percentage of quiescent cells in Fig. 2B). Thus, our model indicated that the switch-like behavior exhibited by p21-deficient cells in response to growth factor stimulation also depended on the positive feedback regulation of CDK2.
Cell Cycle Heterogeneity Is Dependent on WT p21 and SCF/Skp2 Activity.
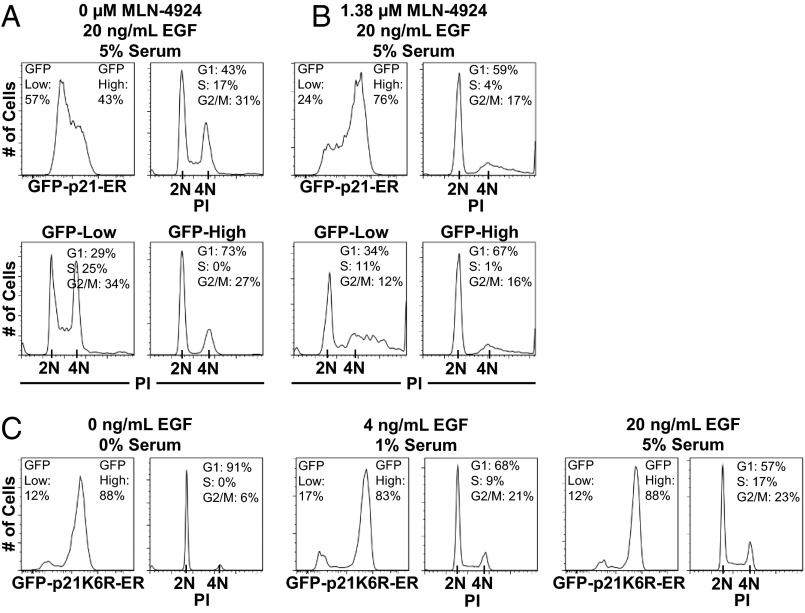
Having proposed this double-negative feedback mechanism, we next sought to establish whether the negative regulation of p21 in our experiments depended on SCF/Skp2. When we inhibited p21 regulation using MLN-4924 (an inhibitor of Cullin-dependent E3 ligases) in p21-deficient cells expressing GFP-p21-ER, flow cytometry analysis of the p21 population distribution showed that a majority of the population had stabilized p21 and become quiescent, even at maximal stimulation (Fig. 4 A and B). We also evaluated expression of GFP-p21K6R-ER, a fusion protein containing a p21K6R mutant that could not be ubiquitinated by SCF/Skp2 at six known lysine targets (26). Across all levels of growth factor stimulation, populations predominantly showed high expression levels of p21K6R (Fig. 4C). Despite this stabilized expression of p21K6R, the cells still responded to varying growth factor stimulation. This result was consistent with previous reports that the mutant is not as active as WT p21 (26, 27). Because the mutant was stabilized at all levels of stimulation but diminished in its ability to inhibit cell cycle activity, the negative regulation of WT p21 (both endogenous p21 and GFP-p21-ER) was likely dependent on ubiquitination by SCF/Skp2 (or other Cullin-dependent complexes) and subsequent proteasomal degradation.
Fig. 4.
Negative regulation of p21 is dependent on SCF/Skp2 and p21 lysine residues targeted for ubiquitination. (A) GFP-p21-ER was expressed in p21−/− cells. Population distributions for GFP-p21-ER were determined by flow cytometry, and GFP-low and -high populations were quantified. Cell cycle phase distributions were determined by PI staining of DNA content (2 N–4 N). (B) Inhibition of SCF/Skp2 by the cullin inhibitor MLN-4924. (C) Expression of GFP-p21K6R-ER, where ubiquitination by SCF/Skp2 has been reduced by mutating six lysine targets.
We then used our CDK2–p21 model to perturb the double-negative feedback regulation mechanism. Stabilizing p21 (setting k2 to zero) (Fig. S4A, Left) caused the system to remain in a quiescent state (high p21/low CDK2 activity), even at high growth factor stimulation (Fig. S4A, Right). This result was consistent with our experimental observation that inhibiting SCF/Skp2 and other Cullin-dependent E3 ubiquitin ligase complexes with MLN-4924 caused populations to remain predominantly quiescent at high growth factor stimulation (Fig. 4 A and B). Because it was not possible to experimentally eliminate the regulation of CDK2 by p21 without also eliminating the regulation of p21 by SCF/Skp2 (26), we used our model (setting k3 to zero) to simulate a p21 mutant unable to inhibit CDK2 but still negatively regulated by CDK2 through SCF/Skp2 (Fig. S4B, Left). This inhibition of p21 regulation caused the CDK2 activity on the steady-state balance plot to remain constant, resulting in monostability at all evaluated growth factor concentrations (Fig. S4B, Right). This result further supported the notion that the double-negative feedback mechanism plays a key role in generating bistability and population heterogeneity.
Discussion
In summary, we report that low basal levels of p21 control growth factor-dependent population heterogeneity in cell cycle activity within an isogenic population of cells (Fig. 5A). Although p21 is perhaps best known as a p53-responsive gene expressed highly in response to DNA damage, our study revealed a physiological role for basal expression levels. Using both live-cell imaging and BrdU staining, we showed that cells expressing p21 exhibited heterogeneity in cycling and quiescent states at intermediate growth factor stimulation—a fraction of the population remained quiescent, whereas the other fraction was cycling. In contrast, cells deficient in p21 did not exhibit this population heterogeneity. In further support of a direct role for p21, ectopic expression at a level approximating the basal endogenous level of p21 expression restored heterogeneity in p21-deficient cells.
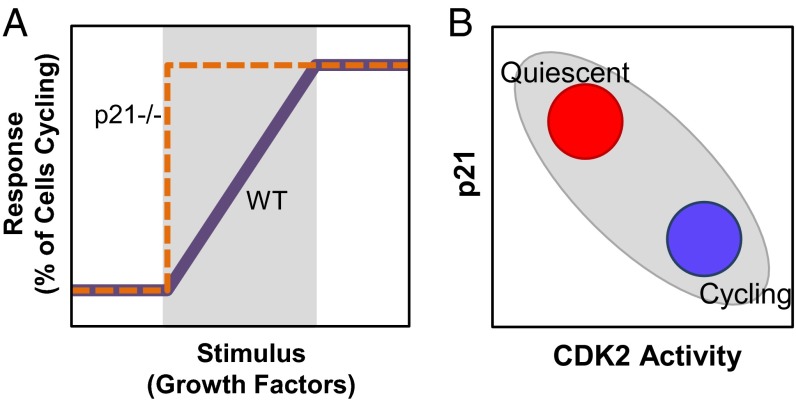
Fig. 5.
A p21–CDK2 axis governs p21-dependent population heterogeneity in quiescent and cycling cellular states. (A) Cells deficient in p21 respond to growth factor stimulation in a more switch-like manner, whereas cells expressing basal levels of p21 respond in a more graded manner. The shaded region represents growth factor levels that support p21-dependent heterogeneity in cell cycle states. (B) Cell states mapped onto a p21–CDK2 axis (shaded region). At a stage before cell cycle entry, cells with high p21 and low CDK2 activity are quiescent (red region), whereas cells with low p21 and high CDK2 activity are cycling (blue region).
Although we have proposed a role for p21 in governing population heterogeneity in response to growth factors, we point out that cells deficient in p21 still responded to changes in growth factor stimulation. The response of these p21-deficient cells, however, was monostable. Compared with WT cells, cells deficient in p21 behaved in a more switch-like manner—predominantly quiescent at low growth factor stimulation and predominantly cycling at intermediate and high growth factor stimulations. This result suggests that mitogenic signaling contributes to the decision to remain quiescent or enter the cell cycle through mechanisms independent of p21—possibly a feedback loop comprised of pRb, E2F, Cyclin E, and CDK2 (17)—but that the presence of p21 causes a decision point to be bistable. This bistability could arise from stochasticity in expression of p21. It has been shown that stochasticity in expression of genes involved in regulatory mechanisms, especially genes that are expressed at a low level, can lead to bistability in a system (14). It is, therefore, possible that stochastic changes in the basal expression level of p21 could lead to the heterogeneity that we observed.
Double-negative feedback regulation between p21 and CDK2 can explain how both cycling and quiescent states can coexist within a single isogenic population. Although we have suggested a role for the SCF/Skp2 E3 ubiquitin ligase in this feedback regulation, we do not discount that other mechanisms that regulate p21 (e.g., CRL4-Cdt2, APC-Cdc20, and possibly other unknown complexes) also contribute to the behavior that we have described. In addition, our experimental results combined with kinetic modeling revealed a p21–CDK2 axis that was predictive of cycling vs. quiescent cellular states (Fig. 5B). We project that this heterogeneity can be mediated by not only p21 but also the CKI proteins p27 and p57, because both are regulated by SCF/Skp2. Because the control scheme has been adopted not only by mammals but also yeast (13, 28), we propose double-negative feedback regulation involving cell cycle activators and inhibitors to be a general strategy for maintaining heterogeneity in cell cycle activity. This mechanism may explain how stem cell populations use p21 to maintain subpopulations of dormant and self-renewing cells (29, 30).
Finally, heterogeneity in cell cycle activity may impact the treatment of tumors, because quiescent cells may evade therapies that target proliferating cells. Thus, therapeutics that inhibit cyclin-dependent kinases (e.g., a small molecule that targets CDK2) could potentially increase heterogeneity, promote quiescent subpopulations, and ultimately show diminished efficacy. Instead, our work suggests that CKIs, such as p21, could be targeted. Such therapeutic agents would be given concurrently with chemotherapy agents, reduce tumor cell heterogeneity, and subsequently increase chemotherapy efficacy.
Materials and Methods
Expression Vectors.
The p21 gene was amplified from human cDNA by PCR. Monomeric GFP (EGFP A207K) was PCR-amplified from pEGFP-N1 (Clontech Laboratories), and ER domain was PCR-amplified from pBabe-Puro-OmoMyc-ER (a gift from Gerard Evan, University of California, San Francisco). Using these DNA fragments, we constructed the gene fusions encoding GFP-p21-ER and GFP-ER; using standard molecular biology techniques, the gene fusions were inserted into plasmids to generate pCru5-GFP-p21-ER and pCru5-GFP-ER, respectively. The fusions were then moved into pCru5-(UUU)GGFP-IRES-Puro (22) using the EcoRI and NsiI restriction sites. The EcoRI-NotI fragment from this vector was then moved into the EcoRI-NotI sites of pCru5-GFP-IRES-mCherry vectors containing various RNA leader sequences and regulatory uORFs (22) to create pCru5-GFP-p21-ER-IRES-Puro and pCru5-GFP-ER-IRES-Puro. For live-cell imaging experiments, RFP (specifically, the fluorescent protein mCherry) was PCR-amplified from pCru5-GFP-IRES-mCherry and substituted for GFP to create pCru5-RFP-p21-ER-IRES-Puro. CSII-EF-DHB-YFP and CSII-EF-H2B-mTurquoise were constructed as previously described (12). GFP fusion constructs were used for flow cytometry experiments, because GFP detection is compatible with detection of PI staining. RFP fusion constructs were used for live-cell imaging experiments, because RFP detection is compatible with detection of the DHB-YFP CDK2 sensor.
Cell Culture.
MCF10A cells were maintained in DMEM/F12 media (11330–032; Life Technologies) supplemented with horse serum (H1138; Sigma-Aldrich), 2 mM glutamine, EGF (SRP3027; Sigma-Aldrich), 10 µg/mL insulin (I1882; Sigma-Aldrich), 500 ng/mL hydrocortisone (H0888; Sigma-Aldrich), 100 ng/mL cholera toxin (C8052; Sigma-Aldrich), 100 U/mL penicillin, and 100 µg/mL streptomycin at 37 °C and 5% (vol/vol) CO2. Full growth medium contained 5% (vol/vol) serum and 20 ng/mL EGF. During routine cell culture and throughout all experiments, cell populations were cultured at subconfluent concentrations to minimize contact inhibition and increased activation of p27.
To make a stable MCF10A WT line expressing the pCru5-IRES-Puro empty vector control and MCF10A p21−/− lines expressing GFP-p21-ER, GFP-ER, RFP-p21-ER, GFP-p21K6R-ER, and the pCru5-IRES-Puro empty vector control, retroviral particles were made by cotransfecting the retroviral expression plasmids with pCL-Ampho (31) in 293T cells using calcium phosphate precipitation (CalPhos Mammalian Transfection Kit; Clontech Laboratories, Inc.). Supernatant containing viral particles was filtered and added to MCF10A cells along with 5 µg/mL polybrene (hexadimethrine bromide) for 18 h. The viral supernatant was tittered, such that each transduced cell received only one copy of the vector. Cells infected with pCru5-GFP-p21-ER, pCru5-GFP-ER, pCru5-RFP-p21-ER, and pCru5-IRES-Puro empty vector were selected with 1 µg/mL puromycin for at least 3 d and supplemented with 1 μg/mL puromycin during routine culture and 1 μg/mL puromycin and 500 nM 4-OHT throughout all experiments. Cultures infected with pCru5-GFP-p21K6R-ER were supplemented with 10 μg/mL blasticidin for at least 3 d to select for stable integration, and cells were maintained with 10 μg/mL blasticidin during routine cell culture and 10 μg/mL blasticidin and 500 nM 4-OHT throughout all experiments. To make MCF10A WT and p21−/− lines that express DHB-YFP and H2B-mTurquoise, lentiviral particles were made by cotransfecting the lentiviral expression plasmids pCSII-EF-DHB-mVenus or pCSII-EF-H2B-mTurquoise (12) with pRSV-Rev, pHCMV-VSVG, and pMdlg in 293T cells using calcium phosphate precipitation. Supernatant containing viral particles was filtered and added to MCF10A cells along with 5 µg/mL polybrene (hexadimethrine bromide) for 18 h. Cells transduced with CSII-EF-H2B-mTurquoise and CSII-EF-DHB-mVenus lentivirus were sorted using a FACSAria (Beckton Dickinson) cell sorter to isolate a population expressing both constructs.
To determine the reversibility of quiescence and p21 stabilization at low growth factor stimulation, MCF10A p21−/− cells expressing GFP-p21-ER or GFP-ER with a vector using the UCU/ACC uORF and RNA leader sequence (pCru5-UCU/ACC-GFP-p21-ER) were induced with 4-OHT for 24 h in media containing 20 ng/mL EGF and 5% (vol/vol) serum. Cells were then cultured with reduced growth factor stimulation (0 ng/mL EGF, 0% serum or 4 ng/mL EGF, 1% serum) for 72 h. Each culture was restored to full growth stimulation [20 ng/mL EGF, 5% (vol/vol) serum] for 72 h. For experiments examining the SCF/Skp2 dependence of p21 degradation, MCF10A p21−/− cells expressing GFP-p21-ER were induced with 4-OHT for 24 h in media containing 20 ng/mL EGF and 5% (vol/vol) serum. Cells were then cultured with no growth factor stimulation (0 ng/mL EGF, 0% serum) for 72 h. Each culture was then restored to full growth stimulation [20 ng/mL EGF, 5% (vol/vol) serum] and concomitantly supplemented with either 0 or 1.38 µM MLN-4924 (A-1139; Active Biochem) cullin inhibitor for 48 h.
Flow Cytometry.
For DNA content analysis by PI staining, cells were trypsinized, rinsed with PBS, added to ice-cold 70% (vol/vol) ethanol, and stored at −20 °C overnight. Each sample was then rinsed with PBS and incubated in a buffer containing PI [PBS with 0.1% (vol/vol) Triton X-100, 0.2 mg/mL RNase A, and 20 μg/mL PI] for 30 min at room temperature.
To measure cell cycling and quiescent states by BrdU incorporation, cells were seeded in six-well plates (353046; Corning) and incubated in complete growth media containing 1 µg/mL puromycin and 500 nM 4-OHT for 24 h. Each well was then washed with PBS and incubated with media containing various concentrations of serum and EGF, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin, 1 µg/mL puromycin, and 500 nM 4-OHT for 72 h; 10 µM BrdU (B23151; Life Technologies) was added to each culture, and cells were incubated for an additional 48 h. Cells were then dissociated with 0.05% trypsin, and DMEM/F12 medium with 1% BSA was used to neutralize the trypsin. Cells were washed with PBS and incubated in 70% (vol/vol) ethanol overnight. After washing with PBS, DNA was denatured by treatment with PBS containing 0.1% Triton-X 100 (PBS-T) supplemented with 4 M HCl for 30 min. The acid was neutralized by washing two times with 0.1 M sodium tetraborate, and samples were incubated with PBS-T containing 1% BSA for 1 h for blocking; 1 µL mouse anti-BrdU antibody conjugated to AlexaFluor488 (clone MoBU1, B35139; Life Technologies) was added to each sample and incubated at 4 °C overnight before washing with PBS-T and incubating with PI staining solution for 30 min.
Cell populations were analyzed on an LSRII flow cytometer (Beckton Dickinson). GFP fluorescence intensity was determined using 488-nm excitation and a 525/50-band pass emission filter (the emission collection was centered at 525 nm with a total range of 50 nm) as well as 405-nm excitation and a 525/50-band pass filter. Flow cytometry data were analyzed using FlowJo (Tree Star).
Live-Cell Imaging.
MCF10A WT and p21−/− cells expressing H2B-mTurquoise, DHB-YFP, and RFP-p21-ER (pCru5-ACC/ACC/ACC-RFP-p21-ER) were seeded in a 96-well plate (3603; Costar) in complete growth media containing 1 µg/mL puromycin and 500 nM 4-OHT and incubated for 24 h. Each well was washed two times with PBS, and 300 µL phenol red-free media containing various concentrations of serum and EGF, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin, 1 µg/mL puromycin, and 500 nM 4-OHT were added. Cells were incubated for 72 h before imaging for 48 h on an IXMicro microscope (Molecular Dynamics) with a heated stage and 5% (vol/vol) CO2. A 10× objective was used, and total exposure time was kept below 900 ms. Images were analyzed using MATLAB (MathWorks).
Immunoblotting.
Immunoblotting was performed according to standard procedures. Lysates were loaded onto a 10% (vol/vol) Mini-Protean TGX Gel (Bio-Rad) and transferred to a PVDF membrane; p21 was detected using a primary anti-p21 mouse monoclonal antibody (2946; Cell Signaling Technology) and a secondary anti-mouse IgG conjugated to HRP (G21040; Molecular Probes). β-Actin was detected using a mouse monoclonal anti–β-actin antibody conjugated to HRP (A00730; Genscript). WesternBright ECL HRP substrate (K-12045-D50; Advansta) was used for HRP detection.
Modeling.
Computational analysis of the CDK2–p21 double-negative regulatory feedback system was performed using MATLAB (MathWorks).
Supplementary Material
Acknowledgments
This work marks the end of C.L.W.'s time at Stanford University. C.L.W. would like to thank Jay Keasling and Matthias Wabl for their tutelage and support along the way, and his students and laboratory members for believing in his laboratory's vision and for doing science the right way. He'd still take reincarnation as a scientist. The authors thank Steve Cappell for providing MLN-4924 and MATLAB code and Mingyu Chung for MATLAB code. Flow cytometry measurements were performed at the Stanford Shared FACS Facility, and sorting was performed on an instrument at the Stanford Shared FACS Facility obtained using National Institutes of Health (NIH) S10 Shared Instrument Grant S10RR025518-01. This study was supported by NIH Grant 5R21AG040360-02 and Ellison Medical Foundation Grant AG-NS-0550-09. K.W.O. was supported by a National Science Foundation Graduate Research Fellowship and a Stanford Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409797111/-/DCSupplemental.
References
- 1.Maheshri N, O’Shea EK. Living with noisy genes: How cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 2.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr Opin Chem Biol. 2009;13(5-6):556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge BB, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335(6064):100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TJ, Yao G, Bennett DC, Nevins JR, You L. Stochastic E2F activation and reconciliation of phenomenological cell-cycle models. PLoS Biol. 2010;8(9):e1000488. doi: 10.1371/journal.pbio.1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 6.Passos JF, et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5(5):e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLoS Comput Biol. 2012;8(4):e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459(7245):428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 10.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 11.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Spencer SL, et al. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155(2):369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Hunter T. Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. Cell Cycle. 2010;9(12):2342–2352. doi: 10.4161/cc.9.12.11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 15.To TL, Maheshri N. Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science. 2010;327(5969):1142–1145. doi: 10.1126/science.1178962. [DOI] [PubMed] [Google Scholar]
- 16.Wong JV, Yao G, Nevins JR, You L. Viral-mediated noisy gene expression reveals biphasic E2f1 response to MYC. Mol Cell. 2011;41(3):275–285. doi: 10.1016/j.molcel.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10(4):476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 18.Yao G, Tan C, West M, Nevins JR, You L. Origin of bistability underlying mammalian cell cycle entry. Mol Syst Biol. 2011;7:485. doi: 10.1038/msb.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn AT, Jones JT, Meyer T. Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle. 2009;8(7):1044–1052. doi: 10.4161/cc.8.7.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westergaard LG, Andersen CY. Epidermal growth factor (EGF) in human preovulatory follicles. Hum Reprod. 1989;4(3):257–260. doi: 10.1093/oxfordjournals.humrep.a136883. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira JP, Noderer WL, Diaz de Arce AJ, Wang CL. Engineering ribosomal leaky scanning and upstream open reading frames for precise control of protein translation. Bioengineered. 2014;5(3):186–192. doi: 10.4161/bioe.27607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira JP, Overton KW, Wang CL. Tuning gene expression with synthetic upstream open reading frames. Proc Natl Acad Sci USA. 2013;110(28):11284–11289. doi: 10.1073/pnas.1305590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dulić V, Stein GH, Far DF, Reed SI. Nuclear accumulation of p21Cip1 at the onset of mitosis: A role at the G2/M-phase transition. Mol Cell Biol. 1998;18(1):546–557. doi: 10.1128/mcb.18.1.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tinkum KL, et al. Bioluminescence imaging captures the expression and dynamics of endogenous p21 promoter activity in living mice and intact cells. Mol Cell Biol. 2011;31(18):3759–3772. doi: 10.1128/MCB.05243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto H, et al. Comparative effects of overexpression of p27Kip1 and p21Cip1/Waf1 on growth and differentiation in human colon carcinoma cells. Oncogene. 1999;18(1):103–115. doi: 10.1038/sj.onc.1202269. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Nacusi L, Sheaff RJ, Liu X. Ubiquitination of p21Cip1/WAF1 by SCFSkp2: Substrate requirement and ubiquitination site selection. Biochemistry. 2005;44(44):14553–14564. doi: 10.1021/bi051071j. [DOI] [PubMed] [Google Scholar]
- 27.Sheaff RJ, et al. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5(2):403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 28.Venta R, Valk E, Kõivomägi M, Loog M. Double-negative feedback between S-phase cyclin-CDK and CKI generates abruptness in the G1/S switch. Front Physiol. 2012;3:459. doi: 10.3389/fphys.2012.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng T, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287(5459):1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 30.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19(6):756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peacock RW, Wang CL. A genetic reporter system to gauge cell proliferation rate. Biotechnol Bioeng. 2011;108(9):2003–2010. doi: 10.1002/bit.23163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.