Abstract
Upregulation of functional voltage-gated Na+ channels (VGSCs) occurs in metastatic human breast cancer (BCa) in vitro and in vivo. The present study aimed to ascertain the specific involvement of the ‘neonatal’ splice variant of Nav1.5 (nNav1.5), thought to be predominant, in the VGSC-dependent invasive behaviour of MDA-MB-231 cells. Functional activity of nNav1.5 was suppressed by two different methods targeting nNav1.5: (i) small interfering RNA (siRNA), and (ii) a polyclonal antibody (NESO-pAb); effects upon migration and invasion were determined. nNav1.5 mRNA, protein and signalling were measured using real-time PCR, Western blotting, and patch clamp recording, respectively. Treatment with the siRNA rapidly reduced (by ~90 %) the level of nNav1.5 (but not adult Nav1.5) mRNA, but the protein reduction was much smaller (~30 %), even after 13 days. Nevertheless, the siRNA reduced peak VGSC current density by 33 %, and significantly increased the cells’ sensitivity to nanomolar tetrodotoxin (TTX). Importantly, the siRNA suppressed in vitro migration by 43 %, and eliminated the normally inhibitory effect of TTX. Migrated MDA-MB-231 cells expressed more nNav1.5 protein at the plasma membrane than non-migrated cells. Furthermore, NESO-pAb reduced migration by up to 42 %, in a dose-dependent manner. NESO-pAb also reduced Matrigel invasion without affecting proliferation. TTX had no effect on cells already treated with NESO-pAb. It was concluded that nNav1.5 is primarily responsible for the VGSC-dependent enhancement of invasive behaviour in MDA-MB-231 cells. Accordingly, targeting nNav1.5 expression/activity may be useful in clinical management of metastatic BCa.
Keywords: Antibody, breast cancer, metastasis, RNAi, voltage-gated Na+ channel
INTRODUCTION
Ion channels are major signalling complexes expressed in many tissues, where they have diverse involvement in cellular activity [1]. Consequently, an increasing body of evidence implicates ion channels in a wide variety of pathophysiological conditions [2, 3], including cancer [4]. In particular, voltage-gated ion channels (activated by a depolarizing shift in membrane potential), including those permeable to K+, Cl− and Na+, are widely expressed in a range of cancer cells [5]. Voltage-gated Na+ channels (VGSCs) have been detected in cell lines of human breast cancer (BCa) [6], prostate cancer (PCa) [7], lymphoma [8], small-cell lung cancer (SCLC) [9, 10], neuroblastoma [11] and melanoma [12]. Importantly, VGSC upregulation has also been found in human BCa, PCa and SCLC in vivo [6, 13-15].
The highly specific VGSC blocker tetrodotoxin (TTX) has been shown to suppress a variety of in vitro cell behaviours associated with the metastatic cascade [16] in BCa, PCa and SCLC cells, including galvanotaxis, endocytic membrane activity, migration and invasion [7, 10, 17-22]. In PCa, TTX suppressed additional metastasis-associated cell behaviours including morphological development and process extension [23], vesicular patterning [24], lateral motility [25], adhesion [26] and gene expression [27]. Furthermore, putative VGSC blockers have been shown to inhibit proliferation of PCa cells [13, 28], and the VGSC-blocking anticonvulsants phenytoin and carbamazepine were found to directly inhibit secretion of prostate-specific antigen (PSA) and interleukin-6 from LNCaP and PC-3 PCa cell lines, respectively [29].
The VGSC proteins are comprised of a core α-subunit, which by itself can generate functional Na+ channels, together with one or more auxiliary, modulatory β-subunits [30]. In the metastatic MDA-MB-231 human BCa cell line, TTX-resistant (IC50 in μM range) functional VGSC activity was detected using the whole-cell patch clamp technique [6, 22]. In this cell line, the predominant isoform, TTX-resistant Nav1.5 alpha-subunit, was expressed ~1800-fold more at mRNA level, compared with the weakly metastatic cell line, MCF-7 [6]. In MDA-MB-231 cells, Nav1.5 constituted over 80 % of the total VGSC α-subunit mRNA level, the remaining 20 % being mainly the TTX-sensitive Nav1.7 [6]. Interestingly, the Nav1.5 was expressed primarily in it’s ‘neonatal’ DI:S3 5′-splice form (nNav1.5), differing from the known DI:S3 3′-splice form at 31 out of 92 nucleotides, resulting in 7 amino acid differences [6]. In the nNav1.5 protein, as with developmentally regulated DI:S3 5′-splice variants of other VGSC subtypes [31], a highly conserved aspartate residue at the extracellular end of DI:S3 was replaced by a positively charged lysine [6, 32]. The DI:S3 5′-splice variant of Nav1.5 has also recently been described in a human neuroblastoma cell line [11].
A novel polyclonal antibody (NESO-pAb) recognising an extracellular epitope (in DI:S3), specific to nNav1.5, but not ‘adult’ Nav1.5 was raised [32]. Immunohistochemistry with NESO-pAb confirmed that nNav1.5 was significantly more abundantly expressed in neonatal than adult heart and brain tissue samples [32]. Immunocytochemistry with NESO-pAb also revealed that nNav1.5 was indeed highly expressed in the plasma membrane of MDA-MB-231 but not MCF-7 cells [6]. Importantly, whole-cell patch clamp recording showed that direct application of NESO-pAb to modified human embryonic kidney (EBNA-293) cells expressing nNav1.5 reversibly blocked functional VGSC activity, whereas EBNA-293 cells expressing ‘adult’ Nav1.5 were ~400-fold less sensitive [32]. EBNA-293 cells are frequently used for transfection studies, and two variants expressing ‘adult’ and ‘neonatal’ forms of Nav1.5 were produced earlier [32].
Although we have previously shown that micromolar concentrations of TTX suppressed in vitro metastatic cell behaviours of BCa [6], the specific VGSC subtype(s) involved was not known. The aim of the present study was to ascertain the extent of involvement of nNav1.5 specifically in upregulating invasive activity in MDA-MB-231 cells. Two independent approaches targeting the functional expression/activity of this isoform were used: (i) RNA interference, and (ii) NESO-pAb. Preliminary findings from this study have been published previously in abstract form [33].
MATERIALS AND METHODS
Cell culture
MDA-MB-231 and PC-3M cells were cultured as described previously [6, 27]. Cells were seeded into Falcon tissue culture dishes (Becton-Dickinson, Oxford, UK) and incubated at 37 °C, 5 % CO2 and 100 % relative humidity.
RNA interference
Two different siRNAs targeting unique nucleotides within the 5′-DI:S3 alternatively spliced exon of nNav1.5 were designed using an online algorithm (Dharmacon, Lafayette, CO), as follows:
siNESO1 target sequence: 5′-UUUGUCGGCUCUUCGAACU-3′
siNESO2 target sequence: 5′-GAGUCCUGAGAGCUCUAAA-3′
Their target specificity was ensured by nucleotide BLAST screening optimised for short sequences [34]. All siRNA duplexes were synthesised by Dharmacon and resuspended and stored according to the manufacturer’s instructions. Cells (70 % confluent in 6-well plates) were transfected for 4 h with siRNA (300 pmol/well) using Oligofectamine (Invitrogen, Paisley, UK), according to the manufacturer’s instructions. mRNA, protein level and functional activity were assayed 3-13 days post-transfection, and compared with mock or control siRNA-treated cells (siControl Non-Targeting siRNA 2). Transfection efficiency was assessed independently using a positive control siRNA (siControl Lamin A/C).
Real-time PCR
Extraction of total RNA, synthesis of cDNA and real-time PCR were performed as described previously [15, 27], with NADH/cytochrome b5 reductase (Cytb5R) measured as a normalising gene. The following primer pairs and annealing temperatures were used:
Cytb5R: 5′-TATACACCCATCTCCAGCGA-3′ and 5′-CATCTCCTCATTCACGAAGC-3′; annealing temperature, 60 °C [35].
Adult Nav1.5: 5′-CATCCTCACCAACTGCGTGT-3′ and 5′-ACATTGCCCAGGTCCACAAA-3′; annealing temperature, 60 °C.
Neonatal Nav1.5: 5′-CATCCTCACCAACTGCGTGT-3′ and 5′-CCTAGTTTTTCTGATACA-3′; annealing temperature, 58 °C.
Nav1.7: 5′-TATGACCATGAATAACCCGC-3′ and 5′-TCAGGTTTCCCATGAACAGC-3′; annealing temperature, 59 °C [35].
Lamin A/C: 5′-CTGCGGCGGGTGGATGCTGAGAAC-3′ and 5′-CCACGGCTGCGCTGCGAGGTAGG-3′; annealing temperature, 67 °C.
The threshold amplification cycles were determined using the Opticon Monitor 2 software (MJ Research, Waltham, MA) and then analysed by the 2−ΔΔCt method [36].
Western blotting
Equivalent amounts of protein lysate (40 μg/well) were resolved by SDS polyacrylamide gel electrophoresis, as described previously [7, 32]. Three primary antibodies were used, at given dilutions, as follows:
Pan-VGSC antibody (1 μg/ml; Upstate);
NESO-pAb antibody (1 μl/ml) [32]; and
Anti-actinin antibody (1 μl/ml; Sigma, Dorset, UK).
The secondary antibodies were peroxidase-conjugated swine anti-rabbit for 1 and 2, and goat anti-mouse for 3 (Dako, Glostrup, Denmark). Blots were developed with ECL (Amersham, Little Chalfont, Buckinghamshire, UK) and visualised by exposure to Super RX100NF film (Fujifilm, London, UK). Densitometric analysis was performed using the Image-Pro Plus software (Media Cybernetics). Signal intensity was normalised to anti-actinin antibody as a loading control/reference, for at least three separate treatments. For each antibody, linearity of signal intensity with respect to increasing protein loading in the range 20-80 μg was ensured using a standard dilution of MDA-MB-231 protein extract.
Migration, proliferation and invasion assays
Cells (2 × 105 /ml and 7.5 × 104 /ml, respectively) were plated onto 12 μm-pore Transwell migration filters in 12-well plates (Corning, NY), or Matrigel-coated invasion chambers in 24-well plates (Becton-Dickinson) according to the manufacturers’ instructions. Cells were incubated with or without TTX (10 μM), and/or NESO-pAb (0.01-20 μg/ml) in a 1-10 % FBS chemotactic gradient. The number of cells migrating over 7 h or invading over 24 h was determined using the thiazolyl blue tetrazolium bromide (MTT) assay [17]. Results were compiled as the mean of at least four repeats. A polyclonal rabbit anti-laminin antibody (10 μl/ml; Serotec, Oxford, UK) and a rabbit IgG fraction from healthy non-immunised rabbits (1 μg/ml; Dako) were used as controls for the NESO-pAb experiments. Proliferation was determined using the MTT assay [17], in at least five different experiments.
Immunocytochemistry and confocal microscopy
Immunocytochemistry was performed as described previously [6, 32], with the following modifications: Cells were labelled with FITC-conjugated concanavalin A (Sigma) as a plasma membrane marker. The secondary antibody was Alexa567-conjugated goat anti-rabbit IgG (Dako).
Samples were viewed using a Leica DM IRBE microscope (with a X100 objective) with a confocal laser scanner (Leica TCS-NT with Ar/Kr laser). FITC and/or Alexa567 were excited with the 488 nm and 568 nm laser lines, respectively. The images (512 × 512 pixels) were obtained simultaneously from two channels using a confocal pinhole of 226.98 μm (Airy 1).
Digital image analysis
Densitometric analysis was performed using the LCS Lite software (Leica), as follows:
Cell surface VGSC expression was determined using the “freeform line profile” function drawn around the cell surface, determined by concanavalin A staining. Measurements were taken from 30 cells per condition, for three repeat treatments.
Protein distribution was determined using the “straight line profile” function drawn across the cytoplasm avoiding the nucleus, as described previously [37-39]. Signal intensity in plasma membrane region, set to cover 1.5 μm inward from the edge of concanavalin A staining, was compared with cytoplasmic signal intensity within the central 30 % of the line profile. Measurements were taken from ≥ 6 cells (randomly chosen) per condition, for three repeat treatments.
For analysing nNav1.5 staining in migrated vs. non-migrated cells, the “histogram profile” function was used to obtain measurements from the whole cell. Measurements were taken from ≥ 15 cells (chosen randomly) per condition, for three repeat treatments.
Electrophysiology
Whole-cell patch clamp recordings were performed 13 days after transfection, when patch pipettes could be successfully sealed upon cell membranes. The procedures used were mostly as described previously [40]. Voltage-activated membrane currents were recorded using an Axopatch 1D patch clamp amplifier (Axon Instruments, Wokingham, UK) compensating for series resistance by ~80 %. Currents were digitised using a Digidata 1200 interface (Axon Instruments), low-pass filtered at 5 kHz, sampled at 50 kHz, and analysed using pClamp 6 software (Axon Instruments). Linear components of leak were subtracted using the leak subtraction facility on the amplifier and/or using the pClamp software.
Two voltage-clamp protocols were used (holding potential = −100 mV), as follows:
Basic current-voltage (I-V) protocol: Cells were depolarised to test potentials within the range −70 to +70 mV in 5 mV steps. The test pulse duration was 60 ms; the interpulse duration was 2 s.
Steady-state inactivation protocol: Prepulses were in the range −130 to −10 mV (10 mV steps) for durations of 1 s. A test pulse of −10 mV was immediately applied for 80 ms; the interpulse duration was 2 s.
Recordings were obtained from a minimum of 20 cells per condition, from at least three repeat treatments. Data from individual dishes were combined to provide an overall mean and standard error (SEM). Conductance-voltage relationships and curve fitting were performed as described previously [41]. TTX dose-repsonse data were fitted using Excel (Micosoft) and Origin (OriginLab, Northampton, MA) software to a double Langmuir adsoption isotherm:
where IC50S and IC50R are the concentrations of TTX for 50 % VGSC blockage, for the TTX-sensitive and TTX-resistant components, respectively; and PTTXs is the proportion of current that is TTX-sensitive [42].
Data analysis
All quantitative data are presented as means ± standard errors, unless stated otherwise. Pairwise statistical significance was determined with Student’s t test, or Mann-Whitney rank sum test, as appropriate. Multiple comparisons were made using ANOVA followed by Newman-Keuls post-hoc analysis. Real-time PCR data were analysed using the 2−ΔΔCT method [36]. Results were considered significant at P < 0.05 (*).
RESULTS
In the present study, the specific involvement of nNav1.5 in upregulating invasive behaviour in MDA-MB-231 cells was tested by two different approaches, aiming to suppress functional nNav1.5 activity with (i) RNA interference, and (ii) NESO-pAb.
siRNA significantly reduced the nNav1.5 mRNA level
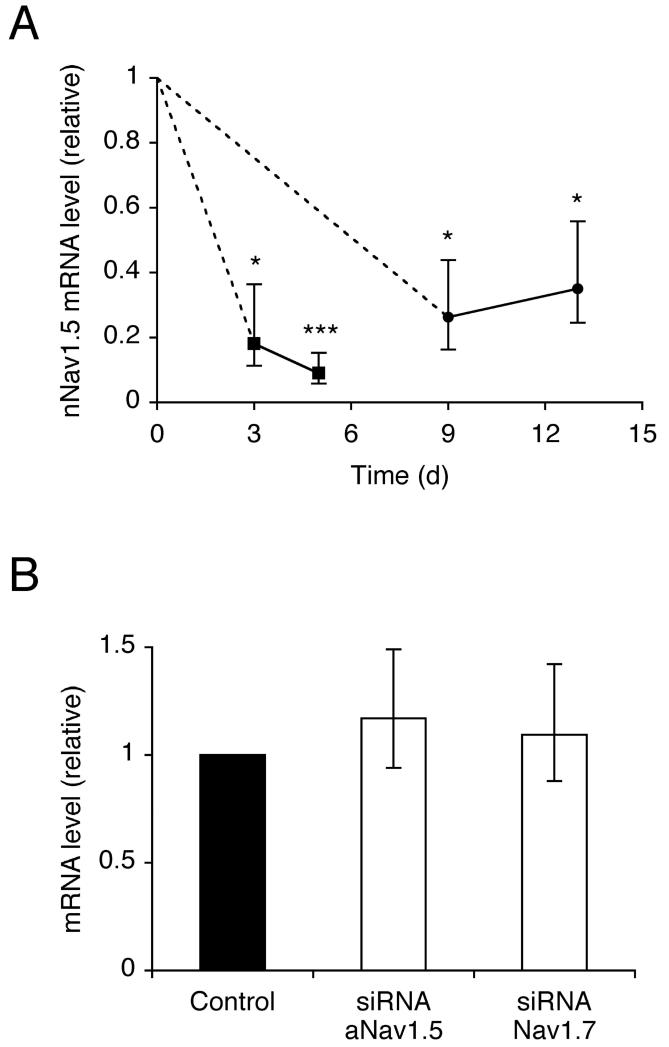
MDA-MB-231 cells were transfected with a positive control siRNA targeting lamin A/C to confirm an efficient transfection protocol. After 3 days, the siRNA significantly reduced the lamin A/C mRNA level by 91 %, compared to mock-transfected cells (P < 0.01; n = 3). Next, cells were transfected independently with one of two siRNAs (siNESO1 and 2) targeting nNav1.5. As both siRNAs produced very similar results, data hereon are presented from either. In siRNA-transfected cells, the nNav1.5 mRNA level was reduced by 82 % after 3 days (P < 0.05; n = 3), increasing to 91 % after 5 days (P < 0.001; n = 3), compared to mock-transfected cells (Figure 1A). In contrast, after 5 days, the mRNA levels of adult Nav1.5 (aNav1.5) and Nav1.7 were unaffected (P = 0.73; n = 3 for both; Figure 1B). After 9 days and 13 days, the reduction in nNav1.5 mRNA level was less marked but still significant, reaching 74 % (P < 0.05; n = 3), and 65 % (P < 0.05; n = 3), respectively (Figure 1A), compared to cells transfected with non-targeting control siRNA (siControl).
Figure 1. RNAi reduced nNav1.5, but not adult Nav1.5, or Nav1.7 mRNA level.
(A) Time course of relative nNav1.5 mRNA level after transfection with siRNA targeting nNav1.5. Squares, data compared with mock-transfected control (no siRNA). Circles, data compared with siControl non-targeting siRNA. (B) Relative level of adult Nav1.5 and Nav1.7 mRNA 5 days after transfection with siControl, or siRNA targeting nNav1.5. VGSC mRNA levels were normalised relative to Cytb5R by the 2−ΔΔCt method. Errors propagated through the 2−ΔΔCt analysis are shown. Significance: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
siRNA reduced the VGSC protein level
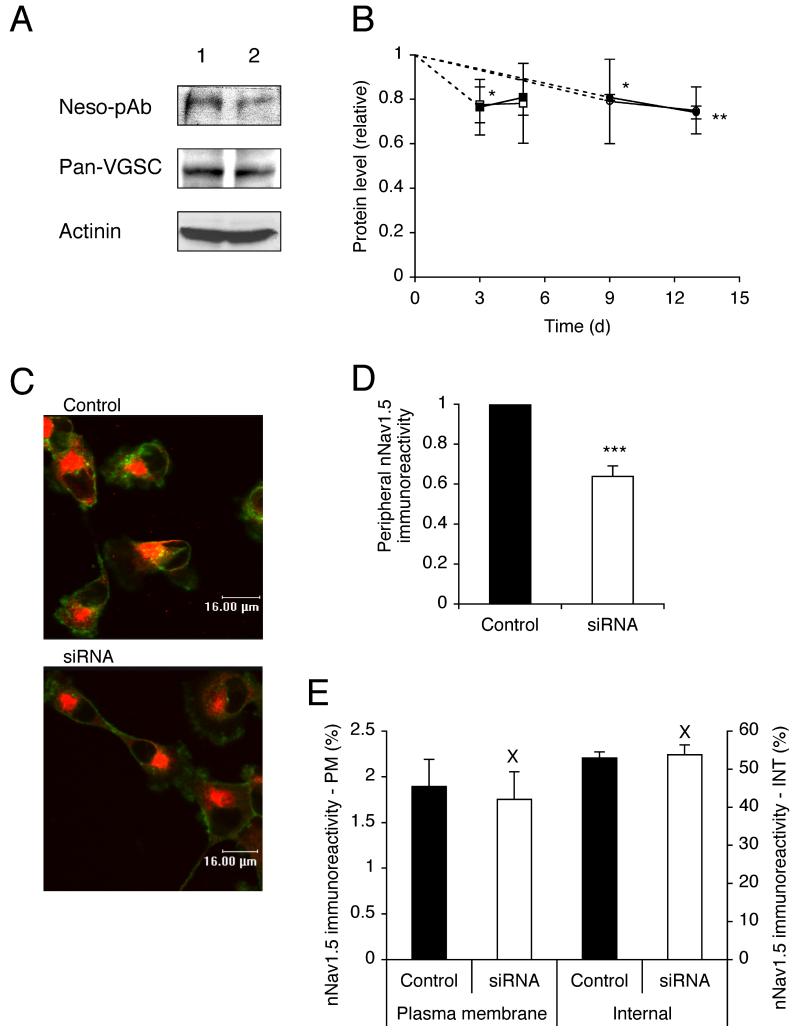
Western blot with pan-VGSC and NESO-pAb antibodies revealed smaller reductions in the total VGSC α-subunit protein level of siRNA-transfected cells. After 3 days, protein was reduced by 24 % (P = 0.23; n = 6), and 23 % (P < 0.05; n = 3), for pan-VGSC and NESO-pAb, respectively, compared to mock-transfected cells (Figure 2A, B). Similarly, after 5 days, protein was reduced by 19 % (P = 0.06; n = 4), and 22 % (P = 0.17; n = 3), for pan-VGSC and NESO-pAb, respectively. The reduction in protein level persisted such that after 9 days, there was a reduction of 19 % (P < 0.05; n = 3), and 21 % (P = 0.19; n = 3), and after 13 days, protein was reduced by 26 % (P < 0.01; n = 3), and 25 % (P = 0.07; n = 3), for pan-VGSC and NESO-pAb, respectively, compared to cells transfected with siControl (Figure 2A, B).
Figure 2. RNAi reduced nNav1.5 protein level.
(A) Western blot with 40 μg of total protein per lane from cells 13 days after treatment with (1) siControl or (2) siRNA targeting nNav1.5. Antibodies used: NESO-pAb, a pan-VGSC antibody, and an actinin antibody as a control for loading. The same membrane was stripped and re-blotted. (B) Time courses of nNav1.5 (light symbols) and total VGSC (dark symbols) protein levels (relative to actinin controls) after transfection with siRNA targeting nNav1.5. Squares, compared with mock-transfected control (no siRNA). Circles, compared with siControl non-targeting siRNA. (C) Typical confocal images of cells double-immunolabelled with NESO-pAb (red) and concanavallin A plasma membrane marker (green) 13 days after transfection with siControl or siRNA targeting nNav1.5. (D) Relative peripheral nNav1.5 protein level in cells 13 days after transfection with siControl or siRNA targeting nNav1.5. (E) nNav1.5 protein distribution (immunoreactivity) along subcellular cross-sections (% of total). Left-hand bars, 1.5 μm sections measured inward from edge of concanavalin A staining; Right-hand bars, middle 30 % of cross-section. PM, plasma membrane; INT, internal. Data are presented as mean and SEM. Significance: (X) P > 0.05, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Consistent with the Western blot data, analysis of confocal images of cells labelled with NESO-pAb revealed that after 13 days, siRNA transfection had significantly reduced the level of nNav1.5 protein at the plasma membrane by 36 % (P < 0.001; n = 90; Figure 2C, D). However, the subcellular (plasma membrane vs. internal/cytoplasmic) distribution of nNav1.5 protein along cellular cross-sections was unchanged by the siRNA treatment. The level of nNav1.5 immunoreactivity in a 1.5 μm section measured inward from the cell edge was assumed to represent the ‘plasma membrane’ fraction; the middle 30 % of the cross-section was assumed to represent the ‘internal’ cytoplasmic fraction [39]. The relative levels of nNav1.5 immunoreactivity for both fractions (expressed as a percentage of the total) were the same in control and siRNA-treated cells (P = 0.74 and P = 0.77, respectively; n = 20 for each; Figure 2E).
In conclusion, the reduction in the total VGSC protein by the siRNA treatment was much smaller and slower than the decrease in the nNav1.5 mRNA level. There was also a reduction in the level of VGSC at the plasma membrane. However, the balance of the subcellular distribution/(re)cycling of protein was not affected.
siRNA reduced VGSC functional activity
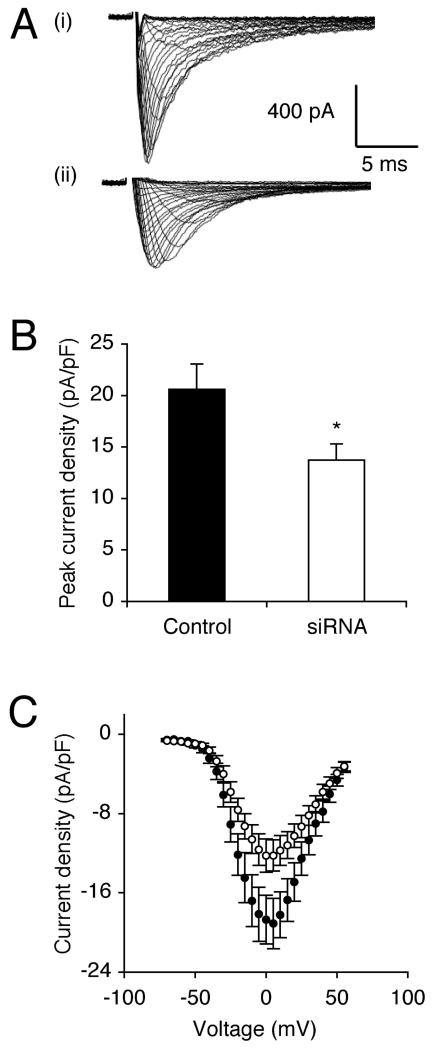
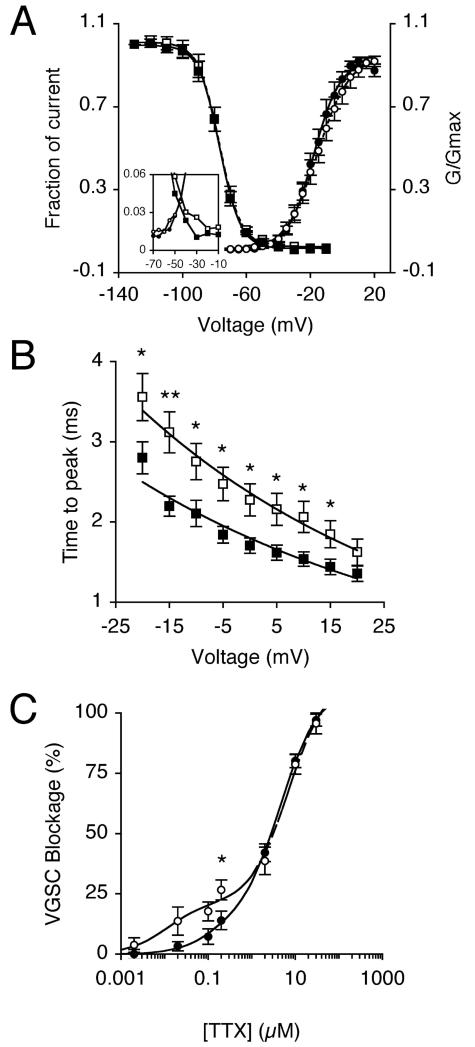
The effect of siRNA on VGSC functional activity was assessed using whole-cell patch clamp electrophysiology, at the same stage, when recordings became possible. Thus, after 13 days, transfection with siRNA significantly reduced peak VGSC current density by 33 %, from 20.6 ± 2.5 pA/pF to 13.7 ± 1.6 pA/pF, compared to siControl (P < 0.05; n > 20 cells of each; Figure 3A-C; Table I). Treatment with siRNA had no effect on activation voltage, voltage for current peak, voltage dependence of activation or steady-state inactivation (Figure 4A; Table I). There was a slight enlargement of the window current in the siRNA-treated cells (Figure 4A inset).
Figure 3. RNAi reduced peak VGSC current density after 13 days.
(A) Typical whole-cell VGSC currents elicited by 60 ms depolarising voltage pulses between −70 mV and +70 mV applied from a holding potential of −100 mV: (i) a siControl-treated cell; (ii) a cell treated with siRNA targeting nNav1.5. (B) Quantitative comparison of peak current densities recorded in siControl-treated cells, and cells treated with siRNA targeting nNav1.5. (C) Mean current-voltage relationships for siControl-treated cells (dark circles), and cells treated with siRNA targeting nNav1.5 (light circles). Data are presented as mean ± SEM. Significance: (*) P < 0.05.
Table I. Effect of siRNA on VGSC characteristics in MDA-MB-231 cells.
| Parametera | Control | siRNA | P |
|---|---|---|---|
| Ip (pA/pF) | 20.6 ± 2.5 | 13.7 ± 1.6 | 0.02* |
| Va (mV) | −52.0 ± 2.4 | −49.6 ± 1.9 | 0.44 |
| Vp (mV) | 1.3 ± 1.8 | 1.9 ± 3.0 | 0.86 |
| Activation V1/2 (mV) | −17.2 ± 1.9 | −16.0 ± 2.1 | 0.68 |
| Activation k (mV) | 7.2 ± 0.5 | 7.9 ± 0.8 | 0.44 |
| Inactivation V1/2 (mV) | −77.9 ± 1.7 | −79.2 ± 2.2 | 0.70 |
| Inactivation k (mV) | −5.3 ± 0.3 | −6.1 ± 0.5 | 0.31 |
| Tp at 0 mV (ms) | 1.7 ± 0.1 | 2.3 ± 0.2 | 0.02* |
| τf at 0 mV (ms) | 1.9 ± 0.3 | 1.9 ± 0.2 | 0.99 |
| τs at 0 mV (ms) | 10.1 ± 0.8 | 11.4 ± 1.1 | 0.33 |
| Block by 200 nM TTX (%) | 14.0 ± 3.9 | 26.7 ± 4.1 | 0.04* |
Abbreviations: Ip, peak current density; Va, activation voltage; Vp, voltage at current peak; V1/2, half-(in)activation voltage; k, slope factor; Tp, time to peak; τf/s, fast/slow time constant of inactivation. Data expressed as mean ± SEM.
Figure 4. RNAi altered VGSC kinetics and TTX sensitivity after 13 days.
(A) Mean availability-voltage (squares) and relative conductance (G/Gmax)-voltage relationships (circles) for siControl-treated cells (dark symbols) and cells treated with siRNA targeting nNav1.5 (light symbols). Control (solid lines) and siRNA data (dotted lines) are fitted with Boltzmann functions. The inset magnifies the voltage range (‘window’) in which current is activated and not fully inactivated. (B) Dependence of time to peak on membrane voltage for siControl-treated cells (dark squares), and cells treated with siRNA targeting nNav1.5 (light squares). Control (solid line) and siRNA data (dashed line) are fitted with single exponential functions. (C) Reduction of VGSC current by TTX for control-treated cells (dark circles), and cells treated with siRNA targeting nNav1.5 (light circles). Control (solid line) and siRNA data (dashed line) are fitted to double Langmuir adsoption isotherms. Data are presented as mean ± SEM. Significance: (*) P < 0.05, (**) P< 0.01.
The time to peak of VGSC current was significantly slower in the voltage range −20 to 15 mV in cells transfected with siRNA (Figure 4B). However, fast (τf) and slow (τs) time constants of inactivation at 0 mV, derived from fitting current inactivations to a double exponential function, were unaffected by the siRNA treatment (Table I). Thus, the siRNA treatment slowed the activation kinetics, consistent with a possible change in the functional VGSC isoform expression profile.
The dose-dependent blockage of VGSC current in MDA-MB-231 cells by TTX was best fitted to a double Langmuir adsorption isotherm, with TTX-sensitive, and TTX-resistant components. The sensitivity of VGSC currents to low concentrations of TTX (< 2 μM) was increased in cells transfected with siRNA, compared to siControl-transfected cells (Figure 4C). The siRNA significantly increased sensitivity to 200 nM TTX, from 14.0 ± 3.9 % to 26.7 ± 4.1 % (P < 0.05; n = 6 cells of each; Table I). We concluded that by downregulating the level of TTX-resistant nNav1.5 with siRNA, the relative proportion of the other TTX-sensitive VGSC isoform(s) was increased. The latter was likely to involve Nav1.7 [6]
Migrated cells expressed more VGSC protein than non-migrated cells
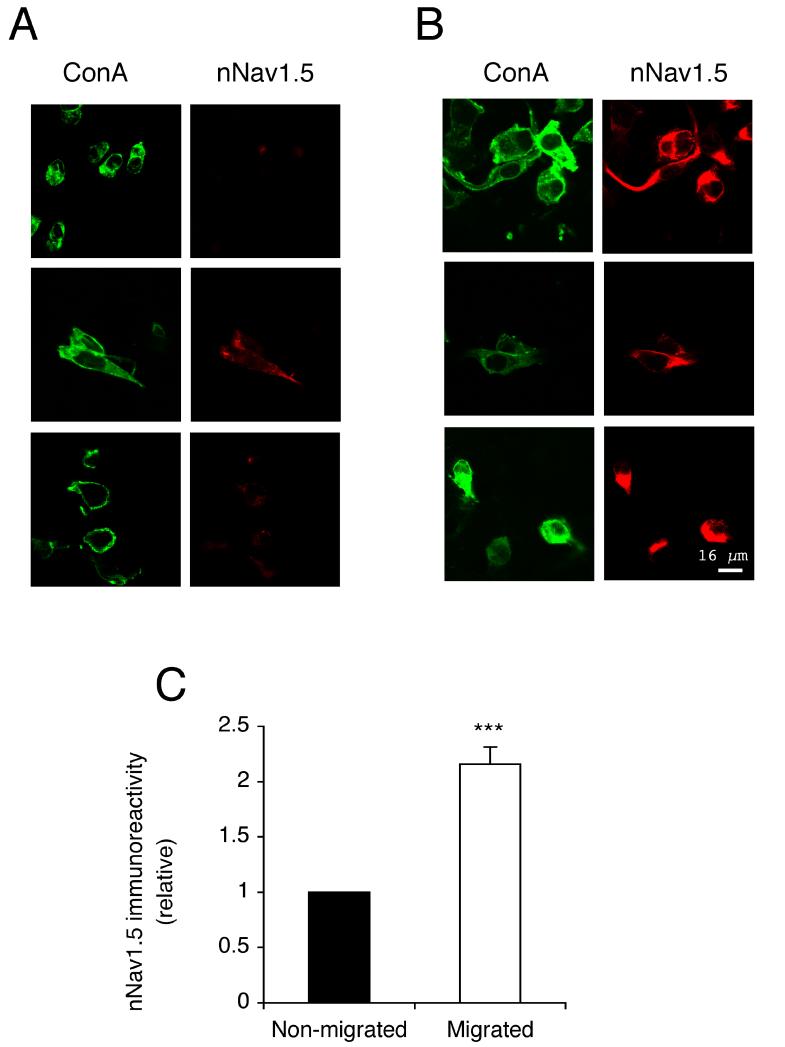
Confocal immunocytochemistry with NESO-pAb was used to compare the nNav1.5 protein expression in plasma membranes of cells that did/did not migrate through the Transwell filters (Figure 5A, B). Cells that migrated through the filter had 2.2-fold more plasma membrane nNav1.5 protein immunoreactivity compared with cells that did not migrate (P < 0.001; n > 45; Figure 5C).
Figure 5. Migrated cells had more nNav1.5 protein at the plasma membrane than non-migrated cells after 16 h.
(A and B) Typical confocal images of non-migrated and migrated cells in Transwell assays, respectively. Cells were non-permeabilized and double-immunolabelled with NESO-pAb (red) and concanavalin A plasma membrane marker (green). (C) Relative nNav1.5 protein level in migrated cells, normalised with respect to the level in non-migrated cells. Significance: (***) P < 0.001.
siRNA and NESO-pAb both suppressed VGSC-dependent migration and invasion
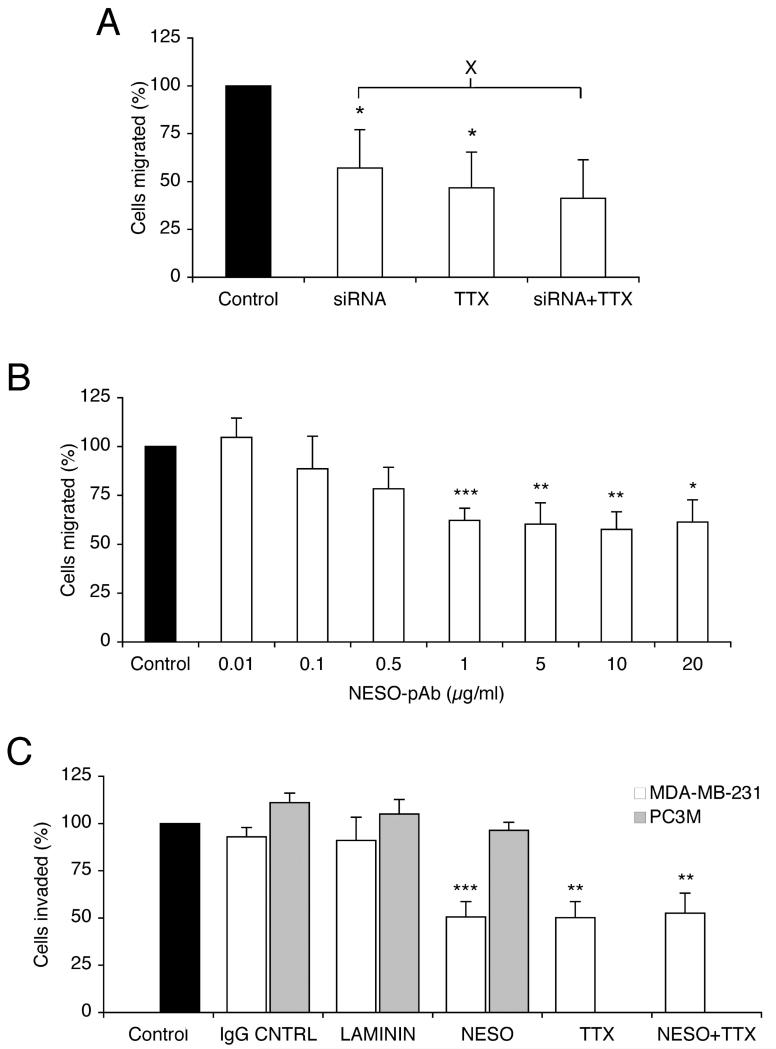
These assays were performed also after 13 days, when the status of the VGSC signalling was known. Thus, transfection with siRNA significantly reduced the number of cells migrating through 12 μm-pore Transwell filters filter by 43 % compared to siControl-transfected cells (P < 0.05; n = 5; Figure 6A). When TTX was applied while the siControl-transfected cells were migrating, their migration was reduced by 53 % (P < 0.05; n = 4), as reported earlier (FRASER). Importantly, TTX applied to siRNA-transfected cells during the assay had no further effect on their migration (P = 0.78; n = 4). Thus, the VGSC-sensitive component of migration was completely suppressed by siRNA targeting nNav1.5.
Figure 6. siRNA and NESO-pAb reduced VGSC-dependent metastatic cell behaviour.
(A) Relative number of cells migrating through a Transwell chamber over 7 h, 13 days after transfection with siControl, or siRNA targeting nNav1.5. Cells were treated with TTX (10 μM), or untreated during the migration assays. (B) Relative number of cells migrating through a Transwell chamber over 7 h, treated with 0.01-20 μg/ml NESO-pAb during the assay. (C) Relative number of cells invading through Matrigel chamber over 24 h. MDA-MB-231 (white bars) and PC-3M (grey bars) cells were treated with IgG (1 μg/ml), anti-laminin (1:100), NESO-pAb (1 μg/ml) with/without TTX (10 μM) during the invasion assay. In both (B) and (C), data shown are with respect to corresponding control, untreated cells (treated as 100 %). Data are presented as mean and SEM. Significance: (X) P > 0.05, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, relative to “control”, unless indicated.
NESO-pAb, applied during Transwell assays, reduced migration in a dose-dependent manner, reaching significance, 38 % reduction at 1 μg/ml (P < 0.001; n = 7; Figure 6B). NESO-pAb (1 μg/ml) also reduced Matrigel invasion of MDA-MB-231 cells by 49 % compared to control (P < 0.001; n = 7), but had no effect on human metastatic PCa PC-3M cells, in which functional Nav1.7 is dominant [35] (J. K. J. Diss, unpublished observation). IgG control and an unrelated anti-laminin antibody had no effect on invasiveness of MDA-MB-231 or PC-3M cells (Figure 6C). TTX (10 μM) reduced invasion of MDA-MB-231 cells by 50 % (P = 0.001; n = 7). Importantly, co-application of NESO-pAb (1 μg/ml) and TTX (10 μM) also reduced cell invasion by 47 % (P < 0.01; n = 7), but had no further effect compared to either NESO-pAb or TTX alone (P = 0.87 and 0.58, respectively; n = 7 for each; Figure 6C). None of the antibody treatments had any effect on proliferation of MDA-MB-231 or PC-3M cells (data not shown).
DISCUSSION
The main findings of this study are as follows: (i) siRNA reduced nNav1.5 mRNA expression in MDA-MB-231 cells rapidly and almost completely. (ii) siRNA-induced protein-level reduction was slower and smaller, leading to decreased VGSC functional activity and increased sensitivity to low concentrations of TTX. (iii) Targeting nNav1.5 with siRNA eliminated the VGSC-dependent component of migration. (iv) Migrated cells expressed more plasma membrane nNav1.5 protein than non-migrated cells. (v) NESO-pAb also reduced migration and eliminated VGSC-dependent invasion. The overall conclusion from this study is that it is indeed nNav1.5 that is primarily responsible for the VGSC-dependent potentiation of invasive behaviour in the MDA-MB-231 BCa cell line. These results agree with, and extend, our previous findings in vitro and in vivo [6].
Effects of RNAi on nNav1.5 mRNA and protein expression
In this study, the unique DI:S3 5′-splice region at exon 6 of nNav1.5 was targeted with siRNA. Although RNAi has previously been used to target ion channels [43-45], including VGSC α-subunit [46, 47], this is the first report where siRNA has been used to specifically target one particular splice variant of a VGSC isoform. There are several potential technical problems associated with the RNAi technique, including off-target effects and poor mRNA/protein-level downregulation [48-50]. In this study, the effects of RNAi were monitored over a 13-day time-course at a hierarchy of levels: mRNA, protein (level and distribution) and functioning; basically the same effects were observed using the two different siRNAs. Although this duration would seem longer than most siRNA experiments, it was necessary (i) to perform the patch clamp recordings to ascertain the level of VGSC signalling, and (ii) to enable correlation of the siRNA effects at the various levels in a systematic approach. The siRNA caused ~90 % reduction of nNav1.5 mRNA after 5 days, beyond which, the level started to partially recover, reaching ~70 % reduction by 13 days. A similar pattern of mRNA silencing has been reported previously [51]. Nevertheless, both adult Nav1.5, and Nav1.7 mRNA levels were not affected by this treatment, confirming the specificity of the siRNA for nNav1.5.
The reduction of nNav1.5 protein level, detected by Western blot using NESO-pAb, was much smaller, becoming significant only after 13 days. A pan-VGSC antibody gave similar results. Confocal immunocytochemistry with NESO-pAb [32] showed that nNav1.5 was present at the cell surface, in agreement with previous observations [6], and that the siRNA reduced the level of nNav1.5 protein expressed at the plasma membrane, without changing the balance of its (re)cycling. At present, the dynamics of VGSC mRNA/protein expression in cancer cells are not known, and this could be compounded by activity-dependent (auto)regulation of VGSCs [39]. The apparent disparity between the time course and extent of target mRNA and protein-level reduction may, in part, be due to large stores of intracellular protein and/or slow turnover rates [52]. Such disparities have been seen before in siRNA experiments. For example, siRNA targeting vitamin K epoxide reductase gene reduced mRNA level after 3 days, but protein was not reduced until after 11 days [51]. Also, in human BCa cells, siRNA-induced protein downregulation persisted for at least 7 days [53]. Given that VGSC protein half-life is 18-26 h [54, 55] and that there was intense intracellular immunoreactivity with NESO-pAb, the slower and smaller reduction in nNav1.5 protein could be due to it being present in large quantity in internal stores and cycling to the plasma membrane persisting.
Involvement of nNav1.5 in the functional VGSC activity of MDA-MD-231 cells
The siRNA treatment reduced the VGSC functional activity in MDA-MB-231 cells by 33 %, in agreement with the molecular data, suggesting a decrease in the availability of VGSC protein at the plasma membrane. Importantly, the siRNA was found also to increase the sensitivity of cells to TTX concentrations of < 2 μM. This implies that the proportion of TTX-sensitive functional VGSCs was increased after the siRNA treatment. This TTX-sensitive component is likely to be Nav1.7, which we have previously shown to constitute ~20 % of the VGSC α-subunit mRNA in MDA-MB-231 cells [6]. This is consistent with the dose-response data (Figure 4C) showing an increase in the level of the TTX-sensitive component up to 20-25 % of total current. On the other hand, the slower time to peak recorded in siRNA-treated cells is unlikely to be due to an increased proportion of Nav1.7, because TTX-sensitive VGSCs tend to have faster kinetics than TTX-resistant VGSCs [56, 57]. This raises the possibility that downregulation of nNav1.5 may have induced secondary, knock-on effects, e.g. alteration of the relative profile of β-subunits, which can have a marked influence on VGSC functional properties [58-60]. Further work is required to determine the functional consequences of the β-subunits and Nav1.7 expression in MDA-MB-231 cells.
A role for nNav1.5 in potentiating metastatic activity
MDA-MB-231 cells were shown to express significantly more nNav1.5 protein at the plasma membrane than weakly metastatic MCF-7 cells [6]. In addition, migrated MDA-MB-231 cells were found to express more plasma membrane nNav1.5 protein than non-migrated cells. Together, these data suggest a correlation between surface expression of nNav1.5 and invasiveness in BCa, as in vivo [6].
In MDA-MB-231 cells, TTX (10 μM) treatment has been shown previously to suppress a variety of in vitro behaviours associated with the metastatic cascade, including galvanotaxis, endocytic membrane activity, migration and invasion, consistent with the involvement of TTX-resistant VGSC activity [6, 22]. The siRNA treatment reduced migration by 43 %, and this was not further suppressed by addition of TTX during the migration assay. Thus, the siRNA almost completely eliminated the VGSC-dependent component of the cells’ invasiveness. Furthermore, NESO-pAb, which can directly inhibit nNav1.5 functional activity [32], was found to reduce both the migration and invasion of MDA-MB-231 cells to a similar extent, without affecting their proliferation, when applied during the assays. Importantly, co-application of TTX did not further reduce the cells’ invasiveness below that of NESO-pAb alone. In addition, NESO-pAb had no effect on the invasion of PC-3M PCa cells, which, like the parent PC-3 cells (34), express primarily Nav1.7 [35] (J. K. J. Diss, unpublished observations), confirming further the specificity of NESO-pAb for nNav1.5. Furthermore, an anti-laminin antibody had no effect on the migration of MDA-MB-231 cells, consistent with the effect of NESO-pAb being specifically upon the nNav1.5 channel. Together, these data imply that nNav1.5 alone is primarily responsible for the VGSC-dependent potentiation of invasiveness in MDA-MB-231 cells. In both approaches, one interesting characteristic was apparent: blocking VGSC activity by only ~30 % was sufficient to eliminate its potentiating effect upon migration or invasion. This would imply that the relationship between VGSC activity and migration/invasion enhancement is rather steep.
Clinical implications
This work has important clinical implications, whereby reduction of nNav1.5 activity, e.g. generally with non-cytotoxic VGSC-blocking drugs [28], could result in suppression of metastatic BCa. More specifically, given that nNav1.5 expression is an oncofetal phenomenon [61] and is not present in normal Nav1.5-expressing adult tissues [6, 32], it appears to have the hallmarks of an ideal target for specific anticancer treatment utilising siRNA [62-64], and/or antibody [65].
ACKNOWLEDGEMENTS
We would like to thank Dr. J. P. Alao and Prof R. C. Coombes for advice on design of initial RNAi experiments.
REFERENCES
- 1.Hille B. Ionic channels of excitable membranes. 2nd edn. Sinauer Associates Inc.; Sunderland (Massachusetts): 1992. [Google Scholar]
- 2.Jurkat-Rott K, Lehmann-Horn F. Human muscle voltage-gated ion channels and hereditary disease. Curr Opin Pharmacol. 2001;1(3):280–287. doi: 10.1016/s1471-4892(01)00050-9. [DOI] [PubMed] [Google Scholar]
- 3.Viswanathan PC, Balser JR. Inherited sodium channelopathies: a continuum of channel dysfunction. Trends Cardiovasc Med. 2004;14(1):28–35. doi: 10.1016/j.tcm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Diss JK, Fraser SP, Djamgoz MB. Voltage-gated Na+ channels: multiplicity of expression, plasticity, functional implications and pathophysiological aspects. Eur Biophys J. 2004;33(3):180–193. doi: 10.1007/s00249-004-0389-0. [DOI] [PubMed] [Google Scholar]
- 5.Fraser SP, Koyuturk M, Djamgoz MB. Ion channel activity and cancer cell proliferation: A short review with particular reference to prostate cancer. In: Rouzaire-Dubois B, Benoit E, Dubois JM, editors. Ion Channels and Physiopathologies of Nerve Conduction and Cell Proliferation. 1st edn. Research Signpost; 2002. [Google Scholar]
- 6.Fraser SP, Diss JK, Chioni AM, et al. Voltage-gated sodium channel expression and potentiation of human breast cancer metastasis. Clin Cancer Res. 2005;11:5381–5389. doi: 10.1158/1078-0432.CCR-05-0327. [DOI] [PubMed] [Google Scholar]
- 7.Laniado ME, Lalani EN, Fraser SP, et al. Expression and functional analysis of voltage-activated Na(+) channels in human prostate cancer cell lines and their contribution to invasion in vitro. Am J Pathol. 1997;150(4):1213–1221. [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser SP, Diss JK, Lloyd LJ, et al. T-lymphocyte invasiveness: control by voltage-gated Na(+) channel activity. FEBS Lett. 2004;569(1-3):191–194. doi: 10.1016/j.febslet.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 9.Blandino JK, Viglione MP, Bradley WA, et al. Voltage-dependent sodium channels in human small-cell lung cancer cells: role in action potentials and inhibition by Lambert-Eaton syndrome IgG. J Membr Biol. 1995;143(2):153–163. doi: 10.1007/BF00234661. [DOI] [PubMed] [Google Scholar]
- 10.Onganer PU, Djamgoz MB. Small-cell lung cancer (human): potentiation of endocytic membrane activity by voltage-gated Na(+) channel expression in vitro. J Membr Biol. 2005;204(2):67–75. doi: 10.1007/s00232-005-0747-6. [DOI] [PubMed] [Google Scholar]
- 11.Ou SW, Kameyama A, Hao LY, et al. Tetrodotoxin-resistant Na+ channels in human neuroblastoma cells are encoded by new variants of Nav1.5/SCN5A. Eur J Neurosci. 2005;22(4):793–801. doi: 10.1111/j.1460-9568.2005.04280.x. [DOI] [PubMed] [Google Scholar]
- 12.Allen DH, Lepple-Wienhues A, Cahalan MD. Ion channel phenotype of melanoma cell lines. J Membr Biol. 1997;155(1):27–34. doi: 10.1007/s002329900155. [DOI] [PubMed] [Google Scholar]
- 13.Abdul M, Hoosein N. Voltage-gated sodium ion channels in prostate cancer: expression and activity. Anticancer Res. 2002;22(3):1727–1730. [PubMed] [Google Scholar]
- 14.Onganer PU, Seckl MJ, Djamgoz MB. Neuronal characteristics of small-cell lung cancer. Br J Cancer. 2005;93:1197–1201. doi: 10.1038/sj.bjc.6602857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diss JK, Stewart D, Pani F, et al. A potential novel marker for human prostate cancer: voltage-gated sodium channel expression in vivo. Prostate Cancer Prostatic Dis. 2005;8(3):266–273. doi: 10.1038/sj.pcan.4500796. [DOI] [PubMed] [Google Scholar]
- 16.Fidler IJ. Timeline: The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 17.Grimes JA, Fraser SP, Stephens GJ, et al. Differential expression of voltage-activated Na(+) currents in two prostatic tumour cell lines: contribution to invasiveness in vitro. FEBS Lett. 1995;369(2-3):290–294. doi: 10.1016/0014-5793(95)00772-2. [DOI] [PubMed] [Google Scholar]
- 18.Djamgoz MBA, Mycielska M, Madeja Z, et al. Directional movement of rat prostate cancer cells in direct-current electric field: involvement of voltage gated Na+ channel activity. J Cell Sci. 2001;114(Pt 14):2697–2705. doi: 10.1242/jcs.114.14.2697. [DOI] [PubMed] [Google Scholar]
- 19.Mycielska ME, Fraser SP, Szatkowski M, et al. Contribution of functional voltage-gated Na(+) channel expression to cell behaviors involved in the metastatic cascade in rat prostate cancer: II. Secretory membrane activity. J Cell Physiol. 2003;195(3):461–469. doi: 10.1002/jcp.10265. [DOI] [PubMed] [Google Scholar]
- 20.Bennett ES, Smith BA, Harper JM. Voltage-gated Na(+) channels confer invasive properties on human prostate cancer cells. Pflugers Arch. 2004;447(6):908–914. doi: 10.1007/s00424-003-1205-x. [DOI] [PubMed] [Google Scholar]
- 21.Smith P, Rhodes NP, Shortland AP, et al. Sodium channel protein expression enhances the invasiveness of rat and human prostate cancer cells. FEBS Lett. 1998;423(1):19–24. doi: 10.1016/s0014-5793(98)00050-7. [DOI] [PubMed] [Google Scholar]
- 22.Roger S, Besson P, Le Guennec JY. Involvement of a novel fast inward sodium current in the invasion capacity of a breast cancer cell line. Biochim Biophys Acta. 2003;1616(2):107–111. doi: 10.1016/j.bbamem.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Fraser SP, Ding Y, Liu A, et al. Tetrodotoxin suppresses morphological enhancement of the metastatic MAT-LyLu rat prostate cancer cell line. Cell Tissue Res. 1999;295(3):505–512. doi: 10.1007/s004410051256. [DOI] [PubMed] [Google Scholar]
- 24.Krasowska M, Grzywna ZJ, Mycielska ME, et al. Patterning of endocytic vesicles and its control by voltage-gated Na(+) channel activity in rat prostate cancer cells: fractal analyses. Eur Biophys J. 2004;33(6):535–542. doi: 10.1007/s00249-004-0394-3. [DOI] [PubMed] [Google Scholar]
- 25.Fraser SP, Salvador V, Manning EA, et al. Contribution of functional voltage-gated Na(+) channel expression to cell behaviors involved in the metastatic cascade in rat prostate cancer: I. lateral motility. J Cell Physiol. 2003;195(3):479–487. doi: 10.1002/jcp.10312. [DOI] [PubMed] [Google Scholar]
- 26.Palmer CP, Mycielska M, Burcu H, et al. A micro-pressure system for measuring cell adhesion: application to cancer cell lines of differing metastatic potential and voltage-gated Na+ channel expression. 2006 doi: 10.1007/s00249-007-0219-2. Manuscript submitted. [DOI] [PubMed] [Google Scholar]
- 27.Mycielska ME, Palmer CP, Brackenbury WJ, et al. Expression of Na+-dependent citrate transport in a strongly metastatic human prostate cancer PC-3M cell line: regulation by voltage-gated Na+ channel activity. J Physiol. 2005;563(Pt 2):393–408. doi: 10.1113/jphysiol.2004.079491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson JD, Hansen TP, Lenkowski PW, et al. Voltage-gated sodium channel blockers as cytostatic inhibitors of the androgen-independent prostate cancer cell line PC-3. Mol Cancer Ther. 2003;2(11):1149–1154. [PubMed] [Google Scholar]
- 29.Abdul M, Hoosein N. Inhibition by anticonvulsants of prostate-specific antigen and interleukin-6 secretion by human prostate cancer cells. Anticancer Res. 2001;21(3B):2045–2048. [PubMed] [Google Scholar]
- 30.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26(1):13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 31.Copley RR. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 2004;20(4):171–176. doi: 10.1016/j.tig.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Chioni AM, Fraser SP, Pani F, et al. A novel polyclonal antibody specific for the Na(v)1.5 voltage-gated Na(+) channel ‘neonatal’ splice form. J Neurosci Methods. 2005;147(2):88–98. doi: 10.1016/j.jneumeth.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Brackenbury WJ, Chioni AM, Djamgoz MB. Further evidence for the neonatal splice variant of Nav1.5 potentiating in vitro metastatic behaviour of MDA-MB-231 human breast cancer cells: application of RNAi and a novel antibody. J Physiol. 2005;568P:PC17. [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Diss JK, Archer SN, Hirano J, et al. Expression profiles of voltage-gated Na(+) channel alpha-subunit genes in rat and human prostate cancer cell lines. Prostate. 2001;48(3):165–178. doi: 10.1002/pros.1095. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Okuse K, Malik-Hall M, Baker MD, et al. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature. 2002;417(6889):653–656. doi: 10.1038/nature00781. [DOI] [PubMed] [Google Scholar]
- 38.Shah BS, Rush AM, Liu S, et al. Contactin associates with sodium channel Nav1.3 in native tissues and increases channel density at the cell surface. J Neurosci. 2004;24(33):7387–7399. doi: 10.1523/JNEUROSCI.0322-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brackenbury WJ, Djamgoz MB. Activity-dependent regulation of voltage-gated Na(+) channel expression in Mat-LyLu rat prostate cancer cell line. J Physiol. 2006 doi: 10.1113/jphysiol.2006.106906. DOI: 10.1113/jphysiol.2006.106906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimes JA, Djamgoz MB. Electrophysiological characterization of voltage-gated Na(+) current expressed in the highly metastatic Mat-LyLu cell line of rat prostate cancer. J Cell Physiol. 1998;175(1):50–58. doi: 10.1002/(SICI)1097-4652(199804)175:1<50::AID-JCP6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 41.Ding Y, Djamgoz MB. Serum concentration modifies amplitude and kinetics of voltage-gated Na+ current in the Mat-LyLu cell line of rat prostate cancer. Int J Biochem Cell Biol. 2004;36(7):1249–1260. doi: 10.1016/j.biocel.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Haufe V, Camacho JA, Dumaine R, et al. Expression pattern of neuronal and skeletal muscle voltage-gated Na+ channels in the developing mouse heart. J Physiol. 2005;564(Pt 3):683–696. doi: 10.1113/jphysiol.2004.079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurney AM, Hunter E. The use of small interfering RNA to elucidate the activity and function of ion channel genes in an intact tissue. J Pharmacol Toxicol Methods. 2005;51(3):253–262. doi: 10.1016/j.vascn.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Rieckhof GE, Yoshihara M, Guan Z, et al. Presynaptic N-type calcium channels regulate synaptic growth. J Biol Chem. 2003;278(42):41099–41108. doi: 10.1074/jbc.M306417200. [DOI] [PubMed] [Google Scholar]
- 45.McCrossan ZA, Lewis A, Panaghie G, et al. MinK-related peptide 2 modulates Kv2.1 and Kv3.1 potassium channels in mammalian brain. J Neurosci. 2003;23(22):8077–8091. doi: 10.1523/JNEUROSCI.23-22-08077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikami M, Yang J. Short hairpin RNA-mediated selective knockdown of NaV1.8 tetrodotoxin-resistant voltage-gated sodium channel in dorsal root ganglion neurons. Anesthesiology. 2005;103(4):828–836. doi: 10.1097/00000542-200510000-00022. [DOI] [PubMed] [Google Scholar]
- 47.Xu X, Shrager P. Dependence of axon initial segment formation on Na+ channel expression. J Neurosci Res. 2005;79(4):428–441. doi: 10.1002/jnr.20378. [DOI] [PubMed] [Google Scholar]
- 48.Whither RNAi? Nat Cell Biol. 2003;5(6):489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 49.Couzin J. Molecular biology. RNAi shows cracks in its armor. Science. 2004;306(5699):1124–1125. doi: 10.1126/science.306.5699.1124. [DOI] [PubMed] [Google Scholar]
- 50.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 51.Li T, Chang CY, Jin DY, et al. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427(6974):541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 52.Zhang W, Lane RD, Mellgren RL. The major calpain isozymes are long-lived proteins. Design of an antisense strategy for calpain depletion in cultured cells. J Biol Chem. 1996;271(31):18825–18830. doi: 10.1074/jbc.271.31.18825. [DOI] [PubMed] [Google Scholar]
- 53.Alao JP, Lam EW, Ali S, et al. Histone deacetylase inhibitor trichostatin A represses estrogen receptor alpha-dependent transcription and promotes proteasomal degradation of cyclin D1 in human breast carcinoma cell lines. Clin Cancer Res. 2004;10(23):8094–8104. doi: 10.1158/1078-0432.CCR-04-1023. [DOI] [PubMed] [Google Scholar]
- 54.Waechter CJ, Schmidt JW, Catterall WA. Glycosylation is required for maintenance of functional sodium channels in neuroblastoma cells. J Biol Chem. 1983;258(8):5117–5123. [PubMed] [Google Scholar]
- 55.Sherman SJ, Chrivia J, Catterall WA. Cyclic adenosine 3′:5′-monophosphate and cytosolic calcium exert opposing effects on biosynthesis of tetrodotoxin-sensitive sodium channels in rat muscle cells. J Neurosci. 1985;5(6):1570–1576. doi: 10.1523/JNEUROSCI.05-06-01570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rola R, Szulczyk B, Szulczyk P, et al. Expression and kinetic properties of Na(+) currents in rat cardiac dorsal root ganglion neurons. Brain Res. 2002;947(1):67–77. doi: 10.1016/s0006-8993(02)02908-6. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida S. Tetrodotoxin-resistant sodium channels. Cell Mol Neurobiol. 1994;14(3):227–244. doi: 10.1007/BF02088322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fahmi AI, Patel M, Stevens EB, et al. The sodium channel beta-subunit SCN3b modulates the kinetics of SCN5a and is expressed heterogeneously in sheep heart. J Physiol. 2001;537(Pt 3):693–700. doi: 10.1111/j.1469-7793.2001.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao YF, Wright SN, Wang GK, et al. Coexpression with beta(1)-subunit modifies the kinetics and fatty acid block of hH1(alpha) Na(+) channels. Am J Physiol Heart Circ Physiol. 2000;279(1):H35–46. doi: 10.1152/ajpheart.2000.279.1.H35. [DOI] [PubMed] [Google Scholar]
- 60.Yu EJ, Ko SH, Lenkowski PW, et al. Distinct domains of the sodium channel beta3 subunit modulate channel gating kinetics and sub-cellular location. Biochem J. 2005;392(Pt 3):519–526. doi: 10.1042/BJ20050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20(56):8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- 62.Rye PD, Stigbrand T. Interfering with cancer: a brief outline of advances in RNA interference in oncology. Tumour Biol. 2004;25(5-6):329–336. doi: 10.1159/000081403. [DOI] [PubMed] [Google Scholar]
- 63.Uprichard SL. The therapeutic potential of RNA interference. FEBS Lett. 2005;579(26):5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karagiannis TC, El-Osta A. RNA interference and potential therapeutic applications of short interfering RNAs. Cancer Gene Ther. 2005;12(10):787–795. doi: 10.1038/sj.cgt.7700857. [DOI] [PubMed] [Google Scholar]
- 65.Willems A, Gauger K, Henrichs C, et al. Antibody therapy for breast cancer. Anticancer Res. 2005;25(3A):1483–1489. [PubMed] [Google Scholar]