Summary
The study of synaptic specificity and plasticity in the Central Nervous System (CNS) is limited by the inability to efficiently visualize synapses in identified neurons using light microscopy. Here we describe Synaptic Tagging with Recombination (STaR), a method for labeling endogenous presynaptic and postsynaptic proteins in a cell-type specific fashion. We modified genomic loci encoding synaptic proteins within Bacterial Artificial Chromosomes such that these proteins, expressed at endogenous levels and with normal spatiotemporal patterns, were labeled in an inducible fashion in specific neurons through targeted expression of site-specific recombinases. Within the Drosophila visual system, the number and distribution of synapses correlate with Electron Microscopy studies. Using two different recombination systems, presynaptic and postsynaptic specializations of synaptic pairs can be co-labeled. STaR also allows synapses within the CNS to be studied in live animals non-invasively. In principle, STaR can be adapted to the mammalian nervous system.
Introduction
The lack of methods to efficiently visualize synapses of identified neurons in the central nervous system (CNS) remains a major obstacle to studying mechanisms of synaptic specificity and plasticity. Due to the cellular complexity of the CNS and the small size of synapses, maps of synaptic connectivity have relied on Serial Section Electron Microscopy (SSEM). A comprehensive map of the synaptic connections between neurons in Caenorhabditis elegans was determined by SSEM in the 1980s (White et al., 1986). In Drosophila melanogaster, detailed maps of synaptic connectivity in the visual system have been determined by several SSEM studies (Meinertzhagen and O'Neil, 1991; Rivera-Alba et al., 2011; Takemura et al., 2013; Takemura et al., 2008). These studies reveal complex and highly specific patterns of connections and provide a foundation for studying the molecular mechanisms of circuit assembly.
SSEM analysis is extremely time-consuming. For instance, Chklovskii and colleagues developed a state-of-the-art semi-automated pipeline to reconstruct a connectome of 379 neurons in the fly visual system (Takemura et al., 2013). In this study, the steps of manually refining the SSEM dataset required ~14400 person-hours in total. As a consequence, assessing variations of synaptic connections among cells of the same cell type and between animals is problematic with Electron Microscopy (EM). In addition, EM analysis of synaptic patterns at multiple developmental stages, in various mutant backgrounds or under different activity-modulated conditions is not feasible in most instances. These limitations have driven researchers to find ways to visualize synapses by light microscopy.
Two approaches have been developed to study synapses using light microscopy. The first involves targeted expression of tagged synaptic proteins to label presynaptic and/or postsynaptic sites (Nonet, 1999; Wagh et al., 2006; Yeh et al., 2005; Zhang et al., 2002). The second relies on visualizing protein interactions across the synaptic cleft. In GFP Reconstitution Across Synaptic Partners (GRASP), these interactions are observed through the reconstitution of GFP fluorescence (Feinberg et al., 2008). In both methods, the modified synaptic proteins are not typically expressed under their endogenous regulatory elements. As a result, the tagged proteins are often over-expressed and may accumulate in inappropriate intracellular locations and thus may not accurately reflect synaptic pairs and the location of synapses (see Results and Discussion). The ideal tool to faithfully label synapses with light microscopy would ensure targeted expression of modified synaptic proteins to only discrete subsets of neurons under the control of their endogenous regulatory mechanisms.
Here we describe Synaptic Tagging with Recombination (STaR), a genetic approach to label synapses in identified neurons with light microscopy. We modified the endogenous genomic loci encoding synaptic proteins within Bacterial Artificial Chromosomes (BACs) to generate inducible presynaptic and postsynaptic markers expressed in a cell-type specific manner, under the control of their endogenous regulatory mechanisms. Here, as a proof-of-principle study, we show that these markers allow the development and distribution of synapses in specific neurons of the fly visual system to be studied in detail in both fixed brain samples and in live animals.
Results
Cell-type Specific Labeling of Presynaptic Sites with the STaR Method
The presynaptic active zones in Drosophila are commonly marked by T-shaped electron dense structures (i.e. T-bars) visualized by Transmission Electron Microscopy (TEM). Similar to the presynaptic ribbons at vertebrate synapses, T-bars are thought to serve as platforms for synaptic vesicle release. They are present at many synapses in Drosophila, including the vast majority, if not all, of the synapses in the visual system, as well as many synapses in the central brain (Butcher et al., 2012; Prokop and Meinertzhagen, 2006; Yasuyama et al., 2003). T-bars are used as the key criterion for synapse identification, as postsynaptic densities are poorly resolved using TEM in flies. T-bars comprise clusters of Bruchpilot (BRP) protein, the single CAST/ERC family member in Drosophila (Fouquet et al., 2009; Kittel et al., 2006; Wagh et al., 2006). By confocal microscopy these clusters appear as fluorescent puncta when stained with BRP antibody, and each punctum correlates with one presynaptic active zone (Fouquet et al., 2009; Hamanaka and Meinertzhagen, 2010). Since BRP protein is broadly expressed in the fly CNS, it is generally not possible to assign BRP puncta to processes of specific cell types, due to the small size of processes and the density of BRP puncta within the neuropil (e.g. see Figure 1B).
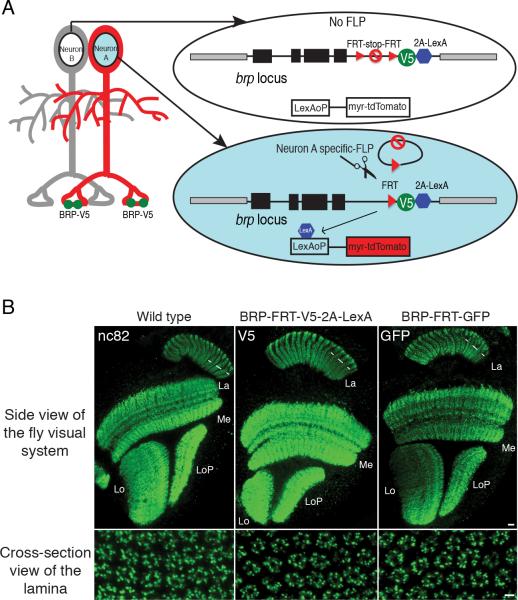
Figure 1. Cell-type Specific Tagging of the Active Zone Protein BRP using STaR.
(A) Schematic diagram for STaR. BRP protein is tagged selectively in Neuron A by targeted expression of FLP recombinase to remove the FRT-Stop-FRT cassette. Black boxes, brp exons; black lines, brp introns; gray boxes, neighboring genes.
(B) The expression pattern of BRP proteins tagged via STaR detected with anti-V5 (middle panels) or with anti-GFP (right panels) is indistinguishable from the endogenous BRP protein detected with an anti-BRP antibody (nc82) (left panels). Scale bars, 10μm (top panels) and 2μm (bottom panels). La, lamina; Me, medulla; Lo, lobula; and LoP, lobula plate.
To visualize presynaptic sites in specific cell types, we devised a genetic strategy, Synaptic Tagging with Recombination (STaR), to label endogenous BRP protein only in processes of identified cells (Figure 1A). We modified the endogenous brp sequence in a BAC by inserting a V5 tag into the brp coding region downstream from transcriptional and translational stop sequences flanked by FRT recombination sites (Golic and Lindquist, 1989; Southern et al., 1991; Venken et al., 2006; Warming et al., 2005). In the absence of FLP recombinase, the BAC encoded BRP protein is not tagged. By contrast, cell-type specific expression of FLP recombinase induces recombination between the two FRT sites to excises the stop sequence, allowing the expression of V5-tagged-BRP only in cells of interest.
To visualize the morphology of cells expressing tagged BRP, and thus to associate BRP puncta with specific neuronal processes, we inserted a 2A-LexA cassette downstream of the V5 tag; the 2A peptide allows for co-translation of LexA which, in turn, drives expression of, for example, myristoylated-tandem-Tomato (myr-tdTomato) from the LexAoP enhancer (Lai and Lee, 2006; Ryan and Drew, 1994). The modified BAC was introduced into flies as a transgene (Groth et al., 2004). When combined with various cell-type specific FLP recombinases, V5-tagged BRP puncta can be detected in identified neurons marking individual presynaptic sites (Figures 1A, 2, 3 and S3). In order to label presynaptic sites in live animals without relying on immuno-staining, we also prepared a GFP-tagged version of the BRP BAC construct with a similar design (see Experimental Procedures and Figure 8).
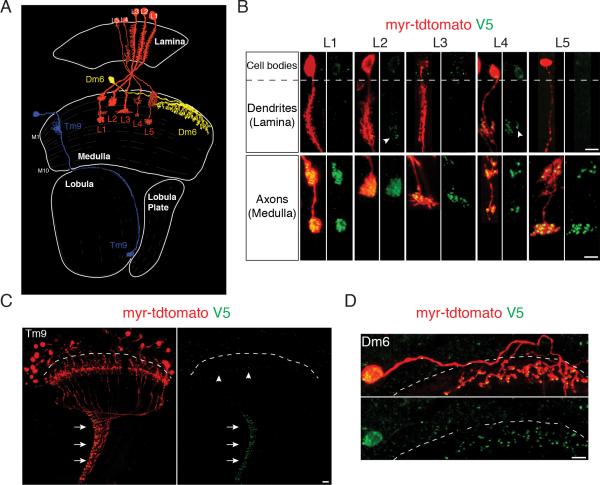
Figure 2. STaR Labeling of Presynaptic Sites in Various Neurons in the Fly Visual System.
(A) Schematic drawing of the Drosophila visual system with neurons shown in this figure: Lamina monopolar neurons L1-L5, Transmedullary neuron Tm9 and Distal medulla neuron Dm6. Adapted from (Fischbach and Dittrich, 1989).
(B-D) Presynaptic sites in various neurons labeled with STaR. Red, myr-tdTomato outlining the neurons expressing the marker; Green, V5 staining labels BRP at presynaptic sites. The dashed lines separate the cortex region comprising the cell bodies (above the line) and the neuropil region comprising neuronal processes (below the line). The V5 antibody shows non-specific background in the cortex, but is specific in the neuropil. Scale bars, 5μm.
(B) Presynaptic sites in L1-L5. Top, L1-L5 dendrites in the lamina neuropil; Bottom, L1-L5 axons in the medulla neuropil. Arrowheads, BRP-V5 puncta in L2 and L4 processes in the lamina.
(C) Presynaptic sites in Tm9. Many Tm9 neurons were labeled. The vast majority of V5 signal was detected at the Tm9 axon terminals in the innermost layer of the lobula neuropil (arrows), while a very small number of puncta was detected in the Tm9 dendrites in the medulla (arrowheads).
(D) Presynaptic sites in Dm6.
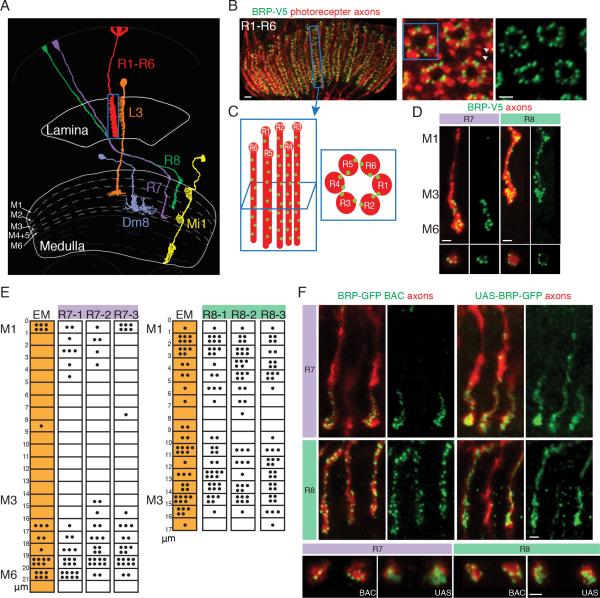
Figure 3. STaR Labeling of Presynaptic Sites Facilitates Quantification in Photoreceptor Neurons.
(A) Schematic drawing of three classes of photoreceptor neurons (R1-R6, R7 and R8) and examples of their postsynaptic partners: L3 for R1-R6, Dm8 for R7 and Mi1 for R8. Only one of the ~750 R1-R6, R7 and R8 neurons is depicted. Adapted from (Fischbach and Dittrich, 1989).
(B) Side view (left panel) and cross-section views (center and right panels) of presynaptic sites in R1-R6 axons in the lamina labeled using STaR. Red, myr-tdTomato labels photoreceptor axons; Green, V5 staining labels presynaptic sites. Arrowheads mark R7 and R8 axons outside of the R1-R6 axon “rosette”. Scale bars, 2μm.
(C) Schematic diagram of one set of R1-R6 axons in a single lamina cartridge from the side view (left) and the cross-section view (right). Red, R1-R6 axons; Green dots, presynaptic sites.
(D) Presynaptic sites in R7 and R8 axons in the medulla labeled with STaR. Top panels, side view; Bottom panels, cross-section view. Red, myr-tdTomato labels R7 and R8 axons, respectively; Green, V5 staining labels BRP at presynaptic sites. Scale bars, 2μm.
(E) The distribution of presynaptic sites in R7 and R8 axons mapped with STaR corresponds well with EM reconstruction results. The numbers on the left represents the distance (in μm) from the top of the medulla (designated “0”). Each black dot represents one presynaptic T-bar (EM data from Takemura et al., 2013; shown as orange shaded columns) or one BRP-V5 punctum (white columns). Each column represents one R7 or R8 axon.
(F). Comparison between STaR labeling of BRP and the Gal4-driven UAS-BRP-GFP marker in R7 and R8 neurons. Only subsets of the axons are labeled with STaR (BRP-GFP BAC) or the UAS-BRP-GFP marker. Top panels, side view; Bottom panels, cross-section view. Red, photoreceptor axons marked with myr-tdTomato (left panels) or 24B10 (right panels); Green, GFP staining. Scale bars, 2μm.
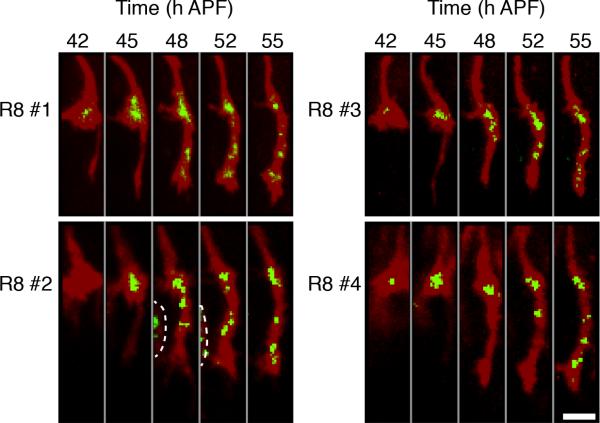
Figure 8. Following Presynaptic Development in R8 Axons in Live Animals using STaR.
Snap-shot images of the axons (red) and the presynaptic sites (green) in four developing R8 neurons in the live pupa labeled with STaR using two photon microscopy. The resolution of these images is lower than the images of the stained samples (Figure 6E), due to the lower intensity of the native fluorescence and the suboptimal orientation of the live pupa (compared to the dissected brain) relatively to the objective lens. Dashed line in R8 #2 separates a neighboring R8 axon. Scale bar, 5μm.
The BAC-encoded modified BRP proteins were expressed in the same manner as the unmodified protein from the endogenous locus. In transgenic flies carrying a BAC encoding the constitutively tagged BRP (i.e. brp-FRT-V5-2A-LexA or brp-FRT-GFP), the tagged BRP expression pattern was indistinguishable from BRP expression pattern in the wild type flies both at adult stage and during development (Figure 1B, S1A and S2A). These BACs also rescued lethality completely in brp mutants. In addition, the BRP expression pattern did not change with gene dosage, as it remained the same in flies carrying one to four copies of the brp gene (Figure S1B and S1C).
BRP Puncta in Light Microscopy Match T-bars Visualized by EM
To address whether the BRP marker reliably marks presynaptic sites, we assessed the distribution of BRP puncta in a variety of cell types in the visual system, using various cell-type specific drivers to express FLP recombinase in identified neurons (Figure 2 and S3). As in EM studies, BRP puncta in lamina monopolar neurons L1-L5 were predominantly localized to their axonal terminals in the medulla neuropil (Figure 2B), while L2 and L4, also, elaborated a small number of presynaptic sites in proximal regions of their processes within the lamina neuropil (arrowheads in Figure 2B) (Meinertzhagen and O'Neil, 1991; Rivera-Alba et al., 2011; Takemura et al., 2013; Takemura et al., 2008). In transmedulla neuron Tm9, the vast majority of BRP puncta were observed at their axon terminals in the lobula neuropil (arrows in Figure 2C), while very few BRP puncta were seen in their dendrites in the medulla (arrowheads in Figure 2C). This distribution is consistent with results from EM analysis (Takemura et al., 2013). In addition, we were able to label the presynaptic sites in large interneurons like Dm6 and Dm8 (Figure 2D and S2D). Determining the distribution of synapses in these neurons is difficult at the EM level, due the large volume of neuropil their branches span. In summary, the BRP marker accurately reflects the distribution of presynaptic sites in specific neurons as determined by EM.
We next assessed whether the BRP marker could be used to quantify the number of presynaptic sites in specific neurons. Initially, we compared the number and distribution of BRP puncta with that of T-bars in photoreceptor neurons, as assessed by EM. The fly compound eye contains some 750 ommatidia, each comprising 8 photoreceptor neurons falling into 3 classes: R1-R6, R7 and R8 (Clandinin and Zipursky, 2002). The R1-R6 neurons terminate within the lamina neuropil and are primarily presynaptic to lamina neurons (Figure 3A) (Meinertzhagen and O'Neil, 1991; Rivera-Alba et al., 2011). R7 and R8 axons innervate the medulla neuropil and are primarily presynaptic to specific medulla neurons (Gao et al., 2008; Takemura et al., 2013; Takemura et al., 2008). To assess the number of BRP puncta in R1-R6, we used a pan-photoreceptor FLP (panPR-FLP) to activate the BRP marker in all photoreceptor neurons and assessed the number and distribution of presynaptic sites in the lamina (Figure 3). Two FLP recombinases, expressed selectively in R7 or R8 neurons respectively, were used to examine the presynaptic sites along the length of R7 or R8 axons in the medulla (Figure 3D).
BRP puncta in different classes of photoreceptor neurons were quantified by taking stacks of cross-section images of the axons (right panels in Figure 3B and bottom panels in Figure 3D). BRP puncta were distributed uniformly along the entire length of R1-R6 axons within the lamina neuropil (Figure 3B and C). The number of BRP puncta was remarkably similar to T-bars assessed using EM (Table 1) (Meinertzhagen and O'Neil, 1991; Meinertzhagen and Sorra, 2001; Rivera-Alba et al., 2011). R7 and R8 axons span the outer 6 and outer 3 layers of the medulla neuropil (i.e. layer M1-M6 and layer M1-M3), respectively. As in the EM reconstruction studies (Takemura et al., 2013; Takemura et al., 2008), each R8 axon elaborates presynaptic sites along its length spanning layers M1-M3. By contrast, the presynaptic sites in R7 axons preferentially localize to the M4-M6 layers (Figure 3D and E). As with R1-R6 neurons, the number and distribution of BRP puncta in R7 and R8 neurons was very similar to that of T-bars assessed by EM (Table 1 and Figure 3E) (Takemura et al., 2013; Takemura et al., 2008). Thus, STaR provides an efficient means of quantifying synapses by light microscopy.
Table 1.
Numbers of presynaptic sites (Mean ± Standard Deviation) in various neurons mapped by EM and the inducible BRP marker. The number of cells quantified for each cell type (n) is shown in the table. For L4, the total number of BRP puncta in a single lamina cartridge innervated by three L4 dendrites is shown here, one from the home cartridge (L4) and two from neighboring cartridges (L4x and L4y).
| Lamina | Neuron | Meinertzhagen and Sorra, 2001 | Rivera-Alba et al., 2011 | BRP Marker (counter #1) | BRP Marker (counter #2) |
| R1-R6 | 283 (n=1) | 264 (n=1) | 262±34 (n=15) | 250±23 (n=15) | |
| L2 | 8 (n=1) | 7 (n=1) | 9±1 (n=25) | 10±2 (n=25) | |
| L4 (L4+L4x+L4y) | 23 (n=1) | 21 (n=1) | 24±3 (n=30) | 26±4 (n=30) | |
| C2 | 16 (n=1) | 8 (n=1) | 9±1 (n=30) | 9±1 (n=30) | |
| Medulla | Neuron | Takemura et al., 2008 | Takemura et al., 2013 | BRP Marker (counter #1) | BRP Marker (counter #2) |
| R7 | 20±2 (n=3) | 26 (n=1) | 27±5 (n=30) | 24±6 (n=30) | |
| R8 | 35±1 (n=3) | 50 (n=1) | 46±5 (n=30) | 47±7 (n=30) | |
| L4 | 22±2 (n=3) | 26 (n=1) | 26±4 (n=30) | 26±5 (n=30) |
Quantification of BRP puncta for several other neuronal classes also correlated well with T-bars analyzed by EM (Table 1) (Meinertzhagen and O'Neil, 1991; Meinertzhagen and Sorra, 2001; Rivera-Alba et al., 2011; Takemura et al., 2008; Takemura et al., 2013). The high correlation between the two methods suggests that the number of presynaptic sites in each cell type is largely stereotyped. In addition, we were able to observe small variations in the number of presynaptic sites among individual neurons of the same class and among individual animals (see Standard Deviations in Table 1 and data not shown). Thus, comparison between STaR and SSEM provides a rapid means of verifying SSEM and conversely the large number of different cells analyzed by STaR permits an assessment of the variability in synapse number not readily achieved by EM.
To assess the importance of using synaptic markers expressed at endogenous levels for mapping synapses, we compared the GFP-tagged BRP STaR marker to the Gal4-driven UAS-BRP-GFP marker in R7 and R8 neurons, respectively (Brand and Perrimon, 1993; Wagh et al., 2006). The protein encoded by UAS-BRP-GFP accumulated in a diffuse pattern within the R7 and R8 axon terminals and thus failed to mark individual presynaptic sites, in sharp contrast to the endogenous STaR marker (Figure 3F and S4). Thus, it is not possible to quantify synapses using a GAL4/UAS version of tagged BRP in these neurons. Similar results were observed with another commonly used presynaptic marker UAS-synaptotagmin-GFP (Figure S4C). Thus, the STaR method, but not GAL4 mediated expression of tagged presynaptic markers, facilitates quantification of synapses within the CNS.
Cell-type Specific Labeling of Postsynaptic Sites with the STaR Method
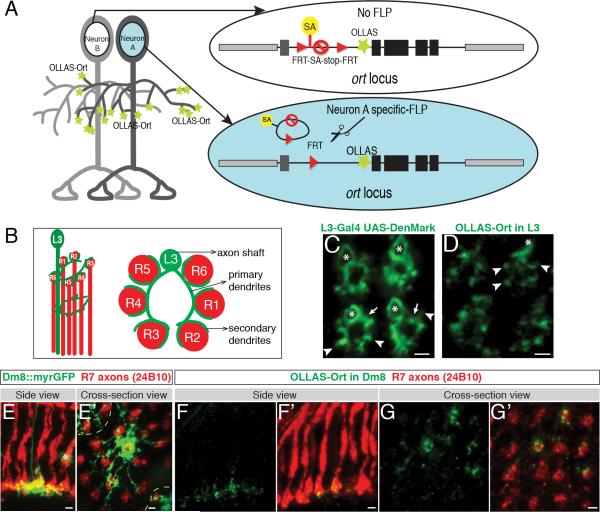
We sought to generate an endogenous marker that selectively marks postsynaptic sites in specific neurons. In contrast to BRP that marks virtually all presynaptic sites, a complementary marker for postsynaptic sites in flies is not known. As an alternative, we chose to tag neurotransmitter receptors in an inducible fashion to demarcate postsynaptic sites. As a proof-of-principle example, we tagged the histamine-gated chloride channel 2 (Ort), the histamine receptor expressed in the postsynaptic partners of the histaminergic photoreceptor neurons, to demonstrate the feasibility of this approach (Gao et al., 2008; Gengs et al., 2002; Pantazis et al., 2008; Stuart et al., 2007).
We modified the endogenous ort sequence in a genomic construct to encode a different inducible epitope tag named OLLAS (Figure 4A) (Park et al., 2008). The OLLAS tag was inserted immediately downstream of the Ort signal sequence (i.e. at the N-terminus of the mature protein). The modified Ort protein is expressed in the same spatiotemporal fashion and at the same level as the unmodified protein from the endogenous locus (Figure S2B and S5). To achieve cell-type-specific expression, an FRT-flanked stop cassette was inserted into the first intron upstream of the translational start. This cassette included a splice acceptor (SA) upstream of the translational and transcriptional stop sequences in the cassette that prevented expression of the ort gene. When combined with a cell-type specific FLP recombinase, the stop cassette was removed, allowing the OLLAS-tagged Ort protein to be expressed in target neurons marking the postsynaptic sites. As insertion of the 2A-LexA sequence at the 3’ end of the ort open reading frame disrupted Ort protein expression, it was not possible to use this strategy to independently assess the morphology of the cell (see Discussion). Nevertheless, by using highly cell-type specific enhancers to drive the FLP recombinase, we were able to reliably label histamine receptors in identified neurons (see Figure 4).
Figure 4. Labeling Postsynaptic Sites in Specific Cell Types using STaR.
(A) Schematic diagram of the cell-type specific postsynaptic marker, the histamine receptor Ort. SA, Splicing Acceptor sequence; OLLAS, an epitope tag; black boxes, ort coding exons; dark gray box, ort non-coding exon; black lines, ort introns; and light gray boxes, neighboring genes.
(B) Schematic drawing of one L3 neuron from the side (left) and in cross-section view (right).
(C) Cross-section view of L3 neurons labeled with the dendritic marker UAS-DenMark.
(D) Postsynaptic sites in L3 neurons labeled with STaR.
In (C) and (D): asterisks, axon shaft; arrows, primary dendrites; and arrowheads, secondary dendrites. Scale bars, 2μm.
(E and E’) Side view (E) and cross-section view (E’) of processes of a single Dm8 neuron labeled with myristoylated GFP (myrGFP). The asterisk in (E) marks processes of a non-Dm8 neuron in the background. The dashed lines in (E’) separate processes of a single Dm8 neuron from processes of its neighbors. Scale bars, 2μm.
(F-G’) Side views (F and F’) and cross-section views (G and G’) of Ort in the Dm8 neurons labeled with STaR. More than one Dm8 neuron is labeled in (G and G’). Red, 24B10 antibody staining labeling axons of R7 and R8 neurons; Green, OLLAS staining. Scale bars, 2μm.
Cell-type specific FLPs were used to selectively activate the Ort marker in L3 neurons (postsynaptic to R1-R6 neurons) and Dm8 neurons (postsynaptic to R7 neurons) (Figure 4B-G) (Gao et al., 2008; Rivera-Alba et al., 2011). L3 neurons extend multiple primary dendrites from the axon shaft with short spike-like secondary dendrites extending between neighboring R1-R6 axons, as shown by the targeted expression of the dendritic marker Denmark via the Gal4-UAS system (Figure 4B and C) (Brand and Perrimon, 1993; Nicolai et al., 2010). OLLAS-tagged Ort in L3 preferentially localized to secondary dendrites (arrowheads in Figure 4D), although some signal was observed along the primary dendrites and the proximal region of the axon shaft (Figure 4D). When we compared the expression pattern of the Ort marker to a general membrane marker (i.e. myr-GFP) expressed in the same L3 neurons, a clear difference in intensity at specific subcellular locations was observed, supporting that the preferential localization of OLLAS-Ort to L3 secondary dendrites is not due to the overall lower expression level of the endogenous construct (Figure S5B).
Dm8 neurons extend a single axon to the M6 layer and arborize extensively within this layer (Figure 4E and E’) (Gao et al., 2008). OLLAS-labeled puncta decorated Dm8 processes and overlapped extensively with the presynaptic R7 terminals (Figure 4F-G’). Taken together, these results demonstrate that the Ort marker is selectively enriched in regions in close proximity to presynaptic terminals of photoreceptor axons.
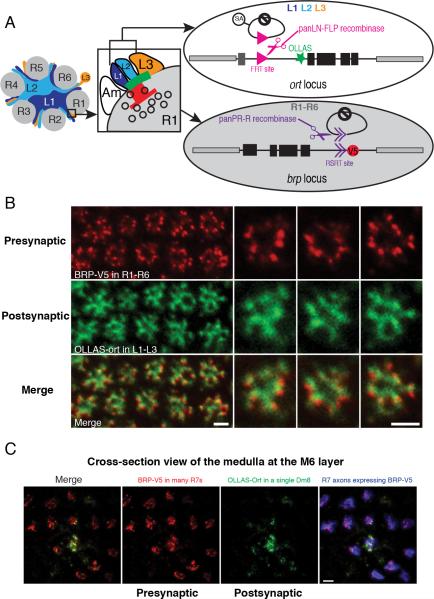
Co-labeling Presynaptic and Postsynaptic Sites with STaR using Two Different Recombination Systems
In order to simultaneously label presynaptic and postsynaptic sites in partner neurons, we incorporated two non-overlapping recombination systems in the same animal (Figure 5) (Nern et al., 2011). The major output synapses of R1-R6 photoreceptor neurons are tetrads (i.e. one presynaptic site opposing four postsynaptic elements); L1 and L2 lamina neurons are postsynaptic at all tetrads and L3 is only postsynaptic at a subset of these with the remainder of the postsynaptic elements provided by amacrine cells or glia (Figure 5A) (Meinertzhagen and O'Neil, 1991; Rivera-Alba et al., 2011). Endogenous Ort is expressed in lamina neurons L1- L3, but neither in L4 nor L5 (Gao et al., 2008). To tag Ort in L1-L3, FLP recombinase was expressed in all lamina neurons. To co-label the presynaptic sites we generated a new version of the BRP marker by replacing the FRT sites with R recombinase recognition sites (RSRT) (Nern et al., 2011). R recombinase was then expressed in all photoreceptor neurons to induce V5 labeling of BRP in R1-R6 neurons (Figure 5A). Combining the two labeling systems in the same animal resulted in matching of V5 puncta with concentrated OLLAS staining in synaptic partners (Figure 5B). This matching was not observed when we co-expressed the BRP marker in R1-R6 neurons and a general membrane marker (myr-tdTomato) in L1-L3 neurons (Figure S6).
Figure 5. Co-labeling of Presynaptic and Postsynaptic Sites in Partner Neurons with Different Recombination Systems using STaR.
(A) Schematic for STaR co-labeling the presynaptic and postsynaptic sites in partner neurons (i.e. R1-R6 neurons synapsing onto L1-L3 neurons). Left: R1-R6 axons and dendrites of L1, L2 and L3 neurons in a single lamina cartridge. Center: a tetrad synapse between a single presynaptic R1 cell and four postsynaptic elements one each from an L1, L2, L3 and amacrine cell (Am). Red, presynaptic T-bar; Green, postsynaptic neurotransmitter receptors. Open circles, synaptic vesicles. Right: the R-RSRT and FLP-FRT recombination systems independently activate the inducible presynaptic marker and postsynaptic marker in R1-R6 neurons and L1-L3 neurons, respectively. Black boxes, coding exons; dark gray box, the non-coding exon; black lines, introns; and light gray boxes, neighboring genes.
(B) Co-labeling presynaptic sites in R1-R6 neurons and postsynaptic sites in L1-L3 neurons using STaR. Each BRP-V5 punctum (red) appears in juxtaposition to concentrated OLLAS-Ort staining (green). Scale bars, 2μm.
(C) Co-labeling presynaptic sites in R7 neurons and postsynaptic sites in Dm8 neurons using STaR. The R7 axons expressing BRP-V5 (red) are simultaneously labeled by myr-tdTomato (blue). Only a subset of R7 neurons is labeled with V5. Scale bar, 2μm.
Using a similar scheme, we labeled V5-tagged BRP in the presynaptic R7 neurons and OLLAS-tagged Ort in the postsynaptic Dm8 neurons in the medulla (Figure 5C). The juxtaposition of V5 puncta and concentrated OLLAS staining shows that STaR allows co-labeling of presynaptic and postsynaptic sites simultaneously in synaptic partners in a complex neuropil.
Formation of Presynaptic Sites in Developing Neurons
The extraordinary density of axonal and dendritic processes of many different neuronal cell types in the developing neuropil hinders the study of synapse formation in the CNS. Here, we took advantage of cell-type specific labeling of presynaptic and postsynaptic sites during development to follow the time course of synapse formation in all three classes of photoreceptor neurons.
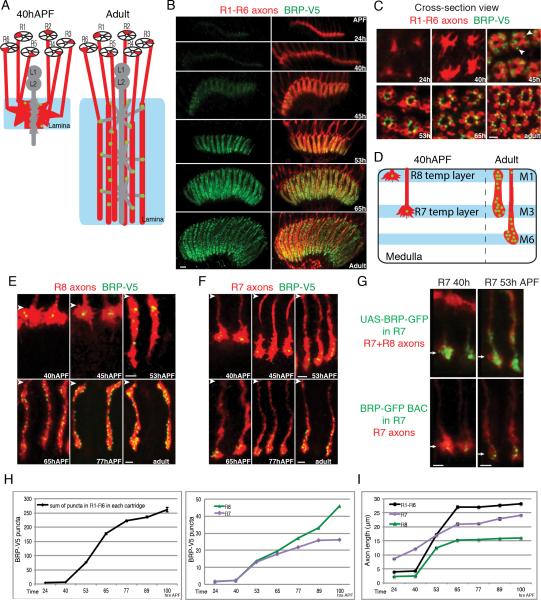
During synapse formation, synaptic proteins are trafficked to specific locations and assembled into presynaptic structures (Owald and Sigrist, 2009). As the tagged BRP protein is expressed via its endogenous regulatory mechanisms, the appearance of BRP puncta provides a marker for active zone assembly (Fouquet et al., 2009). We analyzed presynaptic development in photoreceptor neurons by assessing the spatiotemporal accumulation of BRP puncta within them at different times during development.
R1-R6 neurons extend axons into the lamina during the 3rd instar larval and early pupal stages with their growth cones terminating in the developing lamina (Figure 6A). These growth cones rearrange within the lamina between 30 and 40 hours after puparium formation (hAPF) forming nascent lamina cartridges comprising the growth cones of six R1-R6 axons from six different ommatidia. These axons then extend some 20 microns deeper forming the full depth of the lamina neuropil (Clandinin and Zipursky, 2002). Selective tagging of BRP in photoreceptor neurons revealed a few BRP puncta within R1-R6 growth cones at 40hAPF, just prior to extension (Figure 6B and C). By 45hAPF, R1-R6 axons extend an additional 5 microns as they become morphologically transformed from motile growth cones with filopodia to cylindrically shaped axon terminals. As these axons extend, BRP puncta appear at the borders between neighboring axons suggesting that initial presynaptic sites have formed (arrowheads in Figure 6C). Although terminal extension is complete by 65 hAPF, BRP puncta continue to accumulate through the remaining 30 hours of pupal development (Figure 6B, C, H and I).
Figure 6. STaR Labeling of Presynaptic Sites in the Developing Photoreceptor Axons.
(A) Schematic drawing of R1-R6 axons in the lamina before (left) and after (right) synapse formation.
(B and C) Presynaptic sites in R1-R6 axons labeled with STaR at different developmental times viewed from the side (B) and in cross-section (C) of the lamina. Scale bars, 5μm in (B) and 2μm in (C). Red, R1-R6 axons labeled with myr-tdTomato; Green, V5 staining. Arrowheads point to the initial BRP puncta at 45hAPF.
(D) Schematic drawing of R7 and R8 axons in the medulla before and after synapse formation.
(E and F) Presynaptic sites in R8 (E) and R7 (F) axons labeled with STaR at different developmental times. The arrowheads indicate the top of the medulla neuropil in each image. Scale bars, 2μm.
(G) Comparison between the GFP-tagged BRP endogenous marker and the Gal4-driven UAS-BRP-GFP marker in R7 neurons during development. Arrows point to R7 terminals. Red, photoreceptor axons marked with MAb24B10 (top panels) or myr-tdTomato (bottom panels); Green, GFP staining. Scale bars, 2μm.
(H) Number of BRP-V5 puncta in R1-R6 axons within each lamina cartridge (left) and R7 and R8 axons (right) at different developmental times. The numbers in the left panel refer to the sum of puncta in all 6 axons within the same lamina cartridge. Error bars, Standard Error.
(I) Approximate length of photoreceptor axons at different developmental times. The length is measured from the top of the corresponding neuropil (lamina for R1-R6, medulla for R7 and R8) to the end of the axons marked with myr-tdTomato. Error bars, Standard Error.
A similar time course of BRP accumulation was observed in R7 and R8 terminals. Prior to 40hAPF, R7 and R8 growth cones extend into the developing optic lobes in birth order and terminate in intermediate targets (Figure 6D). R8 growth cones reside at the distal edge of the medulla and R7 growth cones occupy a characteristic position proximal to R8 growth cones. At 40hAPF each R8 growth cone harbors 1-3 bright BRP puncta roughly in the center of the growth cone; a small number of BRP puncta were also seen in R7 growth cones (Figure 6E, F and H). Commencing just after 40hAPF, R7 and R8 growth cones extend into deeper layers of the medulla in a synchronous fashion (Nern et al., 2005; Timofeev et al., 2012; Ting et al., 2005). Coincident with this synchronous extension, BRP puncta begin to accumulate in the newly extended R7 and R8 axon terminals also in a synchronous fashion (Figure 6E, F, H and I). This process is very similar to the presynaptic development in R1-R6 neurons in the lamina.
Together, despite differences in their targeting regions and synaptic partners, the time course of presynaptic development for all three classes of photoreceptor neurons is highly similar: the formation of presynaptic sites initiates in the same time window between 40h-53hAPF and is coincident with the extension of the axons marking the transition from growth cones to synaptic terminals. Importantly, when the UAS-BRP-GFP marker was expressed in R7 neurons via Gal4-mediated expression at these developmental stages, strong GFP signal was observed throughout the terminals at both 40hAPF and 53hAPF, failing to reveal the transition of these growth cones to synaptic terminals (Figure 6G and S4). Thus, STaR facilitates analysis of synaptic development at the level of single identified cells in the context of a complex neuropil.
Accumulation of Neurotransmitter Receptors at Postsynaptic Sites
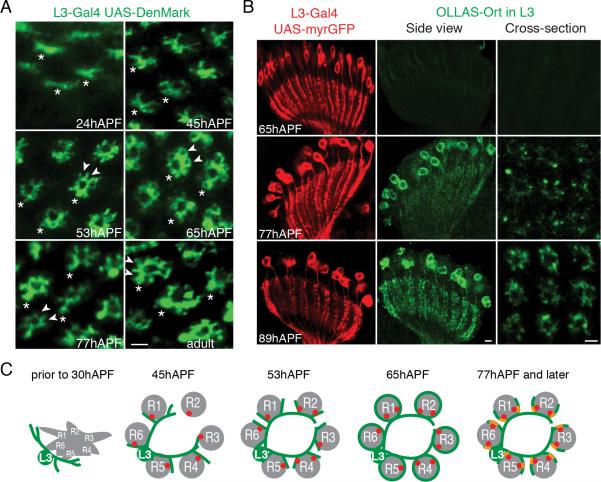
We next followed postsynaptic development of targets of photoreceptor neurons. For this, we focused on L3 neurons in the lamina using targeted expression of Denmark to label L3 dendrites and accumulation of OLLAS-tagged Ort through STaR to label postsynaptic sites. At 24hAPF, filopodia-like dendritic structures extend from the proximal region of the L3 axon (Figure 7A). Between 40-45hAPF, these processes extend into the center of the lamina cartridge between the inner cluster of L1 and L2 axons and the surrounding rosette of R1-R6 axons (Figure 7A and data not shown). By this stage of development, a significant number of presynaptic sites in the R1-R6 axons have formed (Figure 6B, C and H). By 53hAPF, short spike-like secondary dendrites extend out from the primary dendrites and intercalate between the R1-R6 axons. Curiously, these secondary dendrites continue to elongate and surround individual photoreceptor axons at 65hAPF, but later retract (arrowheads in Figure 7A). By 77hAPF, the retraction is complete and the dendrites look morphologically indistinguishable from those of adult L3 neurons. OLLAS-tagged Ort begins to accumulate in L3 dendrites at 77hAPF (Figure 7B), after L3 dendrites have adopted their mature morphology and long after the onset of BRP puncta accumulation in the presynaptic photoreceptor axons.
Figure 7. Accumulation of Neurotransmitter Receptors Labeled with STaR in Developing L3 Neurons.
(A) Cross-section view showing the development of L3 dendrites at different developmental times marked by L3-specific expression of Denmark. Asterisks indicate the axon shafts; arrowheads at 53, 77hAPF and adult stage highlight the cup-shaped staining indicating L3 secondary dendrites only partially surround R cell axons at these times; arrowheads at 65hAPF point to the overgrown secondary dendrites completely surrounding R cell axons (R cell axons are not labeled in these images). Scale bar, 2μm.
(B) Accumulation of Ort in L3 dendrites marked by STaR. Left: Side view showing L3 cell bodies and dendrites marked with myr-GFP. Center: side view showing Ort accumulation in L3 cell bodies and dendrites. Scale bars, 5μm. Right: cross-section view showing Ort accumulation in L3 dendrites. Scale bars, 2μm.
(C) Schematic diagrams summarizing synapse formation between R1-R6 cells and L3 neurons. Presynaptic sites (red dots) form prior to dendritogenesis. Dendrites (green) go through dynamic morphological changes extending beyond their targets followed by retraction to them. These morphological changes occur prior to expression of the neurotransmitter receptors (yellow) within dendrites.
The late localization of Ort to dendrites, after the appearance of BRP puncta also occurred in the other postsynaptic partners of R1-R6 neurons including L1, L2 and Am cells, as well as in the Ort-expressing cells in the medulla postsynaptic to R7 and R8 terminals (Figure S2B). These findings suggest a common timeline for synapse formation in all three classes of photoreceptor neurons (Figure 7C). Presynaptic sites start to form in the photoreceptor axons in close temporal association with dendritogenesis in target neurons. Dendritic processes of postsynaptic neurons go through dynamic morphological changes. It is only after they adopt their mature morphology, some 30 hours after the onset of dendritogenesis, that neurotransmitter receptors become localized to these postsynaptic sites.
Following the Formation of Presynaptic Sites in Live Animals
The ability to image synapses in real-time in live animals is crucial to reveal dynamic changes during synapse formation as well as changes in synaptic organization as a consequence of activity. To test the utility of the STaR strategy in visualizing synapse formation in live animals, we used an R8-specific FLP recombinase to selectively tag BRP with GFP in R8 neurons and followed the dynamics of this marker during the time window of initial synapse formation (i.e. 42-55hAPF) in live, developing pupae with two-photon microscopy (O.A. and S.L.Z., manuscript in preparation). Gal4-mediated expression of UAS-myr-tdTomato was used in parallel to mark R8 axons. After the pupal case surrounding the head was gently removed, each pupa was mounted on a slide with one eye contacting the coverslip. The objective lens was placed against the eye under the coverslip and the dynamics of the developing R8 terminals were visualized directly through the retina in the underlying optic lobe. This is a non-invasive procedure; fertile flies emerge after pupal development with no morphological defects in the visual system.
Consistent with our observation with the fixed samples, after targeting to the distal edge of the medulla, R8 growth cones remain there until 40hAPF with weak GFP signal localized to the center of the growth cone (Figure 8). After 40hAPF, each R8 growth cone starts targeting to the M3 layer first with the rapid extension of a very thin process (Figure 8, red channel, 42-45hAPF). Once this leading process reaches the M3 target layer, it thickens from the top down and the R8 growth cone begins its transformation into a mature axon terminal with the appearance of many BRP puncta along its length (Figure 8B, 45-52hAPF). Live imaging revealed that stable BRP puncta are largely absent from the leading thin process but rapidly populate the thickening terminal. When individual puncta were tracked between successive time points, we found that stable puncta appear from the proximal to distal locations along the length of the extending terminal (data not shown). In summary, STaR allows imaging of synapse formation in specific neurons in real-time within the CNS non-invasively in intact animals.
Discussion
In this study, we describe STaR, a method for marking synapses in defined populations of neurons in both the developing and adult fly nervous system. First, we show that the number and distribution of presynaptic sites labeled by the presynaptic marker correspond well with SSEM studies. Second, by using two independent recombination systems in the same animal, we labeled matching presynaptic and postsynaptic sites simultaneously in synaptic partners. Third, we show that these markers can be used to study synapse formation in identified cells types within the complex CNS in both fixed samples and in live animals.
STaR was inspired by the GRASP method developed by Bargmann and colleagues for studies of neural circuits in C. elegans (Feinberg et al., 2008), and limitations we encountered in using it to study synapse formation in the fly visual system. The initial adaptation of GRASP to Drosophila used the general transmembrane protein CD4 to tether the two split GFP fragments (spGFP1-10 and spGFP11) and was used to detect cell-cell contacts (Gordon and Scott, 2009). We generated synapse-specific GRASP by fusing spGFP1-10 to Neurexin-1 (Nrx-1), a transmembrane protein involved in synapse formation and maturation (Fan et al., 2013; Li et al., 2007; Zeng et al., 2007). This construct (Nrx-1::spGFP1-10) in combination with CD4::spGFP11 detected synaptic connections between neurons in the fly visual system, previously defined by EM reconstruction (e.g. L3-Tm9; L2-L4; R7-Dm8 etc.) (Fan et al., 2013; Takemura et al., 2013) (Y.C. and S.L.Z., unpublished). We also observed GFP signal, however, between pairs of neurons which, though in close association, do not form synaptic contacts (e.g. L1-L2 and L1-L4) (Rivera-Alba et al., 2011)(Y.C. and S.L.Z., unpublished). Thus, signals detected by this method may not strictly reflect synaptic contacts, presumably due to localization artifacts arising from over-expression.
The STaR method facilitates rapid comparisons of the pattern and number of synapses in specific neurons at multiple developmental stages, in various mutant backgrounds or under different activity-modulated conditions. The synaptic markers can be readily integrated into genetic schemes, such as RNA-interference and Mosaic Analysis with a Repressible Cell Marker (MARCM) to study the molecular mechanisms underlying synaptic specificity (Dietzl et al., 2007; Lee and Luo, 2001; Ni et al., 2009). These markers can also detect the modulation of synapse number and structure by activity. Indeed, after exposing fruit flies reared in the darkness to light for 15 minutes, we observed a 15% increase in the number of BRP puncta in R1-R6 photoreceptor axons with STaR (Figure S7), similar to results of previous EM studies on the houseflies Musca domestica (Rybak and Meinertzhagen, 1997). In live animals, the intensity of BRP-GFP fluorescence is lower than GFP or V5 antibody staining on the fixed samples but we did not observe differences in BRP localization. Together with the fact that the number and pattern of V5-tagged BRP puncta correlates very well with EM studies in adult flies, we believe that our fixation and immuno-staining conditions introduced minimal, if any, artifacts, which may have influenced the interpretations of our observations in mature and developing animals.
In many neurons presynaptic sites are relatively sparse (see Table 1 and Figure 2 and 3) and, hence, quantification of BRP puncta is not limited by the resolution of light microscopy (i.e. neighboring presynaptic sites are easily separable). By contrast, for some lamina neuron terminals in the medulla (e.g. L2 in Figure 2B) quantification is hindered by increased density of presynaptic sites even when imaged from the optimal orientation to resolve neighboring puncta. Preliminary studies indicate that STaR is suitable for super-resolution techniques, such as Stochastic Optical Reconstruction Microscopy (STORM), which would facilitate accurate mapping and quantification of the number of presynaptic sites in neurons with particularly high density of synapses (data not shown) (Rust et al., 2006).
STaR can be extended readily to studies in other regions of the fly nervous system. Although the application of the STaR was greatly facilitated by the large collection of cell-type-specific enhancer/promoters available in the fly visual system (Gohl et al., 2011; Jenett et al., 2012; Pfeiffer et al., 2008), similar reagents are rapidly becoming available for many other regions of the fly nervous system and thus labeling presynaptic sites of vast numbers of different neurons will become straight forward. Although the Ort postsynaptic marker is limited to the histaminergic neurons, the same design principle can be applied to other types of neurotransmitter receptors to expand our postsynaptic marker tool kit to label different types of postsynaptic sites. In future studies inclusion of the GAL80 repressor within the stop cassette will facilitate the use of the GAL4-UAS system to label the neurotransmitter receptor-expressing cells without inserting the 2A-LexA cassette to the C-terminus (Ma and Ptashne, 1987), which may disrupt the expression of tagged receptors as it did for Ort.
The STaR strategy can be expanded to generate markers for other types of communication within neural circuits not readily accessible to EM analysis. Gap junctions, for instance, are widely used in the fly nervous system yet are difficult to identify by EM (Shaw et al., 1989; Shimohigashi and Meinertzhagen, 1998). Cell-type specific tagging of gap junction components, the innexin family proteins (Bauer et al., 2005), may provide an entry point to study these structures. In a similar fashion, different neuromodulatory and neuropeptide receptors can be labeled in an inducible fashion. The use of different recombination systems, such as the FLP, R, B2, B3 and KD (Nern et al., 2011), allows different tagged synaptic proteins to be visualized in combination to characterize the molecular properties of identified synapses and other structures in the brain.
While we used BAC recombineering techniques to modify genomic loci of synaptic proteins (Venken et al., 2006), a variety of additional methods are available to generate STaR-based markers, such as Minos-mediated integration cassette (MiMIC) insertions (Venken et al., 2011), and site-specific recombination through cassette exchange (Pecot et al., 2013; Weng et al., 2009). In principle, the STaR method can be extended to the mouse and zebrafish through knock-in and BAC transgenic techniques and through the use of multiple recombination systems (e.g. Cre-Lox and the B3 and KD systems) (Capecchi, 2005; Nern et al., 2011; Stuart et al., 1988).
In summary, we have developed STaR, a cell-type specific synaptic tagging method for light microscopy in fixed and live preparations. We anticipate that this methodology will be useful to investigators examining the architecture, development, dynamics and function of synapses in a wide variety of neural circuits.
Experimental procedures
Generation of the BRP presynaptic marker and Ort postsynaptic marker
See Supplemental Experimental Procedures for detailed information regarding the generation these markers.
Drosophila stocks
Specific fly genotypes in each experiment are described in supplementary methods. Flies were reared at 25° on standard cornmeal/molasses food. Pupal staging was performed by counting the number of hours at 25° after selecting pre-pupa (i.e. 0 hAPF). For instance, 24hrs after the pre-white pupal stage at 25° is 24hAPF.
In addition to the STaR markers presented in this study, we used the following lines: (1) 9-9 Gal4 (L3); (2) Dac-FLP 20 (Pecot et al., 2013); (3) MH56 Gal4 (L3) (Timofeev et al., 2012); (4) Rh4-Gal4 (R7); (5) Rh6-Gal4 (R8) (Tahayato et al., 2003); (6) GMR-FLP (PanPR-FLP) (Pignoni et al., 1997); (7) UAS-FLP (Duffy et al., 1998; Nern et al., 2011); (8) GMR-Gal4 (PanPR-Gal4) (Wernet et al., 2003); (9) UAS-Denmark (Nicolai et al., 2010); (10) UAS-BRP-GFP (Wagh et al., 2006); (11) 20C11(R7)-FLP; (12) Senseless (R8)-FLP; (13) LexAoP-myr-tdTomato in su(Hw)attP2 and su(Hw)attP5; (14) GMR>stop>Gal4; (15) UAS>stop>myr-tdTomato in su(Hw)attP2; (16) UAS-R in attP2 (Nern et al., 2011); (17) UAS-myr-GFP in attP40 (18) UAS-myr-tdTomato in su(Hw)attP2 (Pfeiffer et al., 2010); (19) R27G05-FLP2::PEST in attp40 (PanLN-FLP); (20) R20C11-Gal4 (R7); (21) R24C08-Gal4 (Tm9); (22) R53C12-Gal4 (Mi1); (23) R20G06-Gal4 (Dm6); (24) w; R20C11-AD; R25B02-DBD (C2); (25) w; R26H02-AD; R29G11-DBD (C3); and (26) w; R55F02-AD; R69A01-DBD (Dm8) (Pfeiffer et al., 2008; Tuthill et al., 2013). Stock (4)-(10) were acquired from Bloomington Drosophila Stock Center. Stock (11)-(15) were generated in this study. Stocks (24)-(26) are split GAL4 stocks.
Histology
Histology was performed as described previously with minor modification (Pecot et al., 2013). Fly brains were fixed with PBL (4% paraformaldehyde, 75 mM lysine, 37 mM sodium phosphate buffer pH 7.4) for 25min at room temperature. After multiple rinses in PBS with 0.5% Triton X-100 (PBT), brains were blocked in 10% normal goat serum in PBT (blocking solution) for 1hr. Brains were incubated with primary and secondary antibodies for 2 days each at 4° with multiple rinses in blocking solution in between and afterwards. Brains were mounted in Slow Fade Gold anti-Fade Reagent (Life technologies).
See Supplementary Experimental Procedures for a complete list of primary and secondary antibodies used in this study.
Microscopy and image analysis
Confocal images were acquired with Zeiss LSM780 confocal microscope. The staining patterns were reproducible between samples but overall fluorescence signal and noise unavoidably shows some variation between sections and samples. Some adjustments of laser power, gain and black level settings were therefore made to obtain similar overall fluorescence signals. Single plane or maximum intensity projection confocal images were exported into TIF files using LSM Image Browser (ZEISS).
For quantification of the BRP-V5 puncta, confocal stack images were taken from the optimal orientation (cross-section of the axon axes) to cover the entire axon length. For each cell type, several Z-step values were tested out to find an optimal value (normally between 0.8-1.5μm) to avoid imaging the same puncta redundantly in adjacent sections or missing puncta. The number of BRP-V5 puncta were then scored by two counters (counter #1 and #2 in Table 1) independently by going through the stack images section by section with LSM Image Browser. Counter #2 was an undergraduate student who was unfamiliar with synapses in the fly visual system. Five optic lobes from five animals were imaged and quantified for each cell type. For R1-R6 neurons, three cartridges in each optic lobe were scored, giving a total of 15 cartridges or R1-R6 sets. For L2 neurons, five cells in each optic lobe were scored, giving a total of 25 cells. For the other cell types, six cells in each optic lobe were scored, giving a total of 30 cells for each cell type.
Live imaging of Drosophila pupae
Live Drosophila pupae were staged and mounted on slides at around 36hAPF with one eye contacting the coverslip. Stack images of the R8 axons were taken directly through the retina into the underlying optic lobe every 15 minutes for the next 24hrs with custom 2-photon microscope. These stack images were then reconstructed and aligned to generate maximum-intensity projection images shown in Figure 8. (O.A. and S.L.Z., manuscript in preparation). Detailed information regarding the 2-photon microscope, the imaging set-up and data processing are available upon request.
Supplementary Material
Acknowledgments
We thank Dorian Gunning for help in generating the Ort antibody. We thank Martin Wallner and Richard Ohlsen for recordings in Xenopus oocytes to test different locations for tagging Ort. We thank Koen Venken and Hugo Bellen for providing reagents and advice for BAC recombineering, Tzumin Lee for providing DNA constructs containing LexA::VP16, Gerry Rubin and Barrett Pfeiffer for flies containing various cell-type specific Gal4s and DNA constructs, and Stephan Sigrist for providing brp mutant flies. We thank Mitya Chklovskii, Louis Scheffer, Shinya Takemura, Marta Rivera-Alba and Ian Meinertzhagen for providing the latest results from the fly EM project at Janelia Farm Research Campus. For critical reading of the manuscript, we would also like to thank Ian Meinertzhagen, Kelsey Martin, Eddy De Robertis, Utpal Banerjee, Alvaro Sagasti and members of the S.L.Z. laboratory. This work was supported by UCLA Dissertation Year Fellowship and Howard Hughes Medical Institute. S.L.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauer R, Loer B, Ostrowski K, Martini J, Weimbs A, Lechner H, Hoch M. Intercellular communication: the Drosophila innexin multiprotein family of gap junction proteins. Chemistry & biology. 2005;12:515–526. doi: 10.1016/j.chembiol.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Butcher NJ, Friedrich AB, Lu Z, Tanimoto H, Meinertzhagen IA. Different classes of input and output neurons reveal new features in microglomeruli of the adult Drosophila mushroom body calyx. J Comp Neurol. 2012;520:2185–2201. doi: 10.1002/cne.23037. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nature reviews Genetics. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Making connections in the fly visual system. Neuron. 2002;35:827–841. doi: 10.1016/s0896-6273(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- Fan P, Manoli DS, Ahmed OM, Chen Y, Agarwal N, Kwong S, Cai AG, Neitz J, Renslo A, Baker BS, Shah NM. Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell. 2013;154:89–102. doi: 10.1016/j.cell.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Fischbach KF, Dittrich APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell and Tissue Research258. 1989:441–475. [Google Scholar]
- Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, et al. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengs C, Leung HT, Skingsley DR, Iovchev MI, Yin Z, Semenov EP, Burg MG, Hardie RC, Pak WL. The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA). J Biol Chem. 2002;277:42113–42120. doi: 10.1074/jbc.M207133200. [DOI] [PubMed] [Google Scholar]
- Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nature methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka Y, Meinertzhagen IA. Immunocytochemical localization of synaptic proteins to photoreceptor synapses of Drosophila melanogaster. J Comp Neurol. 2010;518:1133–1155. doi: 10.1002/cne.22268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li J, Ashley J, Budnik V, Bhat MA. Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron. 2007;55:741–755. doi: 10.1016/j.neuron.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, O'Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Sorra KE. Synaptic organization in the fly's optic lamina: few cells, many synapses and divergent microcircuits. Prog Brain Res. 2001;131:53–69. doi: 10.1016/s0079-6123(01)31007-5. [DOI] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Svoboda K, Rubin GM. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc Natl Acad Sci U S A. 2011;108:14198–14203. doi: 10.1073/pnas.1111704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML. Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein-GFP fusions. J Neurosci Methods. 1999;89:33–40. doi: 10.1016/s0165-0270(99)00031-x. [DOI] [PubMed] [Google Scholar]
- Owald D, Sigrist SJ. Assembling the presynaptic active zone. Curr Opin Neurobiol. 2009 doi: 10.1016/j.conb.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Pantazis A, Segaran A, Liu CH, Nikolaev A, Rister J, Thum AS, Roeder T, Semenov E, Juusola M, Hardie RC. Distinct roles for two histamine receptors (hclA and hclB) at the Drosophila photoreceptor synapse. J Neurosci. 2008;28:7250–7259. doi: 10.1523/JNEUROSCI.1654-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Cheong C, Idoyaga J, Kim JY, Choi JH, Do Y, Lee H, Jo JH, Oh YS, Im W, et al. Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. Journal of immunological methods. 2008;331:27–38. doi: 10.1016/j.jim.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot MY, Tadros W, Nern A, Bader M, Chen Y, Zipursky SL. Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron. 2013;77:299–310. doi: 10.1016/j.neuron.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Prokop A, Meinertzhagen IA. Development and structure of synaptic contacts in Drosophila. Semin Cell Dev Biol. 2006;17:20–30. doi: 10.1016/j.semcdb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Rivera-Alba M, Vitaladevuni SN, Mishchenko Y, Lu Z, Takemura SY, Scheffer L, Meinertzhagen IA, Chklovskii DB, de Polavieja GG. Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr Biol. 2011;21:2000–2005. doi: 10.1016/j.cub.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak J, Meinertzhagen IA. The effects of light reversals on photoreceptor synaptogenesis in the fly Musca domestica. Eur J Neurosci. 1997;9:319–333. doi: 10.1111/j.1460-9568.1997.tb01402.x. [DOI] [PubMed] [Google Scholar]
- Shaw SR, Frohlich A, Meinertzhagen IA. Direct connections between the R7/8 and R1-6 photoreceptor subsystems in the dipteran visual system. Cell Tissue Res. 1989;257:295–302. doi: 10.1007/BF00261833. [DOI] [PubMed] [Google Scholar]
- Shimohigashi M, Meinertzhagen IA. The shaking B gene in Drosophila regulates the number of gap junctions between photoreceptor terminals in the lamina. J Neurobiol. 1998;35:105–117. doi: 10.1002/(sici)1097-4695(199804)35:1<105::aid-neu9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. The Journal of general virology. 1991;72(Pt 7):1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- Stuart AE, Borycz J, Meinertzhagen IA. The dynamics of signaling at the histaminergic photoreceptor synapse of arthropods. Prog Neurobiol. 2007;82:202–227. doi: 10.1016/j.pneurobio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Stuart GW, McMurray JV, Westerfield M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- Tahayato A, Sonneville R, Pichaud F, Wernet MF, Papatsenko D, Beaufils P, Cook T, Desplan C. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Developmental cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol. 2008;509:493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeev K, Joly W, Hadjieconomou D, Salecker I. Localized netrins act as positional cues to control layer-specific targeting of photoreceptor axons in Drosophila. Neuron. 2012;75:80–93. doi: 10.1016/j.neuron.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, Nern A, Holtz SL, Rubin GM, Reiser MB. Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron. 2013;79:128–140. doi: 10.1016/j.neuron.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, Bellen HJ. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R, Chen YW, Bushati N, Cliffe A, Cohen SM. Recombinase-mediated cassette exchange provides a versatile platform for gene targeting: knockout of miR-31b. Genetics. 2009;183:399–402. doi: 10.1534/genetics.109.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, Desplan C. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA, Schurmann FW. Synaptic connections of cholinergic antennal lobe relay neurons innervating the lateral horn neuropile in the brain of Drosophila melanogaster. J Comp Neurol. 2003;466:299–315. doi: 10.1002/cne.10867. [DOI] [PubMed] [Google Scholar]
- Yeh E, Kawano T, Weimer RM, Bessereau JL, Zhen M. Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J Neurosci. 2005;25:3833–3841. doi: 10.1523/JNEUROSCI.4978-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sun M, Liu L, Chen F, Wei L, Xie W. Neurexin-1 is required for synapse formation and larvae associative learning in Drosophila. FEBS Lett. 2007;581:2509–2516. doi: 10.1016/j.febslet.2007.04.068. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.