Preface
The discovery that cancer can be governed above and beyond the level of our DNA presents a new era for designing therapies that reverse the epigenetic state of a tumor cell. Understanding how altered chromatin dynamics leads to malignancy is essential for controlling tumor cells while sparing normal cells. Polycomb and trithorax group proteins are evolutionarily conserved and maintain chromatin in the “off” or “on” state, thereby preventing or promoting gene expression, respectively. Recent work highlights the dynamic interplay between these opposing classes of proteins, providing new avenues for understanding how these epigenetic regulators function in tumorigenesis.
Introduction
Tumors are composed of diverse cell types, some of which contribute to resistance to anti-cancer therapy, tumor relapse, and metastasis. Models explaining tumor heterogeneity include the cancer stem cell hypothesis and the clonal evolution model 1–3. Despite ones view, the ultimate goal of cancer therapeutics is to target cells that lead to tumor recurrence—the tumor-propagating cells. Developmental pathways are often inappropriately reactivated in cancer cells, suggesting that tumor-propagating cells have hijacked cellular networks that control the behavior of normal stem cells, empowering them with enhanced proliferative potential 4, 5. In some cancers such as acute myelogenous leukemia (AML), renewal capacity relies on a transcriptional program that is active in embryonic stem cells (rather than adult stem cells) 6. Thus, striking parallels between mechanisms orchestrating normal embryogenesis and those that modulate tumorigenesis could be exploited to treat cancer.
Development of an embryo requires cell fate decisions to establish the lineage of a cell, as well as a system for ‘remembering’ this identity throughout subsequent rounds of division. How this cellular memory is maintained during the life of an organism is a fundamental question in development. This also impinges upon our understanding of cancer, as the cellular memory system that normally governs cell behavior goes awry in cells within a tumor, which cripples the activity of tumor suppressors and spuriously activates proto-oncogenes, endowing cells with relentless proliferative capacity and loss of cell identity.
An understanding of the mechanistic basis of cellular memory began in 1978 when Pam and Ed Lewis described a series of Drosophila melanogaster mutants called Polycomb (Pc) (fig 1). Specific body segments of these mutants had the identity of a completely different, but otherwise normal, body segment. Whereas transcription factors had initially established the proper identity of cells within the region of the embryo that would eventually give rise to the adult segment, cells of Pc mutants failed to transmit this information to daughter cells, producing remarkable phenotypes such as adult animals with legs in the place of antennas 7–9. Pc mutations caused inappropriate reactivation of genes that should have been repressed, such as homeotic genes that dictate segment identity, causing a specific body segment to take on an entirely new character. This failure in the cellular or transcriptional memory system led to the idea that Polycomb group (PcG) proteins repress gene expression by keeping chromatin in a transcriptionally inactive or ‘off’ state 10–14 (box 1).
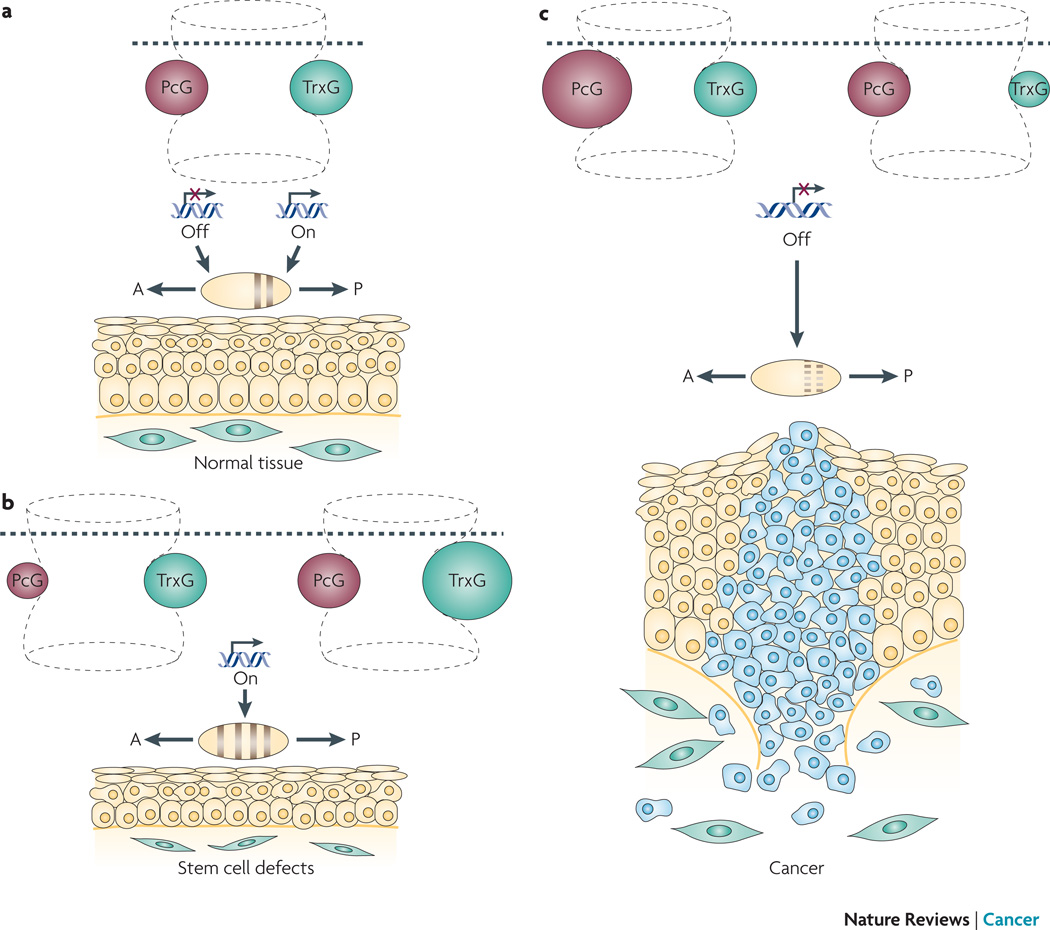
Figure 1. The dynamic hourglass model for PcG-TrxG-mediated interactions.
The hourglass represents developmental time, with the establishment and maintenance of chromatin states being depicted by the upper and lower portions of the hourglass, respectively. The hourglass is ‘flexible,’ as alterations in chromatin structure affects gene expression in a highly dynamic manner. a. The chromatin state established by transcription factors during development (above dotted line) is maintained by an evolutionarily conserved mechanism involving a balance between PcG (pink) and TrxG (green) proteins (below dotted line). PcG and TrxG proteins regulate chromatin to evoke transcriptionally inactive (“off”) or active (“on”) states, respectively, thereby maintaining cellular memory. This PcG-TrxG-modulated cellular memory system maintains the identity of body segments along the anterior (A) - posterior (P) axis of the embryo, and regulates tissue homeostasis in the adult. b. Perturbation of cellular memory due to compromised PcG or excessive TrxG leads to inappropriate segment identity and defects in stem cell renewal. c. Perturbation of cellular memory due to excessive PcG or compromised TrxG leads inappropriate segment identity and cancer.
Chromatin dynamics regulates transcriptional memory and cancer.
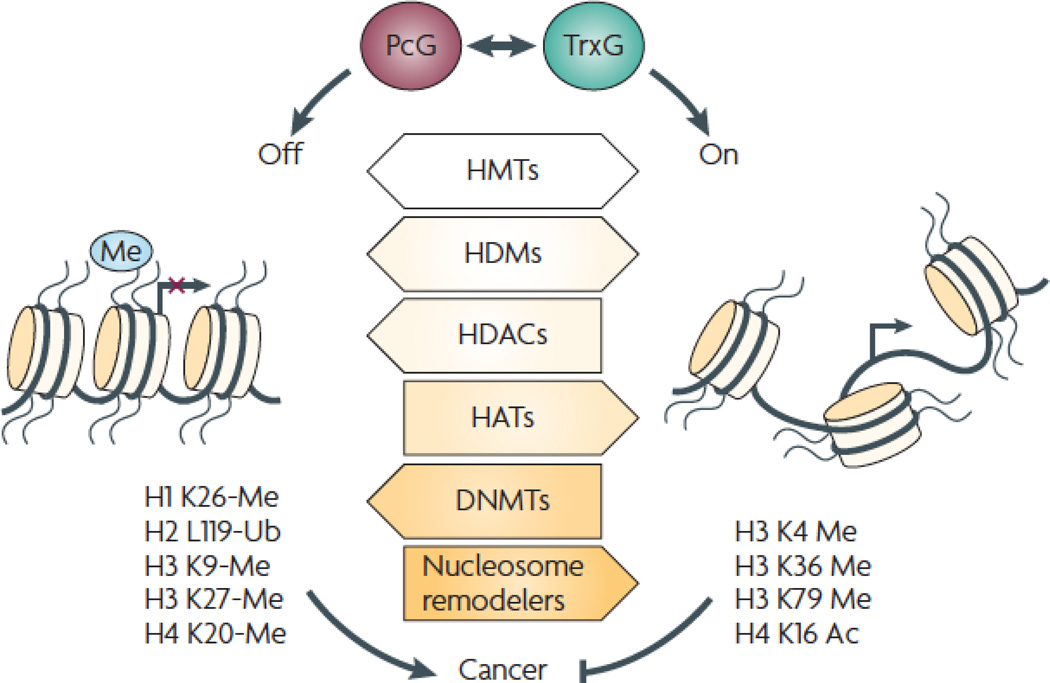
Chromatin structure is regulated by enzyme-containing PcG and TrxG complexes that keep chromatin in a “off” (left) or “on” (right) state, respectively, thereby regulating proteins controlling transcription, DNA replication and DNA repair. Chromatin plays a fundamental role in regulating biological processes that are commonly perturbed in cancer. An alteration in expression of proteins regulating chromatin structure and aberrancy of the specific chromatin marks they evoke, have been extensively documented in human cancer 18, 132–134. An imbalance in chromatin dynamics can lead to cancer by inactivating tumor suppressors, activating oncogenes, or reactivating pathways that inhibit differentiation or favor stem cell renewal. The three general strategies outlined below—histone modification, DNA methylation, and nucleosome remodeling, work together to regulate chromatin structure and to maintain cellular memory, regulating processes that frequently go awry in cancer.
Covalent modification of histones (white arrows): amino terminal histone tails protruding from nucleosomes can be modified at specific residues via methylation, acetylation, phosphorylation, ubiquitination, sumolation, ADP-ribosylation. These modifications are likely to affect the ability of chromatin readers to recognize and to bind to local nucleosomes. Enzymes that covalently modify histone residues include histone methyltransferases (HMTs) that establish the H3K27me3 mark characteristic of repressed genes (left), as well as HMTs that establish the H3K4me3 and H3K36me3 marks characteristic of actively transcribed genes (right). Histone demethylases (HDMs) remove these marks. Histone acetylases (HATs) acetylate histone tails, whereas histone deacetylases (HDACs) remove these acetyl groups. Acetylation of histones is a characteristic of the promoters of actively transcribed genes; therefore HDACs and HATs inhibit and activate gene expression, respectively.
DNA methylation (light gray arrow): DNA is methylated at CpG dinucleotides by DNA methyltransferases (DNMTs). DNA hypomethylation was the first epigenetic change to be reported in human cancer, leading to enhanced genomic instability. In contrast, hypermethylation of CpG islands located near promoters causes silencing of tumor suppressor genes such as hypermethylated in cancer 1 (HIC1), INK4/ARF, mutL homolog 1 (MLH1), Ras association domain family member 1 (RASSF1), and von Hippel-Lindau tumor suppressor (VHL)135.
Nucleosome remodelers (dark gray arrow): ATP-dependent chromatin remodeling complexes use the energy obtained from ATP hydrolysis to alter positioning, conformation, and/or composition of nucleosomes, serving as chromatin remodeling engines. Nucleosome dynamics has a dramatic effect on chromatin structure, with relatively nucleosome-free regions being associated with actively transcribed genes.
Subsequent genetic screens in D. melanogaster identified a second category of proteins, the trithorax group (TrxG) proteins, which counteract the action of PcG proteins (fig 1, box 1). This indicated that TrxG proteins function as ‘anti-silencers’ to activate expression of homeotic genes 8, 15, 16. Thus, PcG and TrxG proteins work together to regulate chromatin dynamics and gene expression cascades to retain cellular memory throughout the life of the organism 17. Not only is this PcG-TrxG-regulated cellular memory system evolutionarily conserved, an imbalance in this system in mammals has been associated with compromised stem cell renewal, as well as with an increased risk of cancer (fig 1b, c). Genetic lesions that cause inappropriate PcG and TrxG activity—such as alterations in expression level, mutational activation or inactivation, as well as chromosomal deletion, amplification or translocation—predispose to cancer (table 1). Anti-cancer therapies aimed at reversing perturbations in the PcG-TrxG cellular memory system have shown significant promise in the clinic 18. This Review focuses on recent advances in understanding how PcG-TrxG-mediated chromatin dynamics converge to regulate tumorigenesis.
Table.
| Complex |
Drosophita melanogaster |
Human | Domain | Activity | Examples of alterations in human cancer |
|---|---|---|---|---|---|
| PcG proteins: establishes of the histone code | |||||
| PRC2 | ESC | EED | WD40 repeats | Required for the HMT activity of EZH2 | Ewing’s sarcoma136, oral SCC137, breast cancer131,138,139, hepatocellular carcinoma140,141, prostate cancer131,142,143, melanoma131 and endometrial cancer131 |
| E(Z) | EZH1 and EZH2 | SET | HMT, establishes H3K27me3mark | ||
| SU(Z)12 | SUZ12 | Zinc finger | Required for the HMT activity of EZH2 | ||
| NURF55 | RBBP4 and RBBP7 | Histone binding | Forms minimal nucleosome-binding module with SU(Z)12 | ||
| PcG proteins: interpreters of the histone code | |||||
| PRC1 | PC | CBX2, CBX4 and CBX8 | Chromodomain | Binds H3K27me3 mark | Oesophageal cancer144, medulloblastoma145 leukaemia146, lymphoma147, neuroblastoma148, hepatocellular carcinoma140, 141, bladder cancer140, astrocytoma150 and prostate cancer142 |
| PH | PHC1, PHC2 and PHC3 | Zinc finger and SPM | Required for silencing | ||
| RING | RING1 and RNF2 | RING zinc finger | Ubiquitin ligase | ||
| PSC | BMI1, PCGF2 and ZNF134 | Zinc finger | Protein-protein interaction | ||
| TrxG proteins: histone modifiers | |||||
| MLL/ASH related | TRX | MLL, MLL2, MLL3 and MLL5 | SET and AT hook | HMT, establishes H3K4me3 mark and DNA binding | Leukaemia151–154 |
| ASH1 | ASH1L | SET | HMT, establishes H3K4me3 mark | ||
| ASH2 | ASH2L | ||||
| WDR5 | WD40 repeats | Essential for H3K4me3 | |||
| MEN1 | MEN1 and HCF1 | ||||
| TrxG proteins: nucleosome remodelers | |||||
| SWI-SNF | OSA | ARID1A and ARID1B | Rhabdoid tumours155–157, lung cancer158–161, oral cancer162 HNSCC163, gastric cancer164 and skin cancer165 | ||
| BRM | BRM | SWI-SNF-like helicase | ATPase activity | ||
| BRG1 | Bromodomain | Binds acetylated histones | |||
| MOIRA | SMARCC2 | ||||
| SNR1 | SNF5, ARID4A and ARID4B | Non catalytic core subunit | |||
| ISWI | ISWI | SMARCA1 | SANT and SWI-SNF like helicase | Histone binding and ATPase activity | |
| NURF301 | BPTF | PHD zinc finger and bromodomain | Binds H3K4me3 and binds acetylated histones | ||
| NURF55 | RBBP4andRBBP7 | ||||
| CHD | CHD1 | CHD1 and CHD2 | Chromodomain and SWI-SNF like helicase | ATPase activity | Glioma58, neuroblastoma166–171, meniniqioma172, melanoma173, pheochromocytoma174–177, oligodendroglioma178–180, CML181, non-Hodgkin's lymphoma182, T-ALL183, thyroid cancer184, colon cancer185, cervical cancer186, breast cancer187–189 and ovarian cancer190 |
| MI-2 | CHD3, CHD4 and CHD5 | Chromodomain, SWI-SNF-like helicase and PHD zinc finger | ATPase activity | ||
| KIS | CHD6, CHD7, CHD8, and CHD9 | Chromodomain, SWI-SNF-like helicase and PHD zinc finger | ATPase activity | ||
ARID, AT-rich interactive domain: ASH, absent small or homeotic; CBX, chromobox homologue; CHD, chromodomain helicase DNA binding; CML, chronic myeloid leukaemia; EED, embryonic ectoderm development: EZH, enhancer of zeste homologue; HCF1, host cell factor C1; HMT, histone methyltransferase; HNSCC, head and neck squamous cell carcinoma; KIS, kismet: MEN1, multiple endocrine neoplasia 1; MLL, mixed lineage leukaemia; NURF, nucleosome remodelling factor; PC, polycomb; PcG, polycomb group: PH, polyhomeoiic; PHC, polyhomeotic homologue; PHD, plant homeodomain; PRC, polycomb repressive complex; RBBP, retinoblastoma binding protein; SCC, squamous cell carcinoma; SUZ12, suppressor of zeste homologue: SPM; T-ALL, T cell acute lymphoblastic leukaemia; TrxG, trithorax group; WDR5, WD repeal domain 5.
PcG and TrxG complexes are dynamic regulators of chromatin
Histone modification
Covalent modification of specific residues within amino terminal tails of histones alters chromatin structure and function; the unique combination of modifications has been described as the histone code. Both PcG and TrxG proteins affect histone methylation, for example, but these opposing classes of proteins methylate distinct residues, resulting in different biological outcomes. PcG proteins establish histone modifications that repress transcription, whereas TrxG proteins establish histone modifications that activate transcription (box 1).
There are two core PcG complexes, each of which is made up of multiple subunits (fig 2a; table 1). Polycomb repressive complex 2 (PRC2) establishes the histone code, and polycomb repressive complex 1 (PRC1) interprets this code 19. The mammalian PRC2 complex contains enhancer of zeste homolog 2 (EZH2), embryonic ectoderm development (EED), suppressor of zeste homolog 12 (SUZ12), and retinoblastoma binding protein 4 (RBBP4) or RBBP7 20. EZH2 contains a SET domain and is a histone methyltransferase (HMT) that trimethylates histone H3 at lysine 27, thereby establishing H3K27me3 characteristic of inactive chromatin 21. Although PRC2 had also been reported to establish the H3K9me3 repressive mark 22, this has remained controversial. EED and SUZ12 do not possess HMT activity themselves, but these subunits are required for the HMT activity of EZH2 23. In D. melanogaster, PRC2 binds polycomb repressive elements (PREs) throughout the genome. PREs in mammals have been more difficult to identify, although PREs within the Krysler and Hox loci were recently characterized functionally 24, 25. H3K27me3, but not H3K9me3, is lost in PcG mutants, highlighting the importance of H3K27me3 in maintaining cellular memory.
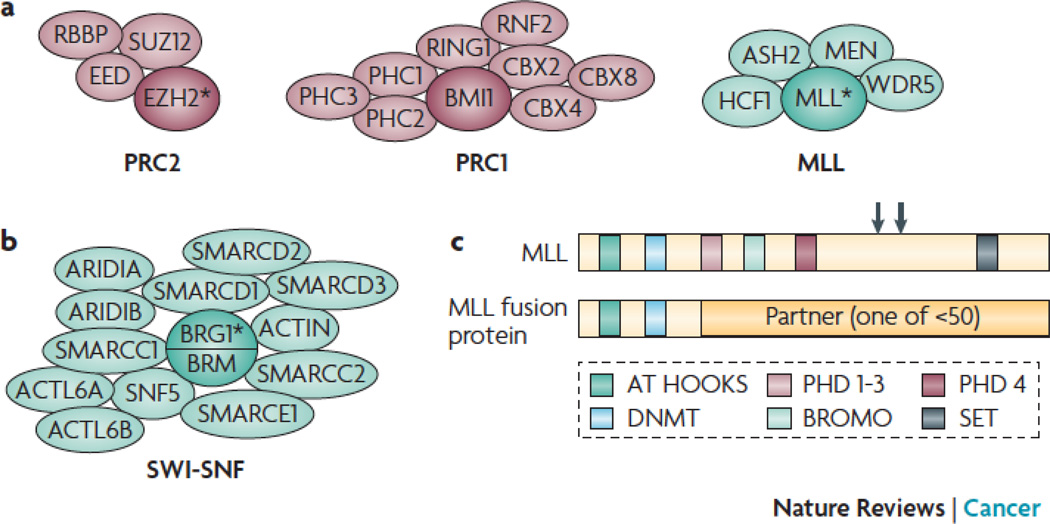
Figure 2. PcG and TrxG-containing complexes.
a. PcG complexes PRC2 and PRC1 (left) and the TrxG MLL complex (right) directly methylate histone tails via the HMT activity of the subunit depicted by an asterisk.
b. Some TrxG complexes such as SWI-SNF mobilize nucleosomes via the ATPase activity of the subunit depicted by an asterisk.
c. Other TrxG proteins such as MLL directly methylate histone tails via the HMT activity encoded by the SET domain (upper diagram). The common breakpoint region (large arrow) and TASPASE cleave sites (small arrows) are shown. Translocations involving MLL (lower diagram) are found in human cancer. These fusions typically retain the MLL N-terminal region, and are devoid of HMT activity.
The repressive histone marks established by PRC2 are recognized by PRC1, which stabilizes the inactive state so that transcriptional memory is maintained. There are multiple forms of mammalian PRC1 complexes, which may contain chromobox homolog 2 (CBX2), CBX4 or CBX8, polyhomeotic homolog 1 (PHC1), PHC2 or PHC3, BMI1 polycomb ring finger oncogene (BMI1), and ring finger protein 1 or 2 (RING1 or RNF2) 26 (fig 2a; table 1). CBX proteins contain a CHROMO domain, which recognizes and binds the H3K27me3 mark established by PRC2. BMI1, RING1, and RNF2 have ring-finger domains, a motif crucial for nuclear localization as well as for the oncogenic activity of BMI1 (discussed later) 27.
A working model for how PcG complexes modulate chromatin is that transcription factors and their associated machinery dictate loci destined for silencing, and PRC2 tags this target by methylating lysine 27 of histone H3. This H3K27me3 mark is recognized and bound by the chromodomain of CBX2, CBX4, or CBX8 within PRC1, which mediates transcriptional silencing by binding methylated histones in the vicinity and ubiquitinating H2A 28, 29. Although in vitro assays have suggested that PRC complexes mediate silencing by physically blocking the binding of transcriptional machinery at promoters 30, PRC complexes are more likely to function by impeding RNA polymerase II (pol II) initiation or elongation. Indeed, several groups have shown that pol II and basal transcription factors are present at PcG-silenced loci 31,32,33. While the hierarchical recruitment model in which PRC2 and PRC1 bind sequentially is attractive, recent evidence indicates that PRC2 and PRC1 bind some loci simultaneously, and that not all PRC2-bound loci are bound by PRC1 34. Furthermore, PRC1 can bind chromatin in the absence of PRC2 35, 36.
Like PcGs, TrxG proteins are also components of multi-subunit complexes, however, their composition is not as well characterized. TrxG proteins can, however, be broadly classified into two categories: the histone modifiers 37 and the nucleosome remodelers 38 (fig 2; table 1). The histone modifiers include HMTs that establish histone marks favoring transcriptional activation. TrxG complexes contain subunits that, like EZH2, are SET domain-containing HMTs, such as the founding member of the D. melanogaster TrxG family trithorax (TRX), and absent, small, or homeotic 1 (ASH1). However, in contrast to EZH2, which methylates histone H3 at lysine 27 to establish a repressive histone mark, TrxG HMTs methylate histone H3 at lysine 4 to establish active chromatin marks 39 (fig 2, fig 3a). The human TRX homolog myeloid-lymphoid or mixed-lineage leukemia (MLL) occupies the promoters of HOX genes, facilitating H3K4 methylation and transcriptional activation 40. Thus, both PcG and TrxG complexes regulate chromatin by directly methylating histones.
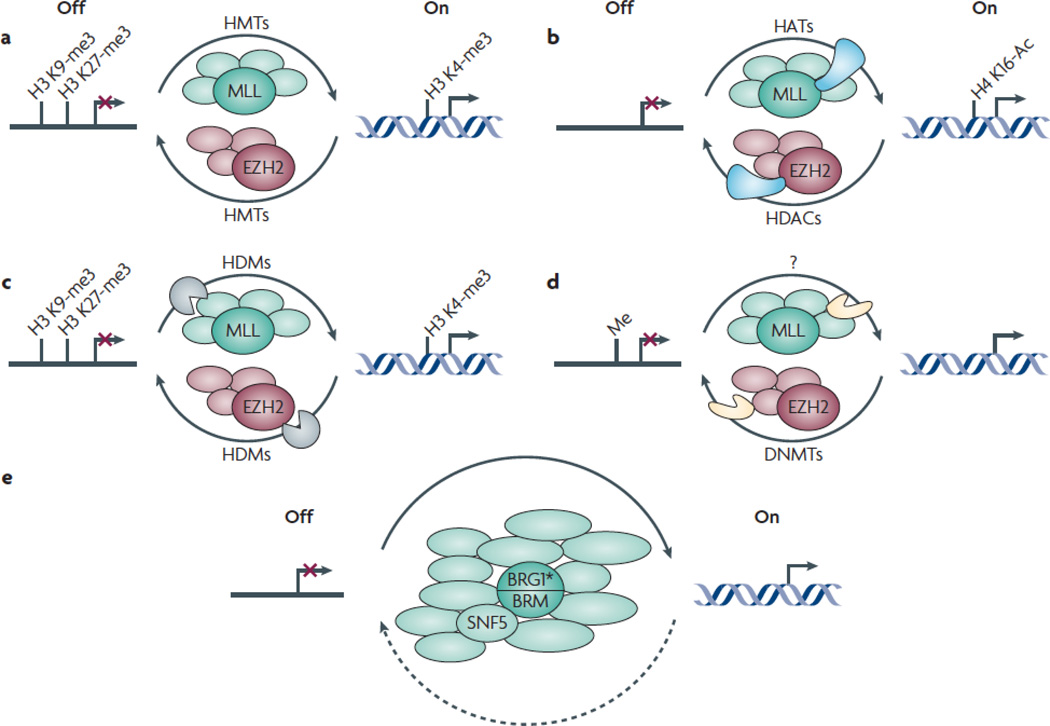
Figure 3. PcG and TrxG complexes affect transcription by modulating enzymatic activities in opposing ways.
PcG and TrxG-mediated regulation involves multiple enzymatic activities. The mechanisms depicted below are not mutually exclusive.
a. PcG and TrxG proteins directly methylate specific histone residues. The PcG and TrxG proteins EZH2 (pink) and MLL (green) are HMTs that establish repressive and activating histone marks, respectively.
b. PcG and TrxG complexes recruit enzymes that modulate acetylation of histone residues (blue). Whereas PcG complexes can associate with HDACs that deacetylate histone residues, TrxG complexes can associate with HATs that acetylate histone residues.
c. PcG and TrxG complexes recruit enzymes (brown) that remove specific methyl marks from histones. Whereas PcG complexes contain HDMs that remove activating marks, TrxG complexes contain HDMs that remove repressive histone marks.
d. PcG and TrxG complexes modulate enzymes (orange) that methylate DNA. Whereas PcG complexes recruit DNMTs that methylate and thereby transcriptionally silence specific genes, TrxG complexes evict DNMTs.
e. TrxG proteins remodel nucleosomes. The TrxG proteins BRG1 and BRM are ATP-dependent nucleosome remodelers (green).
In addition to methylation, PcG and TrxG complexes either directly or indirectly recruit proteins that facilitate other covalent histone modifications, such as acetylation (fig 3b). Histone acetylation status is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Acetylation of histones near promoters is associated with transcriptional activation; therefore, HATs and HDACs favor transcriptional activation and repression, respectively. PcG and TrxG complexes recruit these enzymes (fig 3b). For example, the HDACs sirtuin 1 (SIRT1) and HDAC2 have been purified within PRC complexes 20. By contrast, the HAT MYST1, which acetylates H4K16, a mark of actively transcribed genes, has been purified within MLL-containing complexes 41. Although ASH1 complexes are not fully characterized, ASH1 interacts with the HAT CREB binding protein (CBP, also known as CREBBP), which is a transcriptional co-activator 42. Importantly, acetylation prevents the establishment of H3K27me3. Thus, TrxG proteins facilitate transcriptional activation by establishing activating histone methylation and acetylation marks, while simultaneously preventing PcG complexes from depositing inactivating histone methylation marks. These complexes link histone methylation and histone acetylation. PcG and TrxG complexes are also able to recruit histone demethylases (HDMs), indicating that the reversal of PcG- and TrxG-mediated histone methylation regulates chromatin dynamics and transcription (fig 3c). In addition, PcG complexes can recruit DNA methyltransferases (DNMTs) (fig 3d), indicating that repressive methylation of histones is intimately linked to methylation of DNA—a mechanism by which tumor suppressor genes can be inactivated in human cancer (discussed below). PcG-TrxG not only regulates transcriptional on and off states, it is an adaptable system that allows for graded responses between these extremes. In this dynamic fashion, the bivalent chromatin state of pluripotent embryonic stem cells in which both activating and repressive histone marks are present 43 can be fine-tuned as differentiation proceeds, as was shown in cells of the ectodermal lineage 44. Thus, although PcG and TrxG proteins can directly methylate histones, they are also able to recruit additional players that greatly expand their repertoire of covalent histone and DNA modifying activities.
Nucleosome remodeling
The nucleosome repositioning class of TrxG complexes contain ATP-dependent chromatin remodeling that – along with their associated cofactors – modulate nucleosome structure 45, 46 (fig 2b, table 1). One of the first hints that TrxG proteins modulate chromatin dynamics by affecting nucleosomes was the realization that D. melanogaster BRM was homologous to Saccharomyces cerevisiae (S. cerevisiae) SWI2-SNF2 47. The observation that S. cerevisiae SWI2-SNF2 mutant phenotypes are repressed by mutations in nuclear histones firmly established a functional connection between SWI2-SNF2 and nucleosomes 48, 49. Subsequent work revealed that SWI2-SNF2 complexes consist of multiple subunits and modulate nucleosome position and or composition.
ATP-dependent chromatin remodeling enzymes contain ATPase subunits belonging to the SNF2 superfamily; the sequence of the ATPase domain further subdivides this superfamily 50. Mammalian SNF2 families are also characterized by domains homologous to D. melanogaster proteins: SWI-SNF subunits have a BROMO domain related to BRM, ISWI subunits have a SANT domain related to ISWI, and the CHROMO domain helicase DNA binding (CHD) family is characterized by CHROMO domains related to CHD1, Mi-2 and Kismet (table 1). Mammalian ATP-dependent nucleosome remodeling proteins enhance gene expression by facilitating transcriptional elongation by (pol II)51. Although nucleosome remodeling proteins were first associated with transcriptional activation, these complexes can also function as transcriptional repressors (fig 3e) 52. Models proposed for the mechanism by which nucleosome remodeling proteins affect gene expression include chromatin looping, nucleosome sliding, nucleosome eviction, and exchange of variant histones; these events facilitate binding of transcription factors 53 and basal transcription machinery 54. The histone modifying and the nucleosome remodeling classes of TrxG proteins are not mutually exclusive; TrxG proteins that are HMTs can recruit nucleosome remodeling TrxG proteins. For example, MLL has been purified from a complex containing WD repeat domain 5 (WDR5), a PHD-containing protein required for H3K4me3 that recruits the nucleosome remodeling factor (NURF) complex, indicating that that H3K4me3-mediated transcriptional activation is linked with nucleosome remodeling 55. Thus, the interplay between PcG and TrxG complexes integrate histone modifications, DNA methylation and nucleosome remodeling, affecting chromatin structure and gene expression in a complex and dynamic fashion. The varied composition of the complexes may reflect dynamic changes during development or within tissue-specific contexts, adding further diversity to the PcG-TrxG-regulated cellular memory system.
PcG-TrxG-mediated regulation of tumorigenesis
Gain of PcG and loss of TrxG is a common theme in human cancer, demonstrating the oncogenic and tumor suppressive role, respectively, of these complexes (fig 1c; table 1). For example, BMI1 is an oncogene 56, 57, whereas CHD5 is a tumor suppressor 58. Perturbations of other TrxG proteins such as MLL, however, cause malignancy through gain-of-function mechanisms. Mechanisms leading to deregulation of PcG proteins in cancer highlight the role of cell fate transcription factors and long non-coding RNAs, as recently reviewed 14. Many tumors have reactivated expression of stem cell-associated genes, such as HOX genes—the best characterized targets of PcG and TrxG proteins—consistent with the idea that cellular memory is compromised. PcG and TrxG complexes function reciprocally to regulate transcriptional programs affecting senescence, the cell cycle, apoptosis and genomic stability.
Cellular senescence
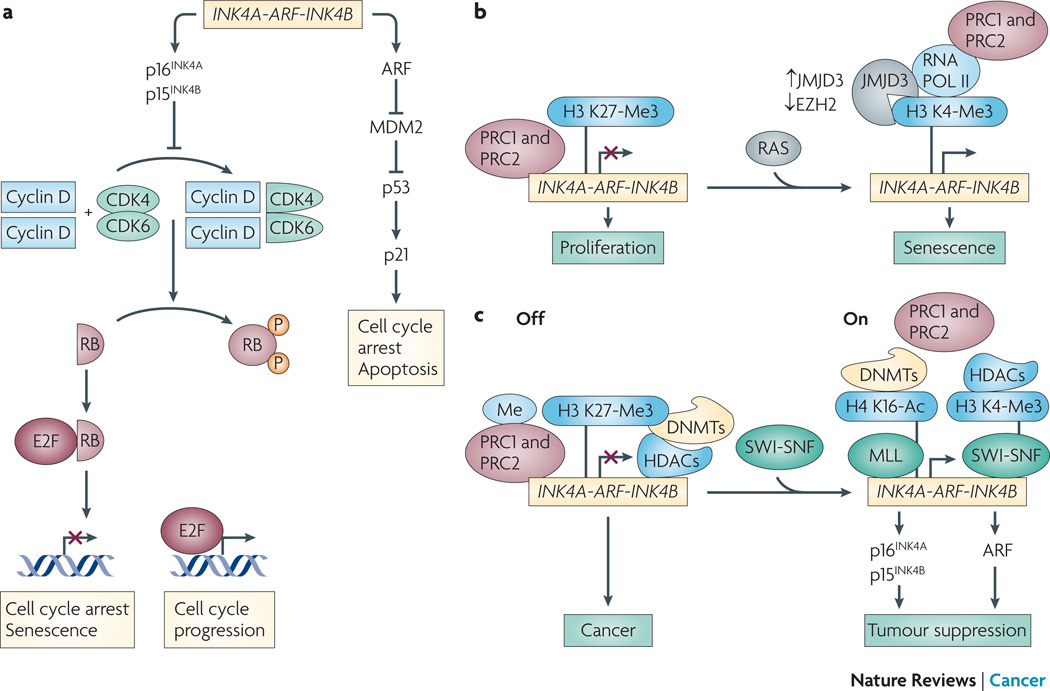
BMI1 was the first PcG protein implicated in cancer. It was initially identified as an oncogene that cooperates with MYC in lymphomagenesis 56, 57, and subsequent studies found it robustly expressed in human cancers (table 1). BMI1 facilitates tumorigenesis by mediating escape from cellular senescence, a state of cell cycle withdrawal that provides a potent tumor suppressive mechanism in vivo 59. The mechanism whereby BMI1 promotes evasion of senescence involves transcriptional silencing of the cyclin-dependent kinase inhibitor 2A (CDKN2A, which encodes both p16INK4A and p14ARF) and CDKN2B (which encodes p15INK4B) loci, mapping to human chromosome 9p21 20, 60. Silencing of CDKN2A and CDKN2B simultaneously shuts down expression of three tumor suppressors 61, 62 (figure 4a). INK4A (also known as p16) and INK4B (also known as p15) are CKIs that prevent CDK4-dependent phosphorylation of RB, thereby preventing E2F-mediated cell cycle progression. BMI1 coordinately represses CDKN2A and CDKN2B loci via CDC6 loading onto a cis-acting replication origin several thousand nucleotides upstream of the transcriptional start site 63. This is the first example of a replication-coupled transcriptional repression mechanism in mammals. Repression of CDKN2A and CDKN2B loci is a recurring theme in tumorigenesis: mouse models with deletions of this locus are tumor prone, and this locus is frequently inactivated in human cancers by deletion, mutation, or DNA hypermethylation 64.
Figure 4. PcG and TrxG proteins regulate the INK4a-ARF-INK4b in a dynamic and reversible manner.
a. The INK4/ARF locus (Ink4/Arf locus in mouse) encodes three tumor suppressors, p15INK4b, ARF (also called p19Arf in mouse) and p16INK4a. p16INK4a induces cell cycle arrest/senescence by inhibiting E2F-mediated transcription of target genes necessary for cell cycle progression. ARF induces cell cycle arrest/apoptosis by facilitating p53 activity via inhibition of MDM2-mediated degradation of p53. p16INK4a/RB- and ARF/p53-mediated tumor suppressive pathways play a nodal role in tumorigenesis. PcG and TrxG proteins function as repressors and activators of the INK4/ARF locus, respectively. b. Normal proliferating cells keep senescence in check by repressing the INK4a-ARFINK4b locus. PcG complexes PRC1 and PRC2 bind INK4a-ARF-INK4b under proliferating conditions, thereby inhibiting senescence. Expression of activated RAS results in recruitment of JMJD3 and pol II, and eviction of PRC proteins. c. TrxG-mediated chromatin dynamics can override PcG-mediated repression in cancer cells. PcG proteins PRC1 and PRC2 are localized at the INK4/ARF locus. This coincides with the recruitment of HDACs, DNMTs, and DNA methylation at the repressed locus in cancer cells. SWI/SNF recruitment to INK4/ARF causes displacement of PcG proteins, DNMTs, and HDACs. MLL is recruited, and the locus is marked with active chromatin marks such as H4K16Ac and H3K4me3, as well as decreased DNA methylation. This coincides with transcriptional activation and activation of p16INK4a-RB- and ARF-p53-mediated tumor suppression. Based on findings from 73.
Senescence limits proliferation, therefore proliferating cells must inhibit senescence to maintain proliferative potential; PcG-mediated transcriptional repression of CDKN2A plays a pivotal role in senescence inhibition (fig 4b) 65. Renewal capacity of both neural and hematopoietic stem cells is dramatically compromised in Bmi1−/− mice 66, 67, concomitant with homeotic phenotypes such as the 2nd cervical vertebrae having the identity of the 3rd, reminiscent of loss of cellular memory observed in D. melanogaster PcG mutants (fig. 1b) 68. These phenotypes are partially rescued by depletion of p16INK4A, underscoring the critical role of BMI1-mediated repression of this senescence inducer. Renewal capacity is also maintained in MEFs via PcG-mediated repression. Activated oncogenes such as H-RAS or BRAF—signals that cause senescence of proliferating MEFs—result in reversal of PcG-mediated repression of p16INK4A 69, 70. Oncogene-induced senescence coincides with loss of PcG protein (EZH2, SUZ12, BMI1 and CBX8) binding at the CDKN2A and CDKN2B loci, as well as transcriptional repression of EZH2, and coincident transcriptional activation and recruitment of the H3K27me3 HDM, lysine (K)-specific demethylase 6 (KDM6B, also known as jumonji domain containing 3, JMJD3). The repressive H3K27me3 marks are replaced with activating ones (H3K4me3), pol II is recruited and p16INK4A expression is induced, culminating in senescence. The presence of AP-1 consensus binding sites in the JMJD3 promoter suggests that this transcription factor plays a role in BRAF-mediated transcriptional activation of JMJD3 70. This indicates that the ability of normal primary cells to resist oncogene-mediated transformation depends on a switch between PcG-mediated repression and TrxG-mediated activation.
PcG-mediated repression is also reversible in cancer cells, as TrxG proteins can override PcG-mediated repression, leading to reactivation of tumor suppressors. SWI-SNF chromatin remodeling complexes contain an ATPase subunit (either BRM or BRG1) as well as the non-catalytic subunit (SNF5) that is common to various SWI-SNF-like complexes (fig 2) 71, 72. Loss of SNF5, BRM, and BRG1 has each been associated with human cancer (table 1). Malignant rhabdoid tumors (MRTs)—a rare but extremely aggressive form of childhood cancer—are caused by bi-allelic deletion or truncating mutations of SMARCB1 (which encodes SNF5).
Reintroduction of SNF5 into human MRT cells (which are deficient for this non-catalytic subunit of SWI-SNF) can alleviate PcG-mediated repression and reverse the tumorigenic phenotype (fig 4c) 73. Expression of SNF5 in MRT cells induces cell cycle arrest and senescence 74, 75, increasing expression of p16INK4A, increasing hypophosphorylated Rb, and compromising expression of E2F target genes 76, 77. SNF5 recruits the catalytic BRG1 subunit to the CDKN2A and CDKN2B promoters, facilitating binding of pol II. SNF5-mediated recruitment of BRG1 coincides with removal of PRC1 and PRC2 repressive complexes, as well as dissociation of DNMT3B and hence decreased DNA methylation 73. MLL is simultaneously recruited, repressive histone marks are replaced with active ones, and senescence is induced. SNF5–mediated senescence requires the ATPase activity of BRG1. This demonstrates that contrary to previous reports obtained in vitro 78, SWI-SNF activity can override PcG-mediated silencing, exemplifying the dynamic interplay between PcG and TrxG complexes.
The above studies in MRT cells showed that SNF5 induces senescence by activating p16INK4A-RB-mediated pathways. Although Smarcb1 (which encodes SNF5)+/− mice develop spontaneous MRTs of the brain 79–81, tumorigenesis was not increased in either Rb1 or Cdkn2aINK4A deficient backgrounds 82, 83. Similarly, although Smarca4 (which encodes BRG1)+/− mice develop mammary gland carcinomas 84, tumorigenesis was not increased in an Rb1+/− background 85. Although these findings implicate SWI-SNF proteins in tumor suppression in vivo, they do not support the findings that p16INK4A-Rb-mediated pathways are involved.
In contrast, evidence for a genetic connection between SNF5 and RB-regulated pathways was gained when it was found that inactivation of cyclin D1 inhibits tumorigenesis of Smarcb1+/− mice, placing cyclin D1 downstream of SNF5 86. To determine whether SWI-SNF complexes inhibit tumorigenesis through RB-related proteins, tumor incidence was monitored in Smarcb1+/− mice that also carried the T121 transgene—a truncated version of SV40 T-antigen that inhibits each of the three Rb family pocket proteins (Rb, p107, and p130) 85. Mice expressing the T121 transgene under control of the lymphotrophic papovavirus (LPV) promoter develop choroid plexus carcinomas (CPCs), a tumor type that also develops in patients with inactivating mutations in SNF5. Although CPC development is not enhanced in Smarcb1+/−; LPVT121 mice and the wild type Smarcb1 locus was retained, these mice develop MRTs with increased penetrance and decreased latency. Thus, SNF5 and RB family members cooperate to suppress cancer in a cell type that leads to MRTs, but not in a cell type that leads to CPCs 85.
TrxG proteins of the nucleosome remodeling class also regulate senescence by transcriptional modulation of CDKN2A. CHD5 was shown to be a tumor suppressor that regulates expression of p16INK4A and p19ARF 58. Mammalian CHD proteins are homologues of the D. melanogaster TrxG proteins Mi-2 and kismet (KIS), and based on structural homology to these TrxG proteins, CHD5 is proposed to function as a nucleosome remodeler 87. CHD3 and CHD4 (the CHD proteins most similar to CHD5) are homologs of D. melanogaster Mi-2α and Mi-2β, respectively; these proteins define the nucleosome remodeling and histone deacetylation (NURD) complex. The function of CHD5 in cancer is striking. Compromised CHD5—either by heterozygous deletion of the region of the chromosome encompassing Chd5 using chromosome engineering or by specific depletion of Chd5 in MEFs using RNAi—compromises p16INK4A and p19ARF expression and enhances proliferation 58. Whereas expression of oncogenic Ras (H-RasG12V) in wild type MEFs induces senescence and prevents tumor formation, Ras-mediated senescence is bypassed in MEFs in which Chd5 has been knocked down, leading to robust tumour formation in vivo 58. Conversely, gain of the chromosomal region encompassing Chd5 augments expression of p16INK4A and p19ARF, causing reduced proliferation and senescence in MEFs, as well as homeotic defects and stem cell depletion in vivo 58; (A.A.M. personal observations).
As was the case for Bmi1−/− mice, phenotypes resulting from gain of the chromosome region encompassing Chd5 were alleviated when levels of p16INK4A and p19ARF were reduced 58. The reciprocal nature of the cancer, homeotic and stem cell phenotypes of mouse models with altered dosage of Chd5 with those of Bmi1 58, 88, support the idea that CHD5 and BMI1 have opposing roles in vivo (fig 1). Recent work in D. melanogaster indicates that KIS counteracts PcG-mediated repression by recruiting TRX and ASH1 to chromatin, reversing the H3K27me3 repressive mark and facilitating transcriptional elongation by pol II 89. Whether CHD5 functions to counteract PcG-mediated repression awaits further study. These observations provide yet another example of the dynamic interplay between PcG-TrxG proteins in cancer and underscore the critical role that gene dosage plays in this process.
Cell cycle
Whereas the findings discussed above indicate that TrxG proteins are tumor suppressors, the role of MLL in cancer is more complex. MLL interacts with a number of proteins, including PcG (BMI1, HPC2), TrxG (SNF5, WDR5), and their accessory proteins (RBBP5, histone deacetylase, histone acetylase p300/CBP. Thus, translocations involving human chromosome 11q23—the most frequent genetic lesion in leukaemia 90, 91 that result in MLL fusions—likely disrupt PcG-TrxG-mediated chromatin dynamics. This idea is supported by the observation that MLL fusions lose their HMT domain yet retain the PcG-interacting regions 92. MLL is normally expressed in a cell cycle-dependent manner; it is dynamically regulated by SCFSKP2 and anaphase promoting complex (APC)CDC20-mediated degradation, resulting in two peaks of expression that facilitate G1-S and G2-M transitions 93. In contrast to wild type MLL, MLL fusions are insensitive to cell cycle-coupled degradation, resulting in constitutive levels of MLL throughout the cell cycle and inappropriate expression of target genes 94.
In addition to being tightly coupled with cell cycle transitions, MLL is cleaved by the endopeptidase taspase 1 (TASP1) to generate amino- and carboxy-terminal peptides that heterodimerize to form functional MLL 95–97. MLL is not cleaved in Tasp1−/− cells, compromising HMT activity 98. The HMT activity of MLL appears critical for its ability to transactivate its targets, as MLL increases H3K4me3 and activates transcription of E2F-regulated genes required for cell cycle progression 99. The co-regulator host cell factor C1 (HCF1) interacts with E2F1 and recruits MLL, thereby serving as a switch during the transition between G1- and S-phase. Mature MLL transcriptionally modulates both cyclins and CKIs; in the absence of TASP1-mediated cleavage of MLL, expression of cyclins A, E, and B are reduced and p16INK4A and p19ARF are induced, inhibiting cell cycle progression 98. Thus, MLL is intimately associated with the cell cycle on two different levels: it directly regulates expression of key cell cycle regulators, and its level oscillates with cell cycle transitions.
The current view is that the oncogenic activity of MLL fusions are through a gain-of-function mechanism. Mll-deficient mouse models are inviable 100, 101. Mll+/− mice are not tumor prone, but have aberrant expression of Hox genes and homeotic defects such as cervical vertebrae with an identity that is inappropriate for that portion of the skeleton, reminiscent of the phenotypes observed in D. melanogaster. On the other hand, leukemia cells with MLL fusions have expression patterns typical of embryonic stem cells, consistent with the idea that cellular identity has been lost.
How can MLL fusions activate gene expression when the HMT-encoding C-terminus is lacking? 102,90,103,91. Intriguingly, the MLL fusion partner AF10 interacts with DOT1-like histone H3 methyltransferase (DOT1L) 104, a non-SET domain-containing HMT that trimethylates H3K79, a mark of actively transcribed genes 105–107. An artificial MLL-DOT1L fusion not known to occur in patients transforms myeloid cells in a manner similar to the tumor-derived MLL-AF10 fusion, and transformation activity is dependent upon DOT1L-mediated H3K79me3 and enhanced HOXA9 expression 104. The finding that MLL-DOT1L and MLL-AF10 fusions have similar transactivation activities suggested a model in which the MLL-AF10 fusion causes abnormal recruitment of DOT1L, changing H3K4me3 to H3K79me3 at specific target loci, including HOXA9 104. DOT1L activity is required for both establishment and maintenance of the transformed state in leukaemia cells with the MLL-AF10 fusion, suggesting that targeting this HMT may pose a new therapeutic strategy for leukemias caused by this MLL fusion. It is possible that other MLL fusions are also capable of recruiting “moonlighting” HMTs, an idea that requires further study.
Other MLL fusions can also recruit additional proteins that facilitate malignancy. The MLL fusion partner AF9 interacts with YEATs domain containing 4 (YEATS4), a protein aberrantly upregulated in neuroblastoma 108 and required for repression of p53 109. The ability of AF9 to recruit YEATS4 is retained in the MLL-AF9 fusion, providing a mechanism for aberrant inactivation of p53. The finding that YEATS4 can recruit SNF5, suggests that this MLL-AF9-YEATS4 triad promotes tumorigenesis by recruiting SWI-SNF complexes 110. Aberrant recruitment of SWI-SNF complexes may also be a strategy used by the MLL-ENL fusion protein, as ENL has been purified from complexes that include SWI-SNF subunits 111. In support of this model, MLL-ENL recruits SWI-SNF to the HOXA7 promoter resulting in increased expression of HOXA7—a protein essential for MLL-ENL-mediated oncogenesis 103.
MLL fusions have also been found in complexes containing JMJD3, suggesting that MLL fusions recruit machinery that removes repressive histone marks 112, 113. These findings exemplify the different means by which leukemogenic TrxG fusions exploit various components of the epigenetic machinery. Thus, MLL fusions drive lymphomagenesis through a gain-of-function mechanism involving misregulation of MLL during the cell cycle or aberrant reactivation of specific developmental programs through opportunistic recruitment of PcG-TrxG-associated machinery.
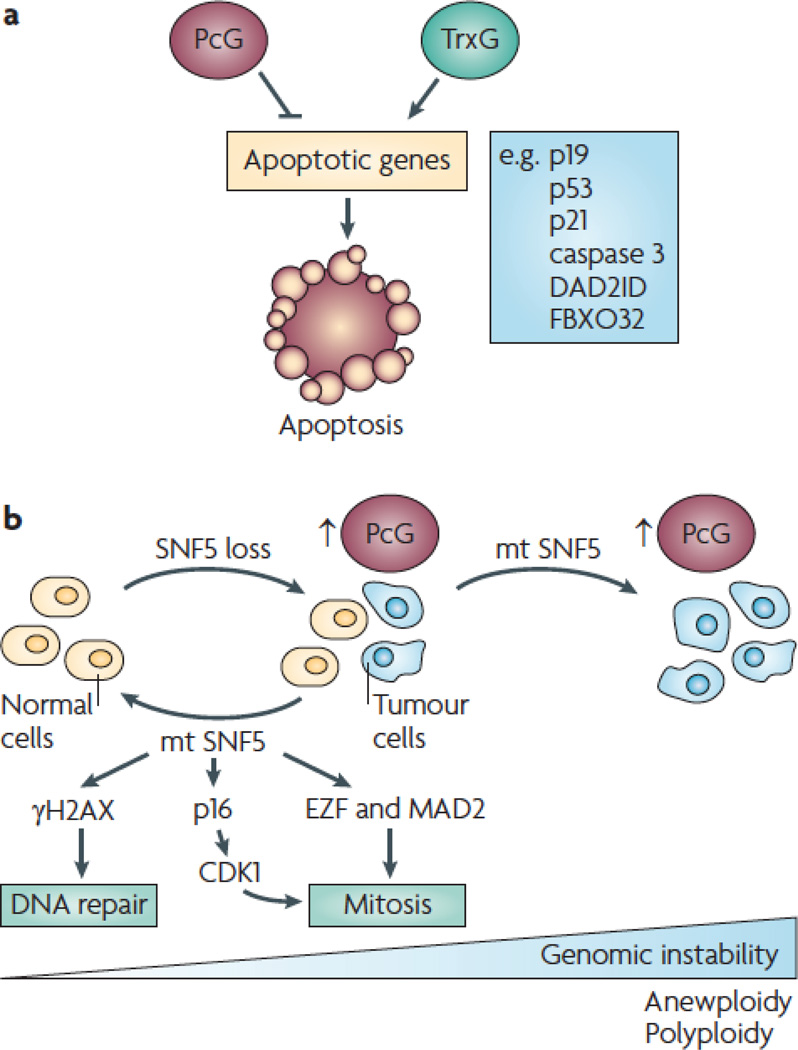
Apoptosis
PcG-TrG-mediated chromatin dynamics affects expression of genes that induce apoptosis (fig 5a). The general theme is that PcG proteins inhibit- whereas TrxG proteins induce-, apoptosis. For example, the ability of PcG and TrxG to work in opposing ways to regulate the CDKN2A locus discussed above, has a dramatic affect on ARF expression. As an inhibitor of MDM2-mediated ubiquitylation and degradation of p53, ARF facilitates apoptosis; therefore, PcG and TrxG repress and evoke apoptosis, respectively.
Figure 5. PcG and TrxG complexes modulate apoptosis and genomic stability.
a. PcG and TrxG proteins repress- and activate expression of genes required for apoptosis. b. Loss of the TrxG protein SNF5 leads to genomic instability and cancer. Re-expression of wild type SNF5 in reestablishes a diploid population of cells, whereas expression of tumor-derived SNF5 exacerbates the polyploidy and aneuploid population of cells.
Loss of PcG repression causes excessive p19ARF-p53-mediated apoptosis. Indeed, Bmi1−/− mice have increased apoptosis in lymphoid tissues, a phenotype that is rescued by depleting p16INK4A and p19ARF or by overexpressing the apoptosis inhibitor Bcl2 61. Apoptosis is also enhanced in Bmi1+/−; EµMyc mice relative to Bmi1+/+; EµMyc mice, compromising lymphomagenesis. Lymphoma development in Bmi1+/−; EµMyc mice requires repression of p19ARF, as apoptosis is compromised in Bmi1+/−; EµMyc; Cdkn2a+/− mice, enhancing lymphomagenesis 61. Gain of PcG-mediated activity has the opposite effect: BMI1 cooperates with oncogenic H-Ras in breast cancer cells by suppressing apoptosis and attenuating p53, p21 and caspase 3, leading to highly malignant tumors that metastasize to the brain 114. BMI1 is essential for the survival and proliferation of both normal stem cells and leukemia-propagating cells; depletion of BMI1 causes apoptosis, as well as senescence 115. These findings underscore the role of PcG-mediated repression in modulating apoptosis via p19ARF-mediated pathways.
PcG proteins can also affect apoptosis by modulating expression of genes besides p19ARF. PRC2-mediated recruitment of DNMTs can silence specific loci, such as those encoding the apoptotic machinery 116. An ‘apoptotic methylation signature’ has been suggested as a strategy for diagnosing cancer 117. EZH2 suppresses expression of the apoptosis mediator DAB2 interacting protein (DAB2IP) in prostate cancer, thereby inhibiting tumour necrosis factor (TNF)-mediated apoptosis 118. Yet, this repression is reversible, as HDAC inhibitors can induce apoptosis 119,120, in some instances sensitizing tumor cells to TNF-related apoptosis inducing ligand (TRAIL)-induced apoptosis 121. The reversibility of this repression is underscored by the ability of the global histone methylation inhibitor DZNep to remove PRC2 components (EZH2, EED, SUZ12) from chromatin, reduce H3K27me3, and activate expression of the apoptosis effector F-box-only protein 32 (FBXO32) in breast cancer but not in normal cells 122.
TrxG proteins also modulate apoptosis. For example, conditional inactivation of SNF5 in MEFs compromises cell survival, induces expression of p53 and p21, leading to apoptosis 82, 123. Whereas p53 deficiency inhibited apoptosis of SNF5−/− MEFs, p53 loss did not reverse their proliferation defect or rescue the embryonic lethality of Smarcb1−/− mice 123. However, tumour onset was extremely rapid when Snf5 and p53 were co-inactivated using a Smarcb1 inverting conditional-p53 conditional-Mx-Cre model, presumably because apoptosis was suppressed 82.
CHD5 also regulates p53-mediated apoptosis. Whereas loss of Chd5 compromises p53 activity and leads to tumorigenesis in vivo, gain of Chd5 increases senescence of MEFs in culture and triggers excessive apoptosis and perinatal lethality in vivo 58. In contrast to the lethal apoptotic phenotype caused by SNF5 deficiency 123, the apoptotic phenotypes caused by increased dosage of the region encompassing Chd5 were rescued in a p53 compromised background 58. It remains to be determined whether p53 deficiency exacerbates tumorigenesis in mice with compromised Chd5.
In contrast to other TrxG proteins that induce apoptosis, some MLL fusions inhibit apoptosis. This further supports the idea that MLL fusions work by a gain of function mechanism.
DNA damage response and genomic integrity
SWI-SNF complexes suppress cancer by maintaining the genome 77 (fig 5b). Proliferation of human MRT cells could still be inhibited by expression of a tumor-derived SNF5 mutant, indicating that loss of cell cycle control was not the only thing responsible for tumorigenesis 124. A subset of SNF5 deficient tumor cells grown in culture had multi-lobed nuclei and were polyploid or aneuploid, indicating that the mitotic checkpoint was defective 124. Wild type SNF5 inhibited proliferation of these abnormal cells, thereby establishing a population of cells that were essentially all diploid. By contrast, expression of tumor-derived SNF5 mutants exacerbated genomic instability. Although wild type SNF5 inhibited proliferation of aneuploid cells, this was not the case when a version of CDK4 that could not be inhibited by p16INK4A was simultaneously expressed, indicating that p16INK4A-mediated pathways are essential for SNF5 to re-establish genomic integrity 124. MEFs in which SNF5 was conditionally ablated were hypersensitive to DNA damage caused by cross-linking (UV irradiation) and double strand breaks (doxorubicin) 123. MEFs in which SNF5 was ablated had altered nuclear structure and extra centrosomes, indicating that SNF5 deficiency leads to cytokinesis failure, polyploidy, and genomic instability (fig 5b). E2F1 and its downstream target, Mad2—a protein implicated in mitotic defects and aneuploidy 125—were downregulated by SNF5 expression 124. Further, SWI-SNF complexes facilitate double-strand break repair by inducing the activity of gamma-H2AX at DNA lesions 126. Yet, results of a separate study—in which the effects of SNF5 ablation was assessed at an earlier time point—indicated that SNF5 loss does not cause hypersensivity to DNA damage and does not compromise the DNA damage checkpoint 127, in agreement with the finding that human MRT cell lines have a normal cell cycle checkpoint in response to DNA damage 128.
In vivo evidence has also pointed to a role for SWI-SNF in regulating genomic stability. Mammary carcinomas of Smarca4+/− mice are genomically unstable, but in contrast to MRT cells maintained in culture, these tumors are not aneuploid 84. Human MRTs as well as MRTs that develop in Smarcb1+/− mice are diploid and genomically stable 127. This implies that disruption of SWI-SNF-mediated chromatin remodeling activity can substitute for genomic instability in cancer. Thus, perturbation of epigenetic events can contribute to cancer in the absence of genetic lesions 129. These findings support the hypothesis that SWI-SNF proteins maintain genomic stability by coupling cell cycle and DNA damage checkpoints.
Conclusions and future perspectives
The mechanism whereby the PcG-TrxG-regulated cellular memory system goes awry during tumorigenesis is just beginning to emerge, unveiling the dynamic and complex nature of this epigenetic process. The ability to regulate cancer above and beyond the level of the DNA offers hope that cancer-causing perturbations can be reversed. Whereas classic tumor suppressors are lost or inactivated during tumorigenesis, a number of genes encoding TrxG proteins are retained. The unique strategies that these modulators of nucleosome structure utilize in malignant cells to circumvent the gauntlet of growth control mechanisms not only helps to elucidate the fascinating biology underlying chromatin regulation, it suggests that modulation of this system is useful in the clinic. First, aberrant expression of PcG-TrxG or the stem cell-associated transcriptional programmes they regulate may be used for cancer diagnosis. For example, the ability of BMI1 to cooperate with oncogenic H-Ras leads to aggressive and highly metastatic tumors that colonize the brain or lung, therefore BMI1 levels may help to classify tumor subtypes 114. Second, PcG-TrxG status may be used to predict survival or treatment outcome. For example, in breast tumours, increased expression of EZH2 is an especially powerful indicator of poor prognosis when coupled with low expression of p57KIP2 (one of the CKIs that is silenced by EZH2) 130. High expression of EZH2 is also a predictor of poor response to therapies for prostate cancer 131. Third, strategies that reestablish the PcG-TrxG balance may be useful in treating cancer. HDAC- and DNMT inhibitors—anti-cancer treatments currently being used in the clinic—effectively reverse H3K27me3, leading to transcriptional activation of cell cycle regulators 130. Development of new HDAC- and DNMT inhibitors are likely to help in this regard. Importantly, the reversibility of PcG-mediated transcriptional repression has important implications for the feasibility of reactivating loci that have been silenced in cancer. Future work that provides insight into the mechanisms whereby PcG-TrxG-mediated pathways converge to regulate cancer should continue to provide new insight.
Box 1.
Acknowledgements
Thanks to Jim Duffy and Yow-Ning Chang for assistance in preparing this manuscript, and to members of my laboratory for helpful discussions.
Glossary terms
- Cancer stem cell hypothesis
The theory that that tumors are composed of a heterogeneous population of cells, including a subset of relatively rare cancer stem cells or “tumor-propagating” cells essential for malignancy.
- Clonal evolution model
The theory that tumor-propagating cells make up a significant proportion of cells within a tumor, and the malignant state is the consequence of selection of dominant clones within the population.
- Body segments
Transcriptional cascades set up during development of D. melanogaster partition the embryo into 14 sections or “parasegments” that correspond to the final anatomical regions of the adult.
- Chromatin
A highly dynamic structure consisting of DNA plus nucleosomes. Each nucleosome unit (comprised of 147 nucleotides of DNA wrapped around a core of histones consisting of a histone 3-histone 4 tetramer and two histone 2A-histone 2B dimers) is separated by a segment of linker DNA that is bound by histone 1.
- Histone code
The concept that the establishment and interpretation of an intricate pattern of histone tail modifications orchestrate chromatin dynamics and gene expression.
- SET domain
The Su(var)3–9, Enhancer of zeste, Trithorax (SET) domain is a ~130 amino acid motif that provides the HMT activity in a variety of proteins that methylate specific lysine residues of histones as well as other protein substrates.
- Polycomb repressive element (PRE)
Specific DNA sequences identified in D. melanogaster that are recognized and bound by PRC2 complexes.
- CHROMO domain
The CHRromatin Organization Modifier (CHROMO) domain is a ~60 amino acid motif originally discovered in HP1 and PcG proteins from D. melanogaster; they are present in a variety of chromatin regulating proteins.
- Ring-finger domain
The Really Interesting New Gene (RING) domain is an ~40–60 residue Cys4-His-Cys3-containing motif that binds zinc, facilitating interactions with DNA, RNA, proteins, and lipids.
- SWI2-SNF2
A complex identified in yeast that is required for transcriptional activation of ~7% of the genome, including the mating type switch (SWI) and the sucrose non fermenting (SNF) genes for which SWI2-SNF2 are named.
- BROMO domain
A motif that binds acetylated lysines present on histone tails.
- PHD finger
The Plant Homeo Domain (PHD) is 50–80 residue Cys4-His-Cys3-containing motif originally identified in Arabidopsis thalinia that binds specifically modified histones.
- Chromosome engineering
A Cre-loxP-based ES cell technology that enables the generation of mouse models harbouring defined chromosome rearrangements such as deletions, duplications, inversions, and translocations.
References
- 1.Wang JCa, D JE. Cancer stem cells: Lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Clarke MF, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 3.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 4.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nature Reviews Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 5.Hill RP. Identifying cancer stem cells in solid tumors: case not proven. Cancer Res. 2006;66:1891–1895. doi: 10.1158/0008-5472.CAN-05-3450. discussion 1890. [DOI] [PubMed] [Google Scholar]
- 6.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: transregulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 8.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 9.Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- 10.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Kerppola TK. Polycomb group complexes--many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 14. Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. This review highlights the intriguing role of cell fate transcription factors and long non-coding RNAs in regulation of PcG function in cancer.
- 15.Breen TR, Harte PJ. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech Dev. 1991;35:113–127. doi: 10.1016/0925-4773(91)90062-b. [DOI] [PubMed] [Google Scholar]
- 16.Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- 17.Brock HW, van Lohuizen M. The Polycomb group - no longer an exclusive club? Current Opinion in Genetics & Development. 2001;11:175–181. doi: 10.1016/s0959-437x(00)00176-3. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine SS, King IFG, Kingston RE. Division of labor in Polycomb group repression. Trends in Biochemical Sciences. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 21.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 22.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 23.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Sing A, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 25. Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. References 24–25 provide the first functional evidence for the presence of PREs in mammals.
- 26.Levine SS, et al. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.David Allis C, Danny Reinberg TJ. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 28.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 29.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 31.Dellino GI, et al. Polycomb silencing blocks transcription initiation. Mol Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- 32.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 34.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 38.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 40.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 41.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Petruk S, et al. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 44.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 46.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 47.Tamkun JW, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 48.Kruger W, et al. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 49.Recht J, Osley MA. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. Embo J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17:1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 54.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 55. Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. This seminal paper links covalent histone modification to chromatin remodelling.
- 56.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 57.van Lohuizen M, et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 58. Bagchi A, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. This manuscript identified CHD5 as a tumor suppressor, providing the first functional evidence that members of the CHD chromatin remodeling family modulate tumorigenesis.
- 59. Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. This review discusses evidence for cellular senescence as a tumor suppressive mechanism and its utility for monitoring the success of anti-cancer therapies.
- 60.Bruggeman SW, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jacobs JJ, et al. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c- Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 63. Gonzalez S, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. This groundbreaking paper identified a cis-acting element that functions to coordinately regulate expression of the INK4A-INK4B locus, providing the first example of replication-coupled transcriptional regulation in mammals.
- 64.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 65.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4AARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 67.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Lugt NM, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 69.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. References 70–71 provide important mechanistic insight into PcG-TrxG-mediated chromatin dynamics in response to activated oncogenes.
- 71.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 72.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 73. Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARFINK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. This work demonstrates that TrxG chromatin remodeling proteins can override PcG-mediated repression in cancer cells.
- 74.Ae K, et al. Chromatin remodeling factor encoded by ini1 induces G1 arrest and apoptosis in ini1-deficient cells. Oncogene. 2002;21:3112–3120. doi: 10.1038/sj.onc.1205414. [DOI] [PubMed] [Google Scholar]
- 75.Versteege I, Medjkane S, Rouillard D, Delattre O. A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle. Oncogene. 2002;21:6403–6412. doi: 10.1038/sj.onc.1205841. [DOI] [PubMed] [Google Scholar]
- 76.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- 77.Imbalzano AN, Jones SN. Snf5 tumor suppressor couples chromatin remodeling, checkpoint control, and chromosomal stability. Cancer Cell. 2005;7:294–295. doi: 10.1016/j.ccr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 78.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 79.Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guidi CJ, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Isakoff MS, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guidi CJ, et al. Functional interaction of the retinoblastoma and Ini1/Snf5 tumor suppressors in cell growth and pituitary tumorigenesis. Cancer Res. 2006;66:8076–8082. doi: 10.1158/0008-5472.CAN-06-1451. [DOI] [PubMed] [Google Scholar]
- 84.Bultman SJ, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- 85.Chai J, et al. Tumor-specific cooperation of retinoblastoma protein family and Snf5 inactivation. Cancer Res. 2007;67:3002–3009. doi: 10.1158/0008-5472.CAN-06-4207. [DOI] [PubMed] [Google Scholar]
- 86.Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc Natl Acad Sci U S A. 2005;102:12129–12134. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 88.Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 89.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 91.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 92.Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 93.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21:2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liu H, Takeda S, Cheng EH, Hsieh JJ. Biphasic MLL takes helm at cell cycle control: implications in human mixed lineage leukemia. Cell Cycle. 2008;7:428–435. doi: 10.4161/cc.7.4.5426. References 93–94 discovered that MLL levels oscillate as a function of the cell cycle, and that MLL fusion proteins found in human leukemia are resistant to this regulation.
- 95.Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 96.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 97.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takeda S, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–2409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 100.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci U S A. 1998;95:10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yagi H, et al. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92:108–117. [PubMed] [Google Scholar]
- 102.Kroon E, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. Embo J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 105.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 106.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 107.Ng HH, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Munnia A, et al. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene. 2001;20:4853–4863. doi: 10.1038/sj.onc.1204650. [DOI] [PubMed] [Google Scholar]
- 109.Park JH, Roeder RG. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol Cell Biol. 2006;26:4006–4016. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Debernardi S, et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99:275–281. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- 111.Nie Z, et al. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol Cell Biol. 2003;23:2942–2952. doi: 10.1128/MCB.23.8.2942-2952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 113.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 114.Hoenerhoff MJ, et al. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009 doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 116.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 117.Murphy TM, Perry AS, Lawler M. The emergence of DNA methylation as a key modulator of aberrant cell death in prostate cancer. Endocr Relat Cancer. 2008;15:11–25. doi: 10.1677/ERC-07-0208. [DOI] [PubMed] [Google Scholar]
- 118.Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem. 2005;280:22437–22444. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 119.Marchion D, Munster P. Development of histone deacetylase inhibitors for cancer treatment. Expert Rev Anticancer Ther. 2007;7:583–598. doi: 10.1586/14737140.7.4.583. [DOI] [PubMed] [Google Scholar]
- 120.Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin Exp Metastasis. 2008;25:183–189. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- 121.Fulda S. Modulation of TRAIL-induced apoptosis by HDAC inhibitors. Curr Cancer Drug Targets. 2008;8:132–140. doi: 10.2174/156800908783769355. [DOI] [PubMed] [Google Scholar]
- 122.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2- mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–2674. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vries RG, et al. Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev. 2005;19:665–670. doi: 10.1101/gad.335805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 126.Park JH, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. Embo J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. McKenna ES, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol Cell Biol. 2008;28:6223–6233. doi: 10.1128/MCB.00658-08. This manuscript supports the concept that inactivation of TrxG chromatin remodeling proteins can substitute for genomic instability in cancer.
- 128.Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65:10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 129.Sansam CG, Roberts CW. Epigenetics and cancer: altered chromatin remodeling via Snf5 loss leads to aberrant cell cycle regulation. Cell Cycle. 2006;5:621–624. doi: 10.4161/cc.5.6.2579. [DOI] [PubMed] [Google Scholar]
- 130.Yang X, et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 132.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 133.Lin JC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends in Molecular Medicine. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 135.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 136.Richter GH, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kidani K, et al. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol. 2009;45:39–46. doi: 10.1016/j.oraloncology.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 138.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pietersen AM, et al. EZH2 and BMI1 inversely correlate with prognosis and TP53 mutation in breast cancer. Breast Cancer Res. 2008;10:R109. doi: 10.1186/bcr2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sasaki M, et al. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–882. doi: 10.1038/labinvest.2008.52. [DOI] [PubMed] [Google Scholar]
- 141.Yonemitsu Y, et al. Distinct expression of polycomb group proteins EZH2 and BMI1 in hepatocellular carcinoma. Hum Pathol. 2009 doi: 10.1016/j.humpath.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 142.van Leenders GJ, et al. Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur Urol. 2007;52:455–463. doi: 10.1016/j.eururo.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 143.Hoffmann MJ, et al. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B, are associated with DNA methylation changes in prostate cancer. Cancer Biology & Therapy. 2007;6:1403–1412. doi: 10.4161/cbt.6.9.4542. [DOI] [PubMed] [Google Scholar]
- 144.He XT, et al. Association between Bmi1 and clinicopathological status of esophageal squamous cell carcinoma. World Journal of Gastroenterology. 2009;15:2389–2394. doi: 10.3748/wjg.15.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leung C, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- 146.Mohty M, Yong AS, Szydlo RM, Apperley JF, Melo JV. The polycomb group BMI1 gene is a molecular marker for predicting prognosis of chronic myeloid leukemia. Blood. 2007;110:380–383. doi: 10.1182/blood-2006-12-065599. [DOI] [PubMed] [Google Scholar]
- 147.van Galen JC, et al. Expression of the polycomb-group gene BMI1 is related to an unfavourable prognosis in primary nodal DLBCL. J Clin Pathol. 2007;60:167–172. doi: 10.1136/jcp.2006.038752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nowak K, et al. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 2006;34:1745–1754. doi: 10.1093/nar/gkl119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shafaroudi AM, et al. Overexpression of BMI1, a polycomb group repressor protein, in bladder tumors: a preliminary report. Urol J. 2008;5:99–105. [PubMed] [Google Scholar]
- 150.Tirabosco R, De Maglio G, Skrap M, Falconieri G, Pizzolitto S. Expression of the Polycomb-Group protein BMI1 and correlation with p16 in astrocytomas an immunohistochemical study on 80 cases. Pathol Res Pract. 2008;204:625–631. doi: 10.1016/j.prp.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 151.Ziemin-van der Poel S, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci U S A. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gu Y, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 153.Domer PH, et al. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci U S A. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Thirman MJ, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 155.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 156.Sevenet N, et al. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet. 1999;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- 157.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 158.Reisman DN, et al. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21:1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- 159.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 160.Medina PP, et al. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRGI, in lung tumors. Genes Chromosomes & Cancer. 2004;41:170–177. doi: 10.1002/gcc.20068. [DOI] [PubMed] [Google Scholar]
- 161.Medina PP, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 162.Gunduz E, et al. Genetic and epigenetic alterations of BRG1 promote oral cancer development. Int J Oncol. 2005;26:201–210. [PubMed] [Google Scholar]
- 163.Gunduz E, et al. Loss of heterozygosity at the 9p21-24 region and identification of BRM as a candidate tumor suppressor gene in head and neck squamous cell carcinoma. Cancer Invest. 2009;27:661–668. doi: 10.1080/07357900802563010. [DOI] [PubMed] [Google Scholar]
- 164.Yamamichi N, et al. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67:10727–10735. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
- 165.Moloney FJ, et al. Hotspot mutation of Brahma in non-melanoma skin cancer. J Invest Dermatol. 2009;129:1012–1015. doi: 10.1038/jid.2008.319. [DOI] [PubMed] [Google Scholar]
- 166.Brodeur GM, Sekhon G, Goldstein MN. Chromosomal aberrations in human neuroblastomas. Cancer. 1977;40:2256–2263. doi: 10.1002/1097-0142(197711)40:5<2256::aid-cncr2820400536>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 167.White PS, et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2-36.3. Proc Natl Acad Sci U S A. 1995;92:5520–5524. doi: 10.1073/pnas.92.12.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Caron H, et al. Chromosome bands 1p35-36 contain two distinct neuroblastoma tumor suppressor loci, one of which is imprinted. Genes Chromosomes Cancer. 2001;30:168–174. doi: 10.1002/1098-2264(200102)30:2<168::aid-gcc1072>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 169.Godfried MB, et al. Lack of interstitial chromosome 1p deletions in clinically-detected neuroblastoma. Eur J Cancer. 2002;38:1513–1519. doi: 10.1016/s0959-8049(02)00137-5. [DOI] [PubMed] [Google Scholar]
- 170.White PS, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 171.Okawa ER, et al. Expression and sequence analysis of candidates for the 1p36.31 tumor suppressor gene deleted in neuroblastomas. Oncogene. 2008;27:803–810. doi: 10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- 172.Piaskowski S, et al. GADD45A and EPB41 as tumor suppressor genes in meningioma pathogenesis. Cancer Genet Cytogenet. 2005;162:63–67. doi: 10.1016/j.cancergencyto.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 173.Poetsch M, Dittberner T, Woenckhaus C. Microsatellite analysis at 1p36.3 in malignant melanoma of the skin: fine mapping in search of a possible tumour suppressor gene region. Melanoma Res. 2003;13:29–33. doi: 10.1097/00008390-200302000-00006. [DOI] [PubMed] [Google Scholar]
- 174.Moley JF, et al. Consistent association of 1p loss of heterozygosity with pheochromocytomas from patients with multiple endocrine neoplasia type 2 syndromes. Cancer Res. 1992;52:770–774. [PubMed] [Google Scholar]
- 175.Mulligan LM, Gardner E, Smith BA, Mathew CG, Ponder BA. Genetic events in tumour initiation and progression in multiple endocrine neoplasia type 2. Genes Chromosomes Cancer. 1993;6:166–177. doi: 10.1002/gcc.2870060307. [DOI] [PubMed] [Google Scholar]
- 176.Edstrom Elder E, et al. Loss of heterozygosity on the short arm of chromosome 1 in pheochromocytoma and abdominal paraganglioma. World J Surg. 2002;26:965–971. doi: 10.1007/s00268-002-6626-8. [DOI] [PubMed] [Google Scholar]
- 177.Aarts M, et al. Microarray-based CGH of sporadic and syndrome-related pheochromocytomas using a 0.1-0.2 Mb bacterial artificial chromosome array spanning chromosome arm 1p. Genes Chromosomes Cancer. 2006;45:83–93. doi: 10.1002/gcc.20268. [DOI] [PubMed] [Google Scholar]
- 178.Dong Z, et al. Identification of two contiguous minimally deleted regions on chromosome 1p36.31-p36.32 in oligodendroglial tumours. Br J Cancer. 2004;91:1105–1111. doi: 10.1038/sj.bjc.6602093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Felsberg J, et al. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–130. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–1128. [PubMed] [Google Scholar]
- 181.Mori N, et al. Chromosome band 1p36 contains a putative tumor suppressor gene important in the evolution of chronic myelocytic leukemia. Blood. 1998;92:3405–3409. [PubMed] [Google Scholar]
- 182.Melendez B, et al. Coincidental LOH regions in mouse and humans: evidence for novel tumor suppressor loci at 9q22-q34 in non-Hodgkin's lymphomas. Leuk Res. 2003;27:627–633. doi: 10.1016/s0145-2126(02)00278-3. [DOI] [PubMed] [Google Scholar]
- 183.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kleer CG, Bryant BR, Giordano TJ, Sobel M, Merino MJ. Genetic Changes in Chromosomes 1p and 17p in Thyroid Cancer Progression. Endocr Pathol. 2000;11:137–143. doi: 10.1385/ep:11:2:137. [DOI] [PubMed] [Google Scholar]
- 185.Zhou CZ, Qiu GQ, Zhang F, He L, Peng ZH. Loss of heterozygosity on chromosome 1 in sporadic colorectal carcinoma. World J Gastroenterol. 2004;10:1431–1435. doi: 10.3748/wjg.v10.i10.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cheung TH, et al. Clinicopathologic significance of loss of heterozygosity on chromosome 1 in cervical cancer. Gynecol Oncol. 2005;96:510–515. doi: 10.1016/j.ygyno.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 187.Bieche I, et al. Two distinct regions involved in 1p deletion in human primary breast cancer. Cancer Research. 1993;53:1990–1994. [PubMed] [Google Scholar]
- 188.Bieche I, Khodja A, Lidereau R. Deletion mapping of chromosomal region 1p32-pter in primary breast cancer. Genes Chromosomes Cancer. 1999;24:255–263. doi: 10.1002/(sici)1098-2264(199903)24:3<255::aid-gcc11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 189.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 190.Gorringe KL, et al. Mutation and methylation analysis of the chromodomain-helicase-DNA binding 5 gene in ovarian cancer. Neoplasia. 2008;10:1253–1258. doi: 10.1593/neo.08718. [DOI] [PMC free article] [PubMed] [Google Scholar]