Abstract
The cerebral cortex and hippocampus are important for the control of cognitive functions and social behaviors, many of which are sexually dimorphic and tightly regulated by gonadal steroid hormones via activation of their respective nuclear receptors. As different levels of sex steroid hormones are present between the sexes during early development and their receptors act as transcription factors to regulate gene expression, we hypothesize that sexually dimorphic gene expression in the developing mouse cortex and hippocampus might result in sex differences in brain structures and neural circuits governing distinct behaviors between the sexes as adults. To test our hypothesis, we used gene expression microarrays to identify 90 candidate genes differentially expressed in the neonatal cortex/hippocampus between male and female mice, including 55 male-biased and 35 female-biased genes. Among these genes, sexually dimorphic expression of eight sex chromosome genes was confirmed by reverse transcription with quantitative PCR (RT-qPCR), including three located on the X chromosome (Xist, Eif2s3x, and Kdm6a), three on the Y chromosome (Ddx3y, Eif2s3y, and Kdm5d), and two in the pseudoautosomal region of the X and Y chromosomes (Erdr1 and Mid1). In addition, five autosomal genes (Cd151, Dab2, Klk8, Meg3, and Prkdc) were also validated for their sexually dimorphic expression in the neonatal mouse cortex/hippocampus. Gene Ontology annotation analysis suggests that many of these sexually dimorphic genes are involved in histone modifications, cell proliferation/death, androgen/estrogen signaling pathways, and synaptic organization, and these biological processes have been implicated in differential neural development, cognitive function, and neurological diseases between the sexes.
Keywords: sex differences, cortex, hippocampus, gene expression microarray, sex chromosomes, sex-biased genes
1. Introduction
Sex differences in normal brain function and behavior are substantial in many species, including humans and mice (Breedlove and Hampson 2002; Jazin and Cahill 2010; McCarthy and Arnold 2011). Gender differences are also noted in the prevalence and symptomatology of many neurological diseases and mental illnesses such as multiple sclerosis, schizophrenia, and autism (Baron-Cohen et al., 2005; Deng et al., 2010; Greer and McCombe 2011). Elucidation of the neural mechanisms that establish sexual dimorphism in brain structures and behaviors will help us understand the processes regulating sex-specific susceptibility to various diseases and assist in development of new treatments for sex-biased disorders.
In mammals, including humans and rodents, sexual differentiation is initiated by the combination of the two sex chromosomes, X and Y (chromosomal sex), generally XX in females and XY in males. The Sry gene located on the Y chromosome encodes a transcription factor that initiates differentiation of the testes in males. In mice, deletion of Sry gene converts XY males to gonadal females while XX females receiving an Sry transgene inserted onto an autosome develop testes rather than ovaries (Gubbay et al., 1990; Koopman et al., 1990). The developing testes release testosterone (T) during late gestation and immediately after birth, causing a rise in circulating T that is essential for producing sex differences in many behaviors and in the neural structures and circuitry underlying these behaviors (organizational effect) (Motelica-Heino et al., 1988; Phoenix et al., 1959). T acts directly on androgen receptor (AR) and/or indirectly on estrogen receptors (ERs) via locally synthesized estradiol (E2) from T by aromatase to masculinize brain structures and behaviors (Davis et al., 1996; Forger 2009; Hines 2006). AR and ERs are abundantly expressed in the developing mouse cortex and hippocampus (Ivanova and Beyer 2000; Kerr et al., 1995). When activated, these receptors act as transcription factors to modulate gene expression, but their specific downstream target genes that are relevant to sex differences in the cortex and hippocampus remain unclear.
Many cognitive behaviors served by the cerebral cortex and hippocampus, such as learning and memory, show sex differences, and gonadal steroids and their nuclear receptors are important for sexual differentiation of these behaviors (Frick and Gresack 2003; Rizk et al., 2005; Sutcliffe et al., 2007). Associated with differential behavioral phenotypes between males and females, sex differences have been reported in the size and laterality of the mouse and rat hippocampus, which may be tied to a greater rate of neurogenesis in males in the first week after birth, as induced by T (Tabibnia et al., 1999; Zhang et al., 2008). Another reported difference in neuroanatomy is the cortical thickness, with adult male mice possessing a thicker cortex than females due to the effects of T (Markham et al., 2003). In humans, a thicker cortex is also seen in male patients suffering from autism compared to healthy male controls (Carper et al., 2002; Doyle-Thomas et al., 2013). Autism is a pervasive neurodevelopmental disorder characterized by deficits in social behavior and interpersonal communication that is four times more likely to occur in males than in females (Baron-Cohen et al., 2011). Auyeung et al. (2009) found that the more T levels in amniotic fluid of a pregnant woman, the higher her child scored on the tests of autistic traits although none of the children in that study were autistic (Auyeung et al., 2009). Along with the positive relationship between normal fetal T levels (ranging between 0.05 and 2.05 nM) and subclinical autistic traits, the cortical thickness influenced by T might be a potential mechanism underlying gender differences in cognitive functions and neuropsychiatric disorders, such as autism. If these sexual dimorphisms are caused by hormone receptors acting as transcription factors, then we should be able to identify specific gene expressions that create these structural and functional differences in the cortex and hippocampus between the sexes.
Besides gonadal hormones, emerging evidence has shown that brain sexual differentiation is also mediated by the action of genes located on the sex chromosomes. For example, the number of mesencephalic dopaminergic cells dissociated and cultured from the XY mouse embryos prior to gonadal differentiation is greater than that of XX mice (Carruth et al., 2002). In addition, a variety of behaviors are modulated by sex chromosome complement (Bonthuis et al., 2012; Cox and Rissman 2011; Cox and Rissman 2011; Gatewood et al., 2006; Gatewood et al., 2006; Gioiosa et al., 2008; Grgurevic et al., 2012; Park et al., 2008; Park et al., 2008; Quinn et al., 2007). Thus, we hypothesize that in the neonatal male cortex/hippocampus, differential expression of sexually dimorphic genes originating from both sex chromosomes and gonadal sex steroid hormones might lead to the development of distinct neural function and behaviors, as well as to underlying differences in brain structure, between the sexes. To test our hypothesis, we used gene expression microarrays to identify sexually dimorphic candidate genes expressed in the neonatal mouse cortex/hippocampus and confirmed selected candidates with reverse transcription with quantitative PCR (RT-qPCR). Through this analysis, we discovered several sex chromosome and autosomal genes differentially expressed in the neonatal male and female mouse cortex/hippocampus.
2. Results
2.1 Mouse body weight, cortex/hippocampus weight, and RNA yield
For the male and female neonatal mice used in the microarray experiment, there were no sex differences in their body weights (p = 0.912), cortex/hippocampus weights (p = 0.084), or RNA yields (p = 0.526) (Table 1). Similarly, the pups used for the RT-qPCR experiment showed no differences in their body weight (p = 0.571), cortex/hippocampal weight (p = 0.423), or RNA yield (p = 0.571) between the sexes (Table 1).
Table 1.
Body weights of the male and female neonatal mice used in the current study as well as the tissue weights and RNA yields of their cortex/hippocampus.
| Groups | N | Body weight (g) |
Tissue weight (mg) |
RNA yield (µg) |
|---|---|---|---|---|
| Microarray– | ||||
| Females | 18 | 1.27 ± 0.03 | 23.18 ± 1.29 | 46.06 ± 2.65 |
| Males | 21 | 1.26 ± 0.03 | 26.78 ± 1.52 | 50.30 ± 4.07 |
| RT-qPCR– | ||||
| Females | 18 | 1.27 ± 0.03 | 23.19 ± 1.59 | 44.01 ± 3.19 |
| Males | 18 | 1.25 ± 0.03 | 25.00 ± 1.58 | 47.25 ± 4.66 |
Data were expressed as mean ± SEM. N, number of mice
2.2 Microarray screening of sex-biased gene expression in the neonatal mouse cortex/hippocampus
Among 45,281 probes included in the Illumina gene expression microarray (San Diego, CA), 17,270 (38.1%) were detected as being expressed in the neonatal cortex/hippocampus. With the BLAST search of the expressed probe sequences in comparison with the NCBI transcriptome database (RefSeq Release 59), 10,762 (62.3%) matched a single transcript, 3,783 (21.9%) matched multiple transcripts corresponding to a single gene, 774 (4.5%) matched multiple genes, and 1,951 (11.3%) matched nothing in the RefSeq database. A total of 14,545 probes from the first two groups (84.2% of expressed probes) corresponding to 10,489 genes were further tested for sex differences in their expression. Using the false discovery rate (FDR) and 1.2-fold change cutoffs, 95 probes (0.65% of 14,545 expressed probes) were identified as sexually dimorphic; these corresponded to 90 distinct genes (0.86% of 10,489 expressed genes) with more being male-biased (55 genes, 61%) than female-biased (35 genes, 39%) (χ2 = 4.44, df = 1, p = 0.035) (Supplemental Table S1).
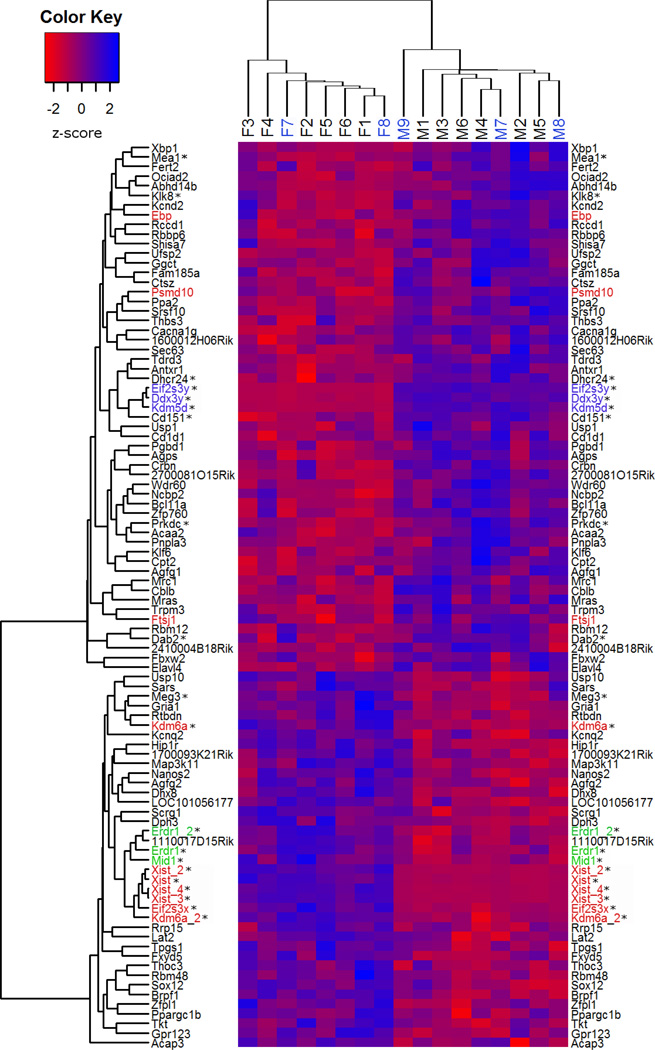
A summary heatmap for hierarchical clustering analysis of samples and sexually dimorphic candidate genes is shown in Figure 1. First, based on their expression patterns, 17 samples were grouped into two clusters (Fig. 1, the top cluster tree). Nine male samples were clustered together (the right half) before joining the females, and vice versa for females (the left half). Additionally, within each of the clusters, the samples used in the first assay (M7–9 or F7, 8) were connected with the same sex samples from the second (M1–6 or F1–6) before they associated with each other, indicating a lack of significant variation between the two assays. The lack of substantial differences between the two assays was also confirmed by principal components analysis (PCA; Supplementary Figure S1). Similar to clustering analysis, male and female samples were consistently mapped closer to the same sex samples before intermingling with the opposite sex in the three PCA plots generated by using expression data of the 95 sexually dimorphic probes (Panel A), the 494 sex chromosome probes (Panel B), and the 5,000 probes with the lowest expression noise (Panel C). Furthermore, within each plot, the samples from the first assay were intermingled with the same sex samples from the second assay, confirming again no significant differences between these two assays.
Fig. 1.
Heatmap showing hierarchical clustering of microarray probes differentially expressed in the neonatal cortex/hippocampus between male and female mice. The rows represent the sexually dimorphic probes with corresponding gene symbols on the left and right sides of the figure. The probes for the sexually dimorphic genes located on the X, Y and X/Y chromosomes are colored red, blue, and green, respectively. The expression level of each probe is displayed on the continuous color scale as shown in the Color Key; blue and red represents the expression level above or below the mean, respectively. The individual samples are shown as columns (1 sample per column). The sample IDs (M1-9 and F1-8) are listed on the top of the columns; the blue ones are from the first microarray assay and the rest are from the second. The dendrogram (clustering) to the left of the heatmap illustrates the similarities between sexually dimorphic probes in terms of their expression levels in different samples; of note are the close clustering of the three Y chromosome genes (Ddx3y, Eif2s3y, and Kdm5d, in blue) up top and the close clustering of the three X (Xist, Eif2s3x, and Kdm6a, in red) and two X/Y (Erdr1 and Mid1, in green) chromosome genes below. Clustering displayed above the sample IDs illustrates the similarities between different samples in terms of how they express the sexually dimorphic genes, exemplified by samples of the same sex grouped together before joining samples of the opposite sex. * indicates a gene that was examined by RT-qPCR.
Clustering analysis assigned 95 sexually dimorphic gene probes into two clusters, male-biased in the top half and female-biased in the bottom half (Fig. 1, the left cluster tree). Three Y chromosome probes, Eif2s3y, Ddx3y, and Kdm5d (in blue), were more closely clustered together than any of the other male-biased genes, and this cluster was next joined by Cd151. Six female-biased probes, including three from the X chromosome (Eif2s3x, Kdm6a, and Xist; in red) were also tightly clustered and then joined by a cluster of three X/Y pseudoautosomal probes (two for Erdr1 and one for Mid1; in green) and a probe matching 1110017D15Rik on chromosome 4.
Among the 90 sexually dimorphic candidate genes expressed in the neonatal mouse cortex/hippocampus, eleven of them were located on the X and Y chromosomes (3.6% of the 304 sex chromosome genes detected as expressed) (Table 2). Three Y chromosome genes, Ddx3y, Eif2s3y, and Kdm5d, showed the most significant sex difference in expression (all p < 0.001, ranked 1st, 2nd, and 4th most significant). Unlike Eif2s3y and Kdm5d expressed exclusively in male samples, Ddx3y expression was detected in both males and females although it was strongly male-biased. Manually searching the sequence similarity of the Ddx3y microarray probe in the BLAST, we found it matches not only Ddx3y, but also several autosomal transcripts, such as Zdhhc3 (43 out of 50 nucleotides matching) and Orc4 (40 out of 50 nucleotides matching) at a threshold lower than that used in the automated search, suggesting that detection of Ddx3y in females is due to cross-reactivity of the probe with other genes. The RT-qPCR measurement with Ddx3y–specific primers confirmed Ddx3y transcript expressed in males only (Fig. 2A). Xist showed female-biased expression with the largest fold change between the sexes, likely due to its high expression; the four distinct microarray probes matching Xist transcripts were expressed in all female, but not male samples, with 37 to 516 times higher expression in females as compared to males. Aside from Xist, six other probes corresponding to five other X chromosome genes were detected with significant sex differences, including three male-biased (Ebp, Ftsj1, and Psmd10) and two female-biased genes (Eif2s3x and Kdm6a). Kdm6a had two probes both showing female-biased expression with 1.34 (9th most significant) and 1.44 (11th most significant) times higher in females than males, respectively. Eif2s3x ranked 8th in terms of statistical significance, and its expression was 1.41 times higher in females than males.
Table 2.
List of sexually dimorphic candidate genes located on sex chromosome genes identified by microarray analysis.
| Symbol | Name | RefSeq Accession |
Greaterin | Chroma | Selected Gene Ontology Annotations |
|---|---|---|---|---|---|
| Eif2s3y* | eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | NM_012011.1 | M | Y | translational initiation, nucleotide binding |
| Ddx3y* | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | NM_012008.2 | M | Y | RNA binding, ATP-dependent helicase activity |
| Kdm5d* | lysine (K)-specific demethylase 5D | NM_011419.3 | M | Y | histone H3-K4 demethylation, DNA binding |
| ErDr1* | erythroid differentiation regulator 1 | NM_133362.2 | F | X|Y | negative regulation of cell proliferation, negative regulation of cell migration, somatic stem cell maintenance |
| Mid1* | midline 1 | NM_010797.2 | F | X| Y | negative regulation of microtubule depolymerization |
| Ebp | phenylalkylamine Ca2+ antagonist (emopamil) binding protein | NM_007898.2 | M | X | cholesterol biosynthetic process, C-8 sterol isomerase activity |
| Eif2s3x* | eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked | NM_012010.3 | F | X | translational initiation, nucleotide binding |
| Ftsj1 | FtsJ homolog 1 (E. coli) | NM_133991.2 | M | X | |
| Kdm6a* | lysine (K)-specific demethylase 6A | NM_00948 3.1 | F | X | neural tube closure, histone demethylase activity (H3-K27 specific) |
| PsMd10 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | NM_016883.3 | M | X | negative regulation of apoptotic process, transcription factor binding, negative regulation of transcription from RNA polymerase II promoter |
| Xist* | inactive X specific transcripts | NR_001463.3 | F | X | dosage compensation by inactivation of X chromosome |
indicates confirmation of sex difference in expression by RT-qPCR.
Chromosome
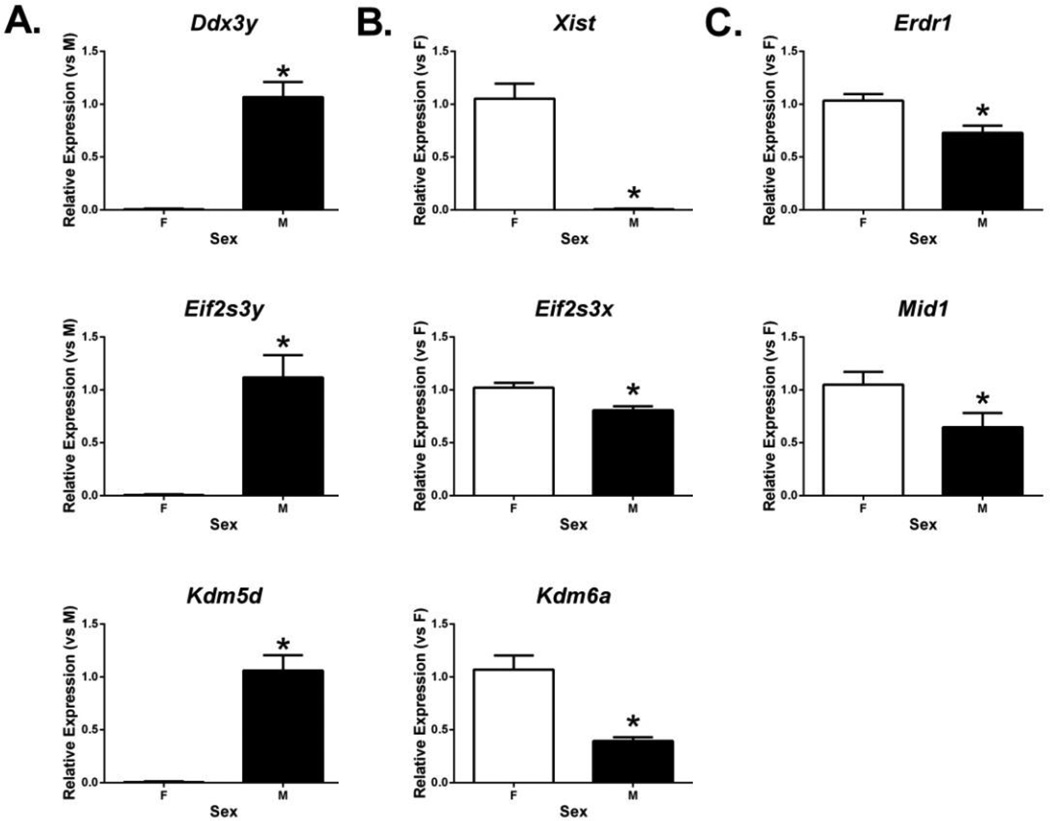
Fig. 2.
Expression of sex chromosome genes in the neonatal female (F) and male (M) mouse cortex/hippocampus measured by RT-qPCR. While three Y chromosome gene expression (A) was quantified relative to expression in males, relative mRNA levels of three X chromosome (B) and two pseudoautosomal (C) genes were calculated as relative to average in females. * indicates significant difference from female expression by two-tailed t-test (p < 0.05). Bars are mean ± SEM.
Three microarray probes matching two sex chromosome genes present in the pseudoautosomal region (PAR) of the X and Y chromosomes were identified as differentially expressed in the male and female mouse cortex/hippocampus: two matching Erdr1 and one matching Mid1. All three probes showed significantly higher expression in females. The two Erdr1 probes ranked 19th and 26th with the fold changes of 1.53 and 1.46, respectively. The Mid1 probe ranked 74th in terms of statistical significance but 18th in terms of fold change, showing expression of 1.56 times higher in females than in males.
Using the Gene Ontology (GO), we examined the functional annotations of these genes and found them involved in a variety of molecular functions (Table 2). For example, Kdm5d and Kdm6a encode enzymes for histone H3 demethylation on K4 and K27, respectively. Eif2s3x and Eif2s3y are involved in translational initiation. Xist involves dosage compensation by silencing one of the two X chromosomes in female cells. Erdr1 negatively regulates cell proliferation, while Psmd10 negatively regulates apoptosis. Ftsj1 showed no GO functional annotation.
2.3 Functional classification of sexually dimorphic candidate genes
Using GO annotation search, we found 26 out of the 90 sexually dimorphic candidate genes with functional linkages to at least one of three biological processes examined (developmental, sex difference, and steroid hormone processes): 20 linked to developmental processes, 6 involved in sex difference processes, and 2 associated with sex hormone processes (Table 3 and Supplemental Table S2). The 20 genes under the category of developmental processes were further explored with their connections to “cell proliferation” (GO: 0008283), “cell death” (GO:0008219), “nervous system development” (GO: 0007399), “cell morphogenesis” (GO:0000902), “cell migration” (GO:0016477), and “synapse organization” (GO:0050808) as well as the descendants of these GO terms (Supplemental Table S2). For example, Klk8 was found to involve “neuron projection morphogenesis” (GO:0048812), a descendant of “cell morphogenesis” (GO:0000902) (Oka et al., 2001). Furthermore, with the other search for “nervous system development” (GO:0007399), Klk8 was also revealed to be involved in “negative regulation of myelination” (GO:0031642) and “negative regulation of axon regeneration” (GO:0048681) (Terayama et al., 2007).
Table 3.
List of sexually dimorphic candidate genes from microarray matching gene ontology (GO) keywords
| Symbol | Name | RefSeq Accession |
Greaterin | Chroma | Gene Ontology Annotations |
|---|---|---|---|---|---|
| 1110017D15Rik | RIKEN cDNA 1110017D15 gene | NM_001048005.1 | F | 4 | spermatogenesis |
| 1600012H06Rik | RIKEN cDNA 1600012H06 gene | NM_001083881.1 | M | 17 | apoptotic process |
| Acaa2 | acetyl-Coenzyme A acyltransferase 2 | NM_177470.3 | M | 18 | negative regulation of mitochondrial outer membrane permeabilization |
| Agfg1 | ArfGAP with FG repeats 1 | NM_010472.2 | M | 1 | spermatogenesis, spermatid nucleus differentiation, acrosome assembly |
| Antxr1 | anthrax toxin receptor 1 | NM_054041.2 | M | 6 | substrate adhesion-dependent cell spreading |
| Bcl11a | B cell CLL/lymphoma 11A (zinc finger protein) | NM_016707.3 | M | 11 | regulation of dendrite development, regulation of neuron projection development, regulation of collateral sprouting |
| Cblb | Casitas B-lineage lymphoma b | NM_001033238.1 | M | 16 | negative regulation of alpha-beta T cell proliferation |
| Cd151* | CD151 antigen | NM_009842.3 | M | 7 | cell migration, T cell proliferation |
| Cd1d1 | CD1d1 antigen | NM_007639.3 | M | 3 | positive regulation of T cell proliferation |
| Dab2* | disabled 2, mitogen- responsive phosphoprotein | NM_001008702.1 | M | 15 | cell morphogenesis involved in differentiation, negative regulation of androgen receptor signaling pathway |
| Dhcr24 | 24-dehydrocholesterol reductase | NM_053272.2 | M | 4 | male genitalia development, negative regulation of cell proliferation, negative regulation of apoptotic process |
| Erdr1* | erythroid differentiation regulator 1 | NM_133362.2 | F | X|Y | negative regulation of cell proliferation, negative regulation of cell migration |
| Fer | fer (fms/fps related) protein kinase, testis specific 2 | NM_001037997.2 | M | 17 | cell proliferation, diapedesis, positive regulation of cell migration |
| Ggct | gamma-glutamyl cyclotransferase | NM_026637.3 | M | 6 | release of cytochrome c from mitochondria |
| Kdm6a* | lysine (K)-specific demethylase 6A | NM_009483.1 | F | X | neural tube closure |
| Klk8* | kallikrein related- peptidase 8 | NM_008940.2 | M | 7 | neuron projection morphogenesis, synapse organization, cell death, negative regulation of myelination, negative regulation of axon regeneration, keratinocyte proliferation |
| Lphn1 | latrophilin 1 | NM)_181039.2 | F | 8 | positive regulation of synapse maturation |
| Map3k11 | mitogen-activated protein kinase kinase kinase 11 | NM_022012.3 | F | 19 | positive regulation of neuron apoptotic process, cell proliferation |
| Mea1 | male enhanced antigen 1 | NM_010787.1 | M | 17 | spermatogenesis |
| Meg3* | maternally expressed 3 | NR_027652.1 | F | 12 | gonadotropin secretion?, negative regulation of cell proliferation? |
| Nanos2 | nanos homolog 2 (Drosophila) | NM_194064.2 | F | 7 | spermatogenesis |
| Ppargc1b | peroxisome proliferative activated receptor, gamma, coactivator 1 beta | NM_133249.2 | F | 18 | intracellular estrogen receptor signaling pathway, estrogen receptor binding |
| Prkdc* | protein kinase, DNA activated, catalytic polypeptide | NM_011159.2 | M | 16 | positive regulation of apoptotic process, germ cell programmed cell death, brain development |
| Psmd10 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | NM_016883.3 | M | X | negative regulation of apoptotic process |
| Sox12 | SRY-box containing gene 12 | NM_011438.2 | F | 2 | spinal cord development |
| Tpgs1 | tubulin polyglutamylase complex subunit 1 | NM_148934.2 | F | 10 | sperm axoneme assembly, spermatogenesis, synaptic transmission |
indicates confirmation of sex difference in expression by RT-qPCR. Underline indicates RT-qPCR failed to confirm sex difference.
indicates an annotation that was present for earlier analysis but has since been removed.
Chromosome
Six candidate genes involved in sex difference processes were similarly analyzed with three GO terms, “sexual reproduction” (GO:0019953), “reproductive system development” (GO:0061458), and “sex differentiation” (GO:0007548) and their descendants (Supplemental Table S2). For example, Prkdc and Dhcr24 were annotated with “germ cell programmed cell death” (GO:0035234) and “male genitalia development” (GO:0030539), which are the descendants of “sexual reproduction” (GO:0019953) and “reproductive system development” (GO:0061458), respectively (Maser et al., 2007; Wechsler et al., 2003). Of the two genes involved in the category of sex hormone processes, Dab2 was linked to "negative regulation of androgen receptor signaling pathway" (GO:0030521) while Ppargc1b was annotated with "intracellular estrogen receptor signaling pathway" (GO:0030520) (Kressler et al., 2002; Zhoul et al., 2005)
2.4 Validation of dimorphic expression of sex chromosome genes in the neonatal cortex/hippocampus
Sixteen out of the 90 sex-biased candidate genes identified by microarray were selected to validate their expression in the neonatal cortex/hippocampus by RT-qPCR, including three on the Y chromosome (Ddx3y, Eif2s3y, and Kdm5d), three on the X chromosome (Xist, Eif2s3x, and Kdm6a), two in the PAR of the X/Y chromosomes (Erdr1 and Mid1), and eight autosomal genes (Cd151, Dab2, Dhcr24, Klk8, Lphn1, Mea1, Meg3, and Prkdc). Eighteen pairs of oligonucleotide primers for these candidate genes and two reference genes were used in RT-qPCR to measure their relative RNA levels (Supplementary Table S3).
Three Y chromosome genes, Ddx3y, Eif2s3y, and Kdm5d, were confirmed to be expressed solely in the male cortex/hippocampus at levels more than 3,000 times higher in males than in females (p <0.001) (Fig. 2A). In contrast, Xist showed expression solely in females with levels over 2,000 times higher in females than in males (p < 0.001) (Fig. 2B, top panel). Unlike Xist, the other two X chromosome genes, Kdm6a and Eif2s3x, were expressed in both sexes, but expression of these genes in females was 2.71 (1.07 ± 0.13 vs. 0.40 ± 0.04, p = 0.001) and 1.28 (1.04 ± 0.07 vs. 0.81 ± 0.05, p = 0.015) times higher than males, respectively (Fig. 2B). Mid1 and Erdr1 are neighboring genes located on the PAR of the X and Y chromosomes as reported by Dal Zotto et al. (1998) and the NCBI Gene database (http://www.ncbi.nlm.nih.gov/gene/); both were confirmed as showing significantly higher gene expression in females, with fold changes of 1.63 (1.05 ± 0.12 vs. 0.65 ± 0.13, p = 0.041) and 1.42 (1.03 ± 0.06 vs. 0.73 ± 0.07, p = 0.002) times greater in females than in males, respectively (Fig. 2C). All of the eight sex chromosome genes tested by RT-qPCR showed significant sex differences consistent with the microarray data.
2.5 Validation of sexually dimorphic expression of autosomal genes by RT-qPCR
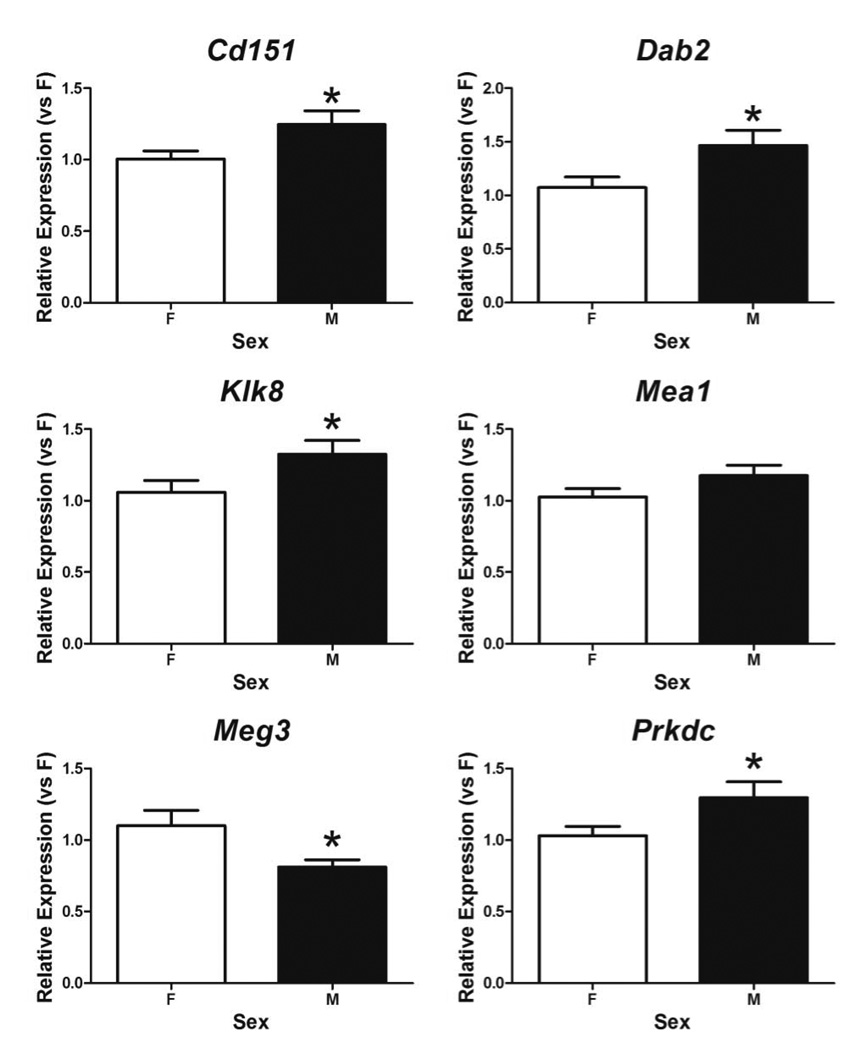
Eight autosomal candidate genes listed on Table 3 were selected to validate their sexually dimorphic expression in the neonatal cortex/hippocampus by RT-qPCR. Five genes were confirmed as sexually dimorphic; of those five, four genes were male-biased (Cd151, Dab2, Klk8, and Prkdc) and one was female-biased (Meg3) (Fig. 3). Similar to microarray data (1.34 fold higher in males than in females), Klk8 expression was 1.25 times greater in males (1.33 ±0.10 vs. 1.06 ± 0.08, p = 0.043) (Fig. 3). Relative mRNA levels of Cd151, Dab2, and Prkdc in the neonatal mouse cortex/hippocampus also showed significant male-biased expression with increased fold changes of 1.24 (1.25 ± 0.10 vs. 1.00 ± 0.06, p = 0.041), 1.36 (1.48 ± 0.15 vs. 1.08 ± 0.10, p = 0.038), and 1.26 (1.30 ± 0.11 vs. 1.03 ± 0.07, p = 0.044) in males as compared to females. Meg3 showed a significant female bias with expression 1.36 times higher in the female cortex/hippocampus than in the male (1.10 ± 0.11 vs. 0.80 ± 0.05, p = 0.024) (Fig. 3).
Fig. 3.
Expression of autosomal genes in the neonatal female (F) and male (M) mouse cortex/hippocampus measured by RT-qPCR. Relative mRNA levels were calculated as relative to expression in females. * indicates significant difference from female expression by two-tailed t-test (p < 0.05). Bars are mean ± SEM.
Expression of the remaining three candidate genes, Dhcr24, Lphn1, and Mea1, was not significantly different between the sexes in RT-qPCR. Dhcr24 and Mea1 showed male-bias and Lphn1 displayed female-bias by microarray. Mea1 showed a trend of male-bias (1.18 ± 0.07 vs. 1.03 ± 0.06, p = 0.113) (Fig. 3). Dhcr24 (F vs. M: 1.03 ± 0.07 vs. 1.00 ± 0.06, p = 0.743) and Lphn1 (F vs. M: 1.02 ± 0.06 vs. 1.11 ± 0.08, p = 0.38) showed essentially even expression between the sexes in the neonatal mouse cortex/hippocampus.
3. Discussion
An earlier microarray study reported 51 genes differentially expressed in the embryonic CD1 mouse brain (head) between the sexes prior to gonadal differentiation with 35 genes over-expressed in females and 16 genes in males (Dewing et al., 2003). Using a similar approach, Yang and colleagues later conducted a comprehensive analysis of gene expression differences between the sexes in multiple somatic tissues, including the brain, of 334 adult mice derived from an F2 intercross between two inbred strains, C57BL/6J and C3H/HeJ (Yang et al., 2006). They reported 355 female-biased and 257 male-biased genes in the whole brain, and these sexually dimorphic genes were highly enriched on the X and Y chromosomes. In the present study, we have observed more male-biased genes (55 or 61%) than female-biased genes (35 or 39%) by microarray analysis. Some discrepancies among these microarray data might be due to differences in mouse strains, mouse ages, brain regions, and microarray designs, but they all demonstrate that the most sexually dimorphic genes are located on the sex chromosomes, including X-specific transcripts (Xist), DEAD box polypeptide 3, Y-linked (Ddx3y; formerly Dby), and eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked (Eif2s3y).
Among the 90 candidate genes identified by microarrays in the current study, we confirmed sexually dimorphic expression of three Y-linked genes, Ddx3y, Eif2s3y, and Kdm5d (formerly Smcy or Jarid1d), in the neonatal mouse cortex/hippocampus using RT-qPCR (Fig. 2A). These genes are localized to the short arm of mouse Y chromosome, and deletion of this region is frequently found in infertile male patients with azoospermia, suggesting the critical roles of these genes in spermatogenesis (Ma et al., 2000; Mazeyrat et al., 2001; Yamauchi et al., 2009). This region is part of the non-recombining region of the Y chromosome (NRY), so the genes on this region, including Ddx3y, Eif2s3y, Kdm5d, Sry, Ubely1, Usp9y, and Uty, are present and thus expressed exclusively in males. Male-predominant expression of Ddx3y, Eif2s3y, and Kdm5d in the mouse brain has been confirmed by our data and a previous study by Xu et al. (2002). Xu and his colleagues have found Ddx3y, Eif2s3y, and Kdm5d as well as the other three NRY genes, Ubely1, Usp9y, and Uty, exclusively expressed in the male mouse brain at different ages, even before gonadal differentiation (Xu et al., 2002). The authors have further used the four core genotypes (FCG) mouse model, which produces XX and XY gonadal males (XXSry and XY−Sry), and XX and XY gonadal females (XX and XY−), to evaluate the effect of genotype (XX and XY), gonadal type, and their interaction on Y-linked gene expression. They observed a similar expression pattern of these Y-linked gene transcripts in the XY- females and XY−Sry males on the day of birth. These findings suggest that expression of Ddx3y, Eif2s3y, and Kdm5d and the other NRY genes are independent of testicular secretions.
Ddx3y encodes an ATP-dependent RNA helicase with a conserved DEAD (Asp-Glu-Ala-Asp) box motif. The specific function of Ddx3y in the brain remains unclear, but the DEAD box proteins have been implicated in RNA metabolism, including secondary structure alteration, splicing, spliceosome assembly, and translation initiation (Abdelhaleem 2005). Eif2s3y is a Y chromosomal homolog of the X-linked Eif2s3x, and both encodes the subunit 3 of the translation elongation and initiation factor 2 (eIF2), a component of the 40S ribosomal complex (Xu et al., 2006). In the presence of GTP, eIF2 forms a ternary complex with initiator tRNA Met-tRNAi and then recruits the 40S ribosomal complex, which determines the rate of protein translation (Gebauer and Hentze 2004; Proud 2005). The function of Eif2s3y in the brain is unknown, but the introduction of Eif2s3y restores normal spermatogonial proliferation and progression in Sxrb mice with partial deletion of the mouse Y short arm (Mazeyrat et al., 2001). Kdm5d encodes for a histone demethylase that specifically demethylates di- and tri-methylated lysine 4 of histone H3 (H3K4). Trimethyl H3K4 generally located near the promoter of genes is associated with an increased rate of transcription and dimethyl H3K4 is often localized in the coding regions of active genes (Bernstein et al., 2002; Santos-Rosa et al., 2002).
Three X chromosome genes, Eif2s3x, Kdm6a, and Xist, were confirmed for their female-biased expression in the neonatal mouse cortex/hippocampus (Fig. 2B). Xist gene encodes a large untranslated RNA transcribed from the inactive X chromosome in females, and thus this gene is normally expressed in female tissues only (Dewing et al., 2003; Shinozaki et al., 1999). Xist is responsible for transcriptional silencing of one of the two X chromosomes to equalize the dosage of X-linked genes between the sexes (X-chromosome inactivation). Skewed X-chromosome inactivation has been implicated in a variety of X-linked neurological diseases and mental illness, such as developmental delay and autism-predisposing chromosomal aberration (Migeon 2006; Qureshi and Mehler 2010; Willard 1996).
While most genes on the inactive X chromosomes are silenced via X inactivation, 3.3% of mouse X-linked genes escape X inactivation and are transcribed from both the active and inactive X chromosomes (Yang et al., 2010). Eif2s3x and Kdm6a were previously reported to escape X inactivation in the neonatal mouse brain (Wang et al., 2010). Consistent with their escaping status, we have observed that Eif2s3x and Kdm6a showed 1.28- and 2.71-fold higher expression in females than in males, respectively (Fig. 2B). In addition, with the fold changes in expression less or larger than 2, we speculate that transcription rates from the two alleles of Eif2s3x and Kdm6a in the neonatal female cortex/hippocampus might not be equal. Kdm6a protein contains a Jumonji-C domain, a structural motif shared by many histone demethylases (Klose and Zhang 2007; Sengoku and Yokoyama 2011; Xu and Andreassi 2011). Kdm6a catalyzes the removal of methylation at lysine 27 of histone H3 (H3K27) (Agger et al., 2007; Hong et al., 2007; Lan et al., 2007). Hypermethylation of H3K27 is a repressive chromatin modification, so Kdm6a might act as an activator of gene expression. A recent study demonstrates that Kdm6a is specifically recruited to Rhox6 and Rhox9 genes in female, but not male, ES cells, resulting in removal of the repressive histone mark, H3K27 trimethylation and increased expression of these two genes (Berletch et al., 2013). In humans, KDM6A point mutations have been identified to associate with Kabuki syndrome characterized by a unique facial appearance, growth retardation, skeletal abnormalities, and intellectual disability (Lederer et al., 2012; Miyake et al., 2013). These findings indicate that sex chromosome genes, like Kdm6a, might directly control sexually dimorphic expression of downstream genes in females via regulation of histone methylation.
Two sexually dimorphic genes, Erdr1 and Mid1, are located in the PAR of the X/Y chromosomes as reported in the literature (Dal Zotto et al., 1998; Trent et al., 2013; Tsakiridis et al., 2007) and the NCBI Gene database (http://www.ncbi.nlm.nih.gov/gene/), and both showed increased expression in the neonatal cortex/hippocampus of female mice by RT-qPCR (Fig. 2C). Erdr1 has been previously demonstrated to be female-biased in both mouse brain and ovary (Nef et al., 2005; Xu et al., 2012), which agrees with our finding. Besides hemoglobin synthesis-inducing activity in erythroleukemia cell lines, Erdr1 protein, erythroid differentiation regulator 1, has been shown to affect apoptosis and stem cell differentiation in hematopoietic stem cells and in keratinocytes (Deneault et al., 2009; Kim et al., 2011). Although its molecular function in brain is not yet clear, Erdr1 expression is altered in multiple mouse models of depression (Hoyle et al., 2011).
Mid1 (midline 1) encodes a protein belonging to the tripartite motif (TRIM) subfamily of zinc finger proteins, with the presence of a RING finger, one or two B-boxes, and a coiled-coil domain (Reymond et al., 2001). Mid1 has been established as showing female bias and escaping X-inactivation (Dal Zotto et al., 1998; Yang et al., 2010). Mid1 protein associates with the microtubules and acts as an E3 ubiquitin ligase that regulates phosphatase 2A degradation (Collison et al., 2013; Du et al., 2013; Trockenbacher et al., 2001). Mid1-null mice show hypoplasia of the vermis, the anterior portion of the medial cerebellum, correlating with impairments in motor coordination and learning (Lancioni et al., 2010); this suggests that Mid1 is important for normal development of the cerebellum. In humans, MID1 has been identified as responsible for the X-linked form of Opitz G/BBB syndrome (Dal Zotto et al., 1998). Since these two genes are neighbors, our results may also suggest an important role of this region on the same chromosome in sexual differentiation of the neonatal mouse cortex/hippocampus.
In C57BL/6 mice, the Mid1 gene (referred to by Fxy) was shown to span the PAR boundary with its first three exons in the X-unique region and the rest 7 exons in the PAR (Palmer et al., 1997). The first three exons are required for Mid1 gene transcription and escape from X inactivation; Mid1 is expressed from both X chromosomes in females. In contrast, with the absence of the 5’ exons of the Mid1 gene on the Y chromosome, Mid1 is transcribed from the only X chromosome in males, resulting in female-biased expression of Mid1. On the other hand, the Erdr1 gene is located 3' of Mid1 and resides entirely within the PAR. Unlike Mid1, Erdr1 is logically expressed from both sex chromosomes. Interestingly, the Erdr1 gene on the X chromosome is only 8,272 bp (170,010,526-170,018,797 bp) as opposed to the one on Y chromosome where it is 31,024 bp (90,785,442-90,816,465 bp) (http://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&dopt=full_report&list_uids=170942), which might be involved in regulation of its differential expression between the sexes. In addition, Erdr1 expression was up-regulated in the male muscle of AR-knockout mice (MacLean et al., 2008), suggesting that female-biased expression of Erdr1 in the mouse cortex/hippocampus might be due to suppression by perinatal T via activation of AR.
A recent microarray study examining the effects of X-chromosome number on gene expression in the whole brains of mouse embryos with four different genotypes, normal XX females (XX), 1X females, (XY*X ), 2X males (XXY*), and 1X males (XY*), also confirmed sexually dimorphic expression of many of the preceding genes (Wolstenholme et al., 2013). It demonstrated higher expression of Eif2s3x, Kdm6a, and Xist in 2X mice compared to 1X regardless of their gonads, consistent with their status of escaping from X inactivation. On the other hand, Y-linked genes, such as Ddx3y, Eif2s3y, and Kdm5d, were exclusively expressed in the males because of the presence of the Y chromosome. Interestingly, unlike our data, dimorphic expression of two pseudoautosomal genes, Erdr1 and Mid1, was not observed in this study, which could be caused by a rearrangement of the PARs of the Y* chromosome that contains Y-chromosome genes, an inverted duplication of a partial PAR, and a small amount of X-chromosome with the centromere (Burgoyne et al., 1998).
In the present study, we have confirmed sexually dimorphic expression of 5 autosomal genes in the neonatal mouse cortex/hippocampus (Fig. 5). Cd151 encodes a member of the tetraspanin family of membrane proteins with four transmembrane domains (Haeuw et al., 2011). Tetraspanins are able to associate with each other or a number of other transmembrane proteins to form tetraspanin-enriched microdomains (TEMs) on the plasma membrane, which organize the plasma membrane and intracellular membranes to selectively concentrate specific membrane proteins and membrane-peripheral signaling molecules (Bailey et al., 2011; Zoller 2009). In humans, CD151 is identified as a positive effecter of metastasis because CD151 overexpression is observed to associate with a poor prognosis in breast, pancreatic, colorectal, and lung cancers (Lee et al., 2013). Along with these observations, sexually dimorphic expression of Cd151 in the neonatal cortex/hippocampus suggests that it may control sex differences in cell proliferation, dynamics, and adhesion during early development (Haeuw et al., 2011).
Dab2 gene located on mouse chromosome 15 encodes a novel phosphoprotein of 82 kDa, which belongs to the Disabled (Dab) gene family. Dab2 is important for cell positioning and formation of visceral endoderm during embryogenesis, and homozygous Dab2 knockout mice have embryonic lethal phenotype with defective cell positioning and formation of the visceral endoderm (Yang et al., 2002). Dab2 is dynamically expressed in the developing central nervous system, primarily in non-neuronal cells within circumventricular organs, secretory cells, immature neuroepithelial cells, endothelial cells, and mononuclear phagocytes, but lacking in neurons, neuroglial cells and in mature microglia (Cheung et al., 2008). Although its role in neuronal development is not yet fully understood, Dab2 inhibits nerve growth factor-mediated neurite outgrowth in vitro (Huang et al., 2007). In addition, Dab2 can modulate AR-mediated cell growth in both normal and malignant prostatic epithelial cells via interaction with c-Src protein (Zhoul et al., 2005). Based on these findings, we speculate that the masculine increase in Dab2 expression in the neonatal male cortex/hippocampus might be responsible for homeostasis of cell proliferation and neurite outgrowth in response to the perinatal rise in T during the critical period.
Klk8, kallikrein related-peptidase 8, is a protein coding gene lying on mouse chromosome 7. Klk8 plays a critical role in hippocampal function. Klk8 knockout mice show impaired spatial memory associated with aberrant synapse formation in their hippocampus (Hirata et al., 2001; Terayama et al., 2007). In the present study, Klk8 was found to be a male-biased gene expressed in the neonatal cortex/hippocampus (Fig. 5), which might be elicited by the perinatal rise in circulating T released by the developing testes. Our hypothesis is supported by a microarray study showing higher expression of Klk8 in the adult female mouse hypothalamus than males (Xu et al., 2012). With higher levels of circulating E2 in adult female mice than males, activation of Klk8 gene might be regulated by E2. In the neonatal male cortex/hippocampus, the masculine increase in Klk8 expression might be similarly regulated by E2 that is locally converted from T by aromatase to masculinize brain structures and behaviors.
Prkdc gene, located on mouse chromosome 16, encodes the catalytic subunit of the DNA-dependent protein kinase (DNA-PK), which plays an important role in DNA double-strand break repair. DNA-PK plays an important role in neuron survival. A significant increase in dying neurons was observed in the cerebral cortex of the mice with a loss of DNA-PK activity, associated with nuclear changes, DNA fragmentation and increased caspase-3 activity (Vemuri et al., 2001). In addition, cultured hippocampal neurons from the mice lacking DNA-PK activity are hypersensitive to apoptosis in response to DNA damage, oxidative stress and excitotoxicity induced by exposing them to topoisomerase inhibitors, amyloid beta peptide (Aβ) and glutamate (Culmsee et al., 2001). Knockout of Prkdc gene results in increased germ cell apoptosis and testicular degeneration in mice (Maser et al., 2007). Double knockout of Prkdc and Atm (ataxia telangiectasia mutated) in mice increases apoptosis of postmitotic neurons (Gladdy et al., 2006), indicating that Prkdc might also be involved in neuronal proliferation during neurodevelopment. Since the hormonal control of cell death is as one of the best-established mechanisms that create sexually dimorphic structures in a variety regions of the brain and spinal cord, such as the principal nucleus of the bed nucleus of the stria terminalis (BNSTp), anteroventral periventricular nucleus (AVPV), and spinal nucleus of the bulbocavernosus (SNB) (Forger 2009), a masculine Prkdc expression in the neonatal cortex/hippocampus might be responsible for regulating neuronal death to change cell number in these brain regions and affects behavior.
Meg3 (maternally expressed gene 3) is the only female-biased, autosomal gene verified by RT-qPCR to express in the cortex/hippocampus of neonatal mice. Meg3 is an imprinted gene, and its transcript is a noncoding RNA highly expressed in the developing and adult mouse neocortex, hypothalamus, hippocampus, and amygdala (Balik et al., 2013). The human homolog of the mouse Meg3 gene, MEG3, is found to express in the brain and pituitary (Gejman et al., 2008). MEG3 expression is diminished in gonadotroph-derived pituitary adenomas, suggesting that Meg3 has an antiproliferative function (Zhou et al., 2012). MEG3 may act as a tumor suppressor by activating p53 to induce expression of p53 target genes in vitro (Zhou et al., 2012). Meg3 knockout mouse embryos showed increased cortical microvascular density in association with increased expression of the genes involving the VEGF and Notch signaling pathways in their brains as compared to wild-type controls (Gordon et al., 2010). Based on its function of tumor suppression and inhibiting angiogenesis, Meg3 may play an important role in sexual differentiation via the control of cell proliferation and vascularization in the brain.
In summary, we have not only replicated sex differences in transcription of several well characterized genes located on the sex chromosomes (Ddx3y, Eif2s3x, Eif2s3y, Kdm5d, Kdm6a, and Xist), but also identified two pseudoautosomal genes (Erdr1 and Mid1) and five autosomal genes (Cd151, Dab2, Klk8, Meg3, and Prkdc) that have been rarely reported as sexually dimorphic in the literature. In general, such sex differences in gene expression might be elicited by the direct actions of sex chromosome genes and/or by the effect of the sex steroid hormones released by the developing testes after gonadal differentiation. Together, our findings add to the theory that gene expression differences might contribute to brain organization in the cortex/hippocampus, which could ultimately lead to changes in social and cognitive behavior.
4. Experimental Procedures
4.1 Animals
Adult male and female C57BL/6J mice (Mus musculus) were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in the California State University, Long Beach (CSULB) Animal Care Facility. Animals were kept on a 12:12 light:dark cycle (lights on at 0600 h). Food (Teklad Mouse/Rat Diet #7012; Harlan Laboratories, Inc., Indianapolis, IN) and water were provided ad libitum. Adult female mice were individually paired with a fertile male, which was removed during late pregnancy. On the day of birth, male and female pups (n= 24–27 per sex) were sacrificed by rapid decapitation. In male mice, a pigment spot was visible on the scrotum, whereas female pups lacked pigmentation in the anogenital region (Wolterink-Donselaar et al., 2009). Their brains were immediately removed from the skulls and coronally blocked between 2.55 mm and 4.95 mm from the most rostral end (Paxinos et al., 2007). The top half of this brain block, including the cerebral cortex and hippocampus, was dissected out, frozen on dry ice, and then stored at −70°C until processed for RNA extraction. Besides examining the presence of pigmentation (males) in their anogenital regions, the tails of these mice were also collected for sex genotyping to confirm their sexes. All experimental procedures were approved by CSULB Institutional Animal Care and Use Committee (IACUC) and performed according to Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines.
4.2 Sex genotyping
Genomic DNA was extracted from the mouse tails using Sigma REDExtract-N-Amp™ Tissue PCR Kit (St. Louis, MO). As described previously (Gatewood et al., 2006; Tsai et al., 2009; Turner et al., 2000), tail DNA was amplified by PCR for the YMT2/B sequence (Ssty1), a member of the Ssty family present on the long arm of Y chromosome, and the myogenin sequence (Myog). PCR products were separated on a 1.5% agarose gel and stained with ethidium bromide. The presence of Ssty1 and Myog was used as the indicator of male genotype and the positive control, respectively.
4.3 RNA Extraction
Total RNA was extracted from the mouse cortex/hippocampus using RNeasy® Lipid Tissue kits (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. RNA quality and concentration of individual samples was determined using Bio-Rad SmartSpec™ Plus spectrophotometer (Hercules, CA) as the samples were diluted (1:50) in 10 mM Tris (pH = 7.5). Absorbance at 260 nm (A260), 280 nm (A280), and 320 nm (A320) was measured. RNA concentration was calculated with A260, and purity was determined by the A260/A280 ratio (between 1.9 and 2.1).
4.4 Gene expression microarray screening of sex-biased genes
To mitigate the possibility of female embryos being affected by T from neighboring males in utero, the RNA samples used for microarray were from the litters with the prevalent sex; male samples from the litters with the majority of male pups, and female samples from those with the majority of females. Two microarray assays were performed with the pooled cortex/hippocampus RNA samples. In the first assay, RNA of 9 male and 6 female mice (3 littermates of the same sex per sample) was mixed to create three male (M7–9) and two female (F7 and F8) pooled samples. The second assay was performed with six male (M1–6) and six female (F1–6) pooled samples, each of which was composed of RNA from two non-littermates of the same sex (Fig. 1).
To evaluate purity, pooled RNA samples were run on an agarose gel and visualized by ethidium bromide staining, and the 28S/18S rRNA ratio calculated. The RNA samples were amplified, labeled, and hybridized to the Illumina MouseWG-6 Expression BeadChip (San Diego, CA), performed by Expression Analysis® (Durham, NC). Arrays were then scanned by the Illumina BeadArray Reader and processed by BeadStudio software to produce summary data, which showed no difference in the average numbers of genes detected as expressed in the first and the second microarray assays with the p-value thresholds of 0.01 (10168.8 ± 63.0 vs. 10118.0 ± 60.1, p = 0.316) or 0.05 (12427.2 ± 88.2 vs. 12340.8 ± 69.6, p = 0.25). Our microarray data have been deposited in the NIH Gene Expression Omnibus (GEO) database (GSE54283, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54283).
4.5 Microarray data analysis
To calculate the fold changes of expression, raw data were first normalized by background subtraction (the median of the negative control probes). Negative values were scrunched into the range 1 to 2 with the same ordering. Normalized data were further processed by quantile normalization (Dunning et al., 2008). Microarray probes were filtered to match current NCBI RefSeq sequences by performing automated BLAST with BioPython (Cock et al., 2009). A probe sequence had to match at least 48 of its 50 nucleotides to be considered effective, and probes matching nothing in the RefSeq database or multiple gene symbols were excluded from further analysis. Filtered probes were tested for expression with significantly higher signal than background (determined by a set of negative control probes) using a one-tailed, one-sample t -test. Probes with significantly higher signals than background (p < 0.01) in at least six of the seventeen total samples were considered expressed. Expressed probes were further tested for a significant difference in expression between the sexes by two-tailed, two-sample t -test on log-transformed expression measurements. Raw p-values were adjusted by false discovery rate (FDR), and an FDR cutoff of 0.05 was used to determine significance (Benjamini and Hochberg 1995). Probes were also filtered for a fold change of at least 1.2 between the sexes. Analysis of the microarray was performed in Python using statistical tools from the SciPy toolkit and storing the data in a MySQL database (Oliphant 2007).
Hierarchical cluster analysis was performed in R with average linkage and a correlation metric to reveal the close relationships among the samples and sexually dimorphic candidate genes. Heatmaps were generated in R using the heatplot function (Eisen et al., 1998). PCA was also performed in Matlab to examine the relationship between the samples based on expression levels of three sets of expressed microarray probes: (1) 95 sexually dimorphic gene probes, (2) 494 X and Y chromosome probes, and (3) 5,000 probes with the smallest mean bead standard errors (noises).
The list of sexually dimorphic candidate genes was searched for the given gene ontology (GO) keywords (Supplementary Table S2) by custom code written in Python to identify annotations of genes with either the listed keywords or any of their descendants in the ontology tree. Annotations were downloaded from Mouse Genome Informatics (MGI) (ftp://ftp.informatics.jax.org/pub/reports/index.html#go) and tree structure was downloaded from the Gene Ontology project (http://geneontology.org/GO.downloads.ontology.shtml).
4.6 cDNA synthesis and PCR primer testing
The cDNA samples (n=18 per sex) were prepared from 1 µg extracted RNA in 20-µl reactions using the Bio-Rad iScript™ cDNA Synthesis Kit. The reverse transcription was performed at 25 C for 5 min, 42 °C for 30 min, and 85 °C for 5 min in a Bio-Rad MyCycler™ (Hercules, CA) thermocycler.
We first looked for PCR primers of selected genes in the literature and in PrimerBank (http://pga.mgh.harvard.edu/primerbank/), and new primers were designed if they were not found. Primers were designed and then checked for suitability using the Primer3 program (http://primer3.wi.mit.edu/), followed by checked for sequence specificity using NCBI’s Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) against RefSeq RNA. All primers found suitable for quantification along with the annealing temperature used and size of the resulting product are listed in Supplementary Table S3.
Primers purchased from Eurofins MWG Operon (Huntsville, AL) were tested in RT-PCR assays using GoTaq® Hot Start Green Master Mix (Promega, Madison, WI) according to the manufacturers’ protocol for 40 cycles. Initial tests used an annealing temperature of 2 °C below the lower melting temperatures of the primers as indicated by Eurofins MWG Operon. PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. All primers used in this study produce single PCR products with the correct size in the positive control as calculated, but no product in the negative control.
Dilution cDNA stocks were prepared with 8 serial dilutions ranging from 1:8 to 1:1,024. For each qPCR reaction, 4 µl of cDNA, 1 ul each of forward and reverse primers (840 nM), and 6 ul of ABsolute™ QPCR SYBR® Green Mix (Waltham, MA) were mixed and performed on an Agilent Stratagene® MX3000P™ qPCR system with MxPro QPCR software (Santa Clara, CA). qPCR amplification were conducted with an initial 15 min denaturation/polymerase-activation step at 95°C, 40 cycles of 15 s at 95°C, 1 min at the annealing temperature, and 30 s at 72°C, and a dissociation step with 1 min at 95°C, 30 s at 60°C, and 30 s at 95°C. Cycle of threshold (Ct) were analyzed by linear regression with log base 2 of the dilution as the predictor and Ct of the reaction as the response. All primers we used were suitable for quantification with the average R2 of the regression of 0.98, Ct values at the 1:16 dilution above 30, and the dissociation curve with single peaks.
4.7 Validation of dimorphic expression of sex-biased candidate genes by RT-qPCR
RT-qPCR for gene expression quantification was conducted under the same conditions as described in the primer dilution tests above, with 4 µl of 1:16 diluted cDNA of the male and female neonatal cortex/hippocampus (n=18 per sex). These samples were randomly selected from all litters; 17 male samples used for RT-qPCR came from male-biased litters, compared to one male from a female-dominated litter. Thirteen female pups used were from female-biased litters, compared to five from male-biased litters. Two replicate reactions for each sample were run and their Ct values averaged before further analysis. For a given sample unnormalized relative expression values for gene expression were calculated using Pfaffl’s method against a female baseline for all cases except Y chromosome genes; for Y chromosome genes, a male baseline was used (Pfaffl 2001). Six reference/housekeeping genes were tested to determine suitability for normalization of relative gene expression. Relative expression values of the target gene were divided by the geometric mean of the relative expression values of the two most stable reference genes (Actb and Rpl13a) to yield normalized relative expression values (Vandesompele et al., 2002). Values more than two standard deviations from their group mean were trimmed to two standard deviations from the group mean in the same direction.
4.8 Statistical Analyses
Aside from the microarray data, the data from individual groups were first tested for normality using Anderson-Darling test, and if passed, the data were subjected to two-tailed, two-sample t -test. In the case of significant deviations from normality, measurements were log transformed, followed by t -test. If transformation still failed to normalize the data, the data were subjected to non-parametric Mann-Whitney test.
Supplementary Material
Highlights.
Microarray analysis identified 90 sex-biased genes in the mouse cortex/hippocampus.
Three Y chromosome genes, Ddx3y, Eif2s3y, and Kdm5d, show male-specific expression.
Xist, Eif2s3x, and Kdm6a are female-based, X-linked genes in the neonatal brain.
Expression of two pseudoautosomal genes, Erdr1 and Mid1, is female-biased.
Cd151, Dab2, Klk8, Meg3, and Prkdc are novel sexually dimorphic autosomal genes.
Acknowledgement
We thank Kathy Trang, Thomas Mota, and Courtney Donovan for their technical assistance. We are also grateful to the reviewers for their valuable comments and suggestions which helped improve and clarify this manuscript. This work was supported by CSUPERB Faculty-Student Collaborative Research Seed Grant and National Institutes of Health Grants, SC3GM102051 and R25GM0500089.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelhaleem M. RNA helicases: Regulators of differentiation. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Auyeung B, Baron-Cohen S, Ashwin E, Knickmeyer R, Taylor K, Hackett G. Fetal testosterone and autistic traits. Br J Psychol. 2009;100:1–22. doi: 10.1348/000712608X311731. [DOI] [PubMed] [Google Scholar]
- Bailey RL, Herbert JM, Khan K, Heath VL, Bicknell R, Tomlinson MG. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans. 2011;39:1667–1673. doi: 10.1042/BST20110745. [DOI] [PubMed] [Google Scholar]
- Balik V, Srovnal J, Sulla I, Kalita O, Foltanova T, Vaverka M, Hrabalek L, Hajduch M. MEG3: A novel long noncoding potentially tumour-suppressing RNA in meningiomas. J Neurooncol. 2013;112:1–8. doi: 10.1007/s11060-012-1038-6. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S, Lombardo MV, Auyeung B, Ashwin E, Chakrabarti B, Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol. 2011;9:e1001081. doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Cohen S, Knickmeyer RC, Belmonte MK. Sex differences in the brain: Implications for explaining autism. Science. 2005;310:819–823. doi: 10.1126/science.1115455. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013;9:e1003489. doi: 10.1371/journal.pgen.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonthuis PJ, Cox KH, Rissman EF. X-chromosome dosage affects male sexual behavior. Horm Behav. 2012;61:565–572. doi: 10.1016/j.yhbeh.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedlove SM, Hampson E. Sexual differentiation of the brain and behavior. In: Becker JB, Breedlove SM, Crews D, McCarthy MM, editors. Behavioral Endocrinology. MIT Press; 2002. p. 75. [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Perry J, Palmer SJ, Ashworth A. The Y* rearrangement in mice: New insights into a perplexing PAR. Cytogenet Cell Genet. 1998;80:37–40. doi: 10.1159/000014954. [DOI] [PubMed] [Google Scholar]
- Carper RA, Moses P, Tigue ZD, Courchesne E. Cerebral lobes in autism: Early hyperplasia and abnormal age effects. Neuroimage. 2002;16:1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- Carruth LL, Reisert I, Arnold AP. Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci. 2002;5:933–934. doi: 10.1038/nn922. [DOI] [PubMed] [Google Scholar]
- Cheung KK, Mok SC, Rezaie P, Chan WY. Dynamic expression of Dab2 in the mouse embryonic central nervous system. BMC Dev Biol. 2008;8 doi: 10.1186/1471-213X-8-76. 76-213X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B. Biopython: Freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison A, Hatchwell L, Verrills N, Wark PA, Siqueira de AP, Tooze M, Carpenter H, Don AS, Morris JC, Zimmermann N, Bartlett NW, Rothenberg ME, Johnston SL, Foster PS, Mattes J. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat Med. 2013;19:232–237. doi: 10.1038/nm.3049. [DOI] [PubMed] [Google Scholar]
- Cox KH, Rissman EF. Sex differences in juvenile mouse social behavior are influenced by sex chromosomes and social context. Genes Brain Behav. 2011;10:465–472. doi: 10.1111/j.1601-183X.2011.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culmsee C, Bondada S, Mattson MP. Hippocampal neurons of mice deficient in DNA-dependent protein kinase exhibit increased vulnerability to DNA damage, oxidative stress and excitotoxicity. Brain Res Mol Brain Res. 2001;87:257–262. doi: 10.1016/s0169-328x(01)00008-0. [DOI] [PubMed] [Google Scholar]
- Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, Valsecchi V, Montini E, Yen C, Chapman V, Kalcheva I. The mouse Mid1 gene: Implications for the pathogenesis of opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet. 1998;7:489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- Davis EC, Popper P, Gorski RA. The role of apoptosis in sexual differentiation of the rat sexually dimorphic nucleus of the preoptic area. Brain Res. 1996;734:10–18. [PubMed] [Google Scholar]
- Deneault E, Cellot S, Faubert A, Laverdure JP, Fréchette M, Chagraoui J, Mayotte N, Sauvageau M, Ting SB, Sauvageau G. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137:369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Sobell JL, Knowles JA. Epigenetic alterations in schizophrenia. FOCUS: The Journal of Lifelong Learning in Psychiatry. 2010;8:358–365. [Google Scholar]
- Dewing P, Shi T, Horvath S, Vilain E. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res Mol Brain Res. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- Doyle-Thomas KA, Duerden EG, Taylor MJ, Lerch JP, Soorya LV, Wang AT, Fan J, Hollander E, Anagnostou E. Effects of age and symptomatology on cortical thickness in autism spectrum disorders. Res Autism Spectr Disord. 2013;7:141–150. doi: 10.1016/j.rasd.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Huang Y, Zaghlula M, Walters E, Cox TC, Massiah MA. The MID1 E3 ligase catalyzes the polyubiquitination of Alpha4 (alpha4), a regulatory subunit of protein phosphatase 2A (PP2A): Novel insights into MID1-mediated regulation of PP2A. J Biol Chem. 2013;288:21341–21350. doi: 10.1074/jbc.M113.481093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning MJ, Barbosa-Morais NL, Lynch AG, Tavare S, Ritchie ME. Statistical issues in the analysis of illumina data. BMC Bioinformatics. 2008;9:85. doi: 10.1186/1471-2105-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger NG. Control of cell number in the sexually dimorphic brain and spinal cord. J Neuroendocrinol. 2009;21:393–399. doi: 10.1111/j.1365-2826.2009.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick KM, Gresack JE. Sex differences in the behavioral response to spatial and object novelty in adult C57BL/6 mice. Behav Neurosci. 2003;117:1283–1291. doi: 10.1037/0735-7044.117.6.1283. [DOI] [PubMed] [Google Scholar]
- Gatewood JD, Wills A, Shetty S, Xu J, Arnold AP, Burgoyne PS, Rissman EF. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J Neurosci. 2006;26:2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2008;93:4119–4125. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioiosa L, Chen X, Watkins R, Klanfer N, Bryant CD, Evans CJ, Arnold AP. Sex chromosome complement affects nociception in tests of acute and chronic exposure to morphine in mice. Horm Behav. 2008;53:124–130. doi: 10.1016/j.yhbeh.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladdy RA, Nutter LMJ, Kunath T, Danska JS, Guidos CJ. p53-independent apoptosis disrupts early organogenesis in embryos lacking both ataxia-telangiectasia mutated and prkdc. Molecular Cancer Research. 2006;4:311–318. doi: 10.1158/1541-7786.MCR-05-0258. [DOI] [PubMed] [Google Scholar]
- Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer JM, McCombe PA. Role of gender in multiple sclerosis: Clinical effects and potential molecular mechanisms. J Neuroimmunol. 2011;234:7–18. doi: 10.1016/j.jneuroim.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Grgurevic N, Büdefeld T, Spanic T, Tobet SA, Majdic G. Evidence that sex chromosome genes affect sexual differentiation of female sexual behavior. Horm Behav. 2012;61:719–724. doi: 10.1016/j.yhbeh.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Haeuw JF, Goetsch L, Bailly C, Corvaia N. Tetraspanin CD151 as a target for antibody-based cancer immunotherapy. Biochem Soc Trans. 2011;39:553–558. doi: 10.1042/BST0390553. [DOI] [PubMed] [Google Scholar]
- Hines M. Prenatal testosterone and gender-related behaviour. Eur J Endocrinol. 2006;155(Suppl 1):S115–S121. doi: 10.1530/eje.1.02236. [DOI] [PubMed] [Google Scholar]
- Hirata A, Yoshida S, Inoue N, Matsumoto-Miyai K, Ninomiya A, Taniguchi M, Matsuyama T, Kato K, Iizasa H, Kataoka Y. Abnormalities of synapses and neurons in the hippocampus of neuropsin-deficient mice. Molecular and Cellular Neuroscience. 2001;17:600–610. doi: 10.1006/mcne.2000.0945. [DOI] [PubMed] [Google Scholar]
- Hong SH, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proceedings of the National Academy of Sciences. 2007;104:18439. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle D, Juhasz G, Aso E, Chase D, Del Rio J, Fabre V, Hamon M, Lanfumey L, Lesch K, Maldonado R. Shared changes in gene expression in frontal cortex of four genetically modified mouse models of depression. European Neuropsychopharmacology. 2011;21:3–10. doi: 10.1016/j.euroneuro.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Huang CH, Cheng JC, Chen JC, Tseng CP. Evaluation of the role of disabled-2 in nerve growth factor-mediated neurite outgrowth and cellular signalling. Cell Signal. 2007;19:1339–1347. doi: 10.1016/j.cellsig.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Ivanova T, Beyer C. Ontogenetic expression and sex differences of aromatase and estrogen receptor-α/β mRNA in the mouse hippocampus. Cell Tissue Res. 2000;300:231–237. doi: 10.1007/s004410000199. [DOI] [PubMed] [Google Scholar]
- Jazin E, Cahill L. Sex differences in molecular neuroscience: From fruit flies to humans. Nature Reviews Neuroscience. 2010;11:9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- Kerr J, Allore R, Beck S, Handa R. Distribution and hormonal regulation of androgen receptor (AR) and AR messenger ribonucleic acid in the rat hippocampus. Endocrinology. 1995;136:3213–3221. doi: 10.1210/endo.136.8.7628354. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Song SB, Yang Y, Eun YS, Cho BK, Park HJ, Cho DH. Erythroid differentiation regulator 1 (Erdr1) is a proapototic factor in human keratinocytes. Exp Dermatol. 2011;20:920–925. doi: 10.1111/j.1600-0625.2011.01354.x. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lancioni A, Pizzo M, Fontanella B, Ferrentino R, Napolitano LM, De Leonibus E, Meroni G. Lack of Mid1, the mouse ortholog of the opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J Neurosci. 2010;30:2880–2887. doi: 10.1523/JNEUROSCI.4196-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, Maystadt I, Dallapiccola B, Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Suh YL, Park TI, Do IG, Seol HJ, Nam DH, Kim ST. Prognostic significance of tetraspanin CD151 in newly diagnosed glioblastomas. J Surg Oncol. 2013;107:646–652. doi: 10.1002/jso.23249. [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Bhasin S. The role of Y chromosome deletions in male infertility. Eur J Endocrinol. 2000;142:418–430. doi: 10.1530/eje.0.1420418. [DOI] [PubMed] [Google Scholar]
- MacLean HE, Chiu WS, Notini AJ, Axell AM, Davey RA, McManus JF, Ma C, Plant DR, Lynch GS, Zajac JD. Impaired skeletal muscle development and function in male, but not female, genomic androgen receptor knockout mice. FASEB J. 2008;22:2676–2689. doi: 10.1096/fj.08-105726. [DOI] [PubMed] [Google Scholar]
- Markham JA, Jurgens HA, Auger CJ, De Vries GJ, Arnold AP, Juraska JM. Sex differences in mouse cortical thickness are independent of the complement of sex chromosomes. Neuroscience. 2003;116:71–75. doi: 10.1016/s0306-4522(02)00554-7. [DOI] [PubMed] [Google Scholar]
- Maser RS, Wong K, Sahin E, Xia H, Naylor M, Hedberg HM, Artandi SE, DePinho RA. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Molecular and Cellular Biology. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR. The role of X inactivation and cellular mosaicism in women’s health and sex-specific diseases. JAMA. 2006;295:1428–1433. doi: 10.1001/jama.295.12.1428. [DOI] [PubMed] [Google Scholar]
- Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, Tsurusaki Y, Nakashima M, Saitsu H, Niikawa N, Matsumoto N. KDM6A point mutations cause kabuki syndrome. Hum Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- Motelica-Heino I, Castanier M, Corbier P, Edwards DA, Roffi J. Testosterone levels in plasma and testes of neonatal mice. J Steroid Biochem. 1988;31:283–286. doi: 10.1016/0022-4731(88)90351-2. [DOI] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Oka T, Akisada M, Okabe A, Sakurai K, Shiosaka S, Kato K. Extracellular serine protease neuropsin (KLK8) modulates neurite outgrowth and fasciculation of mouse hippocampal neurons in culture. Neurosci Lett. 2001;321:141–144. doi: 10.1016/s0304-3940(01)02470-3. [DOI] [PubMed] [Google Scholar]
- Oliphant TE. Python for scientific computing. Computing in Science & Engineering. 2007;9:10–20. [Google Scholar]
- Palmer S, Perry J, Kipling D, Ashworth A. A gene spans the pseudoautosomal boundary in mice. Proc Natl Acad Sci U S A. 1997;94:12030–12035. doi: 10.1073/pnas.94.22.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Burns-Cusato M, Dominguez-Salazar E, Riggan A, Shetty S, Arnold AP, Rissman EF. Effects of sex chromosome aneuploidy on male sexual behavior. Genes Brain Behav. 2008;7:609–617. doi: 10.1111/j.1601-183X.2008.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Halliday G, Watson C, Koutcherov Y, Wang H. Atlas of the Developing Mouse Brain at E17.5, P0, and P6. Burlington, MA 01803, USA: Academic Press; 2007. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45–e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Hitchcott PK, Umeda EA, Arnold AP, Taylor JR. Sex chromosome complement regulates habit formation. Nat Neurosci. 2007;10:1398–1400. doi: 10.1038/nn1994. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Genetic and epigenetic underpinnings of sex differences in the brain and in neurological and psychiatric disease susceptibility. Prog Brain Res. 2010;186:77–95. doi: 10.1016/B978-0-444-53630-3.00006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk A, Robertson J, Raber J. Behavioral performance of tfm mice supports the beneficial role of androgen receptors in spatial learning and memory. Brain Res. 2005;1034:132–138. doi: 10.1016/j.brainres.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, ellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sengoku T, Yokoyama S. Structural basis for histone H3 lys 27 demethylation by UTX/KDM6A. Genes Dev. 2011;25:2266–2277. doi: 10.1101/gad.172296.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki A, Arahata K, Tsukahara T. Changes in pre-mRNA splicing factors during neural differentiation in P19 embryonal carcinoma cells. Int J Biochem Cell Biol. 1999;31:1279–1287. doi: 10.1016/s1357-2725(99)00101-6. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Marshall KM, Neill JC. Influence of gender on working and spatial memory in the novel object recognition task in the rat. Behav Brain Res. 2007;177:117–125. doi: 10.1016/j.bbr.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Tabibnia G, Cooke BM, Breedlove SM. Sex difference and laterality in the volume of mouse dentate gyrus granule cell layer. Brain Res. 1999;827:41–45. doi: 10.1016/s0006-8993(99)01262-7. [DOI] [PubMed] [Google Scholar]
- Terayama R, Bando Y, Murakami K, Kato K, Kishibe M, Yoshida S. Neuropsin promotes oligodendrocyte death, demyelination and axonal degeneration after spinal cord injury. Neuroscience. 2007;148:175–187. doi: 10.1016/j.neuroscience.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Trent S, Dean R, Veit B, Cassano T, Bedse G, Ojarikre OA, Humby T, Davies W. Biological mechanisms associated with increased perseveration and hyperactivity in a genetic mouse model of neurodevelopmental disorder. Psychoneuroendocrinology. 2013;38:1370–1380. doi: 10.1016/j.psyneuen.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers HH, Schneider R, Schweiger S. MID1, mutated in opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet. 2001;29:287–294. doi: 10.1038/ng762. [DOI] [PubMed] [Google Scholar]
- Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiridis A, Tzouanacou E, Larralde O, Watts TM, Wilson V, Forrester L, Brickman JM. A novel triple fusion reporter system for use in gene trap mutagenesis. Genesis. 2007;45:353–360. doi: 10.1002/dvg.20301. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Benavente R, Offenberg HH, Heyting C, Burgoyne PS. Analysis of male meiotic” sex body” proteins during XY female meiosis provides new insights into their functions. Chromosoma. 2000;109:426–432. doi: 10.1007/s004120000097. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri MC, Schiller E, Naegele JR. Elevated DNA double strand breaks and apoptosis in the CNS of scid mutant mice. Cell Death Differ. 2001;8:245–255. doi: 10.1038/sj.cdd.4400806. [DOI] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG. Paternally biased X inactivation in mouse neonatal brain. Genome Biol. 2010;11:R79. doi: 10.1186/gb-2010-11-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler A, Brafman A, Shafir M, Heverin M, Gottlieb H, Damari G, Gozlan-Kelner S, Spivak I, Moshkin O, Fridman E. Generation of viable cholesterol-free mice. Science. 2003;302:2087–2087. doi: 10.1126/science.1090776. [DOI] [PubMed] [Google Scholar]
- Willard HF. X chromosome inactivation and X-linked mental retardation. Am J Med Genet. 1996;64:21–26. doi: 10.1002/(SICI)1096-8628(19960712)64:1<21::AID-AJMG2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Wolstenholme JT, Rissman EF, Bekiranov S. Sexual differentiation in the developing mouse brain: Contributions of sex chromosome genes. Genes Brain Behav. 2013;12:166–180. doi: 10.1111/gbb.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink-Donselaar IG, Meerding JM, Fernandes C. A method for gender determination in newborn dark pigmented mice. Lab Anim (NY) 2009;38:35–38. doi: 10.1038/laban0109-35. [DOI] [PubMed] [Google Scholar]
- Xu J, Andreassi M. Reversible histone methylation regulates brain gene expression and behavior. Horm Behav. 2011;59:383–392. doi: 10.1016/j.yhbeh.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr Patterns. 2006;6:146–155. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Xu X, Coats JK, Yang CF, Wang A, Ahmed OM, Alvarado M, Izumi T, Shah NM. Modular genetic control of sexually dimorphic behaviors. Cell. 2012;148:596–607. doi: 10.1016/j.cell.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Wong SJ, Ojarikre OA, Burgoyne PS, Ward MA. Live offspring from mice lacking the Y chromosome long arm gene complement. Biol Reprod. 2009;81:353–361. doi: 10.1095/biolreprod.109.076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Smith ER, Roland IH, Sheng Z, He J, Martin WD, Hamilton TC, Lambeth JD, Xu XX. Disabled-2 is essential for endodermal cell positioning and structure formation during mouse embryogenesis. Dev Biol. 2002;251:27–44. doi: 10.1006/dbio.2002.0810. [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Konkle AT, Zup SL, McCarthy MM. Impact of sex and hormones on new cells in the developing rat hippocampus: A novel source of sex dimorphism? Eur J Neurosci. 2008;27:791–800. doi: 10.1111/j.1460-9568.2008.06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: A tumor suppressor. J Mol Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhoul J, Hernandez G, Tu SW, Huang CL, Tseng CP, Hsieh JT. The role of DOC-2/DAB2 in modulating androgen receptor-mediated cell growth via the nongenomic c-src-mediated pathway in normal prostatic epithelium and cancer. Cancer Res. 2005;65:9906–9913. doi: 10.1158/0008-5472.CAN-05-1481. [DOI] [PubMed] [Google Scholar]
- Zoller M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.