Abstract
During the past 20 years, autophagy signaling has entered the main stage of the cell biological theater. Autophagy represents an intracellular degradation process that is involved in both the bulk recycling of cytoplasmic components and the selective removal of organelles, protein aggregates, or intracellular pathogens. The understanding of autophagy has been greatly facilitated by the characterization of the molecular machinery governing this process. In yeast, initiation of autophagy is controlled by the Atg1 kinase complex, which is composed of the Ser/Thr kinase Atg1, the adaptor protein Atg13, and the ternary complex of Atg17-Atg31-Atg29. In vertebrates, the orthologous ULK1 kinase complex contains the Ser/Thr kinase ULK1 and the accessory proteins ATG13, RB1CC1, and ATG101. Among these components, Atg1/ULK1 have gained major attention in the past, i.e., for the identification of upstream regulatory kinases, the characterization of downstream substrates controlling the autophagic flux, or as a druggable target for the modulation of autophagy. However, accumulating data indicate that the function of Atg13/ATG13 has been likely underestimated so far. In addition to ensuring proper Atg1/ULK1 recruitment and activity, this adaptor molecule has been implicated in ULK1-independent autophagy processes. Furthermore, recent data have identified additional binding partners of Atg13/ATG13 besides the components of the Atg1/ULK1 complex, e.g., Atg8 family proteins or acidic phospholipids. Therefore, in this review we will center the spotlight on Atg13/ATG13 and summarize the role that Atg13/ATG13 assumes in the autophagy stage play.
Keywords: ATG101, ATG13, RB1CC1, ULK1, autophagy
Atg13: An Essential Autophagy-Related Protein in Yeast
The vesicular trafficking pathway for the lysosomal degradation of cellular components was discovered in the early 1960s by Christian de Duve, who also introduced the terms “autophagy” and “autophagic vacuole.”1 It took 3 more decades until the first regulating genes could be identified in Saccharomyces cerevisiae.2 Another decade later, Daniel J Klionsky and colleagues integrated the various autophagy-defective yeast strains that had been isolated until then under a single unified gene nomenclature, termed autophagy‐related (ATG).3 The yeast ATG gene family has been growing ever since, with 38 members so far.4 Many of these genes additionally regulate the cytoplasm-to-vacuole targeting (Cvt) pathway, an autophagy-related process that is responsible for the selective delivery of acid hydrolases to the yeast vacuole. Some Atgs, like Atg11, Atg20, and Snx4/Atg24, are primarily involved in the Cvt pathway, while others, such as Atg17, Atg29, and Atg31, are autophagy-specific. Still others, such as Atg32 and Atg36, are essential for the selective degradation of mitochondria (mitophagy) or peroxisomes (pexophagy).
Upon starvation, the respective Atg proteins are recruited in a concerted manner to the single site of autophagosome formation, the so-called phagophore assembly site (PAS), which is located in close proximity to the yeast vacuole.5 Several observations by various groups have led to the following preliminary picture of PAS organization: The serine/threonine kinase Atg1 interacts with different sets of Atg proteins—with Atg13, Atg17, Atg29, and Atg316-9 when regulating autophagy and with Atg11, Atg13, Atg20, and Snx4/Atg2410-12 when regulating the Cvt pathway (reviewed in refs. 13 and 14).
According to these observations, Atg13 is involved in both pathways, whereas the constitutively assembled ternary complex of Atg17, Atg29, and Atg31 is exclusively involved in autophagy. The Atg17-Atg31-Atg29 complex localizes at the PAS and functions as a landing platform for further downstream factors.15-17 Upon starvation, Atg1 is one of the first proteins recruited to the PAS. The interaction between Atg1 and the Atg17-Atg31-Atg29 complex is mediated by the bridging molecule Atg13, which directly interacts with both Atg1 and Atg17.18,19 The following enhancement of Atg1 kinase activity by Atg13 and Atg17 is mandatory for the initiation of proper autophagosome formation, although Atg1 kinase activity seems dispensable for the recruitment of further downstream Atg proteins to the site of autophagosome generation.16,18-21 Accordingly, it has been suggested that Atg1 fulfills a structural role during early steps of PAS organization, and that Atg1 kinase activity regulates the dynamics of Atg protein movement at the PAS.20 Along these lines, Suzuki et al. reported that Atg13 and Atg17 stay at a microdomain termed VICS, where the vacuole and phagophore (PG) are in contact, whereas Atg1 can be detected both at this site and the expanding PG.22 Furthermore, Atg1 kinase activity is essential for PG expansion.22 Finally, Yeh et al. found that Atg13 is able to promote the formation of Atg1 dimers and that Atg1 self-interaction subsequently leads to transphosphorylation of Atg1 at S226 in the activation loop.23 Interestingly, this dimerization does not require Atg17, indicating that Atg17 promotes Atg1 kinase activity in a different manner.
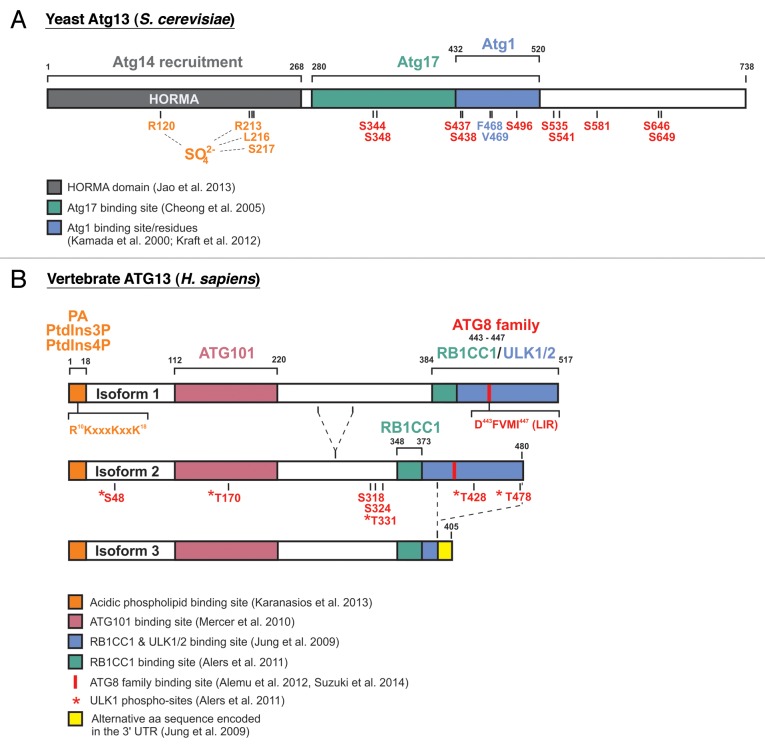
An important step toward our understanding of the molecular details at the phagophore assembly site has been recently made by Ragusa et al., who solved the crystal structure of the Atg17-Atg31-Atg29 complex.24 Their findings can be summarized as follows: The interaction of 2 Atg17 molecules is essential for the formation of a huge hexameric (3:3) complex of 2 Atg17-Atg31-Atg29 heterotrimers. The dimerization is mediated via 4 highly conserved hydrophobic residues located in the C terminus of Atg17. Although the Atg17-dimer possesses a characteristic “double-crescent shaped” curvature, it is not able to bind liposomes of any size.24 Surprisingly, the C-terminal domain of Atg1 shows lipid-tethering and curvature-sensing properties instead.24 It thus has been termed the “early autophagy targeting/tethering” (EAT) domain. Notably, the EAT domain in Atg1 also comprises the Atg13 binding site mapped by Yeh et al.,23 indicating that the C terminus of Atg1 mediates both membrane recruitment and Atg13 interaction in yeast. Vice versa, the binding sites for Atg1 and Atg17 in yeast Atg13 are both located in a central unstructured region, comprising amino acids 280–520 of the full-length (738 amino acid) protein.8,18,25,26 The simultaneous mutation of F468 and V469 of Atg13 to alanine is sufficient to abolish the interaction with Atg1 (Fig. 1A).27
Figure 1. Schematic representation of yeast and vertebrate Atg13/ATG13 proteins. (A) Yeast Atg13 contains an N-terminal HORMA domain and a central Atg1-Atg17-binding region. Jao et al. reported that the HORMA domain is required for the recruitment of Atg14.26 Within this domain, they identified R120, R213, L216, and S217 (amino acid positions of the S. cerevisiae protein; originally the crystal structure of L. thermotolerans was solved with the corresponding amino acid positions R118, R205, L208 and S209) to be involved in binding of a SO42--ion from the crystallization medium, and they hypothesized that the 2 arginine residues might comprise a biological phosphate sensor.26 Kamada et al. mapped the Atg1-binding region to amino acids 432–520 within yeast Atg13.8 In 2012, Kraft et al. reported that mutation of F468 and V469 to alanine is sufficient to abolish Atg1 binding.27 The Atg17-binding site was mapped to amino acids 280–520 by Cheong et al.18 In 2010, Kamada et al. reported the identification and/or prediction of 8 TORC1-sites (S348, S437, S438, S496, S535, S541, S646, and S649).28 Residues S344, S437 and S581 were suggested as PKA-sites.29 (B) The human ATG13 isoforms 1, 2 and 3 are depicted. ATG13 directly interacts with both RB1CC1 and ULK1/2 via its C-terminal region. The binding site has been roughly mapped to residues 384–517 of isoform 1 by Jung et al.30 Our group was able to narrow down the RB1CC1-binding site in ATG13 to residues 348–373 of the full-length avian ATG13 protein, which corresponds to the human isoform 2.31 Terje Johansen’s group reported the existence of a LIR motif within the C-terminal part (amino acids 443–447 in isoform 1).32 Notably, isoform 3 lacks this LIR motif and ULK1-binding capacity, but not the RB1CC1-binding site identified by our group. Mercer et al. mapped the ATG101-binding region to amino acids 112–220,33 and recently Karanasios et al. reported that ATG13 contains an N-terminal membrane binding site with specificity for the acidic phospholipids phosphatidic acid (PA), PtdIns3P and PtdIns4P.34 Apparently, a conserved cluster of 1 arginine (R10) and 3 lysine residues (K11, K15 and K18) mediates this lipid binding capacity.34 Our group was able to identify 5 ULK1-dependent phospho-sites in vitro (corresponding to S48, T170, T331, T428 and T478 in human isoform 2), but their in vivo relevance awaits further clarification.31 Next to these sites, only 2 additional phospho-acceptor sites have been reported so far (corresponding to S318 and S324 in human isoform 2).35-44
In 2013, Jao et al. were able to solve the crystal structure of the N-terminal region of yeast Atg13 (amino acids 1–268).26 Interestingly, this region adopts a HORMA (Hop1, Rev1, and Mad2) fold that shows remarkable structural similarity to the spindle checkpoint protein Mad2,26,45 which is able to switch between 2 completely different conformations.46 Furthermore, the HORMA domain of Atg13 contains a positively charged sulfate-binding pocket that may serve as a binding site for phosphate-containing molecules, such as phosphoproteins or lipids (Fig. 1A). It is tempting to speculate that Atg13 may function as a molecular switch that changes conformation after binding a specific phosphorylated substrate or phospholipid. However, to date it is completely unknown if Atg13 is indeed able to adopt an additional conformation as well, which then would be unable to bind phosphoryl groups.
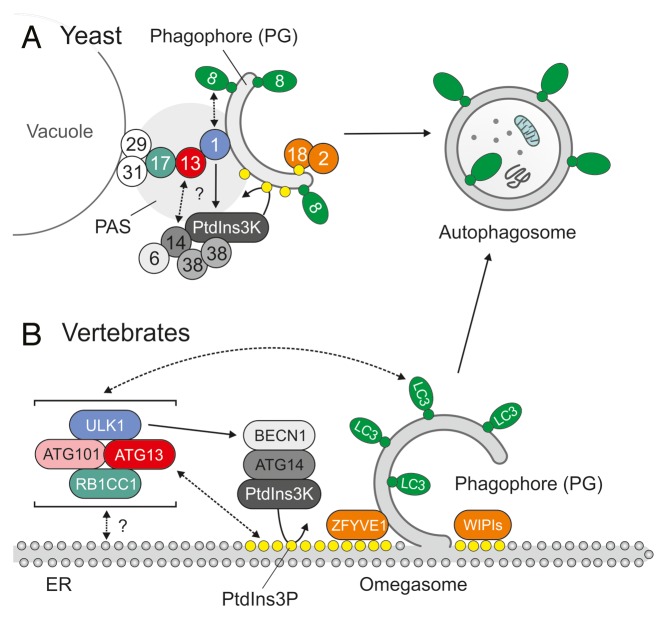
Although the HORMA domain is not essential for the initial recruitment of Atg13 to the PAS, it is crucial for autophagy induction and the subsequent recruitment of Atg14,26 which is a component of another multiprotein‐complex that is important for the formation of autophagosomes. In yeast, the class III phosphatidylinositol 3‐kinase (PtdIns3K) complex that includes Vps34 (vacuolar protein sorting 34) functions in both autophagy and the sorting of vacuolar proteins, and 2 separate Vps34-containing subcomplexes (complex I and II) have been identified to mediate these functions.47 Atg14 is a component of the autophagy-regulating complex I. Although the Atg13 HORMA domain is important for Atg14 recruitment, a direct interaction between the Atg14-containing complex I and the Atg1-Atg13-Atg17 complex has not been reported yet. The membrane lipid phosphatidylinositol 3-phosphate (PtdIns3P) which is generated by the yeast PtdIns3K complexes is an important key factor in autophagosomal membrane trafficking that recruits further autophagy promoting factors such as the Atg2-Atg18 complex to the site of autophagosome generation (Fig. 2A).48
Figure 2. Initiation of autophagosome generation in yeast and vertebrates. (A) In yeast, autophagosomes originate from a single location near the vacuole, the so-called phagophore assembly site (PAS). The pre-assembled Atg17-Atg31-Atg29 complex resides at the PAS and functions as a platform for further downstream factors.15-17 In the initial stage of autophagosome generation, the serine/threonine kinase Atg1 is recruited to the PAS via the adaptor protein Atg13, which is largely dephosphorylated upon autophagy induction and subsequently interacts both with Atg1 and Atg17.18,19 The strong enhancement of Atg1 kinase activity observed during autophagy induction requires both Atg13 and Atg17 and is essential for further progression of autophagosome generation.8 The N-terminal HORMA domain of Atg13 is crucial for the subsequent recruitment of the Atg14-containing class III PtdIns3K complex I.26 The locally restricted production of the membrane lipid PtdIns3P (yellow dots) by PtdIns3K directs additional factors such as the Atg2-Atg18 complex to the PAS and leads to further expansion of the phagophore (PG).48 (B) Although the origin of autophagosomes is more ambiguous in vertebrates, the endoplasmic reticulum (ER) has been proposed as a major site for autophagosome generation. In possible contrast to yeast, the interactions between mammalian ATG13 and the respective counterparts of Atg1 (ULK1) and Atg17 (RB1CC1) are constitutive and all 3 proteins are part of a large complex that is recruited to the equivalent of the PAS upon autophagy induction. The complex additionally includes ATG101, which protects ATG13 from proteasomal degradation and has no corresponding counterpart in yeast.33,49 Upon autophagy induction, the cytosolic ULK1-ATG13-RB1CC1-ATG101 complex redistributes to the ER. There, the class III PtdIns3K, which includes ATG14 and BECN1, is activated, leading to the production of PtdIns3P and the nucleation of membrane compartments termed omegasomes.34 As in yeast, the locally restricted generation of PtdIns3P is a crucial event that recruits further downstream effectors such as ZFYVE1/DFCP1 or members of the WIPI family to the site of autophagosome generation.50,51 The subsequent lipidation of ubiquitin-like molecules of the ATG8 family, such as the microtubule-associated protein 1 light chain 3 (LC3), with phosphatidylethanolamine (green dots) at the omegasome finally leads to the formation and elongation of the phagophore. For reasons of simplicity, ATG9-containing compartments and the Atg12/ATG12–Atg5/ATG5-Atg16/ATG16L1 complex are omitted in (A and B).
Previous findings support the following model for the regulation of autophagy initiation in yeast: Under nutrient-rich conditions Atg13 is kept in a hyperphosphorylated state by the nutrient-sensing serine/threonine kinase TOR (target of rapamycin) complex 1 (TORC1) and by PKA (cAMP-dependent protein kinase). While TORC1-dependent phosphorylation prevents the interaction between Atg1 and Atg13 and the subsequent enhancement of Atg1 activity, PKA phosphorylation interferes with the recruitment of Atg1 to the PAS, at least partially by inhibiting the interaction with Atg17.8,29 Indeed, most of the known TORC1- and PKA-phosphorylation sites in Atg13 are located in the region that mediates Atg1 and Atg17 interaction (Fig. 1A).8,24,26,29 Notably, the expression of an Atg13 mutant in which all 8 TORC1-dependent phosphorylation sites have been mutated to alanine is sufficient to partially induce autophagy under normal growth conditions in yeast.28 Dephosphorylation of Atg13 triggers both the dimerization of Atg1 and the bridging of Atg1 and Atg17, finally leading to the formation of a large Atg1-Atg13-Atg17-Atg31-Atg29 complex at the PAS.15,19,24 The initial translocation of Atg13 to the PAS is not mediated by the N-terminal HORMA domain26 and most likely depends on its Atg17-binding capacity. Atg13 induces Atg1 self-dimerization, enhances its kinase activity and promotes phagophore nucleation.23 Furthermore, since the N-terminal HORMA domain of Atg13 is essential for the recruitment of Atg14 to the PAS, Atg13 most likely represents an essential link between the 2 major autophagy initiation complexes, i.e., the Atg1-Atg13-Atg17-Atg31-Atg29 complex and the PtdIns3K complex Atg6-Atg14-Atg38-Vps34-Vps15 (Fig. 2A).
This attractive model is partially questioned by results recently published by Kraft et al.27 These authors could confirm the direct interaction between Atg1 and Atg13 and the stimulating effect of Atg13 on Atg1 kinase activity, but they did not observe any significant difference in Atg1-Atg13 binding affinity under nutrient-rich vs. nitrogen-starved conditions.27 Although the phosphorylation state of Atg13 was not explicitly analyzed, the authors observed an altered migrational behavior of Atg13 in SDS-PAGE under nutrient-rich conditions, and under these conditions Atg13 still interacted with Atg1. Furthermore, an Atg1 binding-deficient variant of Atg13 could not support the Cvt pathway, indirectly indicating that the interaction of wild-type proteins likely occurs under nutrient-rich conditions. How can these 2 models be integrated? A first approach to answering this question was recently made by Miller-Fleming et al.52 First, there exist several experimental variables which have to be considered, including Atg13 expression levels, cell lysis, and protein purification methods, and the affinity of the applied antibodies.52 Second, the classical model and the observation by Kraft et al. might be combined by the notion that the important step for Atg1 activation is Atg13 dephosphorylation per se rather than the sole association of Atg13 with Atg1.52 In other words, Atg1 activation in yeast is apparently not exclusively controlled by regulated Atg13 binding, but rather involves additional levels of control, e.g., the affinity of the Atg1-Atg13 interaction, conformational alterations, or recruitment of additional factors regulated by the Atg13 phospho-status. Nevertheless, future studies will be needed to reveal the exact nature of the phosphorylation-dependent regulation of the Atg1-Atg13 interaction and/or Atg1 activity.
Quest for the Vertebrate Ortholog of Atg13
Most basic components of the yeast molecular machinery of nonselective autophagy are remarkably conserved in metazoan species (e.g., Atg3, Atg5, Atg8, and Atg12) and homologs could be easily identified in their genome.53 Various autophagy-related genes even split into whole families of orthologs during evolution (e.g., Atg1, Atg4, Atg8, Atg16, and Atg18). In contrast, several autophagy-related genes seem to be absent from vertebrate and mammalian genomes—especially those involved in selective autophagic mechanisms such as the Cvt pathway (e.g., Atg11, Atg19, and Atg20), mitophagy (e.g., Atg11, Atg32, and Atg33) and pexophagy (e.g., Atg11, Pichia pastoris Atg30 and Atg36) even though similar types of selective autophagy occur in higher eukaryotes.
The Caenorhabditis elegans homolog of Atg1 named UNC-51 (uncoordinated-51) was first cloned in 1994,54 the 2 mammalian homologs ULK1 (unc-51 like autophagy activating kinase 1) and ULK2 followed in 1999.55-57 However, nobody was able to clearly identify a respective mammalian homolog of Atg13 or Atg17. It took more than 10 years to stop the discussion of whether higher eukaryotes possess direct homologs of both proteins or not. It had first been proposed by Meijer et al. that the product of the automatically annotated gene KIAA0652 could be the long missing homolog of yeast Atg13,53 although its primary sequence is very weakly conserved and could only be identified after an iterative PSI-BLAST search. In 2009, Chan et al. were able to confirm that the product of this gene is indeed involved in autophagy induction.58 In 2008, Hara et al. provided convincing evidence that RB1CC1/FIP200 (RB1-inducible coiled-coil 1) might be the functional counterpart of yeast Atg17, although there is no significant sequence homology to Atg17.59,60 Interestingly, the extreme C terminus of RB1CC1 displays significant similarity to Atg11,61 which is the counterpart of Atg17 in the yeast Cvt pathway. Thus, it seems that mammals possess at least 2 different Atg1 homologs (of which ULK1 and ULK2 are involved in autophagy), a weakly conserved ATG13 as well as a functional counterpart of Atg17 (RB1CC1)—all of which are involved in early stages of autophagy induction.
Another important step toward our understanding of autophagy initiation in metazoans has been the simultaneous observation by several groups that ATG13 is part of a huge and stable protein complex of approximately 3 MDa.30,49,60,62,63 The complex additionally includes ULK1 (or alternatively ULK2), RB1CC1 and a protein named ATG101. The latter protein has no obvious homolog in S. cerevisiae, directly binds to the middle region of ATG13 (Fig. 1B) and seems to prevent its proteasomal degradation.33,49 The binding sites for ULK1 and RB1CC1 have been mapped to the extreme C terminus of ATG13 (Fig. 1B).30,31
Under nutrient-rich conditions, MTOR (mechanistic target of rapamycin) complex I (MTORC1) associates with the ULK1-ATG13-RB1CC1-ATG101 complex, via direct interaction between ULK1 and RPTOR,63 and negatively regulates ULK1 kinase activity through direct MTORC1-dependent phosphorylation of ULK1 and ATG13.30,49,62,63 When MTORC1 activity is inhibited, e.g., under nutrient starvation conditions or after rapamycin treatment, ULK1 is rapidly dephosphorylated, leading to a prominent increase in enzymatic activity. Subsequently, ULK1 auto-phosphorylates, and phosphorylates both ATG13 and RB1CC1. Our group was able to identify 5 residues in human ATG13 that are directly phosphorylated by ULK1 in vitro (Fig. 1B; Table 1).31 The functional relevance of direct ULK1-dependent phosphorylation of ATG13 and RB1CC1, however, has still to be confirmed in vivo.31 Additionally, Joo et al. reported that S318 (human isoform 2, see below) is another ULK1-dependent phospho-site in ATG13 (Fig. 1B; Table 1).35 They observed that ULK1-dependent phosphorylation of ATG13 leads to its dissociation from ULK1 (see below).35 In general, global phosphorylation of ULK1 and ATG13 is decreased under starvation conditions, while RB1CC1 phosphorylation is decreased under nutrient-rich conditions.13,30,62,63 In other words, it appears that the phosphorylation-status of ULK1 and ATG13 primarily depends on the upstream regulatory kinase MTOR (and on AMPK [AMP-activated protein kinase] in the case of ULK1), whereas the phosphorylation-status of RB1CC1 mainly depends on ULK1. Noteworthy, while the mutation of 8 TORC1-dependent phosphorylation sites in yeast Atg13 is sufficient to induce autophagy no corresponding MTORC1-dependent phosphorylation site has been identified in mammalian ATG13 so far.36 In line with this and in contrast to the situation in yeast, the interaction between ATG13 and both Atg1 homologs ULK1 and ULK2 is constitutive and is neither affected by the phosphorylation status of ATG13 nor by nutrient availability.30,49,62,63 Finally, Shang et al. observed that ATG13 phosphorylation levels are low under nutrient-rich conditions and remain largely unaltered following starvation.36 Using SILAC, the authors identified only 1 phospho-residue, i.e., S324 (human isoform 2), which was also identified by several large-scale phospho-proteomic approaches.37-44
Table 1. Summary of known Atg13/ATG13 phosphorylation sites.
| Yeast Atg13 (S. cerevisiae) | |||||
|---|---|---|---|---|---|
| Kinase | Residue | Sequence | Identified using | Remarks | Ref. |
| PKA | S344 | RRSLS | R-R-x-pS/T consensus site | PKA in vitro kinase assay, Atg13-3SA mutation | 29 |
| S437 | RRHSS | ||||
| S581 | RRNSL | ||||
| TOR | S437 | RRHSS | MS/MS | TORC1 in vitro kinase assay, Atg13-8SA mutation | 28 |
| S438 | RRHSS | ||||
| S646 | SSISP | ||||
| S649 | SPRSI | ||||
| S348 | LSLSP | S-x-pS-P consensus site, similarity to S646 | |||
| S535 | DSHSP | ||||
| S541 | PSISP | ||||
| S496 | ISDSL | similarity to S438 | |||
| Vertebrate ATG13 (H. sapiens) | |||||
| Kinase |
Residue (isoform 2) |
Sequence | Identified using | Remarks | Ref. |
| ULK1 | S48 | PTGSD | MS/MS | ULK1 in vitro kinase assay | 31 |
| T170 | GFQTV | MS/MS | |||
| T331 | VLETI | MS/MS | |||
| T428 | DLGTF | MS/MS | |||
| T478 | FVETL | MS/MS | |||
| S318 | VSNSS | SILAC-based MS/MS | ULK1 in vitro kinase assay, anti-pS318 antibody | 35 | |
| ? | S324 | GRASP | SILAC-based MS/MS | 36 | |
| S324 | GRASP | several phosphoproteome analyses | 37–44 | ||
In yeast, Atg13 could be identified both as a direct PKA and TORC1 substrate. Stephan et al. were able to identify 3 PKA consensus sites in Atg13 and subsequently confirmed the direct phosphorylation by PKA in an in vitro kinase assay.29 Using mass spectrometry (MS/MS), Kamada et al. were able to identify 4 TORC1-dependent in vitro phosphorylation sites in Atg13.28 Four other sites have been predicted, based on their similarity to S649 and S438, respectively.28 Mutation of all 8 serine residues in yeast Atg13 to alanine almost completely abolishes phosphorylation by TORC1. For human ATG13, S324 and S318 are the only in vivo phosphorylation sites reported so far. S324 could be identified by mass spectrometry (MS/MS), both in a SILAC-based approach and several global phospho-proteome analyses.36-44 S318 has been identified as a direct ULK1-dependent in vitro phosphorylation site.35 Its relevance could be subsequently confirmed in vivo using a phospho-specific antibody. Our group was able to identify 5 ULK1-dependent in vitro phosphorylation sites in human ATG13.31 Numbering of amino acids for human ATG13 refers to human isoform 2.
The Atg13/ATG13 homologs of Caenorhabditis elegans (EPG-1/ATG-13) and Drosophila melanogaster (Atg13) both directly interact with the corresponding Atg1 homolog UNC-51 and Atg1, respectively.64,65 Interestingly, while in yeast Atg13 phosphorylation is strongest under normal growth conditions, Drosophila Atg13 is hyperphosphorylated in starved animals, and as in higher metazoans, its interaction with Atg1 is neither affected by the phosphorylation status nor by nutrient availability.64 The knockout of Atg13 in Drosophila larval fat body cells leads to a complete block of autophagosome generation in response to nutrient starvation or rapamycin treatment.64 A loss of function mutation of EPG-1/ATG-13 causes accumulation of PGL granules in somatic cells and shortens the life time of C. elegans under nutrient deprivation conditions.65 Notably, no Drosophila or C. elegans homolog of Atg17 or RB1CC1 has been described so far.
In addition to its central role during autophagy induction, neuronal functions have been attributed to UNC-51/ULK1. In C. elegans, the Atg1 ortholog UNC-51 has been named according to the uncoordinated movement of the mutant strain, and UNC-51 is central for axonal elongation.54,66 Later it was demonstrated that VAB-8 and UNC-14 are substrates of UNC-51 which likely mediate its function in axon outgrowth.67,68 In Drosophila, Atg1 associates with and phosphorylates UNC-76, a protein that participates in axonal transport processes.69 Finally, ULK1 and ULK2 have been implicated in axon outgrowth and branching.70-72 Furthermore, ULK1 interacts with the synaptic proteins SYNGAP1 and SDCBP/syntenin-1, and the latter is directly phosphorylated by ULK1.73,74 However, the role of the additional components of the ULK1 complex in general and of ATG13 in particular for this neuron-specific function remains rather elusive. At least locomotion and axon guidance appear to be unaffected by mutation of EPG-1/ATG-13 in C. elegans.65 Furthermore, despite the evident function of autophagy for neuron homeostasis, it is unknown whether the function of ULK1 (and possibly its associated components) for axon elongation/outgrowth during neuronal development can be connected to its pro-autophagic activity or whether it represents a completely independent mode of action.75 It is tempting to speculate that different adaptor proteins direct ULK1 activity during different physiological processes, e.g., ATG13 during initiation of autophagy. This would resemble the situation of MTOR, whose substrate binding specificity is mainly controlled by RPTOR and RICTOR in MTORC1 and MTORC2, respectively.76 Much more work has to be performed in order to clarify the potential functions of ATG13 outside autophagy signaling.
In yeast, all autophagosomes originate from a single location, the PAS. In higher eukaryotes, several organelles have been proposed as a membrane source (which is also the case in yeast) and as the site of autophagosome formation. The endoplasmic reticulum (ER) and mitochondria, however, represent the most likely and best characterized sources for the autophagosomal membrane.77 In 2008, a PtdIns3P-enriched microdomain in the endoplasmic reticulum (ER) was identified by Axe et al. and proposed as a major site of autophagosome generation.50 Newly generated autophagosomes seem to bud from these ring-shaped structures, termed omegasomes (Fig. 2B).78-80
The recruitment of the ULK1/2-ATG13-RB1CC1-ATG101 complex to the ER is the first event in omegasome formation, and ULK1 kinase activity is essential for the subsequent recruitment of the ATG14-containing class III PtdIns3K complex.51 The locally restricted activity of PtdIns3K leads to the enrichment of PtdIns3P in the ER and finally to the recruitment of PtdIns3P-binding proteins, such as ZFYVE1/DFCP1 (zinc finger, FYVE domain containing 1) or members of the WIPI (WD repeat domain, phosphoinositide-interacting 1) family to the site of autophagosome generation.50,51 In accordance with this picture, Karanasios et al. recently found that about 80% of the newly formed omegasomes in starved HEK293 cells colocalize with punctate structures that contain ATG13, ULK1, and ATG101.34 Interestingly, the single ATG13-positive dot-like structure that associates with each ring-shaped omegasome grows in parallel and fades away after several minutes. Usually, newly formed autophagosomes bud from the ER within 1 min after ATG13 disappearance.34 Although the spatio-temporal orchestration and kinetics of ATG protein recruitment has been analyzed in great detail, it is still completely unknown as to which molecular events trigger the initial recruitment of ATG13 to the omegasome and what leads to its disappearance at later stages of autophagosome maturation.
An interesting observation in this regard was reported by the same authors, who identified a basic region in the extreme N terminus of ATG13 that is able to bind acidic phospholipids in vitro, mainly phosphatidic acid, PtdIns3P, and PtdIns4P.34 The mutation of 4 basic residues, which are highly conserved among higher eukaryotes, greatly abolishes phospholipid binding, without affecting the interaction of ATG13 with ATG101 or ULK1 (Fig. 1B).34 Since the PtdIns3P-generating PtdIns3K complex is recruited at later stages, it is unlikely that the initial recruitment of ATG13 to omegasomes is due to its lipid-binding capacity.34,51 Possibly, other components of the ULK1 complex also participate in the initial recruitment. For example, the C-terminal domain of ULK1 has also been reported to direct membrane binding,58 resembling the membrane-tethering capabilities of the yeast Atg1 EAT domain described above. Nevertheless, it is conceivable that ATG13 represents another PtdIns3P binding factor that might establish a positive feedback loop which leads to sustained ULK1 recruitment and prolonged PtdIns3K activation.34 Furthermore, one might speculate that PtdIns3P dephosphorylation in turn contributes to ATG13 dissociation at a later stage.
The N-terminal HORMA domain in yeast Atg13 is essential for autophagy induction and responsible for the subsequent recruitment of the Atg14-containing PtdIns3K complex to the site of autophagosome generation, although a direct interaction could not be observed yet. Likewise, there is no evidence for a direct interaction between the ULK1/2-ATG13-RB1CC1-ATG101 complex and the mammalian PIK3C3/VPS34-PIK3R4/VPS15-BECN1-ATG14 class III PtdIns3K complex.61 Nevertheless, the interaction might be indirect and mediated by a yet unknown protein that interacts with members of both complexes. Notably in this regard, TAB2 (TGF-β activated kinase 1/MAPK37 binding protein 2) has been recently identified both as an ATG13- and BECN1-interacting protein,82 providing a potential link between both complexes. Furthermore, the 2 class III PtdIns3K complex components AMBRA1 and BECN1 both serve as ULK1 substrates.83,84
In addition, ATG13 directly interacts with several mammalian homologs of Atg8,32 termed LC3A/B/C, GABARAP, and GABARAPL1/L2. Alemu et al. reported that ATG13 most strongly interacts with members of the GABARAP subfamily, which are involved in later steps of autophagosome maturation.85 The interaction is mediated via a single and highly conserved LIR (LC3-interacting region) motif consisting of 5 amino acids (DFVMI) in the C-terminal half of human ATG13 (Fig. 1B; Table 2).32 Interestingly, ULK1/2 and RB1CC1 also interact with LC3 as well as various other members of the GABARAP family.27,32,61,86 In contrast, Suzuki et al. recently reported X-ray crystallographic structures of LC3A, LC3B, and LC3C in complex with ATG13 LIR peptides.81 They also report that the LC3 isoforms exhibit positive 2-hybrid interactions only with ATG13, but not with ULK1, RB1CC1, or ATG101.81 Furthermore, the authors describe that mutation of residue K49 of LC3 isoforms to alanine increases ATG13 binding, whereas mutation of K51 to alanine decreases ATG13 binding. Interestingly, both mutants do not support autophagosome formation. Suzuki et al. propose a model in which a specific affinity range for the binding of LC3 isoforms to the ATG13 LIR is mandatory for proper autophagosome formation, with either too high or too low affinity negatively affecting this process.81 Notably, the observed effect was especially prominent under nutrient-rich conditions, indicating that the affinity-dependency is in effect for selective autophagy.81 Nevertheless, future studies will be needed to reveal the molecular details of when and how ATG13 binds to the different mammalian ATG8 proteins and what the physiological relevance of these associations is.
Table 2. Summary of Atg13/ATG13 interaction partners and binding sites.
| Yeast Atg13 (S. cerevisiae) | |||||
|---|---|---|---|---|---|
| Binding partner or ligand | Binding site | Residues | Remarks | Ref. | |
| Start | end | ||||
| Atg1 | 432 | 520 | 8 | ||
| F468/V469 | Mutation prevents binding | 27 | |||
| Atg17 | 280 | 520 | 18 | ||
| Sulfate (and phosphoryl-groups?) |
R120 | Direct interaction (side chain; R118 in L. thermotolerans) | 26 | ||
| L216/S217 | Direct interaction (backbone; L208/S209 in L. thermotolerans) | ||||
| R213 | Generation of basic pocket (R205 in L. thermotolerans) | ||||
| Vertebrate ATG13 (H. sapiens) | |||||
| Binding partner or ligand | Binding site(isoform 2) | Residues | Remarks | Ref. | |
| Start | end | ||||
| ULK1-RB1CC1 | 347 | 480 | Mapped for human ATG13 (isoform 1 aa 384–517) | 30 | |
| RB1CC1 | 348 | 373 | Mapped for avian ATG13, which corresponds to human isoform 2 (isoform 1 aa 385–410) | 31 | |
| ATG101 | 112 | 220 | 33 | ||
| ATG8 family (preference for GABARAP subfamily) | 406 | 410 | D406FVMI410 | LIR motif (minimal; isoform 1 aa 443–447) | 32 |
| ATG8 family (LC3 subfamily) | 404 | 410 | H404DDFVMI410 | LIR motif (isoform 1 aa 441–447) |
81 |
| Acidic phospholipids (PA, PtdIns3P, PtdIns4P) | 1 | 18 | R10KxxxKxxK18 | 34 | |
| TAB2 | ? | ? | 82 | ||
This table summarizes the binding sites and interaction motifs mapped for yeast and vertebrate Atg13/ATG13, as depicted in Figure 1. Please note that the RB1CC1 binding site in ATG13 reported by Alers et al. has been mapped for the full-length avian ATG13 protein,31 which corresponds to human isoform 2, while the ULK1/RB1CC1 binding site in human ATG13 reported by Jung et al. has been mapped for the 37 amino acids-larger human isoform 1.30 Numbering of amino acids for vertebrate ATG13 refers to human isoform 2.
As described above, ATG13 directly interacts with both RB1CC1 and ULK1/2 via its C-terminal region. Jung et al. have roughly mapped the binding site to residues 384–517 of human ATG13 (isoform 1; as described below, these residues correspond to residues 347–480 of human isoform 2).30 However, since all 3 proteins are part of a common protein complex, it is very unlikely that RB1CC1 and ULK1/2 compete for the interaction with ATG13.30,62,63 Our group was able to further narrow down the RB1CC1-binding site in ATG13 to residues 348–373 of the full-length avian ATG13 protein,31 which corresponds to the human isoform 2 (Fig. 1B; Table 2). Meanwhile, we could confirm these results for the human ATG13 protein and found that the interaction with ULK1 and ULK2 is indeed mediated by a different, nonoverlapping region in the extreme C terminus of ATG13 (our unpublished data).
Alternative Splicing
Although vertebrate genomes have only one ATG13 gene, the alternative splicing of the ATG13 pre-mRNA results in the generation of several ATG13 splice variants. According to Jung et al. and the UniProt database, there is evidence for at least 5 different human ATG13 isoforms at the protein level (ref. 30 and UniProt entry O75143). The longest human isoform 1 (O75143-1) comprises 517 amino acids, while the shorter isoforms 2, 4, and 5 lack various exons in the middle and N-terminal region of ATG13. Interestingly, human isoform 3 (O75143-3) additionally lacks the last 75 C-terminal amino acids. Its mRNA instead makes up a part of the 3′ UTR and thus encodes for an alternative 15 amino acid sequence at the C terminus that completely differs from the other isoforms (Fig. 1B).30 Notably, ATG13 isoform 3 misses both the LIR motif, which is responsible for GABARAP interaction,32 as well as the ULK1 binding site. Isoform 3 is hence unable to interact with ULK1.30 Our group was able to identify at least 7 different ATG13 splice variants at the mRNA level in avian DT40 cells.31 Although there is no evidence for an isoform that resembles human isoform 3 in DT40 cells, we could identify several isoforms incapable of RB1CC1 interaction. The RB1CC1 binding site could be narrowed down to the 26 amino acids encoded by avian exon 12, which are located in the C-terminal region mapped as the RB1CC1/ULK1 binding site in human ATG13 by Jung et al. (Fig. 1B; Table 2).30,31 Notably, only those isoforms able to interact with RB1CC1 were able to rescue the autophagy-defective phenotype of ATG13-deficient DT40 cells.31 Interestingly, while the human ATG13 isoform 3 is unable to interact with ULK1, it still comprises the RB1CC1 binding site mapped for the avian ATG13 protein (ref. 31 and unpublished data; Fig. 1B).
Does Atg13/ATG13 have Functions Outside the Atg1/ULK1 Complex during Autophagy Induction?
Based on the observations described above, the primary and evolutionarily conserved function of Atg13/ATG13 seems to be closely related to its ability to bind Atg1/ULK1 and Atg17/RB1CC1, thereby recruiting Atg1/ULK1 to the site of autophagosome formation and enhancing Atg1/ULK1 kinase activity.8,23,28,62,64 However, there are several lines of evidence that Atg13/ATG13 might have additional functions during autophagy induction.
First, Kraft et al. could show that the autophagy-defective phenotype of yeast ATG13 knockout strains can be partially rescued by an Atg13 mutant that is defective in Atg1 interaction.27 Although autophagy seems to be induced primarily by dephosphorylation of TORC1-dependent sites in Atg13 and subsequent association of Atg13 and Atg1,23,28 the authors conclude that Atg13 might possess an additional function in yeast autophagy that is independent of Atg13-Atg1 complex formation.
Second, Joo et al. could demonstrate that ULK1 (but not ULK2) directly phosphorylates ATG13 at S318 and that this phosphorylation induces the dissociation from ULK1 and relocalization of ATG13 to depolarized mitochondria.35 Furthermore, overexpression of a nonphosphorylatable ATG13-S318A mutant was able to inhibit the selective clearance of damaged mitochondria. These results indicate that ATG13 possesses an additional function in mitophagy that requires previous phosphorylation by ULK1 and involves an alternative and yet unkown mode of action at depolarized mitochondria.35
Third, our group was able to generate ATG13-deficient and ULK1/ULK2 double-deficient DT40 cells.31 The phenotype of both vertebrate cell lines is remarkably different: While ATG13−/− cells are completely autophagy defective, ULK1−/− ULK2−/− cells do not display any impairment of the autophagic pathway, indicating that ATG13 plays an essential role in autophagy induction, while ULK1/2 kinase activity is dispensable in this cell line. Nevertheless, the direct interaction with RB1CC1 is essential for ATG13 function in DT40 cells.31
Conclusion
Yeast Atg13 is an adaptor protein that is hyperphosphorylated by TORC1 and PKA under normal growth conditions.8,28,29 Under starvation conditions Atg13 is dephosphorylated and able to interact with both Atg1 and Atg17 via its middle region.8,28 Dephosphorylation of Atg13 induces the recruitment of Atg1 to the PAS, via simultaneous interaction with Atg1 and the Atg17-Atg31-Atg29 complex, induces dimerization and activation of Atg1, and thereby locally restricts Atg1 activity to the site of autophagosome generation.23,27 Furthermore, the N-terminal HORMA domain of Atg13 is essential for the subsequent recruitment of the Atg14-containing PtdIns3K complex to the PAS.25,26 The locally restricted generation of PtdIns3P recruits further downstream factors to nascent autophagosomes (Fig. 2A). Since the phosphorylation status of Atg13 affects the localization and activity of Atg1, yeast Atg13 may hence best be described as a molecular switch that transduces nutrient-regulated TORC1 and PKA signaling to the autophagic machinery. The structure of the Atg13 N-terminal region implies an additional regulatory mechanism: First, it displays remarkable structural homology to the HORMA domain of Mad2, which exists in 2 distinct conformations. Second, it comprises a sulfate-binding pocket. This may indicate that the N terminus of Atg13 is able to undergo conformational changes upon phosphorylation and may switch between a phospho-binding and nonphospho-binding state.25,26 Although this is an intriguing hypothesis, it still has to be confirmed in vivo.
The mammalian homolog of Atg13 shares weak sequence identity with the yeast protein. In possible contrast to yeast, the interactions between mammalian ATG13 and the respective counterparts of Atg1 (ULK1) and Atg17 (RB1CC1) are constitutive and not differentially regulated by nutrient availability. All 3 proteins are part of a large stable protein complex that localizes to phagophore assembly sites upon autophagy induction. It is not clear which molecular events trigger the translocation of the ULK1-ATG13-RB1CC1 complex to the site of autophagosome generation (Fig. 2B). Furthermore, although human ATG13 is a highly phosphorylated protein that runs as a smeared band in western blot, much less is known about the kinases that phosphorylate ATG13 in vivo (Table 1) or about how its phosphorylation status is linked to autophagy induction mechanistically. Because the N-terminal region of human ATG13 has not been crystalized yet, it is also not known whether it adopts a similar fold as yeast Atg13.
The ability to interact with Atg1/ULK1 and Atg17/RB1CC1 is a central feature of Atg13/ATG13. Accordingly, Atg13/ATG13 is an important component of the autophagic machinery, and this is confirmed by several Atg13/ATG13 knockdown, loss-of-function, and knockout studies (summarized in Table 3). However, future studies will have to reveal whether the view of Atg13/ATG13 as a sole companion of Atg1/ULK1 is an undervaluation of its pro-autophagic role. The experiments described above indeed indicate that Atg13/ATG13 has additional functions that are independent of a direct interaction or permanent colocalization with Atg1 or ULK1, respectively. In this regard, Atg13/ATG13 might step out of the shadow of Atg1/ULK1.
Table 3. Summary of Atg13/ATG13 knockdown, loss-of-function, and knockout phenotypes.
| Species | Cell type/Cell line | Effect on autophagy induction | Additional phenotypes | Ref. | |||
|---|---|---|---|---|---|---|---|
| Stimulus | Assay | Phenotype | |||||
| Saccharomyces cerevisiae | - | KO | Nitrogen-starvation | Pho8Δ60 alkaline phosphatase activity | Blocked | 6 | |
| Reduced viability | |||||||
| - | KO | Nitrogen-starvation | Pho8Δ60 alkaline phosphatase activity | Blocked | 12 | ||
| Autophagic body accumulation in atg13Δ pep4Δ mutants (EM) | Blocked | ||||||
| - | KO | Nitrogen-starvation | Pho8Δ60 alkaline phosphatase activity | Blocked | 27 | ||
| Arabidopsis thaliana | - | KO | Nitrogen starvation | Growth of seedlings | Reduced | 87 | |
| Vacuolar breakdown of GFP-ATG8a (WB) | Blocked | ||||||
| Detection of GFP-ATG8a positive autophagic bodies (FM) | Blocked | ||||||
| Short-day photoperiod | Leaf senescence | Accelerated | |||||
| Fixed-C starvation | Seedling survival | Reduced | |||||
| Caenorhabditis elegans | somatic cells | LOF | - | Accumulation of PGL granules | 65 | ||
| - | Food deprivation | Shortened life time | |||||
| Drosophila melanogaster | Larval fat body | KO | Starvation in 20% sucrose solution | mCherry-Atg8a dot formation (FM) | Blocked | 64 | |
| Endog. Ref(2)P/SQSTM1 and GFP-Ref(2)P degradation (FM) | Blocked | ||||||
| Rapamycin treatment (TOR inhibition) | mCherry-Atg8a dot formation (FM) | Blocked | |||||
| Gallus gallus | DT40 | KO | Starvation in EBSS | endog. LC3-II accumulation (WB) | Blocked | 31 | |
| mCitrine-LC3 dot formation (FM) | Blocked | ||||||
| Autophagosome formation (EM) | Blocked | ||||||
| Mus musculus | MEFs | KD | Starvation in amino acid-free medium | GFP-LC3 dot formation (FM) | Blocked | 62 | |
| GFP-LC3 lipidation (WB) | Blocked | ||||||
| Reduction of ULK1 protein level and increased mobility in WB (loss of auto-phosphorylation?) | |||||||
| MEFs | KO | Starvation in amino acid-free medium | WIPI2 dot formation (IF) | Blocked | 34 | ||
| ATG16 dot formation (IF) | Blocked | ||||||
| PP242 treatment (MTOR inhibition) | WIPI2 dot formation (IF) | Blocked | |||||
| ATG16 dot formation (IF) | Blocked | ||||||
| MEFs | KO | Reduction of ULK1 protein level (WB) and reduced ULK1 phosphorylation at S757 (WB) | 36 | ||||
| Homo sapiens | HEK293 | KD | Rapamycin treatment (MTOR inhibition) | Endog. LC3-II accumulation (WB) | Reduced | 30 | |
| SQSTM1/p62 degradation (WB) | Reduced | ||||||
| HeLa | KD | Endog. LC3 dot staining (IF) | Reduced | ||||
| HeLa | KD | Reduction of ULK1 protein level (WB) | |||||
| HEK293 | KD | Starvation in EBSS | GFP-LC3 dot formation (FM) | Reduced | 58 | ||
| GFP-LC3 lipidation (WB) | Blocked | ||||||
| Endog. LC3-II accumulation (WB) | Reduced | ||||||
| HEK293 | KD | Starvation in amino acid- and serum-free medium | GST-BHMT fragmentation (WB) | Reduced | 33 | ||
| Endog. LC3-II accumulation (WB) | Reduced | ||||||
| HeLa | KD | Starvation in amino acid- and serum-free medium | GFP-LC3 dot formation (FM) | Blocked | 63 | ||
| Starvation in amino acid- and serum-free medium | Autophagosome formation (EM) | Blocked | |||||
| Reduction of ULK1 and RB1CC1 protein levels (WB) | |||||||
| U2OS | KD | Strong reduction of ULK1 protein level (WB) | 36 | ||||
This table summarizes the reported effects of Atg13/ATG13 knockdown (KD), loss-of-function mutations (LOF), and knockout (KO) in various species and cell lines. EM, electron microscopy; FM, fluorescence microscopy; IF, immunofluorescence; WB, western blot.
Disclosure of Potential Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
Related research of BS is supported by grants from the Deutsche Forschungsgemeinschaft (STO 864/3-1) and from the Research Committee of the Medical Faculty of the Heinrich-Heine-University Düsseldorf (58/2013).
Note Added in Proof
During preparation/publication of this review article, Fujioka et al. reported the X-ray crystallographic analysis of the interaction of yeast Atg13 with Atg1 and Atg17 (Nat Struct Mol Biol 2014 May 4. 10.1038/nsmb.2822). Atg13 binds tandem microtubule interacting and transport (tMIT) domains in Atg1 via a 2-part MIT interacting motif (residues 460–521). The authors suggest that this mode of interaction is conserved in mammals. Additionally, the Atg17-binding region was mapped to amino acids 424–436 of Atg13. Finally, the authors propose that starvation-induced dephosphorylation of specific serine residues in Atg13 enhances the interaction with both Atg1 and Atg17.
Glossary
Abbreviations:
- ATG
autophagy-related gene
- LIR
LC3-interacting region
- MTOR
mechanistic target of rapamycin
- PAS
phagophore assembly site
- RB1CC1/FIP200
RB1-inducible coiled-coil 1
- ULK1/2
UNC-51 like autophagy activating kinase 1/2
References
- 1.Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–3. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 2.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 4.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–13. doi: 10.1016/S0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 7.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356:405–10. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamata T, Kamada Y, Suzuki K, Kuboshima N, Akimatsu H, Ota S, Ohsumi M, Ohsumi Y. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005;338:1884–9. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–96. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott SV, Nice DC, 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–9. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–61. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–5. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 16.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–50. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 18.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–53. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–38. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–44. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 23.Yeh YY, Shah KH, Herman PK. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem. 2011;286:28931–9. doi: 10.1074/jbc.M111.250324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–12. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jao CC, Ragusa MJ, Stanley RE, Hurley JH. What the N-terminal domain of Atg13 looks like and what it does: a HORMA fold required for PtdIns 3-kinase recruitment. Autophagy. 2013;9:1112–4. doi: 10.4161/auto.24896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci U S A. 2013;110:5486–91. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–54. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alers S, Löffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, et al. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy. 2011;7:1423–33. doi: 10.4161/auto.7.12.18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Øvervatn A, Kirkin V, Johansen T. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287:39275–90. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–62. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 34.Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, Ktistakis NT. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci. 2013;126:5224–38. doi: 10.1242/jcs.132415. [DOI] [PubMed] [Google Scholar]
- 35.Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell. 2011;43:572–85. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–93. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber TB, Mäusbacher N, Kéri G, Cox J, Daub H. An integrated phosphoproteomics work flow reveals extensive network regulation in early lysophosphatidic acid signaling. Mol Cell Proteomics. 2010;9:1047–62. doi: 10.1074/mcp.M900486-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–29. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villén J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–93. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiśniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J Proteome Res. 2010;9:3280–9. doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 43.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–6. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanivan S, Gnad F, Wickström SA, Geiger T, Macek B, Cox J, Fässler R, Mann M. Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J Proteome Res. 2008;7:5314–26. doi: 10.1021/pr800599n. [DOI] [PubMed] [Google Scholar]
- 45.Aravind L, Koonin EV. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23:284–6. doi: 10.1016/S0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- 46.Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–9. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 50.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–76. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller-Fleming L, Cheong H, Antas P, Klionsky DJ. Detection of Saccharomyces cerevisiae Atg13 by western blot. Autophagy. 2014;10:514–7. doi: 10.4161/auto.27707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 54.Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Müller F, Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 55.Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics. 1998;51:76–85. doi: 10.1006/geno.1998.5340. [DOI] [PubMed] [Google Scholar]
- 56.Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, Shirasawa T, Muramatsu M. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun. 1998;246:222–7. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- 57.Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–9. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 58.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–71. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–7. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 60.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–14. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian E, Wang F, Han J, Zhang H. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 2009;5:608–15. doi: 10.4161/auto.5.5.8624. [DOI] [PubMed] [Google Scholar]
- 66.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai T, Garriga G. The conserved kinase UNC-51 acts with VAB-8 and UNC-14 to regulate axon outgrowth in C. elegans. Development. 2004;131:5991–6000. doi: 10.1242/dev.01457. [DOI] [PubMed] [Google Scholar]
- 68.Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801–11. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- 69.Toda H, Mochizuki H, Flores R, 3rd, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292–307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee EJ, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7:689–95. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–46. doi: 10.1016/S0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 72.Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T, Tomoda T, Tani T, Wooten MW, Wang F. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci U S A. 2007;104:5842–7. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajesh S, Bago R, Odintsova E, Muratov G, Baldwin G, Sridhar P, Rajesh S, Overduin M, Berditchevski F. Binding to syntenin-1 protein defines a new mode of ubiquitin-based interactions regulated by phosphorylation. J Biol Chem. 2011;286:39606–14. doi: 10.1074/jbc.M111.262402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomoda T, Kim JH, Zhan C, Hatten ME. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18:541–58. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–37. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McEwan DG, Dikic I. Not all autophagy membranes are created equal. Cell. 2010;141:564–6. doi: 10.1016/j.cell.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 78.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 79.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki H, Tabata K, Morita E, Kawasaki M, Kato R, Dobson RC, Yoshimori T, Wakatsuki S. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure. 2014;22:47–58. doi: 10.1016/j.str.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 82.Takaesu G, Kobayashi T, Yoshimura A. TGFβ-activated kinase 1 (TAK1)-binding proteins (TAB) 2 and 3 negatively regulate autophagy. J Biochem. 2012;151:157–66. doi: 10.1093/jb/mvr123. [DOI] [PubMed] [Google Scholar]
- 83.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res. 2000;85:1–12. doi: 10.1016/S0169-328X(00)00218-7. [DOI] [PubMed] [Google Scholar]
- 87.Suttangkakul A, Li F, Chung T, Vierstra RD. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–79. doi: 10.1105/tpc.111.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]