Abstract
Microorganisms evolve via mechanisms spanning sexual/parasexual reproduction, mutators, aneuploidy, Hsp90, and even prions. Mechanisms that may seem detrimental can be repurposed to generate diversity. Here we show the human fungal pathogen Mucor circinelloides develops spontaneous resistance to the antifungal drug FK506 (tacrolimus) via two distinct mechanisms. One involves Mendelian mutations that confer stable drug resistance; the other occurs via an epigenetic RNA interference (RNAi)-mediated pathway resulting in unstable drug resistance. The peptidyl-prolyl isomerase FKBP12 interacts with FK506 forming a complex that inhibits the protein phosphatase calcineurin1. Calcineurin inhibition by FK506 blocks M. circinelloides transition to hyphae and enforces yeast growth2. Mutations in the fkbA gene encoding FKBP12 or the calcineurin cnbR or cnaA genes confer FK506 resistance (FK506R) and restore hyphal growth. In parallel, RNAi is spontaneously triggered to silence the FKBP12 fkbA gene, giving rise to drug-resistant epimutants. FK506R epimutants readily reverted to the drug-sensitive wild-type (WT) phenotype when grown without drug. The establishment of these epimutants is accompanied by generation of abundant fkbA small RNA (sRNA) and requires the RNAi pathway as well as other factors that constrain or reverse the epimutant state. Silencing involves generation of a double-stranded RNA (dsRNA) trigger intermediate from the fkbA mature mRNA to produce antisense fkbA RNA. This study uncovers a novel epigenetic RNAi-based epimutation mechanism controlling phenotypic plasticity, with possible implications for antimicrobial drug resistance and RNAi-regulatory mechanisms in fungi and other eukaryotes.
The pathogenic fungus M. circinelloides grows as hyphae aerobically, and as yeast in anaerobic/high CO2 conditions3. FKBP12 is a prolyl-isomerase conserved throughout eukaryotes that interacts with FK506 and rapamycin and mediates their antifungal activity in M. circinelloides4 and other fungi. The FKBP12-FK506 and FKBP12-rapamycin complexes inhibit the protein phosphatase calcineurin and the Torkinase, respectively1,5. FK506 inhibition of calcineurin blocks hyphal growth of M. circinelloides and enforces yeast phase growth2 (Fig. 1a, Extended Data Fig. 1a). Exposure to FK506 yields at moderate frequency (~1 × 10-6) drug-resistant isolates exhibiting hyphal growth emerging from the yeast colony periphery (Extended Data Fig. 1a). A subset of FK506R isolates harbor mutations in the fkbA gene encoding FKBP12 or the calcineurin A or B subunit genes cnaA and cnbR (45/64 isolates [~70%], Supplementary Table 1)2.
Figure 1. RNAi-dependent epimutations confer FK506-resistance in M. circinelloides.
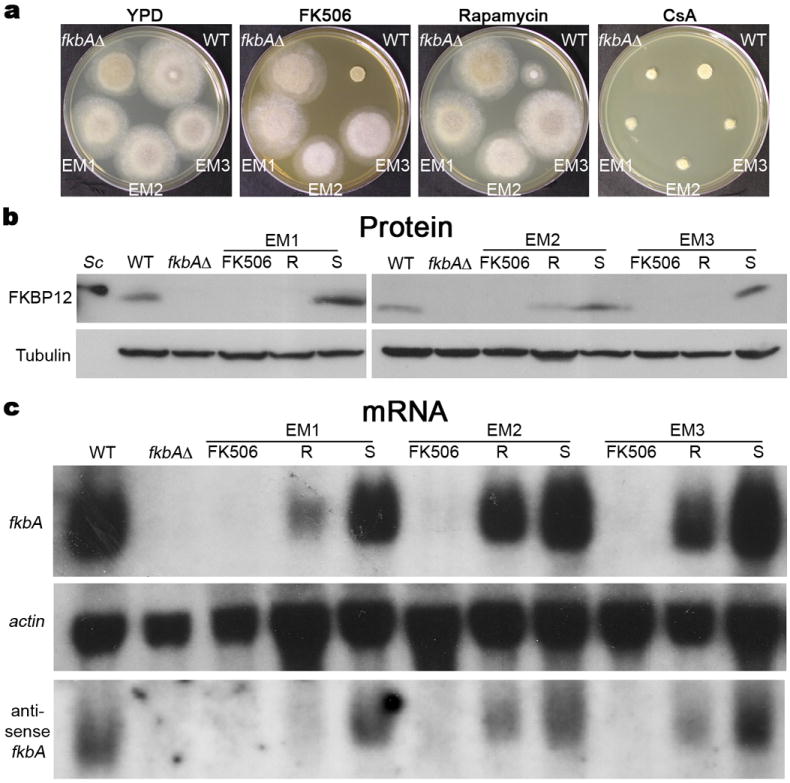
a, WT, fkbA mutant (fkbAΔ), and epimutant strains were grown on YPD media alone or supplemented with FK506, rapamycin, or cyclosporin A (CsA). Images representative of several independent experiments. b-c, The epimutant strains EM1, EM2, and EM3 were grown on YPD media with FK506 (FK506 lanes), or YPD drug-free media (R lanes). The reverted strains EM1-S, EM2-S, and EM3-S (S lanes) were grown in YPD media and whole cell protein and RNA extracts were prepared. b, Equivalent protein amounts (120 μg) were resolved by SDS-PAGE and analyzed by western blot with an anti-S. cerevisiae-FKBP12 antibody. AnS. cerevisiae extract (Sc) was included as control for antibody specificity; tubulin served as loading control. Images representative of seven independent experiments. c, 50 μg total mRNA was analyzed by northern blot employing probes specific for fkbA, act1 (loading control), and antisense fkbA mRNAs. Images representative of six independent experiments for the fkbA and act1 probes, and two for the antisense fkbA probe.
However, several FK506R isolates (17/64, ~27%) harbored no mutations in the fkbA or calcineurin target genes. These isolates exhibited resistance to FK506 and rapamycin, but not to cyclosporin A (which similar to FK506 enforces largely yeast growth, Fig. 1a) or other drugs (nystatin, amphotericin B, not shown), arguing against multidrug resistance mechanisms. These unusual drug-resistant isolates also reverted frequently within several generations of vegetative growth on drug-free media and were restored to a WT phenotype (yeast growth on FK506) (Extended Data Figure 1b, Extended Data Fig. 1c). Expression analyses revealed a complete loss of fkbA mRNA and FKBP12 protein in these drug-resistant isolates when grown in media containing FK506. In contrast, mRNA and protein levels were reduced but detectable in some resistant isolates (R) when grown in drug-free media (Fig. 1b-c), and were restored to WT levels in revertant isolates that became FK506-sensitive (FK506S) (S) following passage in drug-free media (Fig. 1b-c).
Because drug resistance was reversible and an active RNAi pathway is present in the organism6,7 we entertained the hypothesis that drug resistance is RNAi-mediated. Remarkably, sRNAs complementary to fkbA were detected in these unusual drug-resistant isolates (Fig. 2a, Extended Data Fig. 2a), suggesting a new role for RNAi in development of transient resistance to antifungal drug exposure. Consistent with the expression analyses, the sRNA signal was highly abundant during growth with FK506 (100%), less abundant when the isolates were grown in drug-free media (~62 and 25% for EM2 and EM3 respectively, Fig. 2a), and lost when drug resistance reverted (0-0.02%). The fkbA silencing was not associated with DNA methylation in M. circinelloides (Extended Data Fig. 2b). We term these unusual drug-resistant isolates epimutants, by analogy with studies in fungi8,9, plants10, and animals11-13, in which epimutations have been described as silencing of genes that are usually active or vice versa11.
Figure 2. Epimutant strains express abundant sRNA antisense to fkbA.

a, sRNA were extracted from WT, epimutants (FK506 and R lanes), and reverted strains (S lanes) after growth in YPD media alone (R, S lanes) or with FK506 (FK506 lanes). sRNAs (25μg) were analyzed by sRNA blot employing an antisense-specific probe for the fkbA gene or a probe for 5S rRNA (loading control). Images representative of three independent experiments. b, The presence of sense and antisense fkbA sRNA was analyzed by high-throughput sequencing in WT, epimutants, and two revertant strains (EM1-S, EM3-S). sRNA amount is expressed in reads per million, and they are distributed along the fkbA ORF (bottom) c, Analysis of size and first nucleotide (inset) of antisense sRNAs. The representation corresponds to data obtained from isolate EM1-R. Similar results were observed for the EM2-R and EM3-R epimutants.
Our first hypothesis was that RNAi could be triggered via dsRNA production from the overlap in the 3’ regions between fkbA and its convergently transcribed neighboring gene patA (Extended Data Fig. 3 and Supplementary Table 2). But patAΔ mutants did not show any effect on the frequency of fkbA epimutational silencing (Table 1, Supplementary Table 1, Extended Data Fig. 4). 3’ RACE assays confirmed the mRNA from fkbA gene and the pyrG marker replacing patA were not overlapping (Supplementary Table 2). Thus, expression of patA to generate overlapping RNA molecules is not necessary for fkbA silencing.
Table 1.
Frequency of epimutants/mutants in the WT, patA, and RNAimutant strains.
| Strain | Background | Total analyzed | Mutations in fkbA | Mutations in cnaA/cnbR | No mutation found | Epimutants # | Epimutants % | p-value* |
|---|---|---|---|---|---|---|---|---|
| NRRL3631 | Wild type | 33 | 22 | 0 | 11 | 10 | 30.3 | |
| R7B | leuA- | 31 | 22 | 1 | 8 | 7 | 22.6 | |
| MU434 | patAΔ∷pyrGleuA- | 33 | 28 | 0 | 5 | 5 | 15.2 | 0.531 |
| MU435 | patAΔ∷pyrGleuA- | 33 | 26 | 0 | 7 | 7 | 21.2 | 1 |
| MU406 | dcl1Δ∷pyrGleuA- | 25 | 23 | 1 | 1 | 1 | 4 | 0.06 |
| MU407 | dcl1Δ∷pyrGleuA- | 21 | 18 | 3 | 0 | 0 | <4.8 | 0.033 |
| MU410 | dcl2Δ∷pyrGleuA- | 34 | 31 | 2 | 1 | 0 | <2.9 | 0.004 |
| MU413 | ago1Δ∷pyrGleuA- | 26 | 25 | 1 | 0 | 0 | <3.8 | 0.012 |
| MU426 | ago1Δ∷pyrGleuA- | 27 | 27 | 0 | 0 | 0 | <3.7 | 0.012 |
| MU416 | ago2Δ∷pyrGleuA- | 23 | 16 | 3 | 4 | 4 | 17.4 | 0.741 |
| MU414 | ago3Δ∷pyrGleuA- | 27 | 22 | 1 | 4 | 4 | 14.8 | 0.518 |
| MU419 | rdrp1Δ∷pyrGleuA- | 32 | 6 | 0 | 26 | 26 | 81.3 | 0.000004 |
| MU420 | rdrp2Δ∷pyrGleuA- | 25 | 23 | 2 | 0 | 0 | <4.0 | 0.013 |
| MU428 | rdrp2Δ∷pyrGleuA- | 27 | 26 | 1 | 0 | 0 | <3.7 | 0.012 |
p-values were obtained based on a Fisher Exact Probability Test for a 2x2 Contingency Table, comparing each of the mutant strains individually versus the R7Bstrain.
As fkbA is a highly expressed gene (Fig. 1c), and a high RNA turnover rate has been implicated in production of aberrant RNA and triggering of silencing14-17, we speculated that an fkbA antisense RNA may be generated by an RNA-dependent RNA polymerase (RdRP)18. Northern blot analysis with an fkbA antisense-specific probe revealed an antisense fkbA mRNA in all strains with robust fkbA expression (Fig. 1c). This fkbA antisense RNA is perfectly complementary to the intron spliced mature fkbA sense mRNA, and is 5’ capped and poly-adenylated (Extended Data Fig. 5). Thus, the antisense RNA is generated from the mature fkbA mRNA. The fkbA antisense RNA is expressed in the WT strain (Fig. 1c), however RNAi is activated in only a subset of cells selected with FK506. The in vivo efficiency of dsRNA formation or its transport to the cytoplasm may limit sRNA levels restricting silencing to fewer cells. Only traces of antisense sRNA complementary to fkbA (16 reads per million) were detected in the WT strain by high-throughput sequencing (Extended Data Fig. 6), supporting this hypothesis.
To test if mutations may have occurred to promote the formation of epimutations, the reverted and now sensitive epimutant strains were exposed to a second round of FK506 to isolate mutants/epimutants. The frequency of epimutation versus mutation to FK506R in the reverted epimutant strains was similar or even lower than in the parental WT strain (Extended Data Fig. 7). Thus, the underlying mechanism appears solely epigenetic and does not require any genetic change in the genome to promote epimutation.
High-throughput sequencing demonstrated abundant sRNAs complementary to the fkbA mRNA (antisense) as well as sense sRNA in three epimutant resistant isolates, but these were barely detectable in the WT strain or in the two corresponding FK506S revertants analyzed (Fig. 2b). Some sRNA sequences spanned exon-exon junctions (Fig. 2b, Extended Data Fig. 8a), indicating that the source and target of the sRNA is mature mRNA. Most of these sRNAs average 21-24 nt, with a bias towards 5’ terminal uridine (Fig. 2c), features typical of sRNA that interact with Argonaute. No further loci were detected exhibiting the same pattern of sRNA production: high level in epimutants, and very low levels in WT and reverted strains (Extended Data Fig. 8b). Silencing pathways can operate constitutively on M. circinelloides endogenous genes during normal growth7. In contrast, the FK506-selected silencing of the fkbA gene may not affect other loci. However, no other phenotypes were selected and sRNAs are often lost without selection (Fig. 2a, Extended Data Figs. 1b and 8b).
To test if silencing was driven and enhanced by a genome-wide increase in stress-activated RNAi, the WT strain was exposed to stress conditions prior to FK506 exposure (Supplementary Table 3). However, analysis of recovered FK506R isolates did not show an increase (or decrease) of epimutation frequency in fkbA (Supplementary Table 4). Thus, either stress fails to activate RNAi or we have not identified the precise activating stress. The calcineurin inhibitor CsA also did not increase epimutations conferring FK506 resistance; thus inhibition of calcineurin (by CsA or FK506) does not activate RNAi.
To investigate if sRNAs generated against fkbA are produced by canonical RNA silencing, we screened for epimutants in mutants lacking RNAi pathway components. No epimutants were found in dcl2, ago1, or rdrp2 mutants (Table 1, Extended Data Fig. 9a), showing that Dcl2 (Dicer), Ago1 (Argonaute), and RdRP2 are necessary for endogenous silencing of fkbA. Dcl2 and Ago1 are essential for both transgene-induced and endogenous silencing in M. circinelloides7,19,20. RdRP2 is essential to amplify RNAi signals for transgene-induced silencing18, but was thought to play a role secondary to RdRP1 for endogenous RNAi silencing7. Instead, we find RdRP2 is crucial, suggesting different pathways for endogenous silencing operate in M. circinelloides, which may be correlated with the four different known classes of sRNAs7. Surprisingly, we found only one epimutant in two independent dcl1 mutants (1/46 FK506R isolates, 2.2%, Table 1, Supplementary Table 1 and Extended Data Fig. 9a). Dcl1 is not known to play any essential role in transgene-inducedor endogenous silencing7,21, but does provide an auxiliary role in mutants lacking Dcl219. The paucity of fkbA epimutants in the dcl2 and dcl1 mutant backgrounds suggests that both Dcl1 and Dcl2 are involved in epimutational fkbA silencing. ago2 and ago3 mutants exhibited a WT frequency of silencing (15-17%, Table 1, Extended Data Figs. 9a and 2a), thus Ago2 and Ago3 are dispensable for epimutation20. These results reveal establishment of epimutants in M. circinelloides depends upon Dcl2, Dcl1, Ago1, and RdRP2. Except for Dcl1, these genes are involved in biogenesis of the known class I type of sRNAs7. Thus, although epimutant genetic requirements could suggest a distinct sRNA class is involved (given the role of Dcl1), we cannot exclude that fkbA silencing is due to an atypical class I sRNA.
Unexpectedly, our studies revealed a novel role for RdRP1 in constraining epimutational silencing. RdRP1 is required for RNA silencing of exogenous sense transgenes in M. circinelloides. When DNA alleles producing dsRNA are introduced, this by passes RdRP1 to evoke gene silencing18. As noted above, RdRP1 has a major role in endogenous silencing7. RdRP1 is hypothesized to be central for activating RNAi by generating dsRNA from the single-stranded RNA (ssRNA) precursor, both from sense transgenes or mRNA. In this model RdRP1 should be essential for triggering fkbA silencing because it involves an fkbA dsRNA (Fig. 1c, Extended Data Fig. 5). Surprisingly, the rdrp1 mutant showed an elevated silencing rate of ~80% (~22% in WT, Table 1, Extended Data Figs. 9a and 2a), and the epimutants isolated in this mutant did not revert on drug free media (EM4, Extended Data Fig. 1b). In Caenorhabditis elegans the rrf-3 mutant lacking one of several RdRPs has a similar enhanced RNAi phenotype22,23. RRF-3 has also been proposed to generate dsRNA from mRNA templates24, hence RdRP1 and RRF-3 could serve analogous roles. Furthermore, the exosome and RNAi pathways in Schizosaccharomyces pombe compete in their degradation activities. Both mechanisms share common targets25 and in the rdrp1 mutant sRNA formation is abolished, and mRNAs are primarily directed to the exosome26. In Mucor, RdRP1 could have an opposing role to RNAi, promoting assembly of exosome machinery on specific mRNA targets and thereby avoiding activation of RNAi under normal conditions, as was suggested previously7.
To test which RdRP might generate dsRNA from fkbA mRNA, we tested for fkbA antisense RNA in silencing mutants (Extended Data Fig. 9b). All strains analyzed expressed the antisense RNA at similar levels, even in the rdrp1 and rdrp2 mutants. The two RdRP polymerases could play a redundant role in generating fkbA antisense RNA (rdrp1 rdrp2 double mutants appear inviable, precluding analysis, not shown), or other rdrp genes may participate.
Together these results provide evidence for a different route processing endogenous sRNA, wherein RdRP2, Ago1, and both Dcl proteins are necessary to silence mRNA expression via epimutation, and RdRP1 plays an unexpected role constraining epimutational silencing. We consider two possible models. In the first, sRNAs are produced constitutively and stochastically at low levels against the entire genome or some designated loci, allowing adaptation to environments through an RNAi-based pathway. In the second model, some mechanism activates RNAi under adverse/novel physiological conditions, facilitating genomic and phenotypic plasticity. Either could explain the broad range of environments in which M. circinelloides grows, and the limited antifungal drug susceptibility.
Previous studies showed the M. circinelloides species complex includes three distinct subspecies: M. circinelloides f. lusitanicus (Mcl), M. circinelloides f. circinelloides (Mcc), and M. circinelloides f. griseocyanus (Mcg)27. Mating barriers and phylogenetic separation provide evidence these three lineages are different enough to represent distinct species. To generalize our findings, two Mcc strains [Mucho, 1006PhL28], and an Mcg strain [ATCC1207a] were tested in addition to the Mcl strains. Only the Mcc strains grew as yeast in the presence of FK506 (Extended Data Fig. 10a), enabling the recovery of FK506R isolates. The two exhibited different patterns of genomic plasticity; 2.5-fold more FK506R isolates were recovered from 1006PhL than from Mucho (Supplementary Table 5), and appeared earlier (5-7 days for 1006PhL, 5-15 days for Mucho). Epimutants silencing fkbA occurred in the pathogenic isolate 1006PhL at a surprisingly more elevated rate (90%) than in Mucho (<7.7%) or the Mcl strains (~20-30%) (Supplementary Table 5 and Extended Data Fig. 10b). Mcc is the most common Mucor species associated with human infection. The enhanced ability to activate RNAi exhibited by the 1006PhL virulent isolate suggests RNAi may enable this fungal pathogen to readily adapt both in nature and the host. Further studies are required to elucidate whether our observations are generally applicable, or if not, how specificity is brought about.
This study underscores the ability of M. circinelloides to adapt to the environment through two different routes of phenotypic variation, one stable (mutation) and one transient (epimutation). This plasticity evokes a broader phenotypic repertoire including the ability to reverse epimutations when selective pressures are relaxed. This is the first known example of epimutations involving an endogenous gene in fungi identified in a standard genetic screen, however given the ubiquity of RNAi it is unlikely to be unique. While this example involves resistance to an antifungal drug in a human fungal pathogen, these findings could have implications beyond novel modes of transient antimicrobial resistance for the broader evolutionary trajectory of this and other eukaryotes with active RNAi pathways.
METHODS
Strains and growth conditions
The leuA leucine auxotrophic strain R7B, derived from M. circinelloides f. lusitanicus CBS277.49 (syn. Mucor racemosus ATCC1216b), was used as the WT strain to compare with the patA and silencing mutant strains, as they were generated in the R7B background18-21. The strains were grown at room temperature (~26°C) on yeast extract peptone dextrose agar (YPD, 10 g/L yeast extract, 20 g/L 4 peptone, 20 g/L dextrose, 2% agar), MMC medium pH=4.5 (1% casamino acids, 0.05% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose), or YNB minimal medium29 pH 4.5, supplemented with 1 μg/mL of FK506 (Prograf), 100 ng/mL of rapamycin, or 100 μg/mL of CsA when needed. The cultures were routinely incubated for 48hr except when noted otherwise or for the isolation of FK506-resistance patches, when the cultures were incubated for as long as 4 weeks in some cases. The media to test the different stress conditions was prepared by adding the specific compounds at the concentrations indicated (see Supplementary Table 3) except for trisporic acid, which was sprayed over the spores (approximately 100 μg of trisporoid as suggested by previous studies30). All of the compounds were added to YPD medium unless otherwise specified.
FK506 resistant strains isolation
FK506R isolates were obtained after growing the different strains on YPD containing 1 μg/mL of FK506 for three days to three-four weeks at room temperature, until patches with hyphal growth were observed. Each isolate was derived from an independent subculture grown on a different petri dish. The isolates were analyzed after at least three generations of vegetative growth in the presence of FK506 to ensure a high proportion of the nuclei in the mycelium syncytium were mutant or silenced.
DNA/RNA extraction and analysis
The isolates were grown on MMC media pH=4.5, supplemented with 1 μg/mL of FK506 when necessary, prior to DNA or RNA purification. The FKBP12 gene fkbA and calcineurin genes cnaA, cnaB, cnaC, and cnbR were sequenced from DNA purified with CTAB and chloroform extraction from lyophilized mycelia. The FK506-resistant isolates obtained in a mutant background were verified by junction PCR for the deletion of the proper gene. Small and total RNAs were extracted using Trizol as described previously6 from frozen mycelia in liquid nitrogen. 25-35 μg of sRNAs were separated by electrophoresis on 15% TBE-Urea gels (Invitrogen), electrotransferred to Hybond N+ filters at 400 mA for 1 hour in 0.5X TBE, and cross-linked by irradiation with ultraviolet irradiation (2x 1.2 Q 105 mJ/cm2). Prehybridization and hybridization were carried out with ultrasensitive hybridization buffer UltraHyb (Ambion). The fkbA antisense-specific and 5S rRNA riboprobes were prepared by in vitro transcription using the Maxiscript transcription kit (Ambion) following supplier-recommended protocols. Riboprobes were treated as described previously6 to result in an average size of 50 nt.
Protein cell extracts and analysis
Whole proteincell extracts were prepared from mycelia grown on MMC media pH=4.5 and frozen in liquid nitrogen as described previously31 with some modifications. The sample was ground to a powder with a mortar and pestle and transferred to TSA extraction buffer (10 mM Tris-HCl pH=8.0 and 0.15 M NaCl) with protease inhibitors (complete Mini from Roche), and 1 mM of Benzamidine. After 30 minutes incubation on ice and 30 minutes centrifugation at top speed at 4°C, the amount of protein in the supernatant was quantified by Bradford assay (BioRad). Protein (100-120 μg) was loaded in 4-20% polyacrylamide gels for western blot analysis. Antiserum against ScFKBP12 raised in rabbits32 was used to detect FKBP12 in Mucor, and this sera was previously shown to be cross-reactive4. Mouse monoclonal antibodies against α-Tubulin (Sigma, T5168) served as the loading control.
Disruption of patA
A disruption allele containing the pyrG gene as a selectable marker flanked by sequences 5’ and 3’ of the patA ORF was generated via overlap PCR and used to generate patA null mutants by gene replacement. Primers used for the construction of the disruption allele and the confirmation of the deletion are listed in Supplementary Table 6. Strain MU402 (leuA-, pyrG-) was transformed with the patAΔ∷pyrG disruption cassette as described previously33 by electroporation of protoplasts obtained from spores after treatment with chitosanase and lysing enzymes. 5’ and 3’ junction PCRs were used to identify transformants in which homologous replacement of the patA locus had occurred and confirmed with both spanning and ORF specific PCR analysis.
3’ and 5’ RACE assays
Total RNA for 3’ and 5’ RACE assays was extracted following the protocol for filamentous fungi supplied with the RN easy Plant Mini Kit (QIAGEN). The First Choice RLM-RACE kit (Ambion) was utilized for 3’ and 5’ UTR amplification and the gene-specific primers used are listed in Supplementary Table 6. Sequences were aligned with Serial-Cloner software.
High-throughput sRNA sequencing
sRNA libraries were prepared following the instructions of the TruSeq Small RNA Sample Prep Kit (Illumina). sRNA-enriched samples extracted with the mirVana miRNA isolation kit (Ambion) as previously described7 were size-fractioned by electrophoresis and the 18-24 nt fraction purified from the gel was used as input. The library was sequenced by the High-Throughput Sequencing Facility (HTSF), UNC Chapel Hill, using the Illumina platform (HiSeq 2000).
Raw sRNA sequences were processed using fastx_clipper <http://hannonlab.cshl.edu/fastx_toolkit/> to remove adapter sequences. The adapter sequences trimmed from each sample are listed in Supplementary Table 7. Because samples were multiplexed for sequencing, each sample was trimmed using a multiplex index specific sequence. The adapter sequences that were trimmed are based on those supplied by the manufacturer (Oligonucleotide sequences © 2007-2011 Illumina, Inc. All rights reserved). Trimmed reads were then mapped to the Mucor circinelloides CBS277.49 v2.0 genome34 using TopHat35 (TopHat version 2.0.5, Bowtie version 2.0.0.7, Samtools version 0.1.18.0) and the basic library data is represented in Supplementary Table 8. The downloaded genome annotation file was modified to include the coordinates of the 3’ UTRs for the patA and fkbA genes as determined by 3’ RACE (see Supplementary Table 2); this modified annotation file served as the input for TopHat.
Both parts of Figure 2c represented the antisense reads of fkbA from the EM1-R samples, and show plus strand reads that map entirely within the fkbA gene (1803250-1803768 on scaffold_03). Figure 2b presents all reads that map to the fkbA gene, and the read counts were normalized to the total number of mapped reads in the sample.
Extended Data Fig. 8a shows a map for all of the reads in the EM1-R sample, emanating from the region of the exon 1 – exon 2 boundary of the fkbA gene (i.e. reads completely within the range 1803668-1803774 on scaffold_03). Only reads with at least five copies in the sample are shown.
Extended Data Fig. 6 consists of the sequence logos36 for all fkbA antisense reads (plus strand in the range 1803250-1803768 on scaffold_03) in the WT sample, at each read length observed.
Figures 2b, 2c, Extended Data Fig. 6, and 8a were generated using custom software, based on a number of existing software libraries36-41. The analysis and figures can readily be reproduced by downloading the source code from <https://bitbucket.org/granek/mucor_srna>. Raw data from the high-throughput sRNA sequencing of WT, epimutant, and revertant strains have been deposited in NCBI’s Gene Expression Omnibus42 and are accessible through GEO Series accession number GSE56353 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56353).
To search for genes with sRNA patterns correlated with fkbA (Extended Data Fig. 8b), antisense read counts were determined for each annotated gene using htseq-count43, counts were normalized using the R package DESeq238, and genes with counts so low (mean normalized count across samples less than 10) that any true differences between samples are confounded by Poisson noise were excluded from consideration. The similarity to fkbA was determined by computing the Kendall rank correlation coefficient for each annotated gene in the genome. The fifty genes with the highest correlation coefficient (i.e. most closely correlated with fkbA) were plotted using R44 for comparison to fkbA.
GeneBank accession numbers
Sequences for the fkbA and calcineurin genes from the WT strain NRRL3631 and the epimutant strains (EM1, EM2, and EM3) were deposited in GenBank with accession numbers WT fkbA: KF203228, EM1 fkbA: KF203229, EM2 fkbA: KF203230, EM3 fkbA: KF203231, WT cnaA: KJ668831, WT cnaB: KJ668832, WT cnaC: KJ668833, WT cnbR:KJ668834, EM1 cnaA: KJ668835, EM1 cnaB: KJ668836, EM1 cnaC: KJ668837, EM1 cnbR: KJ668838, EM2 cnaA: KJ668839, EM2 cnaB: KJ668840, EM2 cnaC: KJ668841, EM2 cnbR: KJ668842, EM3 cnaA: KJ668843, EM3 cnaB: KJ668844, EM3 cnaC: KJ668845, and EM3 cnbR: KJ668846.
p-value calculation
p-values listed in Table 1 are based on a Fisher Exact Probability Test for a 2×2 Contingency Table, comparing each of the patA and silencing mutant strains individually versus the R7B WT strain, from which all of the mutants were generated.
Extended Data
Extended Data Figure 1. M. circinelloides can develop resistance to FK506 by two mechanisms, one stable (mutations) and one transient (epimutations).
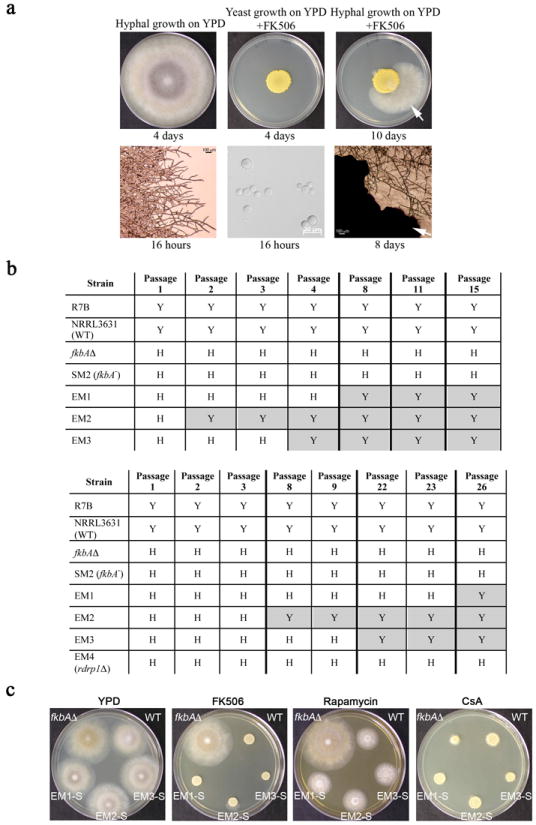
a, The WT strain (NRRL3631) grows as hyphae (white, upper left panel) on YPD and as a yeast (yellow, upper center panel) on YPD containing 1 μg/mL of FK506. An FK506-resistant patch that emerged from the southeastern edge of the yeast patch is shown after 10 days of incubation (arrow, upper right panel). Microscopic images corresponding to the culture plates at the top were taken at different incubation periods as indicated and are shown in the lower panels. For yeast growth, cells from the colony were dispersed in water on a microscope slide. The black patch (arrow) in the microscopic image in the lower right panel corresponds to the edge of the compact yeast colony. Images are representative of all of the FK506 resistant isolates obtained (see Supplementary Tables 1, 3, and 5) b, Epimutant strains revert during passage on drug-free media. Y=yeast, H=hyphal. Shaded areas indicate reversion of epimutants to the WT phenotype (yeast growth on YPD supplemented with 1 μg/mL FK506). SM2 strain4 harbors an A-to-G substitution (A316G) in the acceptor splice site of intron 2. Darker vertical bars indicate intervals in which some passages are not depicted. c, Reverted epimutant strains from (b) lost their resistance to FK506 and rapamycin (central panels) and remained sensitive to CsA whose mechanism of action does not involve FKBP12 (right panel). The images were taken after 48 hours of incubation at room temperature (~26°C) on YPD or YPD media supplemented with the different drugs. Images are representative of two independent experiments. EM1-S, EM2-S, EM3-S=Epimutants 1, 2, and 3 reverted to restore FK506-sensitivity. fkbAΔ=fkbA null mutant.
Extended Data Figure 2. Epimutations are generated by the RNAi pathway, and are not associated with DNA methylation in M. circinelloides.
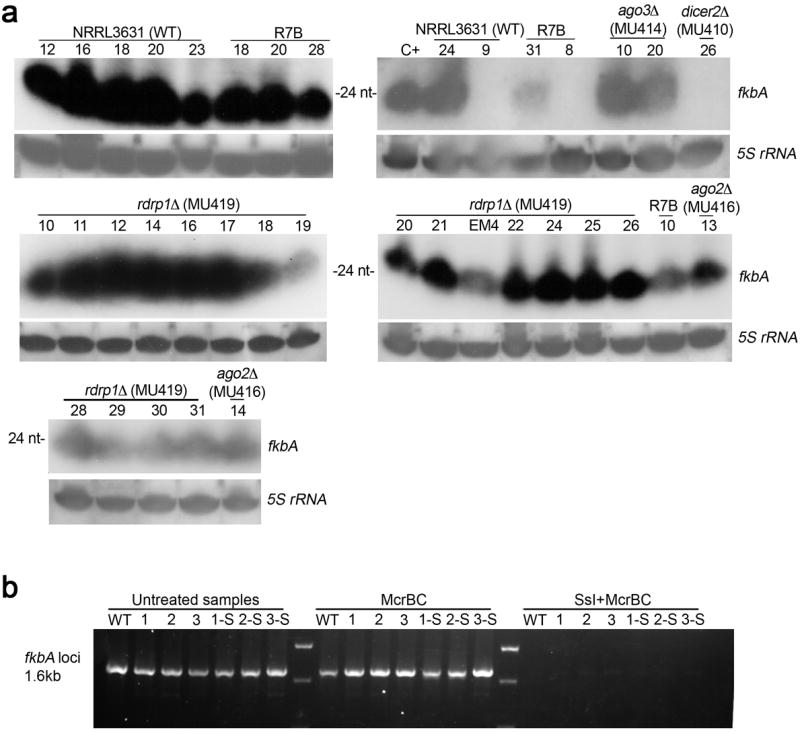
a, Confirmation of the presence of sRNAs in all of the remaining epimutant isolates from the different strains lacking mutations in the fkbA and calcineurin genes (cnaA, cnaB, cnaC, and cnbR) not shown in Figure 2a or Extended Data Fig. 9. The numbers of the isolates correspond to those in Supplementary Table 1. sRNA blots were hybridized with an antisense-specific probe to detect fkbA sRNA. 5S rRNA served as a loading control. Abundant sRNAs were detected in all of the strains with the exception of three of the isolates (NRRL3631 isolate 9, R7B isolate 8, and MU410 isolate 26). These isolates also do not show any mutations in the genes analyzed (fkbA, cnaA, cnaB, cnaC, cnbR) and the mechanism by which they have developed FK506-resistance remains to be established. The image of the blot in which these three isolates were included is representative of two independent experiments. All of the other blots were generated only once, because a positive signal indicates the presence of sRNA. b, Genomic DNA (~40 μg) from the WT strain (NRRL3631), the three epimutants (1=EM1, 2=EM2, and 3=EM3), and the three reverted strains (1-S=EM1-S, 2-S=EM2-S, and 3-S=EM3-S) was treated with the methylated-DNA-specific restriction enzyme McrBC (NEB) with or without previous treatment with the CpG methyltransferase SsI (NEB), following the manufacturer’s protocols. PCR amplification of the fkbA locus (~1.6 kb = 732 bp 5’ upstream fkbA, 457 bp fkbA ORF and 435 bp 3’ downstream fkbA) was carried out using 100 ng of purified DNA. PCR amplification after McrBC treatment yielded similar levels of product as the untreated samples. Virtually no product was obtained by PCR amplification in any of the samples after treatment with the CpG methyltransferase SsI followed by McrBC treatment, indicating that McrBC digested the newly methylated DNA, preventing its amplification. These results indicate that RNAi silencing does not involve DNA methylation of the fkbA locus in M. circinelloides. Image is representative of two independent experiments.
Extended Data Figure 3. mRNAs from fkbA and its neighboring gene patA overlap in their 3’ regions by 92 bp.
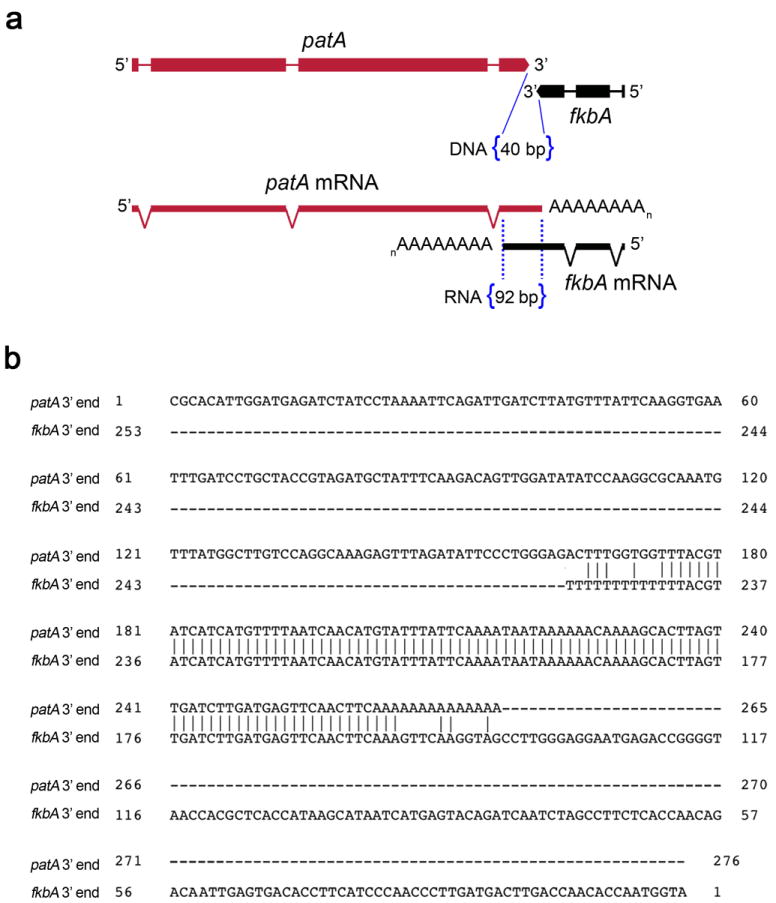
a, The fkbA and patA genes are convergently oriented. The intergenic region is only 40 bp, and the mRNA overlap of their 3’ UTR regions spans 92 nucleotides. b, Alignment of the overlapping fkbA and patA 3’ regions based on 3’ RACE analysis. The direction of the transcripts are the same as in the upper figure, where the patA transcript is 5’ to 3’ end (top sequence) and the fkbA transcript is in the opposite orientation (3’ to 5’, bottom sequence). The polyA tails of both mRNA are shown.
Extended Data Figure 4. patA expression to generate overlapping RNA molecules is not necessary for fkbA silencing.
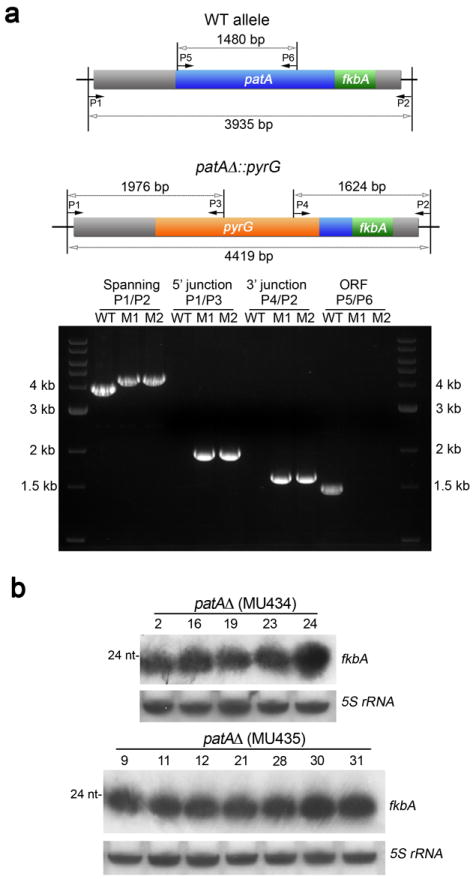
a, Two independent patA null mutants (M1=MU434 and M2=MU435) were generated by homologous recombination, employing pyrG as the selectable marker. The patA ORF was replaced with the pyrG gene after electroporation of protoplasts with a gene deletion cassette, generated by overlap PCR, containing the selectable marker pyrG flanked by 5’ upstream and 3’ downstream sequences flanking the patA ORF. Almost 400 bp from the 3’ end of patA were preserved to keep intact the 3’UTR of fkbA. PCRs from 5’ and 3’ junctions (P1/P3 and P4/P2 respectively), the patA ORF (P5/P6), and spanning the patA and fkbA loci (P1/P2) were performed to confirm the deletion of the patA ORF and correct insertion of the pyrG disruption cassette (bottom). Image is representative of two independent experiments. 3’RACE assays were performed to verify that the pyrG 3’UTR and fkbA 3’UTR do not overlap in the patA null mutants (See Supplementary Table 2). b, Confirmation of the presence of sRNAs in epimutants derived from two independent patA null mutants. The numbers of the isolates correspond to those in Supplementary Table 1. An antisense-specific probe was used to detect fkbA sRNAs by northern blot. 5S rRNA served as a loading control. Abundant sRNAs complementary to fkbA were detected in all of the FK506R strains that lacked Mendelian mutations isolated from the two independent patAΔ. sRNA blots were generated once because apositive signal indicates the presence of sRNA.
Extended Data Figure 5. fkbA antisense RNA is complementary to mature fkbA RNA.
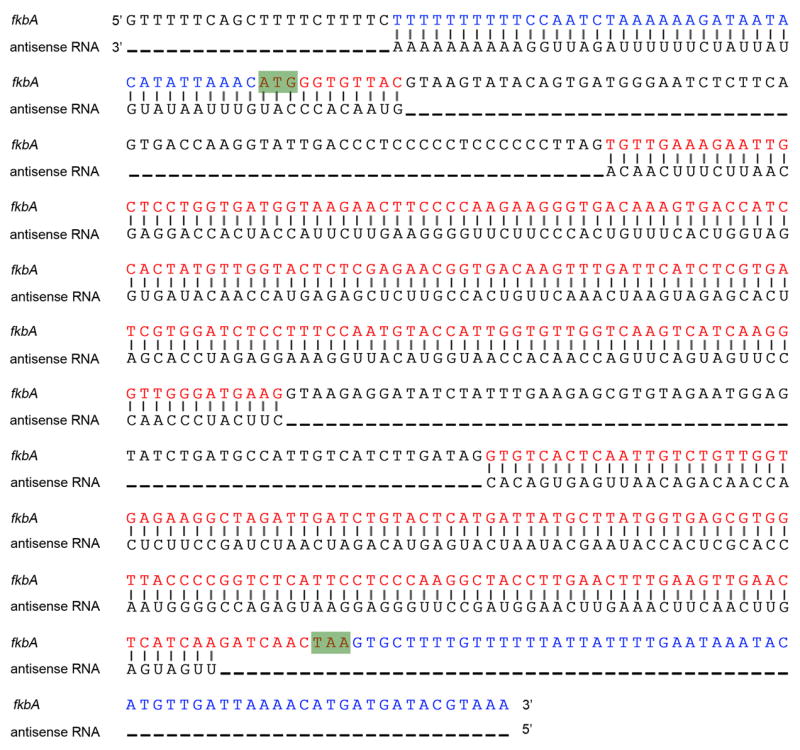
The complete sequence of the antisense RNA was determined based on 5’ and 3’ RACE analyses (bottom sequence) and compared to the fkbA DNA (top sequence). These analyses indicate the antisense RNA is 5’ capped and 3’ poly-adenylated. The fkbA introns were absent in the antisense sequence and the 3’ end matched the beginning of the 5’ UTR found on fkbA mRNA by 5’ RACE analysis, indicating that mature spliced fkbA RNA is used as a template by an RdRP to generate the complementary strand. The antisense RNA 5’ end is located 7 nt upstream of the STOP codon. The 3’ end is located 40 nt upstream of the ATG codon. The fkbA DNA sequence includes the sequenced 5’ and 3’ UTR regions in blue and the introns in black. The fkbA coding region is indicated in red. The ATG start and TAA stop codons are shown in green boxes.
Extended Data Figure 6. Very few sRNAs were detected by high-throughput sequencing in the WT strain.
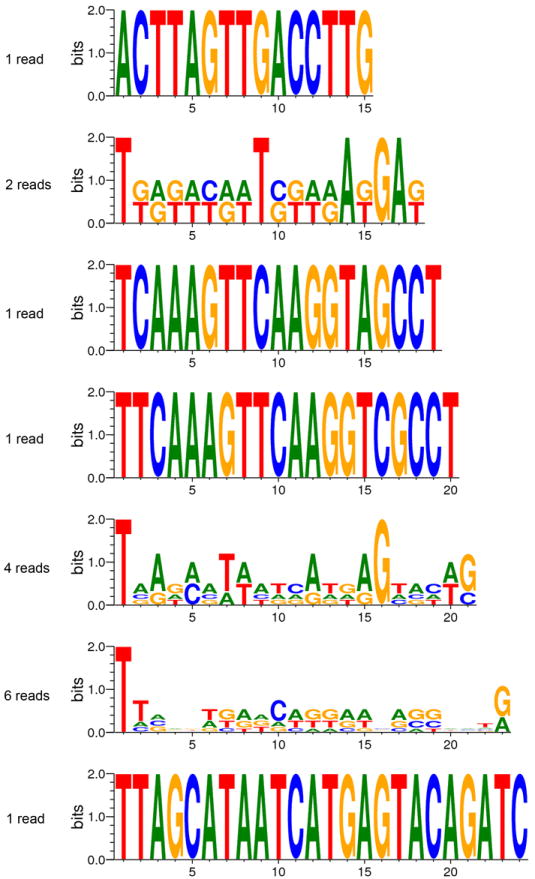
The normalized number of reads of each antisense sRNA complementary to fkbA is extremely low in the WT strain, and below the detection limit of northern blot (numbers expressed in reads per million). Almost all of the antisense sRNA detected have a uridine at the 5’ terminus, features typically found in sRNA that interact with Argonaute proteins, suggesting that they may represent authentic sRNAs but are present at insufficient levels to trigger RNAi silencing. Sense sRNA does not show any bias (data not shown). The height of a letter represents the frequency with which the base is observed in that position, and the total height of the letters in a position indicates how strong the bias is for specific bases in that position. Note that because the total number of fkbA antisense reads in the WT strain is very small, they are not probably representative of the true distribution of sRNAs, so the logos are likely to overestimate any sRNA sequence bias.
Extended Data Figure 7. Reverted strains (EM1-S, EM2-S, and EM3-S) exposed to a second round of FK506 selection undergo epimutations at the same frequency as the WT strain.
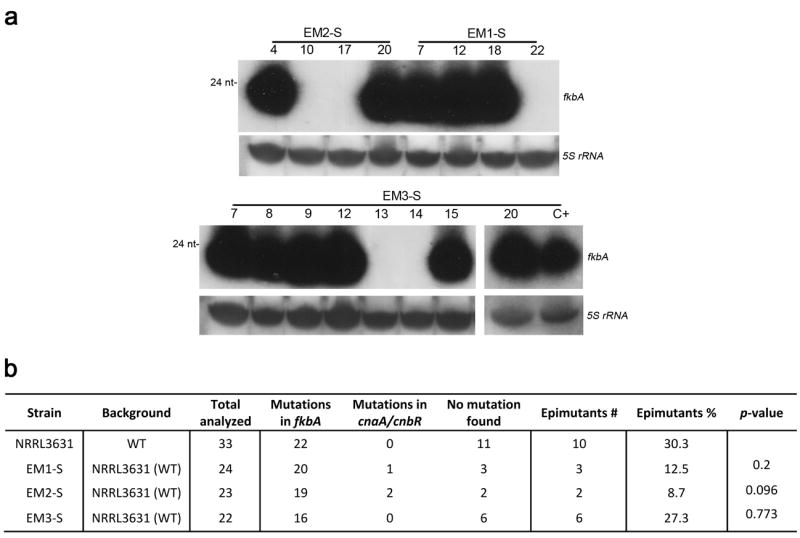
Epimutant strains (EM1, EM2, and EM3) that had reverted to an FK506 sensitive WT phenotype (yeast growth in the presence of FK506) after several passages on drug-free media were exposed a second time to 1 μg/mL FK506 to ascertain if genomic mutations had occurred that enhance epimutant formation. a, The numbers of the isolates correspond to those in Supplementary Table 1. The new FK506R isolates that lacked a Mendelian mutation in any of the target genes showed abundant sRNAs complementary to fkbA based on northern blot of sRNA hybridized with an fkbA antisense-specific probe. 5S rRNA served as a loading control. Images are representative of two independent experiments. EM1-S, EM2-S, EM3-S =Epimutants 1, 2, and 3 reverted to restored FK506-sensitivity. C+=EM1 before reversion of FK506-resistance. b, The frequency of epimutation was similar or lower in the reverted epimutant strains compared to the WT, which argues against mutations that arose promoting epimutation. In addition, because rdrp1 mutations enhance epimutation frequency and stability, the rdrp1 gene was sequenced in EM3 and found to be WT with no mutations.p-values were obtained based on a Fisher Exact Probability Test for a 2×2 Contingency Table, comparing each of the mutant strains individually versus the WT strain NRRL3631.
Extended Data Figure 8. sRNA were detected by high-throughput sequencing in the epimutant strains, but not in the WT and reverted strains. This pattern was not conserved in any other loci in the genome.
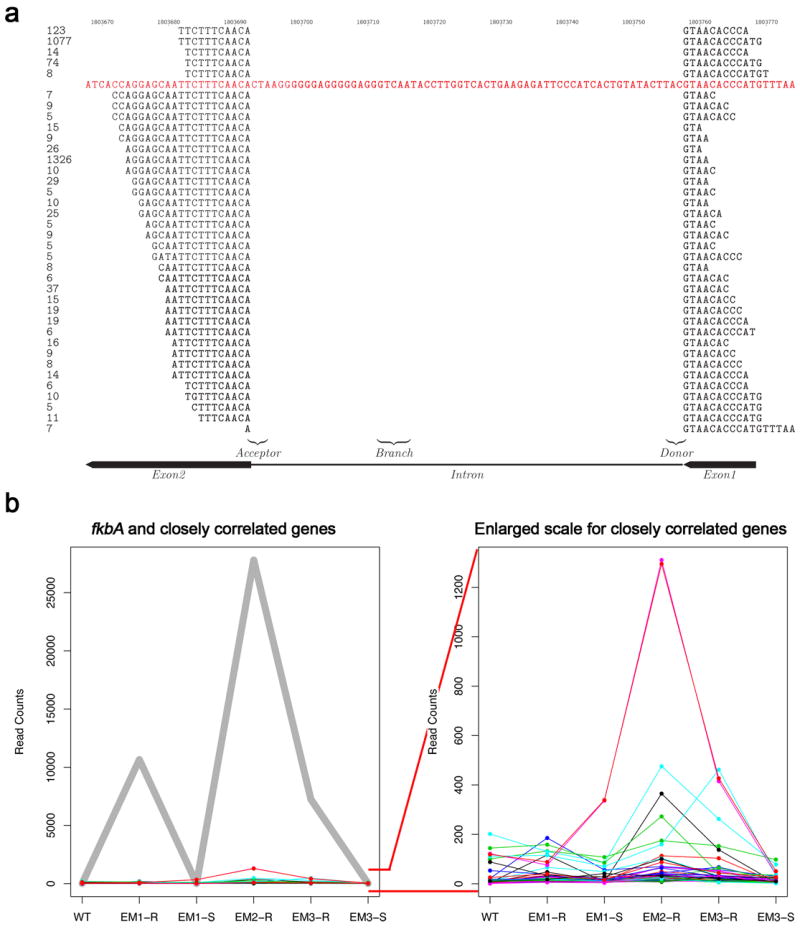
a, sRNAs were found to span exon-exon junctions. Antisense and sense sRNA from EM1 that span intron 1 are shown at the top and bottom respectively. The numbers on the left are the normalized read counts (reads per million) for each specific sRNA. Only sRNAs with 5 or more read counts are shown. The reference sequence is in red and sRNAs spanning the intron are in black. b, Distribution of antisense sRNA for the fkbA gene across the different strains and the 50 genes with sRNA patterns most closely correlated with fkbA. sRNA read counts distribution for the fkbA gene is represented with a heavy gray line (left panel). The 50 genes with the closest correlated pattern are represented with thin colored lines. The bottom part of the left panel has been expanded to elucidate the read count patterns of the correlated genes (right panel). While some of these genes show an apparent similarity to the pattern of sRNA in the fkbA silenced and revertant strains, the levels of read counts are in most cases ~100-fold lower than that for fkbA gene, and were not detected by sRNA blots (data not shown). WT=strain NRRL3631. EM1-R, EM2-R, EM3-R=Epimutants 1, 2, and 3 resistant to FK506. EM1-S and EM3-S=Epimutants 1 and 3 reverted to restored FK506-sensitivity.
Extended Data Figure 9. sRNA and antisense fkbA RNA detection in the different mutant strains lacking RNAi pathway components.
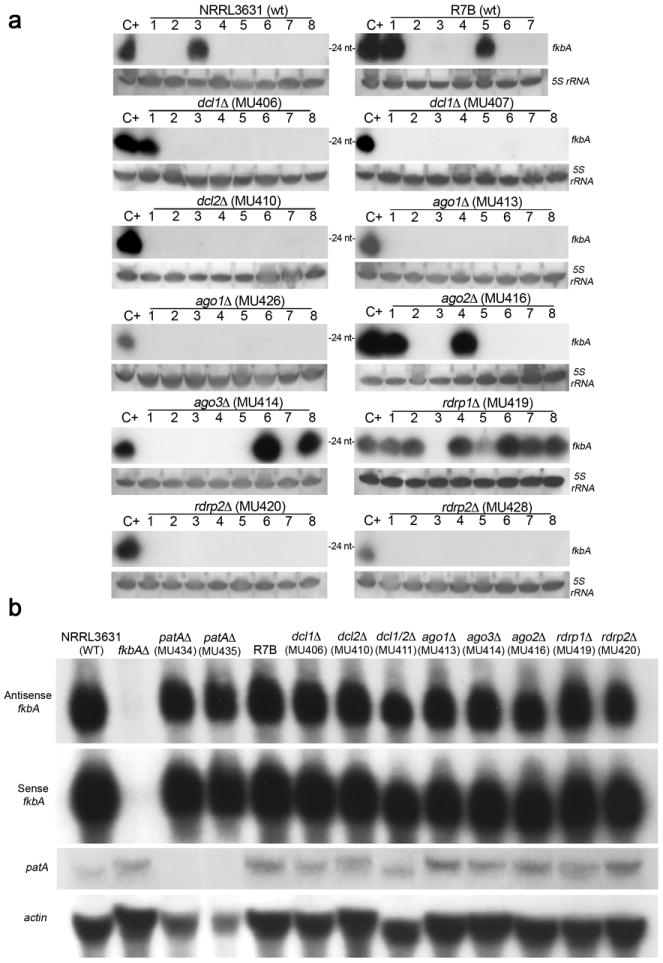
a, The numbers of the isolates correspond to those in Supplementary Table 1. No epimutants (absence of sRNA complementary to fkbA) were found in the dcl2 (MU410), dcl1 (MU407), ago1 (MU413, MU426), or rdrp2 (MU420, MU428) null mutants based on sRNA northern blots hybridized with an fkbA antisense-specific probe. Only one epimutant was recovered in the second independent dcl1 mutant (MU406). We reconfirmed this result (sRNA blot, no fkbA mutation) and also validated the isolate was dcl1Δ by PCR. ago2 (MU416) and ago3 (MU414) null mutants showed a frequency of epimutation similar to the WT strain (R7B). The rdrp1 null mutant (MU419) showed an elevated epimutation frequency. The conclusion that Dcl2, Dcl1, Ago1, and RdRP2 are required for epimutation is supported by the congruence of phenotype, and the analysis of two independent null mutants each for ago1, rdrp2, and dcl1. R7B served as WT for this experiment as all of the RNAi silencing mutants were generated in this background. 5S rRNA served as a loading control. C+=EM1 strain. Images from dicer mutant blots are representative of two independent experiments. The remaining blots were generated once. b, Total RNA was isolated from the WT, fkbA mutant, patA mutant, R7B and the indicated RNAi pathway mutants, and 50 μg of total RNA were used to ensure signals could be detected from all of the mRNA analyzed, as the level of patA expression is low. All of the silencing mutant strains have the same level of expression of the fkbA antisense RNA based on northern blot. Antisense- and sense-specific probes were used to detect antisense and sense fkbA RNA respectively. The northern blot was first probed for antisense RNA to avoid residual signal from fkbA mRNA. patA expression was not affected in any the fkbAΔ or the RNAi mutant strains, but as expected was absent in the two independent patAΔ strains. actin served as a loading control. Images are representative of three independent experiments.
Extended Data Figure 10. M. circinelloides f. circinelloides (Mcc), and M. circinelloides f. griseocyanus (Mcg) strains were tested for generation of FK506R and fkbA silencing.
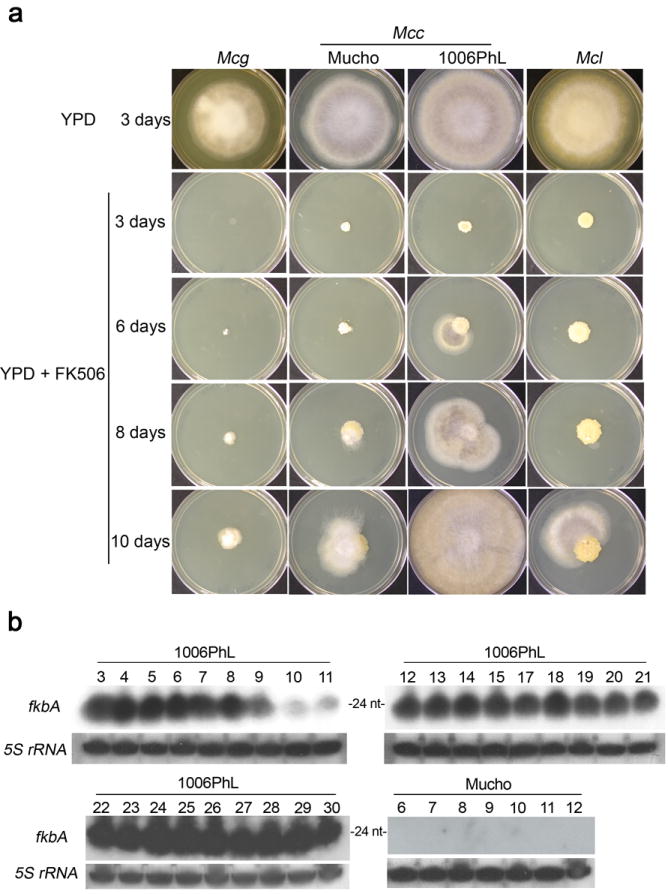
a, The indicated Mucor strains, plus M. circinelloides f. lusitanicus (Mcl) WT strain used as a control, were incubated on YPD media for 3 days (top panel) and on YPD supplemented with 1 μg/mL of FK506 for up to 10 days (lower panels). The Mcl and 1006PhL strains grew as a yeast colony until FK506R sectors started to grow as hyphae. The Mucho strain grew as a yeast colony for several days, and in some of the plates a resistant patch appeared, but after 7-8 days the colonies developed aerial hyphae on top of the yeast colony, producing FK506 sensitive spores that grew as yeast after falling on the media, preventing the development of more FK506R patches. The Mcg strain was more sensitive to FK506 without a visible colony until day 6 when the spores started to germinate as a mixture of yeast and hyphae that did not produce any FK506R growth. Images are representative of ~40 independent colonies from each strain. b, Confirmation of the presence of sRNAs in epimutants derived from one of the two pathogenic M. circinelloides f. circinelloides strains. The numbers of the isolates correspond to those in Supplementary Table 5. An antisense strain-specific probe was used to detect fkbA sRNAs by northern blot from both strains (using 30 μg of sRNA). 5S rRNA served as a loading control. Abundant sRNAs complementary to fkbA were detected in all of the FK506R strains that lacked Mendelian mutations isolated from the 1006PhL strain, but not from the Mucho strain. Images from the lower blots are representative of two independent experiments. Images from the upper blots were generated once since sRNA positive signals were detected from all samples analyzed.
Supplementary Material
Acknowledgments
We thank Rebecca Skalsky and Vikram Ponnusamy for technical support and Johannes Wöstemeyer for trisporic acid. We thank Bryan Cullen, Tom Petes, Blake Billmyre, Marianna Feretzaki, Joanne Kingsbury, and Vikram Ponnusamy for critical reading. This work was supported by NIH grants R37 AI39115-17, R01 AI50438-10, R01 CA154499-04, and the Spanish MICINN BFU2009-07220 and MINECO BFU2012-32246, co-financed by FEDER.
Footnotes
Author contributions
S.C., C.W., S.T-M., R.M.R-V., M.E.C., and J.H designed experiments, interpreted data, and wrote the paper. S.C., C.W., R.J.B., S.C.L., and F.E.N. performed experiments. P.M. sequenced the sRNA library. J.A.G. analyzed deep-sequencing data. S.T-M., R.M.R-V., M.E.C. and J.H. provided materials.
Supplementary Information is available in the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
Sequences for the fkbA gene from WT strain NRRL3631 and epimutant strains (EM1, EM2, EM3) were deposited in GeneBank with accession numbers KF203228, KF203229, KF203230, and KF203231. Raw data from high-throughput sRNA sequencing of WT, epimutant, and revertant strains have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE56353 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56353).
References
- 1.Liu J, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 2.Lee SC, Li A, Calo S, Heitman J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLOS Pathog. 2013;9:e1003625. doi: 10.1371/journal.ppat.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orlowski M. Mucor dimorphism. Microbiol Rev. 1991;55:234–258. doi: 10.1128/mr.55.2.234-258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastidas RJ, Shertz CA, Lee SC, Heitman J, Cardenas ME. Rapamycin exerts antifungal activity in vitro and in vivo against Mucor circinelloides via FKBP12-dependent inhibition of Tor. Eukaryotic Cell. 2012;11:270–281. doi: 10.1128/EC.05284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolas FE, et al. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010;38:5535–5541. doi: 10.1093/nar/gkq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhounim L, Rossignol JL, Faugeron G. Epimutation of repeated genes in Ascobolus immersus. EMBO J. 1992;11:4451–4457. doi: 10.1002/j.1460-2075.1992.tb05546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colot V, Maloisel L, Rossignol JL. Interchromosomal transfer of epigenetic states in Ascobolus: transfer of DNA methylation is mechanistically related to homologous recombination. Cell. 1996;86:855–864. doi: 10.1016/s0092-8674(00)80161-0. [DOI] [PubMed] [Google Scholar]
- 10.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 11.Suter CM, Martin DIK, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nature Genetics. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 12.Chan TL, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nature Genetics. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 13.Hitchins MP, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 14.Baulcombe DC. RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol Biol. 1996;32:79–88. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 15.Wassenegger M, Pelissier T. A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- 16.Elmayan T, Vaucheret H. Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 1996;9:787–797. [Google Scholar]
- 17.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 18.Calo S, Nicolas FE, Vila A, Torres-Martinez S, Ruiz-Vazquez RM. Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Mol Microbiol. 2012;83:379–394. doi: 10.1111/j.1365-2958.2011.07939.x. [DOI] [PubMed] [Google Scholar]
- 19.de Haro JP, et al. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryotic Cell. 2009;8:1486–1497. doi: 10.1128/EC.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervantes M, et al. A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus Mucor circinelloides. PLOS One. 2013;8:e69283. doi: 10.1371/journal.pone.0069283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas FE, de Haro JP, Torres-Martinez S, Ruiz-Vazquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol. 2007;44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 23.Timmons L. Endogenous inhibitors of RNA interference in Caenorhabditis elegans. BioEssays. 2004;26:715–718. doi: 10.1002/bies.20078. [DOI] [PubMed] [Google Scholar]
- 24.Gent JI, et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanaka S, et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li CH, et al. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 2011;7:e1002086. doi: 10.1371/journal.ppat.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SC, et al. Analysis of a foodborne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBio. 2014 doi: 10.1128/mBio.01390-14. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasker BA, Borgia PT. High-frequency heterokaryon formation by Mucor racemosus. J Bacteriol. 1980;141:565–569. doi: 10.1128/jb.141.2.565-569.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wetzel J, Burmester A, Kolbe M, Wostemeyer J. The mating-related loci sexM and sexP of the zygomycetous fungus Mucor mucedo and their transcriptional regulation by trisporoid pheromones. Microbiology. 2012;158:1016–1023. doi: 10.1099/mic.0.054106-0. [DOI] [PubMed] [Google Scholar]
- 31.Murcia-Flores L, Lorca-Pascual JM, Garre V, Torres-Martinez S, Ruiz-Vazquez RM. Non-AUG translation initiation of a fungal RING finger repressor involved in photocarotenogenesis. J Biol Chem. 2007;282:15394–15403. doi: 10.1074/jbc.M610366200. [DOI] [PubMed] [Google Scholar]
- 32.Cardenas ME, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutierrez A, Lopez-Garcia S, Garre V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J Microbiol Methods. 2011;84:442–446. doi: 10.1016/j.mimet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Grigoriev IV, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heger A. pysam: Python interface for the SAM/BAM sequence alignment and mapping format. 2013 http://code.google.com/p/pysam/
- 38.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cock PJ, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- 41.Oliphant TE. Python for scientific computing. Comput Sci Eng. 2007;9:10–20. [Google Scholar]
- 42.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. bioRxiv. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


