Abstract
Low birth weight and intrauterine growth restriction (IUGR) increase the risk of mortality and morbidity during the perinatal period as well as in adulthood. Environmental and genetic factors contribute to IUGR, but the influence of maternal genetic variation on birth weight is largely unknown. We implemented a gene-by-environment study wherein we utilized the growth restrictive effects of high altitude. Multigenerational high-altitude residents (Andeans) are protected from altitude-associated IUGR compared with recent migrants (Europeans). Using a combined cohort of low- and high-altitude European and Andean women, we tested 63 single nucleotide polymorphisms (SNPs) from 16 natural selection-nominated candidate gene regions for associations with infant birth weight. We identified significant SNP associations with birth weight near coding regions for two genes involved in oxygen sensing and vascular control, PRKAA1 and EDNRA, respectively. Next, we identified a significant association for the PRKAA1 SNP with an intermediate phenotype, uterine artery diameter, which has been shown to be related to Andean protection from altitude-associated reductions in fetal growth. To explore potential functional relationships for the effect of maternal SNP genotype on birth weight, we evaluated the relationship between maternal PRKAA1 SNP genotype and gene expression patterns in general and, in particular, of key pathways involved in metabolic homeostasis that have been proposed to play a role in the pathophysiology of IUGR. Our observations suggest that maternal genetic variation within genes that regulate oxygen sensing, metabolic homeostasis, and vascular control influence fetal growth and birth weight outcomes and hence Andean adaptation to high altitude.
Keywords: Andean, genotype-phenotype association study, intrauterine growth restriction, mTOR pathway, natural selection
birth weight affects health from infancy through adulthood, with intrauterine growth restriction (IUGR) being a major contributor to perinatal mortality and morbidity. Despite improvements in health care, IUGR remains a leading cause of infant mortality in developing and developed countries (12). Additionally, low birth weight increases the risk of developing diabetes, hypertension, and heart disease in adulthood (2, 3, 33). Therefore, identifying the genetic and environmental factors contributing to IUGR is of particular interest for improving both perinatal and adult health. While recent studies have helped illuminate the role of maternal genetic variation on infant birth weight (1, 21, 60), much remains to be discovered with respect to the physiological pathways by which such genetic factors operate.
The environmental factors contributing to IUGR include alcohol use and smoking during pregnancy, hypertensive disorders of pregnancy (preeclampsia, eclampsia, and gestational hypertension), and high altitude (>2,500 m) (12). High-altitude studies provide a valuable model for understanding the mechanisms involved in IUGR since chronic hypoxia restricts fetal growth and increases infant morbidity and mortality at high altitude (28, 35) and also in hypertensive disorders of pregnancy. While birth weight decreases with increasing altitude in all populations studied to date, averaging a 102 g decline per 1,000 m altitude gain (22), not all populations are equally affected. Specifically, infants born to women of Andean or Tibetan ancestry have half the birth-weight decline compared with women of European or Han Chinese ancestry living at the same altitudes [167 ± 36 (mean ± SE) vs. 373 ± 48 g, respectively] (24, 40, 51, 66; Moore LG, Niermeyer S, unpublished observations). Furthermore, the percentage of Andean ancestry is significantly correlated with birth weight, suggesting a genetic influence for this phenotype (6, 24, 51). Animal models exhibit a similar trend, with populations endemic to high altitude showing less fetal growth restriction compared with populations native to low altitude (14, 43). Thus, high-altitude populations present a unique opportunity to identify the genetic factors influencing birth weight and increased susceptibility to IUGR.
In previous work, we identified selection-nominated candidate genes for adaptation to high altitude from genome-scan data from Andean and Tibetan populations (7, 8, 39). Specifically, 40 genes or gene regions were identified by one or more tests of natural selection and confirmed in the two Andean genome scans, while 31 genes were identified in Tibetans (7, 8) (Supplemental Table S1).1 To test whether the previously identified selection-nominated candidate genes affect variation in phenotypes related to survival and reproduction at altitude (defined here as giving birth to a normal birth-weight offspring at high altitude), we performed a quantitative association study with 96 single nucleotide polymorphisms (SNPs) located in or near 16 selection-nominated candidate genes to identify genotype associations with birth weight in an independent sample of Andean or European women living at low or high altitude in Bolivia. Significant associations between birth weight and maternal SNPs were identified in or near two genes, namely, endothelin receptor type A (EDNRA) and the alpha-1 catalytic subunit of adenosine monophosphate-activated protein kinase (PRKAA1, also known as AMPKα1). We then compared these maternal genotypes with respect to uterine artery (UA) diameter during pregnancy and the expression patterns of genes in pathways proposed to play a role in fetal growth restriction at high altitude (25, 45, 65). Overall, our results suggest that maternal variation in genes affecting metabolic homeostasis influence physiological responses to pregnancy that are instrumental for fetal growth and therefore may be influential in achieving genetic adaptation to high altitude.
MATERIALS AND METHODS
Study participants.
Recruited for longitudinal studies during pregnancy were 178 healthy Europeans and Andeans residing at two locations in Bolivia, the high-altitude city of La Paz or the neighboring city of El Alto, Bolivia (3,600 and 4,100 m, respectively) and the low-altitude city of Santa Cruz, Bolivia (400 m). Augmenting this longitudinal sample was a cross-sectional cohort of 120 women living in La Paz or El Alto, all but one of whom was Andean. Women were classified as Andean or European by self-identified ancestry as verified from a panel of ancestry-informative gene markers (AIMs) and parental and grandparental surnames as previously described (25). Participant exclusion criteria were maternal smoking, chronic hypertenslon, gestational or other forms of diabetes, absence of birth weight data, or failure for their DNA samples to pass quality control (QC) as described below, leaving a total of 245 participants whose data are reported here (Fig. 1). All high-altitude studies were performed at the Instituto Boliviano de Biología de Altura (Bolivian High Biology Institute) or the Clinica del Sur (Southern Clinic) in La Paz, and all low-altitude studies were conducted at the Clinica Sirianí (Siriani Clinic) in Santa Cruz. All study participants provided informed written consent to study procedures that had been approved by the Colorado Multiple Institutional, the Colegio Médico in Bolivia, the Pennsylvania State University, and the University of Michigan Institutional Review Boards.
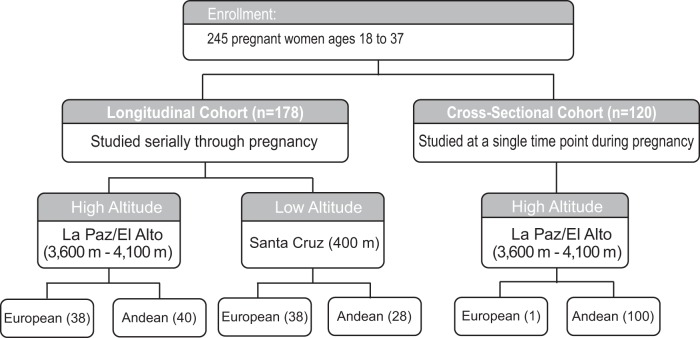
Fig. 1.
Study design. Maternal and infant data were combined from 2 cohorts, a longitudinal cohort and a cross-sectional cohort. For the longitudinal cohort, European and Andean women were studied at pregnancy weeks 20, 36, and 3–4 mo postpartum at low and high altitude. For the cross-sectional cohort, studies were conducted at 1 time point during pregnancy ranging from 17 to 43 wk with a mean of 31.1 (± 5.7). Sample sizes are listed in parentheses.
Phenotype collection.
Each participant provided biographical information (age, altitude of birth, self-identified ancestry, parental and grandparental surnames and ethnicity, body weight before pregnancy, number of previous pregnancies and live births) on a questionnaire that was administered in her native language. Phenotypic studies were conducted in the longitudinal cohorts at both altitudes at pregnancy weeks 20 and 36 and 3–4 mo postpartum for a measurement in the nonpregnant state (25). The cross-sectional cohort was studied once between 17 and 43 wk of pregnancy with an average of 31.1 wk (± 5.7 SE) (10). For both cohorts, height was measured by stadiometer, body weight by balance scale in lightly clothed participants, subscapular and tricep skinfolds by Lange caliper (Beta Technology, Santa Cruz, CA) and summed from the right and left sides for an index of body fat during pregnancy, and blood pressure by arm cuff sphygmomanometer.
Infant sex, birth weight, length, and gestational age data were collected by hospital personnel at the time of delivery or by study personnel for infants born at home. Gestational age was based on the date of the last menstrual period and confirmed by fetal biometry at week 20 or by clinical assessment at delivery.
In the longitudinal cohorts, UA diameter was measured with an ATL3000 ultrasound unit with color imaging and Doppler at pregnancy weeks 20 and 36. Measurements were obtained bilaterally in the longitudinal view via a 4 Mhz curved linear array probe at the point where the UA appears to cross over the external iliac artery. Mean vessel diameter was expressed as (2 × end-diastolic + peak systolic)/3 and averaged from the right and left sides as previously described (25).
Selection of candidate genes and SNPs.
In previous work, we employed population genetic techniques to identify gene regions exhibiting strong signatures of natural selection among Andeans and Tibetans from dense genome scan data (7, 8). In brief, first we scanned across each chromosome to identify gene regions with previously unknown function with respect to altitude adaptation showing evidence of positive selection. We identified extended regions of statistical significance for three test statistics commonly used in population genetic research (locus-specific branch length, lnRH, and Tajima's D), using the hypergeometric distribution calculated for one megabase nonoverlapping windows along each chromosome. The results of this analysis revealed 14 candidate regions for high-altitude adaptation in Tibetans and 37 regions in Andeans. Second, we identified signatures of positive selection in genes with known physiological roles in oxygen homeostasis, including genes in the hypoxia inducible transcription factor (HIF) pathway and genes in or related to the globin family. We considered genes where two or more test statistics were statistically significant in Andeans or Tibetans (see Supplemental Tables S1 and S2 for additional detail). From these two analyses, we selected the 16 candidate genes listed in Table 1 to be assayed in our cohort based on rank, function, and assay design success. The chosen regions included: 1) 14 genes or gene regions with known physiological roles in oxygen homeostasis, including genes in the HIF pathway and the β-globin gene cluster located on chromosome 11 and 2) two genomic regions with unknown function with regard to high-altitude phenotypes that included portions of chromosome 12 [108,830 kilobase (kb) pairs–113,266 kb] and chromosome 15 (40,327–42,697 kb). Hereafter, we refer to these regions by their respective chromosome number (CHR12 or CHR15).
Table 1.
Candidate genes and SNPs
| Gene Symbol or Chr Region | Gene Name | Chr | SNP(s) | SNP Count |
|---|---|---|---|---|
| CDH1 | cadherin 1 | 16 | rs7203337, rs11648220, rs1111722, rs7196626 | 4 |
| CHR12 | NA | 12 | rs659964, rs2339840 | 2 |
| CHR15 | NA | 15 | rs675996, rs11638531, rs7179662, rs9920624 | 4 |
| EDNRA | endothelin receptor type A | 4 | rs11731134, rs7675774, rs12508797 | 3 |
| EGLN1 | Egl nine homolog 1 (C. elegans) | 1 | rs1339896, rs2812385, rs16854779 | 3 |
| EPAS1 | endothelial PAS domain protein 1 | 2 | rs12619696, rs10187368, rs13420857, rs7587138 | 4 |
| Globin Cluster | NA | 11 | rs2736588, rs10082629, rs2499957, rs2500035, rs17250577, rs11037417, rs2499945 | 7 |
| IL6 | interleukin 6 (interferon, β 2) | 7 | rs10271156, rs3094968, rs9639436, rs2905347, rs3094969 | 5 |
| KCNMA1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 10 | rs7918675, rs12783632, rs2637260 | 3 |
| NOS2 | nitric oxide synthase 2, inducible | 17 | rs1009484, rs12948602, rs4795856, | 3 |
| POLR2A | polymerase (RNA) II (DNA directed) polypeptide A, 220 kDa | 17 | rs2302764, rs16956822, rs4511593, rs2269459 | 4 |
| PRKAA1 | protein kinase, AMP-activated, α1 catalytic subunit | 5 | rs3805490, rs6861121, rs9292780, rs10055860, rs9292785, rs1345778 | 6 |
| PSMC3 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 11 | rs11600292 | 1 |
| SATB1 | SATB homeobox 1 | 3 | rs4130090, rs336641, rs1553995, rs1822120, rs7427289 | 5 |
| TGFA | transforming growth factor, α | 2 | rs17581575, rs17615746, rs2902451, rs959827, rs414399, rs11902650 | 6 |
| TNC | tenascin C | 9 | rs4979472, rs2482077, rs10982455 | 3 |
Chr, chromosome; SNP, single nucleotide polymorphism.
For each of the 16 candidate genes, we selected haplotype-tagging (tag) SNPs for Andean (i.e., Quechua and Aymara) haplotypes. To identify tagSNPs in this population, genotype data spanning 500 kb upstream and downstream of the start and end coordinates of each gene were phased with fastPHASE (v1.1) (46), and the resulting haplotypes were inferred with Haploview v4.1 (4). In this way, we included SNPs located within each candidate gene as well as SNPs upstream and downstream as the latter have been shown to regulate adjacent genes (57). All SNPs upstream or downstream of the transcription initiation start site or the termination codon are referred to by the HIF-pathway candidate gene for which they were selected. See Supplemental Table S2 for a listing of each SNP selected.
SNP genotyping.
Peripheral blood was drawn from an antecubital vein into a collection tube containing EDTA and transported on dry ice to our University Park lab. DNA was isolated by the Puregene DNA purification system (Qiagen, Valencia, CA) according to the manufacturer's instructions. Genotyping of 78 AIMs that distinguish West African, European, and Native American parental populations was performed at Prevention Genetics (Marshfield, WI) as previously described (9, 11, 48) and used for calculating individual ancestry estimates using a dihybrid model implemented in the program Maximum Likelihood (17).
Genotyping of candidate SNPs was performed by using the ABI SNPlex genotyping technology (Applied Biosystems, Foster City, CA) and Genemapper v3.5 (Applied Biosystems) software for genotype calling. In total, 82 SNPs produced reliable genotype calls and were subject to QC. SNPs were filtered from our dataset if they exhibited ≥20% missing data in our combined longitudinal and cross-sectional cohort (n = 15 SNPs) or if they violated Hardy-Weinberg equilibrium (P < 0.001, n = 4 SNPs), as such violations were likely to indicate genotyping error. Therefore quantitative association analyses were performed on 63 SNPs distributed across the 16 candidate gene regions (Table 1). We also excluded 53 individuals [30 Andeans (8 low, 22 high altitude) and 23 Europeans (5 low, 18 high altitude)] from our analysis as they exhibited ≥20% missing SNP data or lacked birth weight data. The mean birth weight of infants born to excluded vs. included women did not differ within any altitude and ancestry group.
Association analysis.
We combined European and Andean groups and the longitudinal and cross-sectional cohorts to increase our power to detect associations between selection-nominated candidate genes and birth weight in a cross-population manner. SNP analysis and genetic-model assessment were conducted with PLINK version 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/). We performed standard linear regression on birth weight to calculate regression coefficients and 95% confidence intervals using an additive model of inheritance. SNPs showing significant uncorrected (P < 0.05) birth-weight associations in an additive model next were run with dominant and recessive models of inheritance. Models were adjusted for Native American ancestry proportion (NAAP) as estimated by the AIMs to control for population substructure, altitude of birth (either low or high), gestational age, infant sex, and maternal height as these variables were shown to influence birth weight in our full cohort (63). We applied a Bonferroni correction for multiple testing with a P value cutoff of P < 0.0008 (α = 0.05, 63 tests).
Gene expression analysis.
Forty-six (46) high-altitude Andean women who delivered at term from the cross-sectional cohort were included for gene expression analyses, 27 of whom delivered non-IUGR infants, and 19 delivered IUGR infants. Infants were classified as IUGR if their birth weight was <10th percentile of sea-level values for their gestational age and sex, and non-IUGR if the birth weight was between the 10th and 90th percentiles (62). No babies weighed more than the 90th percentile or were postterm (>42 wk).
Third trimester peripheral blood samples (8 ml) were collected into a BD Vacutainer CPT Cell Preparation tube (Franklin Lakes, NJ) containing sodium citrate and Ficoll Hypaque density fluid. The resultant peripheral blood mononucleated cells (PBMCs) were then resuspended in RNAlater (Ambion, Austin, TX) and stored at −80°C until analysis. Total RNA was prepared with an RNeasy kit (Qiagen, Hilden, Germany) and used for cDNA synthesis with an oligo (dT) primer (Affymetrix, Santa Clara, CA). The gene expression profile for each participant was assessed with the Affymetrix GeneChip Human Gene 1.1 ST microarray, which measures 764,885 probes covering 28,869 genes throughout the genome. Raw chip files were background corrected, log2 transformed, and normalized by robust multiarray average in the Affymetrix Expression Console (Affymetrix) (20). A linear model was then fit to the normalized data to generate a matrix, including an expression value, for each probe.
RNA transcriptional profiles were contrasted by maternal genotypes with Limma and Surrogate Variable Analysis (31, 50) packages in R (44) with gene-chip processing date, gestational age at the time of study, and the presence or absence of hypertensive complications of pregnancy (preeclampsia, gestational hypertension) as covariates. Genes that were differentially expressed between our genotypes of interest at a Benjamini-Hochberg (BH) corrected P value <0.05 to control for multiple comparisons (5) were then analyzed by Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com) software. IPA subdivides input data, in this case a list of differentially expressed genes, on the basis of molecular connectivity (networks), the assumption being that interrelationship likely represents significant biological function. A numerical score is given to indicate the degree of fit between the network and a list of differentially expressed genes; the score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher's exact test. Finally, given the overrepresentation of mechanistic target of rapamycin (mTOR) pathway genes in the list of Andean selection-nominated candidate genes (see below), we generated a single P value for mTOR pathway genes using the Stouffer-Liptak method (32, 53) to evaluate the relationship of SNP genotypes with transcriptional activity within that pathway specifically.
RESULTS
Maternal and infant characteristics.
The Andean women were younger, shorter, weighed less when nonpregnant or during the 3rd trimester, and were of higher parity than the Europeans at high altitude (Table 2). The Andeans were also shorter than the Europeans at low altitude, but nonpregnant body mass index, pregnancy weight gain, and pregnancy skinfolds were similar in all groups. As expected the average NAAP was higher for Andeans than Europeans at high altitude. The cross-sectionally studied Andeans were also predominantly of Native American ancestry and did not differ from the longitudinal cohort of Andean women in any maternal or infant characteristic. At low altitude, NAAP reported for Europeans was considerably higher than at high altitude, but values in the low-altitude group principally reflected Native American ancestry of low-altitude origin.
Table 2.
maternal and infant characteristics for longitudinal and cross-sectional cohorts
| Longitudinal |
|||||||
|---|---|---|---|---|---|---|---|
| High Altitude |
Low Altitude |
||||||
| Variable | Andean | European | P | Andean | European | P | Cross-sectional High-altitude Andean |
| Maternal characteristics | |||||||
| Age, yr | 28 ± 7 (40) | 32 ± 5 (38) | P < 0.01 | 27 ± 5 (28) | 27 ± 6 (38) | NS | 27 ± 6 (100) |
| Nonpregnant height, cm | 150 ± 4 (40) | 161 ± 6 (34) | P < 0.001 | 157 ± 5 (27) | 160 ± 6 (38) | P < 0.05 | 151 ± 6 (100) |
| Nonpregnant weight, kg | 59 ± 10 (40) | 64 ± 11 (28) | P < 0.05 | 60 ± 14 (27) | 65 ± 17 (36) | NS | NR |
| Nonpregnant BMI, kg/m2 | 26 ± 4 (40) | 25 ± 4 (27) | NS | 24 ± 6 (27) | 26 ± 5 (35) | NS | NR |
| 3rd trimester weight, kg | 67 ± 9 (39) | 71 ± 9 (32) | P < 0.05 | 72 ± 11 (23) | 77 ± 14 (31) | NS | 66 ± 10 (100) |
| Pregnancy weight gain, kg | 7.3 ± 5.7 (39) | 8.3 ± 3.9 (24) | NS | 12.4 ± 17.8 (22) | 11.6 ± 19.8 (29) | NS | NR |
| NAAP, % | 85 ± 12 (40) | 20 ± 22 (38) | P < 0.001 | 64 ± 18 (27) | 57 ± 15 (37) | NS | 86 ± 12 (100) |
| Skinfolds, mm | 45 ± 13 (26) | 41 ± 15 (22) | NS | 56 ± 18 (27) | 60 ± 20 (36) | NS | NR |
| Parity, no. of live births | 3 ± 2 (40) | 2 ± 1 (35) | P < 0.01 | 2 ± 2 (28) | 2 ± 2 (38) | NS | 3 ± 2(100) |
| Infant characteristics | |||||||
| Birth weight, g | 3063 ± 421 (40) | 3091.5 ± 493 (38) | NS | 3276 ± 547 (28) | 3199 ± 527 (38) | NS | 3020 ± 606 (100) |
| Adjusted birth weight, g | 3242 ± 145 (40) | 2921 ± 156 (34) | P < 0.001 | 3299 ± 199 (27) | 3205 ± 167 (38) | NS | NA |
| Length, cm | 49 ± 2 (40) | 49 ± 4 (31) | NS | 50 ± 2 (28) | 50 ± 1 (36) | NS | 48 ± 5 (88) |
| Gestational age, wk | 39 ± 2 (38) | 39 ± 2 (38) | NS | 39 ± 2 (27) | 38 ± 2 (38) | NS | 39 ± 2 (100) |
| Female % | 55 [40–70] (40) | 38 [23–55] (36) | NS | 36 [18–53] (28) | 61 [45–76] (38) | NS | 48 [38–58] (100) |
Data are means ± SD, 95% confidence intervals (CI) for proportions in brackets, and samples sizes in parentheses. The single European woman in the cross-sectional cohort residing at high altitude was not included. Birth weight was adjusted using covariance analysis where factors related to birth weight (gestational age, maternal height) were set to the mean value for the 2 groups combined. NAAP, Native American ancestry proportion; no., number; NS, not significant; NR, not reported; NA, not applicable.
The Andean and European infants at high or low altitude were similar with respect to length, gestational age, sex, and birth weight prior to adjustments for covariates. According to multiple linear regression analysis, maternal height (P < 0.05) and gestational age (P < 0.001) were positively associated with birth weight in our longitudinal cohort (Table 3). After adjusting for these covariates by setting them to the mean values for the two ancestry groups combined, we found birth weight to be greater in Andean compared with European infants at high but not low altitude (P < 0.001) (Table 2).
Table 3.
Multiple linear regression analysis of the relationship between gestational age, maternal height, altitude of residence, and infant birth weight for the longitudinal cohort
| Independent Variable | Beta | P |
|---|---|---|
| Gestational age | 85.46 | <0.001 |
| Maternal height | 11.53 | 0.048 |
| Altitude | 177.80 | 0.031 |
Dependent variable (y) = infant birth weight (g). Effects of gestational age, nonpregnant maternal height, and altitude on infant birth weight are shown. Parity and infant sex were not significantly associated with infant birth weight and were excluded from the analysis.
Candidate gene associations with birth weight.
We performed standard linear regression for each SNP to isolate the effect of maternal SNP genotype on infant birth weight after controlling for maternal height, gestational age, altitude of residence, infant sex, and population structure (using NAAP). Two of the 63 SNPs from the 16 natural selection-nominated candidate genes, PRKAA1 SNP rs1345778 and EDNRA SNP rs11731134, were related to infant birth weight in the 245 European or Andean women for the two altitudes combined with additive or dominant models (Table 4). Figure 2 shows the best model for each SNP, and Supplemental Tables S3–S5 provide the full results for additive, dominant, and recessive models.
Table 4.
PRKAA1 and EDNRA SNP associations with infant birth weight and maternal UA diameter
| Additive Model |
Dominant Model |
Recessive Model |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Alleles | Minor Allele | n | Beta | P | Beta | P | Beta | P |
| Associations with birth weight at the two altitudes combined | ||||||||||
| rs1345778 | PRKAA1 | G/T | G | 228 | −225.2 ± 123.2 | 0.0004 | −258.0 ± 136.9 | 0.0003 | −211.4 ± 443.7 | NS |
| rs11731134 | EDNRA | C/G | G | 229 | −186.2 ± 105.8 | 0.0007 | −208.1 ± 124.2 | 0.0012 | −275.9 ± 306.4 | NS |
| Associations with week 36 UA diameter at HA | ||||||||||
| rs1345778 | PRKAA1 | G/T | G | 65 | −0.048 ± 0.03 | 0.0026 | −0.054 ± 0.03 | 0.0034 | −0.072 ± 0.10 | NS |
| rs3805490 | PRKAA1 | A/T | A | 67 | −0.044 ± 0.02 | 0.0001 | −0.061 ± 0.03 | 0.0006 | −0.066 ± 0.04 | 0.0034 |
| rs9292785 | PRKAA1 | C/T | C | 64 | −0.053 ± 0.02 | 0.0001 | −0.066 ± 0.03 | 0.0001 | −0.063 ± 0.06 | NS |
Regression coefficients (beta) ± the 95% CI intervals are shown. UA, uterine artery; HA, high altitude.
Bonferroni threshold of P < 0.0008, α = 0.05 for birth-weight analysis and P < 0.006, α = 0.05 for UA diameter analysis.
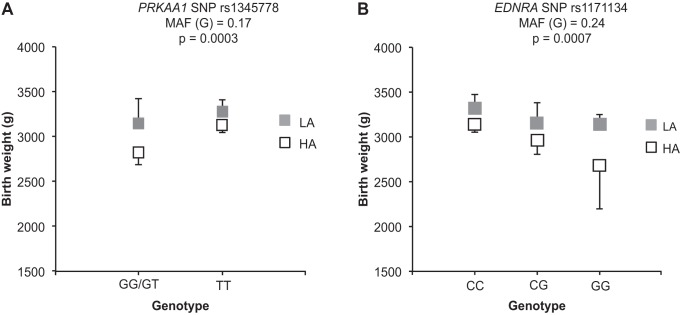
Fig. 2.
Single nucleotide polymorphism (SNP) associations with birth weight at low or at high altitude. Two SNPs were significantly associated with birth weight in additive and dominant models of inheritance in our combined low- and high-altitude cohort. Shown are PRKAA1 SNP rs1345778 using a dominant model (A) and EDNRA SNP rs11731134 using an additive mode (B). For both SNPs, mean birth weight is depicted as filled boxes for low altitude and open boxes for high altitude. The y-axis shows birth weight in grams for the genotypic categories indicated on the x-axis. The 95% confidence intervals (CIs, lines) were calculated for each genotypic category for birth weight. Minor allele frequency (MAF) is reported for the combined high- and low-altitude study participants. LA, low altitude; HA, high altitude.
For PRKAA1 SNP rs1345778, the GG and GT genotypes were associated with lower birth weight, and the TT genotype was associated with higher birth weight (Fig. 2A). Furthermore, the higher birth weight allele, T, was observed at a significantly higher frequency in the Andean (T = 0.88) compared with the European women (T = 0.73), and there were no high-altitude Andean women with the GG genotype (Table 5). For EDNRA SNP rs11731134, infants born to women with the CC or GC genotypes had higher birth weights compared with those born to women of the GG genotype (Fig. 2B). There were no differences in EDNRA SNP rs11731134 allele frequencies between ancestry groups (Table 5).
Table 5.
PRKAA1 rs1345778 and EDNRA rs11731134 allele and genotype frequencies for high-altitude study participants
| Gene | SNP | Alleles | Minor Allele | Ancestral Allele | Allele/Genotype | Andean | European | P | Total |
|---|---|---|---|---|---|---|---|---|---|
| PRKAA1 | rs1345778 | G/T | G | T | G | 0.12 (34) | 0.27 (21) | 0.003 | 0.16 (55) |
| T | 0.88 (240) | 0.73 (57) | 0.84 (297) | ||||||
| GG | 0.00 (0) | 0.08 (3) | 0.02 (3) | ||||||
| GT | 0.25 (34) | 0.38 (15) | 0.28 (49) | ||||||
| TT | 0.75 (103) | 0.54 (21) | 0.70 (124) | ||||||
| EDNRA | rs11731134 | C/G | G | G | C | 0.79 (222) | 0.72 (56) | NS | 0.78 (278) |
| G | 0.21 (58) | 0.28 (22) | 0.22 (80) | ||||||
| CC | 0.61 (86) | 0.54 (21) | 0.60 (107) | ||||||
| CG | 0.36 (50) | 0.36 (14) | 0.36 (64) | ||||||
| GG | 0.03 (4) | 0.10 (4) | 0.04 (8) |
Allele and genotype frequencies are shown with the samples sizes provided in parentheses. The ancestral allele is reported according to dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). PRKAA1 rs1345778, but not EDNRA rs11731134, differ between Andeans and Europeans (χ2 = 8.63). Allele and genotype frequencies are reported for the high-altitude study participants only.
PRKAA1 and EDNRA SNP associations with UA diameter.
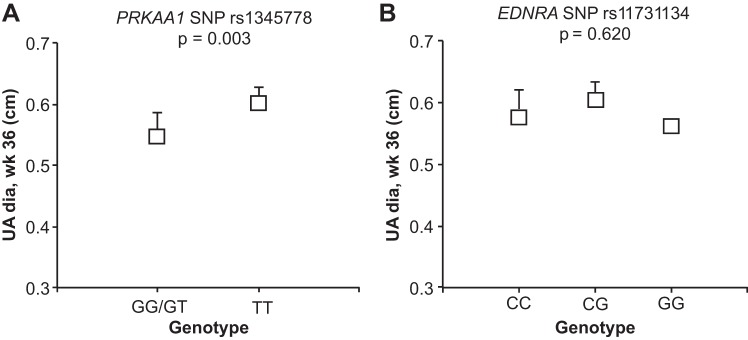
Previously we showed that greater UA blood flow protected Andeans against altitude-associated reductions in fetal growth (25). Since one means by which SNPs in or near EDNRA and PRKAA1 may influence birth weight is via alterations in UA diameter, we explored its associations with ENDRA and PRKAA1 SNP genotypes in our longitudinally studied participants at weeks 20 and 36 of pregnancy at high altitude. Maternal genotypes for PRKAA1 SNPs rs929785 and rs3805490 were associated with UA diameter measured at high altitude for pregnancy week 20 after a Bonferroni correction for multiple tests with a P value cutoff of P < 0.0056 (α = 0.05, 9 tests) was applied using additive and dominant models of inheritance (Supplemental Table S6). Furthermore, maternal genotypes for PRKAA1 SNPs rs929785, rs1345778, and rs3805490 were significantly associated with UA diameter measured at high altitude at 36 wk after the application of a Bonferroni correction for multiple tests with a P value cutoff of P < 0.0056 (α = 0.05, 9 tests) regardless of whether an additive or dominant model of inheritance was employed (Supplemental Table S7). For simplicity and because there were no women with GG genotypes at high altitude (Table 5), the associations between UA diameter measured at 36 wk and PRKAA1 SNP rs1345778 genotype using the dominant model are shown in Fig. 3A. EDNRA SNP rs765774 was associated with UA diameter measured at high altitude for pregnancy week 20 (Supplemental Table S6). There were no associations between UA diameter measured at high altitude for pregnancy week 36 with any of the three EDNRA SNPs considered (rs12508797, rs11731134, rs765774), including the rs11731134 SNP associated with birth weight (Fig. 3B, Supplemental Tables S6 and S7).
Fig. 3.
SNP associations with uterine artery (UA) diameter (dia). Women with the PRKAA1 SNP rs1345778 TT genotype giving birth to heavier birth-weight infants at HA had larger UA diameters measured at week 36 of pregnancy (A), but that pregnancy week 36 UA diameter at HA was not associated with EDNRA rs11731134 genotype (B). For both SNPs, mean UA dia in centimeters is depicted as unfilled boxes with 95% CIs (lines) for the combined Andean and European HA participants of each genotypic category.
PRKAA1 SNP rs1345778 associations with gene transcriptional activity.
To explore the metabolic pathways by which maternal PRKAA1 rs1345778 SNP genotypes might influence fetal growth, we contrasted the transcriptional profiles for the GG/GT vs. TT women. We identified 828 transcripts that differed, 410 of which were upregulated and 418 downregulated, using a BH P < 0.05 to control for multiple comparisons. Using IPA we identified networks implicated by the 780 mapped genes (Supplemental Table S8). Table 6 shows the top networks identified (cellular assembly and organization, cellular function and maintenance, and amino metabolism, lipid metabolism and molecular transport) and the transcripts differing between GG/GT vs. TT genotypes for each network.
Table 6.
Top molecular networks represented by genes that differ in expression by maternal PRKAA1 rs1345778 genotype
| Top Networks (58) | Molecules in Network | Score |
|---|---|---|
| Cellular assembly and organization, cellular function and maintenance, cancer | ATG4C, BMPR1A, Caspase 3/7, Cbp/p300, CD226, CDC42EP5, CYBA, DEFB114, EEA1, ERK1/2, HOOK2, HOOK3, Igf, JAM2, KIAA1279, MAPKAPK5, mir-181, MYOG, Nectin, PARD3, PIGF, PNMA1, PVRL2, RGS5, RHEB, Smad1/5/8, Smad2/3, SPTLC1, TIGIT, TOM1L1, TP53RK, TSHZ3, ZFYVE16, ZMYND11, ZNF638 | 39 |
| Cellular assembly and organization, cellular function and maintenance, connective tissue disorders | ACAN, ADAMTS4, ALOX15, Alpha actinin, Alpha catenin, BCAN, BRK1, CHRNA2, COL1A2, Collagen type I, Collagen type III, CTNNA1, CTTNBP2, EPHA7, ERK, IL3RA, ITGA11, KIF9, LAMTOR3, MAFF, MEP1B, NCKAP1, NRG (family), P4HA1, Ppp2c, PRMT3, Rock, RYK, SELP, STEAP4, TLL1, TMEM59, TNC, VCAN, YTHDC1 | 37 |
| Amino acid metabolism, lipid metabolism, molecular transport | ACSL1, Akt, ASAH2, ATP1B2, C1GALT1C1, CAMLG, CCDC85A, CEP70, CPA1, Cyclin D, DGKD, GAD2, GAD, GDNF, GRB14, HLA-DQ, INSIG2, ITGB8, KLF15, MEF2, mir-133, N-cor, Nfat (family), Nr1 h, PSMB1, PTF1A, RNF139, Rxr, SIM1, SLC32A1, SPOCD1, STMN2, TCL1A, ULBP1, WRB | 34 |
Boldface indicates molecules belonging to each network that were differentially expressed in women with PRKAA1 SNP rs1345778 TT vs. GG/GT genotypes.
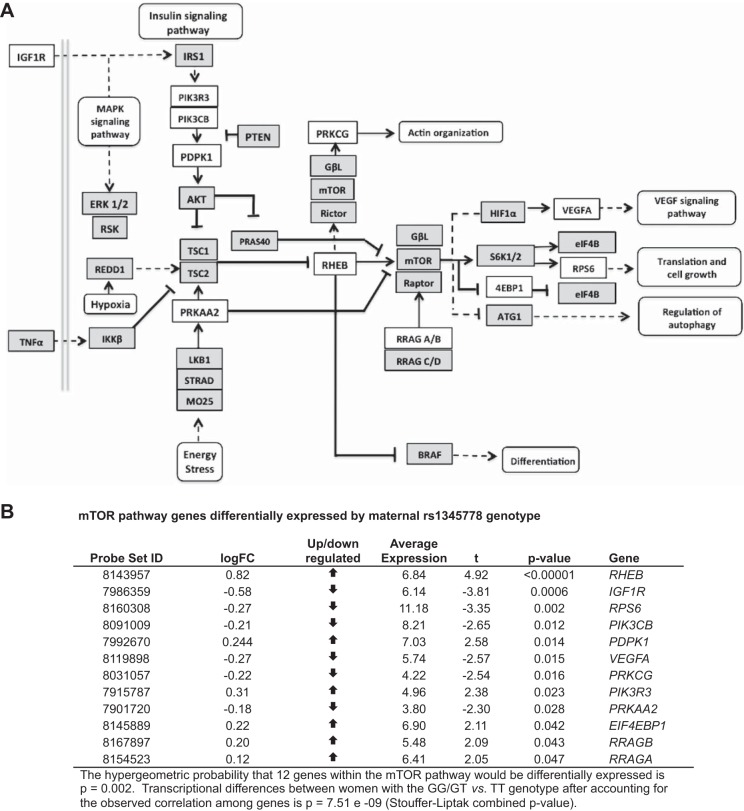
We then asked whether PRKAA1 SNP rs1345778 genotype influenced the expression of genes belonging to the mTOR pathway as defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG). We focused on the mTOR pathway because 1) PRKAA1 is the catalytic subunit of AMPK, which is a regulator of the mTOR pathway, and 2) mTOR genes were overrepresented in the list of Andean selection-nominated candidate genes. Specifically, the probability that six genes, PRKAA1, PRKAA2, MTOR, PIK3CA, TNF, VEGFA, within our list of 40 selection-nominated candidate genes (see Supplemental Table S1) would be part of the 61 gene KEGG mTOR pathway is P = 6.25e-12. The transcriptional activity of 12 mTOR pathway genes (RHEB, IGF1R, RPS6, PIK3CB, PDPK1, VEGFA, PRKCG, PIK3R3, PRKAA2, EIF4EBP1, RRAGA, and RRAGB) differed between women with the GG/GT vs. TT genotype with a nominal P value of P < 0.05; the hypergeometric probability that 12 genes within the mTOR pathway would be differentially expressed due to random chance is P = 0.002 (Fig. 4). Furthermore, after observed correlation among genes were accounted for, transcriptional differences of mTOR pathway genes varied between women with the GG/GT vs. TT genotype at a P = 7.51e-09.
Fig. 4.
mTOR transcriptional activity. Maternal PRKAA1 SNP rs1345778 genotype was associated with the transcriptional activity of 12 genes belonging to the mTOR pathway as defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG). A: transcripts that differed during the 3rd trimester in women with TT vs. GG/GT PRKAA1 SNP rs1345778 genotypes as white rectangles, those that did not differ as shaded rectangles, and functional pathways implicated as rectangles with rounded corners. Specifically, PIK3R3, EIF4EBP1, RHEB, PDPK1, RRAGA, RRAGB transcripts were downregulated, and PIK3CB, PRKAA2, IGF1R, RPS6, VEGFA, PRKCG transcripts were upregulated in women with TT vs. GG/GT rs1345778 genotypes using a nominal P value of <0.05; the hypergeometric probability for 12 genes in the mTOR pathway being differentially expressed is P = 0.002. B: the Affymetrix probe set ID, fold change, direction of change, average expression value, t score, and P value for transcriptional differences for each mTOR pathway gene. The combined P value for transcriptional differences between maternal TT vs. GG/GT rs1345778 genotypes after accounting for the observed correlation among genes is 7.51e-09.
DISCUSSION
We investigated the genotypic effects of 63 SNPs in 16 selection-nominated candidate genes or gene regions on birth weight and the intermediate phenotype, UA diameter, in Andeans and Europeans residing at high and low altitude. We identified a gene-by-environment interaction for two SNPs near the genes PRKAA1 and EDNRA such that 1) the associations between the SNPs and birth weight were observed in Andeans and Europeans across the two altitudes and 2) several SNPs in/near PRKAA1, but not EDNRA, were consistently associated with UA diameter at high altitude. Furthermore, the frequency of the PRKAA1 T allele associated with the advantageous phenotypes (heavier birth weights, larger UA diameters) was present at a higher frequency in the Andeans who, as previously reported, have larger UA diameters and greater blood flow than the Europeans during pregnancy and give birth to heavier birth weight infants at high altitude (24). Pointing to a potential functional link, maternal PRKAA1 SNP genotype is associated with gene expression patterns in general and, in particular, of key pathways involved in metabolic homeostasis that have been proposed to play a role in the pathophysiology of fetal growth restriction.
Our study benefited from certain strengths but also had some weaknesses. The depth and breadth of genotype- and phenotype-related information collected for these participants are a prominent strength of this study. Genotypes included for study belonged to a list of selection-nominated candidate genes that we previously identified using two genome scans in two high-altitude and multiple low-altitude control populations (7, 8). As such, we worked under the assumption that genes belonging to this list were likely linked to phenotypes characteristic of long-term, adapted high-altitude populations. Ninety-six (96) maternal SNPs representing 16 of these candidate genes or gene regions were evaluated for their relationship with infant birth weight, a variable that is important for reproductive success at high altitude and thus highly sensitive to selective pressure. To strengthen the biological interpretation of our observed SNP associations with birth weight, we also assessed the relationship between maternal SNP genotype and an intermediate phenotype, UA diameter, that is important both for maintaining sufficient uteroplacental blood flow to support fetal growth and is related to birth weight at high altitude (23, 25, 67). Our observation that PRKAA1 SNP rs1345778 was associated not only with birth weight but also UA diameter raised our confidence that this genetic variant may be of particular importance for fetal growth. While the number of participants was small relative to that required for genome-wide association studies (34), reported are the largest number of pregnant high- or low-altitude residents to date (n = 245) with genetic, UA diameter, and birth weight data. However, our study findings were limited by SNPlex-assay design constraints, with many assayed SNPs lying outside of the coding regions. Because tagSNPs were chosen, the two SNPs associated with birth weight were in close linkage disequilibrium with the coding regions, but full sequencing data and additional experiments are required to determine the specific gene changes involved and, in particular, whether they affect protein abundance or transcriptional activity and hence provide a gain or loss of function. Another limitation is that spurious associations between a phenotype and a SNP can occur in stratified-population samples, as has been shown for skin color (48). We attempted to control for this by removing the effects of admixture stratification by the inclusion of NAAP as a covariate in our regression analyses. While the AIMs currently available are not able to distinguish Andean from other Native American ancestry, by using the surname and ancestry data provided by questionnaire, we were able to determine that the NAAP detected in the high- and low-altitude Andeans was of high-altitude origin, whereas the NAAP detected in the high- and low-altitude Europeans was of low-altitude origin. Furthermore, we explored the contribution of maternal genetic variation to fetal growth and infant birth weight but did not consider the contribution of paternal or infant genetic variation. Given the strong assortative mating present in this population (see for example Ref. 6), we suspect that genetic traits present at high frequency in the mother are also likely to have been present in the baby. Future studies considering the effects of both maternal and fetal genetic variation on fetal growth and birth weight outcomes will be useful for improving our understanding of the genetic basis of IUGR and for elucidating the complex interplay between maternal and fetal physiology. One challenge posed by performing gene-expression studies in humans, particularly during pregnancy, is the ability to obtain relevant cells or tissue in sufficient quantity. We reasoned that, despite being unable to infer the relationship between maternal genotype and tissue-specific gene expression, the use of PBMCs would allow us to gain insight into the influence of maternal genotype on systemic processes involved in maternal vascular and metabolic adaptation to pregnancy (e.g., inflammation). Additionally, since RNA samples were not available for all women, gene-expression studies were performed on a subset of the total study population, thus limiting our power to detect differences in transcriptional activity between maternal genotypes.
Our studies extend previous reports showing that fetal growth and birth weight are influenced by genetic (fetal, maternal, and paternal) and environmental factors (e.g., high altitude). High altitude profoundly reduces birth weight by slowing fetal growth in the third trimester, rather than shortening gestation (37). Therefore it follows that high altitude presents a unique opportunity to identify the maternal genetic factors influencing birth weight and hence has been the subject of numerous studies over the past 30 years in residents of the Tibetan Plateau, the Andean Altiplano, and the North American Rocky Mountains (16, 23, 24, 40, 41, 66, 67). Historical accounts dating to the time of the Spanish conquest of South America chronicle the difficulty experienced by early Spaniards in reproducing at high altitude. For example, the first child born to Spanish parents to survive in Potosi, Bolivia (4,100 m) occurred 53 years after the arrival of the Spanish to this region (36). Admixed children born to one Spanish parent and one Andean parent, usually an Andean mother and a Spanish father, were not uncommon following the conquest and appeared not to suffer the same difficulties as pregnancies from two European parents. Francisco Pizarro himself had two children with the Inca Quispe Sisa, both of whom were born in Juaja (3,410 m) (15). Today, Han residents of Tibet often descend to low altitude during pregnancy and remain there until the infants are ∼2 yr in age (40, 42). Beyond these anecdotal reports, we and others have shown associations between birth weight and parental ancestry as determined by surname (6, 24), a positive association between NAAP and UA diameter (or blood flow) during pregnancy at high altitude (25), a negative correlation between European ancestry proportion and birth weight in Andean women living at high altitude (68), and that Andeans were able to maintain normal fetal growth compared with Europeans at high altitude likely due, at least in part, to greater UA diameter, blood flow, and uteroplacental O2 delivery during pregnancy (63). Therefore, association studies utilizing the effect of hypobaric hypoxia on birth weight are a powerful method for understanding the maternal genetic contribution to birth weight, the physiological pathways by which hypoxia-associated IUGR occurs, and ultimately the inherited basis of hypoxia-associated IUGR.
PRKAA1 encodes the alpha1 catalytic subunit of AMPK, a heterotrimeric enzyme made up of two alpha, two beta, and three gamma subunits (18). This enzyme belongs to the ancient 5′-adenosine monophosphate-activated protein kinase (AMPK) gene family involved in regulation of cellular ATP (reviewed in Ref. 27). During periods of metabolic stress such as those experienced in hypoxia, AMPK is activated by increases in 5′-adenosine monophosphate (AMP) (19). Our observation that PRKAA1 was one of two genes associated with infant birth weight is interesting in light of AMPK's central role in oxygen sensing and several major metabolic signaling pathways (58). In particular, PRKAA1's gene product is essential for the transcriptional activity of hypoxia-inducible factor-1 (HIF-1), a gene that trans-activates multiple genes in the HIF pathway that are critical for both embryonic vascularization and development (30), as well as intimately involved in physiological responses to hypoxia and pregnancy, which serve to maintain tissue oxygenation and metabolism in the face of limited oxygen supply or increased oxygen demand (47, 61). Another central mechanism of action involves the mTOR pathway, which AMPK activation can inhibit by phosphorylating tuberous sclerosis 1 or 2 (TSC1, TSC2) either indirectly via inhibiting Rheb or directly via phosphorylating Raptor. Highlighting the importance of the mTOR pathway for fetal growth, mTOR activity is reduced in IUGR (45) and elevated in cases of fetal overgrowth (13). Yung et al. (65) have recently reported increased levels of phosphorylated ELF2 in placentas from IUGR pregnancies at sea level as well as at 3,100 m in Colorado, suggesting that both decreased mTOR activity and a longer-term block to translation contribute to IUGR at high altitude. Differences in AMPK levels have not been seen in these studies, which may be because other factors were involved in regulating mTOR, sample sizes were too small (n = 3), or AMPK levels were highly variable due to the rapidly metabolized nature of this substance. Further supporting a link between AMPK and reduced birth weight, we have shown that transcriptional activity of peroxisome proliferator activated receptor gamma, a ligand-activated nuclear receptor and a key regulator of cellular metabolic homeostasis, is impaired during pregnancy at high altitude (26), an effect that may be attributed to enhanced AMPK activity (52). AMPK activation also has vasodilatory effects due in part to increasing endothelial nitric oxide production but also by acting directly on the vascular smooth muscle (29, 59), both of which are increased by chronic hypoxia in isolated mouse uterine arteries (49). In short, our observations that maternal PRKAA1 genotype was associated with infant birth weight, UA diameter, and the transcriptional activity of metabolic pathways involved in cellular metabolism supports the possibility that maternal PRKAA1 SNP genotype influences fetal growth by affecting vascular control and/or metabolic homeostasis. However, additional data are required to determine the specific functional changes responsible for the observed associations.
EDNRA controls the production of endothelin receptor type A and is part of the endothelin system, which also includes endothelin receptor type B (EDNRB) and endothelins 1–3 (EDN1, EDN2, EDN3). EDN1 encodes the peptide ET-1, a potent vasoconstrictor involved in vascular homeostasis (64). EDNRA encodes the EDN1 receptor and is expressed primarily in vascular smooth muscle cells with no known effects beyond vasoconstriction. Animal models have shown that EDNRA protein expression is significantly decreased in the uteroplacental vascular bed and that its antagonism prevents hypoxia-induced IUGR in rats (55, 56). In humans, ET-1 levels decrease during pregnancy, but this normal fall does not occur or is diminished in preeclamptic women (54). In the present study, we did not find maternal EDNRA genotype to be associated with circulating ET-1 levels (data not shown) or to be consistently associated with UA diameter at high altitude, but further study is warranted for evaluating the effects of genotypic variation in EDNRA in reproductively relevant, maternal or placental vascular tissues.
In summary, we have identified two novel loci, PRKAA1 and EDNRA, associated with infant birth weight. Both of these genes are involved in cellular oxygen sensing and delivery as part of the HIF pathway. Our results suggest that maternal variation in genes influencing maternal vascular responses to pregnancy and UA diameter in particular and transcriptional activity more broadly are likely to play key roles in protecting Andeans against altitude-associated IUGR. These results advance our understanding of the genetic mechanisms and physiological pathways contributing to IUGR and adaptation to high altitude but require replication in additional Andean populations to confirm our findings. Further studies also are required to determine the specific gene variants involved and the mechanisms by which genetic variation in or near PRKAA1 and EDNRA could influence uteroplacental blood flow and/or the delivery or metabolism of oxygen and other nutrients required for normal fetal growth. Other kinds of genetic variation, such as those in small interfering RNAs and microRNAs, should also be considered. Continued study of IUGR at high altitude is expected to yield important new discoveries into the pathways governing maternal and fetoplacental physiology as well as shed light on the evolutionary processes that have resulted in high-altitude adaptive phenotypes. Ultimately, such findings promise to lead to novel approaches for diagnosing and treating pregnancy-associated disorders and for improving neonatal health.
GRANTS
This work was funded by the National Institutes of Health (NIH) Grants HLBI-079647 (L. G. Moore), HLBI-060131 (L. G. Moore), TW-001188 (L. G. Moore), HG-002154 (M. D. Shriver); National Science Foundation Grants 0622337 (A. W. Bigham and M. D. Shriver); the Wenner-Gren Foundation Grant 7538 (A. W. Bigham); National Science Foundation Graduate Research Fellowships (A. W. Bigham and M. J. Wilson) and Doctoral Dissertation Research Improvement Grant (C. G. Julian); and an American Heart Association Predoctoral Fellowship (C. G. Julian). C. G. Julian is supported by NIH Building Interdisciplinary Research Careers Women's Health Grant 5 K12 HD-057022-07.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.W.B., C.G.J., M.J.W., E.V., V.A.B., M.D.S., and L.G.M. conception and design of research; A.W.B., C.G.J., M.J.W., V.A.B., and L.G.M. performed experiments; A.W.B., C.G.J., M.J.W., and L.G.M. analyzed data; A.W.B., C.G.J., and L.G.M. interpreted results of experiments; A.W.B., C.G.J., and L.G.M. prepared figures; A.W.B., C.G.J., M.J.W., and L.G.M. drafted manuscript; A.W.B., C.G.J., and L.G.M. edited and revised manuscript; A.W.B., C.G.J., M.J.W., V.A.B., M.D.S., and L.G.M. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank the participants who so graciously volunteered their time to be a part of this study and the numerous physicians, technical staff, and graduate students who helped with its conduct. We also thank Robert Stearman and Brent Pedersen for constructive criticism of earlier drafts of the manuscript and suggestions for the conduct of the statistical analyses.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Adkins RM, Somes G, Morrison JC, Hill JB, Watson EM, Magann EF, Krushkal J. Association of birth weight with polymorphisms in the IGF2, H19, and IGF2R genes. Pediatr Res 68: 429–434, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker DJ, Osmond C, Golding J, Kuh D, Wadsworth ME. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ 298: 564–567, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker DJ, Osmond C, Law CM. The intrauterine and early postnatal origins of cardiovascular disease and chronic bronchitis. J Epidemiol Community Health 43: 237–240, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B 57: 289–300, 1995. [Google Scholar]
- 6.Bennett A, Sain SR, Vargas E, Moore LG. Evidence that parent-of-origin affects birth-weight reductions at high altitude. Am J Hum Biol 20: 592–597, 2008. [DOI] [PubMed] [Google Scholar]
- 7.Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Lopez Herraez D, Brutsaert T, Parra EJ, Moore LG, Shriver MD. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet 6: 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigham AW, Mao X, Mei R, Brutsaert T, Wilson MJ, Julian CG, Parra EJ, Akey JM, Moore LG, Shriver MD. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics 4: 79–90, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York city. Hum Genet 115: 57–68, 2004. [DOI] [PubMed] [Google Scholar]
- 10.Browne VA, Toledo-Jaldin L, Davila RD, Lopez LP, Yamashiro H, Cioffi-Ragan D, Julian CG, Wilson MJ, Bigham AW, Shriver MD, Honigman B, Vargas E, Roach R, Moore LG. High-end arteriolar resistance limits uterine artery blood flow and restricts fetal growth in preeclampsia and gestational hypertension at high altitude. Am J Physiol Regul Integr Comp Physiol 300: R1221–R1229, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brutsaert TD, Parra EJ, Shriver MD, Gamboa A, Leon-Velarde F. Ancestry explains the blunted ventilatory response to sustained hypoxia and lower exercise ventilation of Quechua altitude natives. Am J Physiol Regul Integr Comp Physiol 289: R225–R234, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Creasy RK, Resnik R. Intrauterine growth restriction. In: Creasy and Resnik's Maternal-Fetal Medicine Principles and Practice, edited by Creasy RK, Resnik R, Iams JD, Lockwood CJ, Moore TR, Greene MF. Philadelphia, PA: WB Saunders, 2014, p. 743–755. [Google Scholar]
- 13.Gaccioli F, White V, Capobianco E, Powell TL, Jawerbaum A, Jansson T. Maternal overweight induced by a diet with high content of saturated fat activates placental mTOR and eIF2 alpha signaling and increases fetal growth in rats. Biol Reprod 89: 96, 2013. [DOI] [PubMed] [Google Scholar]
- 14.Giussani DA, Salinas CE, Villena M, Blanco CE. The role of oxygen in prenatal growth: studies in the chick embryo. J Physiol 585: 911–917, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzales GE. Peruvian contributions to the study on human reproduction at high altitude: from the chronicles of the Spanish conquest to the present. Resp Physiol Neurobi 158: 172–179, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Haas JD, Morenoblack G, Frongillo EA, Pabon J, Pareja G, Ybarnegaray J, Hurtado L. Altitude and infant growth in Bolivia - a longitudinal-study. Am J Phys Anthropol 59: 251–262, 1982. [DOI] [PubMed] [Google Scholar]
- 17.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates - disease associations and individual risk of diabetes and gallbladder-disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol 70: 433–441, 1986. [DOI] [PubMed] [Google Scholar]
- 18.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785, 2007. [DOI] [PubMed] [Google Scholar]
- 19.Hardie DG, Carling D. The AMP-activated protein kinase - fuel gauge of the mammalian cell? Eur J Biochem 246: 259–273, 1997. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Ishida M, Monk D, Duncan AJ, Abu-Amero S, Chong J, Ring SM, Pembrey ME, Hindmarsh PC, Whittaker JC, Stanier P, Moore GE. Maternal inheritance of a promoter variant in the imprinted PHLDA2 gene significantly increases birth weight. Am J Hum Genet 90: 715–719, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen GM, Moore LG. The effect of high altitude and other risk factors on birthweight: independent or interactive effects? Am J Public Health 87: 1003–1007, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julian CG, Galan HL, Wilson MJ, Desilva W, Cioffi-Ragan D, Schwartz J, Moore LG. Lower uterine artery blood flow and higher endothelin relative to nitric oxide metabolite levels are associated with reductions in birth weight at high altitude. Am J Physiol Regul Integr Comp Physiol 295: R906–R915, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julian CG, Vargas E, Armaza JF, Wilson MJ, Niermeyer S, Moore LG. High-altitude ancestry protects against hypoxia-associated reductions in fetal growth. Arch Dis Child Fetal Neonatal Ed 92: 372–377, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julian CG, Wilson MJ, Lopez M, Yamashiro H, Tellez W, Rodriguez A, Bigham AW, Shriver MD, Rodriguez C, Vargas E, Moore LG. Augmented uterine artery blood flow and oxygen delivery protect Andeans from altitude-associated reductions in fetal growth. Am J Physiol Regul Integr Comp Physiol 296: R1564–R1575, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julian CG, Yang IV, Browne VA, Vargas E, Rodriguez C, Pedersen BS, Moore LG, Schwartz DA. Inhibition of peroxisome proliferator-activated receptor gamma: a potential link between chronic maternal hypoxia and impaired fetal growth. FASEB J 28: 1268–1279, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp BE, Stapleton D, Campbell DJ, Chen ZP, Murthy S, Walter M, Gupta A, Adams JJ, Katsis F, van Denderen B, Jennings IG, Iseli T, Michell BJ, Witters LA. AMP-activated protein kinase, super metabolic regulator. Biochem Soc Trans 31: 162–168, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Keyes LE, Armaza JF, Niermeyer S, Vargas E, Young DA, Moore LG. Intrauterine growth restriction, preeclampsia, and intrauterine mortality at high altitude in Bolivia. Pediatr Res 54: 20–25, 2003. [DOI] [PubMed] [Google Scholar]
- 29.Lee KY, Choi HC. Acetylcholine-induced AMP-activated protein kinase activation attenuates vasoconstriction through an LKB1-dependent mechanism in rat aorta. Vasc Pharmacol 59: 96–102, 2013. [DOI] [PubMed] [Google Scholar]
- 30.Lee M, Hwang JT, Lee HJ, Jung SN, Kang IS, Chi SG, Kim SS, Ha JH. AMP-activated protein kinase activity is critical for hypoxia-inducible factor-1 transcriptional activity and its target gene expression under hypoxic conditions in DU145 cells. J Biol Chem 278: 39653–39661, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liptak T. On the combination of independent tests. Magyar Tud Akad Mat Kutato Int Kozl 3: 171–197, 1958. [Google Scholar]
- 33.McCance DR, Pettitt DJ, Hanson RL, Jacobsson LT, Knowler WC, Bennett PH. Birth weight and non-insulin dependent diabetes: thrifty genotype, thrifty phenotype, or surviving small baby genotype? BMJ 308: 942–945, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369, 2008. [DOI] [PubMed] [Google Scholar]
- 35.Mccullough RE, Reeves JT, Liljegren RL. Fetal growth retardation and increased infant-mortality at high altitude. Obstet Gynec Surv 32: 596–598, 1977. [DOI] [PubMed] [Google Scholar]
- 36.Monge C. Acclimatization in the Andes. Baltimore, MD: Johns Hopkins University Press, 1948. [Google Scholar]
- 37.Moore LG. Human genetic adaptation to high altitude. High Alt Med Biol 2: 257–279, 2001. [DOI] [PubMed] [Google Scholar]
- 39.Moore LG, Shriver M, Bemis L, Hickler B, Wilson M, Brutsaert T, Parra E, Vargas E. Maternal adaptation to high-altitude pregnancy: an experiment of nature–a review. Placenta 25, Suppl A: S60–S71, 2004. [DOI] [PubMed] [Google Scholar]
- 40.Moore LG, Young D, McCullough RE, Droma T, Zamudio S. Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am J Hum Biol 13: 635–644, 2001. [DOI] [PubMed] [Google Scholar]
- 41.Mortola JP, Frappell PB, Aguero L, Armstrong K. Birth weight and altitude: a study in Peruvian communities. J Pediatr 136: 324–329, 2000. [DOI] [PubMed] [Google Scholar]
- 42.Niermeyer S, Yang P, Shanmina, Drolkar, Zhuang J, Moore LG. Arterial oxygen saturation in Tibetan and Han infants born in Lhasa, Tibet. N Engl J Med 333: 1248–1252, 1995. [DOI] [PubMed] [Google Scholar]
- 43.Parraguez VH, Atlagich M, Diaz R, Bruzzone ME, Behn C, Raggi LA. Effect of hypobaric hypoxia on lamb intrauterine growth: comparison between high- and low-altitude native ewes. Reprod Fertil Dev 17: 497–505, 2005. [DOI] [PubMed] [Google Scholar]
- 44.R Development Core Team R: a language and environment for statistical computing. In: R Foundation for Statistical Computing. Vienna, Austria: 2008. [Google Scholar]
- 45.Roos S, Jansson N, Palmberg I, Saljo K, Powell TL, Jansson T. Mammalian target of rapamycin in the human placenta regulates leucine transport and is down-regulated in restricted fetal growth. J Physiol 582: 449–459, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78: 629–644, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenza GL. Life with oxygen. Science 318: 62–64, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 112: 387–399, 2003. [DOI] [PubMed] [Google Scholar]
- 49.Skeffington KL, Higgins JS, Sferruzzi-Perri AN, Fowden AL, Yung HW, Burton GJ, Moore LG, Giussani DA. Uterine artery (UA) reactivity to AMP-activated protein kinase in control and hypoxic pregnancy. Reprod Sci 31: 378A, 2014. [Google Scholar]
- 50.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. [DOI] [PubMed] [Google Scholar]
- 51.Soria R, Julian CG, Vargas E, Moore LG, Giussani DA. Graduated effects of high-altitude hypoxia and highland ancestry on birth size. Pediatr Res 74: 633–638, 2013. [DOI] [PubMed] [Google Scholar]
- 52.Sozio MS, Lu C, Zeng Y, Liangpunsakul S, Crabb DW. Activated AMPK inhibits PPAR-α and PPAR-γ transcriptional activity in hepatoma cells. Am J Physiol Gastrointest Liver Physiol 301: G739–G747, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stouffer S, Schuman EA, DeVinney LC, Star SA, Williams RM. The American Soldier: Adjustment During Army Life. Princeton, NJ: Princeton University Press, 1949. [Google Scholar]
- 54.Taylor RN, Varma M, Teng NN, Roberts JM. Women with preeclampsia have higher plasma endothelin levels than women with normal pregnancies. J Clin Endocrinol Metab 71: 1675–1677, 1990. [DOI] [PubMed] [Google Scholar]
- 55.Thaete LG, Jilling T, Synowiec S, Khan S, Neerhof MG. Expression of endothelin 1 and its receptors in the hypoxic pregnant rat. Biol Reprod 77: 526–532, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thaete LG, Neerhof MG, Caplan MS. Endothelin receptor A antagonism prevents hypoxia-induced intrauterine growth restriction in the rat. Am J Obstet Gynecol 176: 73–76, 1997. [DOI] [PubMed] [Google Scholar]
- 57.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, Ibrahim M, Omar SA, Lema G, Nyambo TB, Ghori J, Bumpstead S, Pritchard JK, Wray GA, Deloukas P. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39: 31–40, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res 100: 328–341, 2007. [DOI] [PubMed] [Google Scholar]
- 59.Viollet B, Horman S, Leclerc J, Lantier L, Foretz M, Billaud M, Giri S, Andreelli F. AMPK inhibition in health and disease. Crit Rev Biochem Mol Biol 45: 276–295, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, Wang X, Laird N, Zuckerman B, Stubblefield P, Xu X. Polymorphism in maternal LRP8 gene is associated with fetal growth. Am J Hum Genet 78: 770–777, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol 300: C385–C393, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams R, Creasy R, Cunningham G, Hawes WE, Norris FD, Tashiro M. Fetal growth and perinatal viability in California. Obstet Gynecol 58: 624–632, 1982. [PubMed] [Google Scholar]
- 63.Wilson MJ, Lopez M, Vargas M, Julian C, Tellez W, Rodriguez A, Bigham A, Armaza JF, Niermeyer S, Shriver M, Vargas E, Moore LG. Greater uterine artery blood flow during pregnancy in multigenerational (Andean) than shorter-term (European) high-altitude residents. Am J Physiol Regul Integr Comp Physiol 293: R1313–R1324, 2007. [DOI] [PubMed] [Google Scholar]
- 64.Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, Yazaki Y, Goto K, Masaki T. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature 332: 411–415, 1988. [DOI] [PubMed] [Google Scholar]
- 65.Yung HW, Cox M, Tissot van Patot M, Burton GJ. Evidence of endoplasmic reticulum stress and protein synthesis inhibition in the placenta of non-native women at high altitude. FASEB J 26: 1970–1981, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zamudio S, Droma T, Norkyel KY, Acharya G, Zamudio JA, Niermeyer SN, Moore LG. Protection from intrauterine growth-retardation in Tibetans at high-altitude. Am J Phys Anthropol 91: 215–224, 1993. [DOI] [PubMed] [Google Scholar]
- 67.Zamudio S, Palmer SK, Droma T, Stamm E, Coffin C, Moore LG. Effect of altitude on uterine artery blood-flow during normal-pregnancy. J Appl Physiol 79: 7–14, 1995. [DOI] [PubMed] [Google Scholar]
- 68.Zamudio S, Postigo L, Illsley NP, Rodriguez C, Heredia G, Brimacombe M, Echalar L, Torricos T, Tellez W, Maldonado I, Balanza E, Alvarez T, Ameller J, Vargas E. Maternal oxygen delivery is not related to altitude- and ancestry-associated differences in human fetal growth. J Physiol 582: 883–895, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.