The Ustilago maydis Hook protein Hok1 is part of an evolutionarily conserved protein complex that regulates bidirectional early endosome trafficking by controlling attachment of both kinesin-3 and dynein.
Abstract
Bidirectional membrane trafficking along microtubules is mediated by kinesin-1, kinesin-3, and dynein. Several organelle-bound adapters for kinesin-1 and dynein have been reported that orchestrate their opposing activity. However, the coordination of kinesin-3/dynein-mediated transport is not understood. In this paper, we report that a Hook protein, Hok1, is essential for kinesin-3– and dynein-dependent early endosome (EE) motility in the fungus Ustilago maydis. Hok1 binds to EEs via its C-terminal region, where it forms a complex with homologues of human fused toes (FTS) and its interactor FTS- and Hook-interacting protein. A highly conserved N-terminal region is required to bind dynein and kinesin-3 to EEs. To change the direction of EE transport, kinesin-3 is released from organelles, and dynein binds subsequently. A chimaera of human Hook3 and Hok1 rescues the hok1 mutant phenotype, suggesting functional conservation between humans and fungi. We conclude that Hok1 is part of an evolutionarily conserved protein complex that regulates bidirectional EE trafficking by controlling attachment of both kinesin-3 and dynein.
Introduction
The organization of eukaryotic cells depends on bidirectional membrane trafficking along microtubules (MTs; Welte, 2004). Anterograde transport along MTs is mediated predominantly by kinesin-1, the founding member of the kinesin superfamily (Brady, 1985; Vale et al., 1985), and kinesin-3. Kinesins are opposed by cytoplasmic dynein that moves cargo toward MT minus ends (Vale, 2003; Vallee et al., 2004; Hirokawa et al., 2010). Kinesins and dynein bind to the same organelle, where they compete with each other in a “tug-of-war” to determine the direction of transport (Gross, 2004; Welte, 2004; Müller et al., 2008; Soppina et al., 2009; Hendricks et al., 2010; Jolly and Gelfand, 2011). Recent experimental evidence both in vitro (Derr et al., 2012) and in living cells (Kunwar et al., 2011) suggests that additional higher order control modulates the tug-of-war, thereby orchestrating bidirectional transport (Jolly and Gelfand, 2011). Indeed, cargo-bound adapter complexes have been reported that bind and coordinate both kinesin-1 and dynein (Deacon et al., 2005; Caviston and Holzbaur, 2009; Akhmanova and Hammer, 2010; Fridolfsson et al., 2010; Splinter et al., 2010; Mitchell et al., 2012; Fu and Holzbaur, 2013; van Spronsen et al., 2013). However, 8 of the 45 reported kinesins in mice and humans are kinesin-3 motors (Miki et al., 2001), and these motors play pivotal roles in axonal transport (DeGiorgis et al., 2008) and motility of early endosomes (EEs; Hoepfner et al., 2005). Despite this importance, no coordinating adapter is known for kinesin-3 and dynein.
The fungus Ustilago maydis is a genetically tractable organism that shares remarkable conservation with human cells (Steinberg and Perez-Martin, 2008). This includes the use of kinesin-3 and dynein for bidirectional EE motility (Wedlich-Söldner et al., 2002b; Lenz et al., 2006). Anterograde EE transport is driven by three to six kinesin-3 motors, whereas retrograde movement is mediated by single dyneins, which change the direction of EE transport upon binding (Schuster et al., 2011c). Here, we report that a Hook protein, Hok1, is essential for EE motility. Hook proteins have previously been reported as organelle–MT linkers, involved in endocytic protein trafficking, Golgi organization, and cilia formation, but have not been implicated in bidirectional transport (Krämer and Phistry, 1996, 1999; Sunio et al., 1999; Walenta et al., 2001; Ge et al., 2010; Baron Gaillard et al., 2011; Maldonado-Báez et al., 2013). We show that Hok1 forms a complex with homologues of the human fused toes (FTS) protein (Lesche et al., 1997) and its interactor FTS- and Hook-interacting protein (FHIP; Xu et al., 2008). A conserved coiled-coil (CC) domain within the N-terminal region of Hok1 controls both dynein and kinesin-3 attachment to EEs. A change from anterograde to retrograde transport is accompanied by kinesin-3 release. When considered together, our results suggest that Hok1 is part of an adapter complex that orchestrates bidirectional transport by coordinating kinesin-3 and dynein attachment to EEs.
Results
Identification of factors involved in bidirectional EE motility
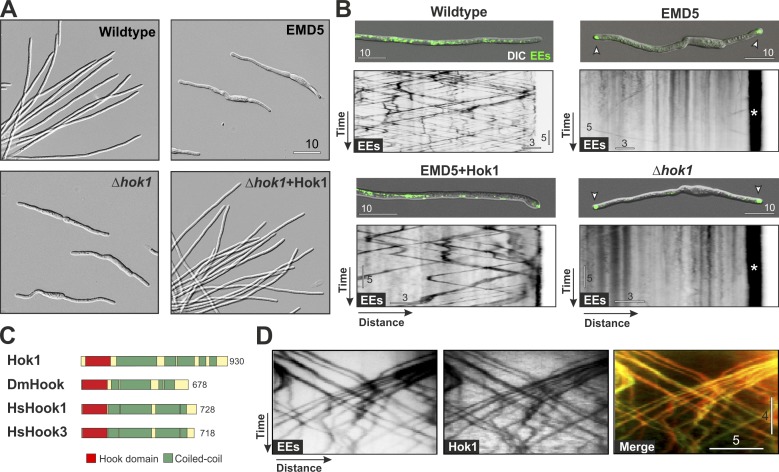
We identified mutants defective in EE motility by screening U. maydis for impaired growth morphology. Normal wild-type hyphae elongate and carry a central nucleus (Fig. S1 A). Dynein and kinesin-3 mediate bidirectional EE motility along MTs (Lenz et al., 2006). These are unipolar near the tip and septum, extending their plus ends to the cell poles, but form bipolar bundles in the center of cells, with their minus ends concentrated near the nucleus (Fig. S1 B; Schuster et al., 2011b). Consequently, inactivation of either dynein or kinesin-3 leads to unidirectional transport mediated by the opposite motor. This, in turn, results in largely immobile clusters of EEs near the cell tip (Fig. S2 A, a dynein heavy chain mutant Dyn2ts; Wedlich-Söldner et al., 2002a) or in subapical regions (Fig. S2 A, kinesin-3 mutant ΔKin3; Lenz et al., 2006). On agar medium plates, colonies of wild type are white and filamentous (Fig. S2 B). Impaired EE motility results in short hyphae and gray colonies (Fig. S2, B and C, ΔKin3 shown as an example). We exploited the appearance of this EE motility–associated morphological phenotype to screen for UV-induced mutants expressing the EE-specific small GTPase Rab5a fused to GFP (Fuchs et al., 2006). We selected strains that (a) appear gray, (b) form short hyphal cells, and (c) show EE motility defects (Fig. S2), identifying an endosome motility defect mutant (EMD5; Figs. 1 A and S2 B, EMD5). Whereas EEs are motile and evenly distributed in wild type (Fig. 1 B and Video 1), EE motility in EMD5 was almost abolished, and organelles clustered at the cell ends (Fig. 1 B, EMD5, arrowheads and asterisk indicate EE clusters). This is reminiscent of a defect in retrograde, dynein-mediated motility (Fig. S2 A).
Figure 1.
Hok1 is required for EE motility. (A) Morphology of wild type and Δhok1 mutants. (B) EE distribution and motility in wild type, EMD5 mutants expressing hok1 (EMD5 + Hok1), and Δhok1 mutants (Δhok1). Deletion or mutation of hok1 abolishes motility and induces EE clusters (arrowheads; asterisk in kymograph). Contrast-inverted kymographs show GFP-Rab5a fluorescence (EEs). See also Video 1. (C) Domain organization of selected Hook proteins. For accession numbers, see Materials and methods. (D) Contrast-inverted kymographs showing bidirectional motility of mCherry-Rab5a–labeled EEs and Hok1-GFP. Hok1 locates on all moving organelles (yellow lines in merged image). See also Video 2. Images in A, B, and D were adjusted in brightness, contrast, and γ settings. Horizontal bars are in micrometers, and vertical bars are in seconds.
A Hook protein is required for EE motility
We performed whole genome sequencing of EMD5 and identified 11 point mutations. Two resided in the open reading frame Um05551.1 (E843 to K and G883 to E; see Materials and methods for accession numbers; Kämper et al., 2006). The predicted protein shares 22.1/38% and 23.0/39.1% sequence identity/similarity with a Hook from Drosophila melanogaster and Hook1 from humans, respectively. In addition, it displays a similar domain architecture (Fig. 1 C), containing an N-terminal Hook superfamily domain (P = 8.9 × 10−38) and an extended CC region (Fig. 1 C). Um05551.1 was named Hok1 following standard nomenclature rules (see gene nomenclature conventions in the Saccharomyces Genome Database). Similar to D. melanogaster, the U. maydis genome encodes only one Hook protein. When expressed in EMD5, Hok1 rescued the mutant growth defect and restored EE motility (Figs. 1 B and S2 D and Video 1). We confirmed the importance of Hok1 for EE motility deleting hok1. This Δhok1 mutant grew identically to EMD5 with short, bipolar cells that contained polar clusters of largely immobile GFP-Rab5a–positive EEs (Fig. 1, A and B, indicated by arrowheads). Introducing a fusion protein of Hok1 and GFP into the Δhok1 mutant restored hyphal growth and EE motility (Figs. 1 A and S2 E). We conclude that Hok1 is required for bidirectional motility of EEs. When expressed in Δhok1, the Hok1-GFP fusion protein was located on both anterograde- and retrograde-moving EEs (Fig. 1 D and Video 2). This suggests that Hok1 controls EE motility while bound to the organelles.
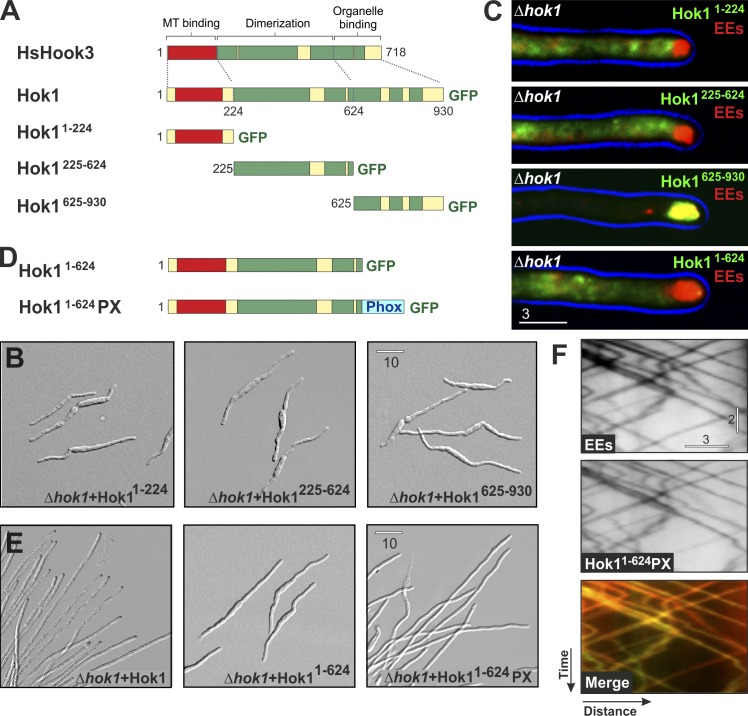
The C-terminal part of Hok1 targets to EEs
Loss-of-function mutations in hok1 were located in a conserved stretch of amino acids near the C terminus. This region targets human Hook3 to the Golgi apparatus (Walenta et al., 2001). The N terminus of human Hook proteins interacts with MTs (Walenta et al., 2001) or, in the case of ZYG-12 from worms, binds the dynein complex (Malone et al., 2003). We therefore divided Hok1 into three analogous regions (Fig. 2 A, N-terminal part, aa 1–224; middle part, aa 225–624; and C-terminal part, aa 625–930) and fused these individual domains to GFP and tested their localization pattern. We expressed the fusion proteins in Δhok1 mutants, containing mCherry-Rab5a, to avoid dimerization or competition with endogenous Hok1. Each fusion protein was expressed (Fig. S3 A), but none of the truncated proteins rescued the Δhok1 phenotype (Fig. 2 B). Neither Hok11–224 nor Hok1225–624 showed specific localization in the cell (Figs. 2 C and S4 A), suggesting that the N-terminal domain of Hok1 has no strong affinity for either EEs or MTs. In contrast, Hok1625–930 localized to the apical EE cluster (Fig. 2 C), indicating that the C-terminal region targets Hok1 to EEs. To determine whether this is the only role of the C-terminal region, we generated a C-terminal–truncated Hok1 protein (Fig. 2 D, Hok11–624). This protein was expressed (Fig. S3 B) but neither bound to EEs (Fig. 2 C) nor complemented Δhok1 (Fig. 2 E). However, when Hok11–624 was fused to the Phox (PX) domain of the endosomal t-SNARE Yup1 (Fig. 2 D; Wedlich-Söldner et al., 2000), the chimeric Hok11–624PX protein was targeted to moving EEs (Fig. 2 F and Video 3) and partially restored growth in Δhok1 (hyphal cell length of Δhok1 + Hok1: 80.47 ± 16.03 µm, n = 283 cells; length of Δhok1 + Hok11–624PX: 45.69 ± 22.9 µm, n = 504 cells; length of Δhok1: 31.40 ± 7.04 µm, n = 268 cells; Δhok1 + Hok11–624PX significantly longer than Δhok1, P < 0.0001; Fig. 2 E). We conclude that the C-terminal region of Hok1 anchors the protein to its cargo, which confirms findings in humans (Walenta et al., 2001). Artificial targeting of the remaining N-terminal part of Hok1 to EEs partially restored the morphology phenotype, suggesting that this part is essential for regulating EE motility.
Figure 2.
The C-terminal region of Hok1 targets to endosomes. (A) The organization of human Hook3, Hok1, and the truncated proteins Hok11–224, Hok1225–624, and Hok1625–930. (B) Morphology of Δhok1 expressing Hok11–224, Hok1225–624, and Hok1625–930. (C) Colocalization of Hok11–224, Hok1225–624, Hok1625–930, and Hok11–624 and mCherry-Rab5a–labeled EEs in Δhok1. The cell edge is indicated in blue. Only the C-terminal fragment of Hok1 localizes to apical EE clusters. (D) Organization of Hok11–624 and Hok11–624PX. The latter carries an EE-targeting PX domain (Wedlich-Söldner et al., 2000). (E) Morphology of control cells and Δhok1 that express Hok1, Hok11–624, and Hok11–624PX. Targeting of truncated Hok1 to EEs partially rescues the growth defect. (F) Bidirectional motility of mCherry-Rab5a–labeled EEs and Hok11–624PX. The upper two kymographs are contrast inverted. Hok11–624PX locates on the moving organelles (yellow lines in merged image). See also Video 3. Images in B, C, E, and F were adjusted in brightness, contrast, and γ settings. Horizontal bars are in micrometers, and the vertical bar is in seconds.
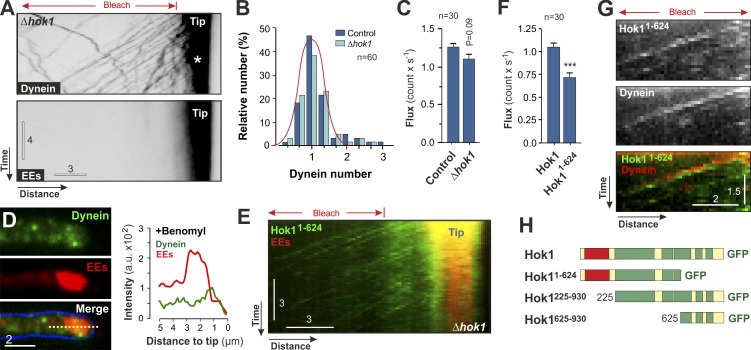
Hok1 mediates binding of dynein to EEs
Δhok1 mutants showed short hyphae and apical EE clustering, reminiscent of dynein mutants (Fig. S2 A; Lenz et al., 2006). This suggests that Hok1 supports retrograde dynein-dependent motility of EEs. To test this idea, we coexpressed a functional fusion protein of the dynein heavy chain Dyn2 with a triple GFP tag (GFP3-Dyn2; Lenz et al., 2006) and mCherry-Rab5a in Δhok1. Consistent with a previous study, we found that dynein accumulated at apical MT plus ends (Schuster et al., 2011a), from where it moved toward MT minus ends located in the subapical region of the cell (Figs. 3 A, asterisk indicates dynein cluster; and S4 B; and Video 4). We used fluorescent nuclear porins as internal calibration standards (Schuster et al., 2011c) and confirmed a previous study that these retrograde signals represent individual dynein motors (Fig. 3 B) that carry EEs to the cell center (Schuster et al., 2011c). In contrast, no dynein was found on anterograde EEs (Fig. S4 C; Schuster et al., 2011c). The retrograde flux of dynein was not significantly different between Δhok1 and control cells (P = 0.09; Fig. 3 C), demonstrating that Hok1 is dispensable for retrograde dynein motility. However, dynein was not able to move EEs to MT minus ends in Δhok1 (Fig. 3 A, EEs), suggesting that Hok1 mediates the association of the motor with organelles. To confirm this, we treated Δhok1 cells with the MT-depolymerizing drug benomyl, a nocodazole-like, fungal-specific benzimidazole carbamate that binds to β-tubulin (Davidse and Flach, 1977; Jung et al., 1992). In contrast to nocodazole, benomyl reversibly disrupts MTs in U. maydis (Fuchs et al., 2005). When MTs were disrupted, apical dynein accumulation disappeared, whereas EE clusters were unaffected, and no colocalization with dynein fluorescence was observed under these conditions (Fig. 3 D). Collectively, these data suggest that dynein is not associated with EEs in the absence of Hok1.
Figure 3.
The N-terminal region of Hok1 mediates dynein binding to EEs. (A) GFP3-labeled dynein heavy chain (dynein) and mCherry-Rab5a–labeled endosomes (EEs) in Δhok1 mutants. Dynein (asterisk) accumulates at the cell tip (tip), from where it leaves without EEs. Cells were photobleached (bleach) to reduce signal interferences, which did not affect EE or motor motility (Schuster et al., 2011c). See also Video 4. (B) Estimated dynein numbers in moving GFP3-Dyn2 signals in control and Δhok1 cells. Estimation used an internal calibration standard, assuming that the GFP3-labeled dynein heavy chain forms dimers (Schuster et al., 2011c). Data represent two experiments. The red line shows a normal distribution curve. (C) Retrograde dynein flux in control and Δhok1 cells at ∼10 µm behind the tip. Bars represent data from two experiments and are means ± SE; sample size is indicated. No significant difference was found, P = 0.09. (D) Images and linescan plot of dynein and EE colocalization in Δhok1 hyphal tips after disruption of the MTs (+benomyl). The dotted line in merged image indicates the region of intensity scan. a.u., arbitrary unit. (E) Motility of Hok11–624 and mCherry-Rab5a–labeled EEs in Δhok1. Hok11–624 moves in retrograde direction, whereas EEs remain stationary. See also Video 5. (F) Retrograde flux of Hok1 and Hok11–624 at ∼10 µm behind the cell tip. Bars represent data from two experiments and are means ± SE; sample size is indicated. Significant difference is indicated: ***, P < 0.0001. (G) Co-migration of Hok11–624 and mCherry3-labeled dynein heavy chain. Kymographs were slightly misaligned to better show colocalization. See also Video 6. (H) Domain organization of proteins used in immunoprecipitation and mass spectrometry experiments. Images in A, D, E, and G were adjusted in brightness, contrast, and γ settings. Horizontal bars are in micrometers, and vertical bars are in seconds.
Our results showed that the N-terminal domain and the CC region of Hok1, when targeted to EEs, are sufficient to restore EE motility, suggesting that Hok11–624 interacts with dynein. We therefore coexpressed Hok11–624 and mCherry-Rab5a in a Δhok1 mutant and, consistent with the lack of the organelle-binding C terminus, found that EEs are immobile (Fig. 3 E, EEs in red). However, Hok11–624 moved in a retrograde direction (Fig. 3, E and F; and Video 5), suggesting that the N-terminal half of Hok1 interacts with dynein independently of its association with EEs. Indeed, Hok11–624 and mCherry3-labeled dynein heavy chain traveled together in Δhok1 cells (Fig. 3 G and Video 6), suggesting that the dynein–dynactin complex interacts with Hok1. To investigate this potential interaction further, we performed immunoprecipitation experiments followed by mass spectrometry, using truncated Hok11–624 and the entire Hok1. Neither Hok11–624 nor the entire Hok1 revealed any interaction with dynein–dynactin or any other motor protein. Instead, Hok1 interacted with the U. maydis proteins um00451 and um10821 (8–26 peptides; see Materials and methods for accession numbers). Both proteins show significant amino acid sequence similarity with human FTS (17.1% identity/27.4% similarity; Lesche et al., 1997) and human FHIP (18.9% identity/27.5% similarity; Xu et al., 2008), respectively, and share a similar domain structure (Fig. S4 D, U. maydis FTS named Fts1 and U. maydis FHIP named Fhp1). Both proteins also bound to Hok1225–930 (8–19 peptides; Fig. 3 H) but only weakly to Hok1625–930 (two to five peptides) and Hok11–624 (less than two peptides; Fig. 3 H), indicating that Hok1 binding requires both the C terminus and the middle region. Consistent with binding to Hok1 in vitro, we found that fluorescent Fts1-GFP and Fhp1-GFP localized to mCherry-Rab5a–labeled EEs (Fig. S4 E). We conclude that Hok1, Fts1, and Fhp1 form a complex, which was previously reported in humans (Xu et al., 2008). Whether Fts1 or Fhp1 mediate interaction with dynein on organelles remains to be investigated.
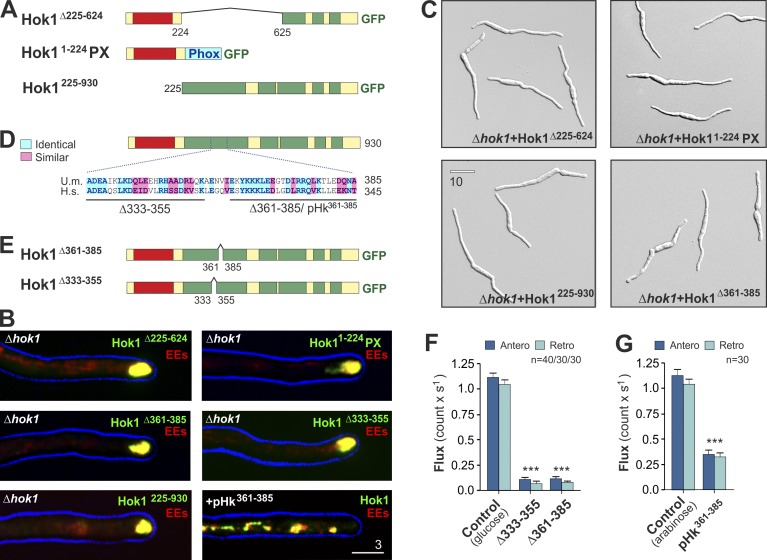
A conserved region within the extended CC is crucial for dynein recruitment to EEs
We asked whether the N-terminal or middle part of Hok1 is sufficient to bind dynein and restore filamentous growth of Δhok1. We generated mutant proteins, missing either the middle region (Hok1Δ225–624) or consisting of the N-terminal 224 aa fused to the PX domain of Yup1 (Hok11–224PX). In addition, we deleted the N-terminal region (Hok1225–930; Fig. 4 A). All proteins localized to EEs (Fig. 4 B), but none were able to restore filamentous growth of Δhok1 mutants (Fig. 4 C). Thus, both regions of Hok1 cooperate in dynein binding and motility control. All Hook proteins contain an extended CC domain of 260–280 aa, adjacent to the N-terminal Hook superfamily domain (Fig. 1 C; Walenta et al., 2001). Hok1 shares sequence identity with the human orthologues Hook1 and Hook3 (∼23.6 and ∼23.7%, respectively). Sequence conservation was highest within a central part of the CC region (50.9% identity and 75.7% similarity between Hok1 and Hook3 in a central stretch of 53 aa; Hook1: 37.7% identity and 69.8% similarity; Fig. 4 D). Within this conserved region, 18 of 25 identical residues are charged, suggesting that they are surface exposed and so could interact with a binding partner. We tested for the importance of this region by introducing short deletions into the conserved region (Fig. 4 D). The resulting mutant proteins Hok1Δ361–385 and Hok1Δ333–355 were expressed in Δhok1 mutants (Figs. 4 E and S3, B and C). Both localized to polar EE clusters (Fig. 4 B), but neither was able to restore EE motility or cell growth (Figs. 4, C and F; and S5 A). This demonstrates that the highly conserved CC region is essential to the function of Hok1. As human Hook3 forms homodimers (Krämer and Phistry, 1996; Xu et al., 2008), we tested whether deletions in the CC region prevent dimerization, thereby inactivating Hok1. Native gel electrophoresis did not reveal differences between Hok1, Hok1Δ361–385, and Hok1Δ333–355 (Fig. S5 B), suggesting that small truncations do not alter protein tertiary structure. Thus, we consider it likely that the conserved region of Hok1 interacts with a functionally important binding partner.
Figure 4.
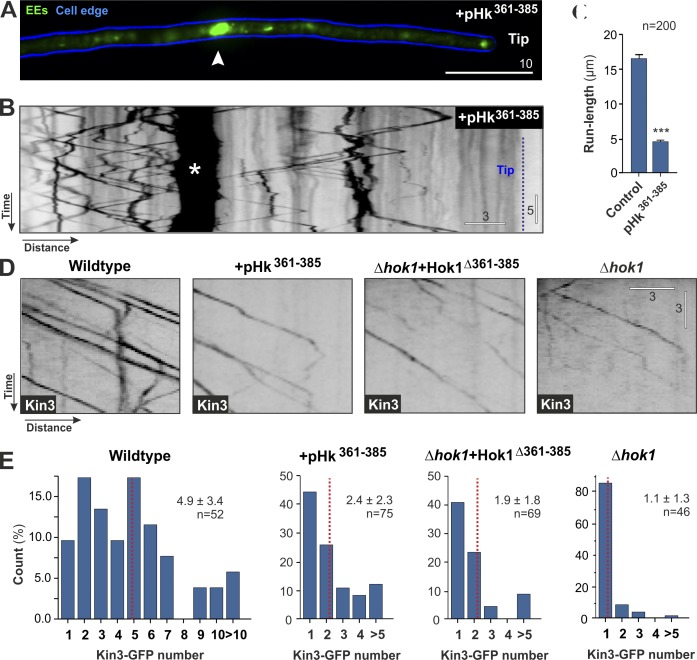
A conserved region in the first CC is essential for Hok1 function. (A) Organization of truncated Hok1Δ225–624, Hok11–224PX, which carries an EE-targeting PX domain, and Hok1225–930. (B) Colocalization of truncated Hok1 proteins and mCherrry-Rab5a–labeled EEs in Δhok1 and colocalization of Hok1-GFP and EEs in cells expressing the peptide pHk361–385. Cell edges are indicated in blue. All constructs localize to EEs without rescuing Δhok1. (C) Morphology of Δhok1 mutants expressing Hok1Δ225–624, Hok11–224PX, Hok1225–930, and Hok1Δ361–385. Neither rescues the morphology defect. (D) Localization of a highly conserved region within the first CC of Hok1 and human Hook3. Identical amino acids are shown in light blue, and similar amino acids are shown in pink. The alignment was generated in ClustalW. H.s., Homo sapiens; U.m., U. maydis. (E) Organization of the truncated proteins Hok1Δ361–385 and Hok1Δ333–355. (F) Anterograde and retrograde flux of mCherry-Rab5a–labeled EEs in control cells and Hok1Δ361–385 and Hok1Δ333–355 (Δ333–355 and Δ361–385) at ∼10 µm behind the cell tip. Bars represent data from two experiments and are means ± SE; sample size is indicated. (G) Anterograde and retrograde flux of mCherry-Rab5a–labeled EEs in control cells and cells expressing the peptide pHk361–385 (see D for sequence). Bars represent data from two experiments and are means ± SE; sample size is indicated. Images in B and C were adjusted in brightness, contrast, and γ settings. Significant difference is indicated: ***, P < 0.0001. Bars are in micrometers.
Hok1 mediates kinesin-3 attachment to the cargo
We reported previously inhibitory peptides, which interfere with protein–protein interaction in vivo (Schuster et al., 2011a). We designed an inhibitory peptide pHk361–385 covering the region E361 to A385 (Fig. 4 D, pHk361–385). We placed this under the control of strong inducible crg promoter (Bottin et al., 1996) and expressed it in cells that contain Hok1-GFP– and mCherry-Rab5a–labeled EEs. After peptide expression for 2 h, both anterograde and retrograde EE motility was lowered significantly (Fig. 4 G, pHk361–385). The motility inhibition was not caused by release of Hok1 from the EEs (Fig. 4 B, +pHk361–385), suggesting that the peptide instead inhibits Hok1 activity. This result confirms a pivotal role for the conserved region within the first CC.
Interestingly, the reduction in EE motility occasionally coincided with formation of EE clusters in subapical cell parts (Fig. 5, A [arrowhead] and B [asterisk]; and Video 7). Here, EE motility is predominantly mediated by kinesin-3 (Fig. S1 B; Schuster et al., 2011b). We noticed that the anterograde run length of kinesin-3–driven EEs was reduced significantly when the inhibitory peptide was expressed (Fig. 5, B and C). An extended run length of cargo requires multiple kinesin motors (Klumpp and Lipowsky, 2005; Beeg et al., 2008), suggesting that inhibition of Hok1 by pHk361–385 reduced the number of kinesin-3 on EEs. We tested this by tagging kinesin-3 with GFP and quantifying the number of EE-associated motors, using fluorescent nuclear porins as internal calibration standards. Consistent with Schuster et al. (2011c), we estimated that approximately five kinesin-3 motors (range of 1–18) bind to a single EE in wild-type cells (Fig. 5, D and E, wild type). In contrast, in the presence of the pHk361–385 peptide, the number of EE-attached kinesin-3 motors was reduced significantly (two to three motors; P < 0.0001; Fig. 5, D and E, +pHk361–385). A similar reduction was found when Hok1Δ361–385 was expressed in Δhok1 (approximately two motors; Fig. 5, D and E, Δhok1 + Hok1Δ361–385), and only one motor was associated with EEs when hok1 was deleted (Fig. 5, D and E, Δhok1). These results demonstrate that Hok1, in particular, the conserved region within the CC, participates in attachment of kinesin-3 to EEs. On the other hand, Hok1 still localizes to EEs in the absence of kinesin-3 (Fig. S5 C). Thus, we conclude that Hok1 is an adapter for kinesin-3 cargo attachment. However, the interaction is most likely indirect or transient because immunoprecipitation and mass spectrometry analysis did not reveal a significant interaction between kinesin-3 and Hok1.
Figure 5.
Hok1 is required for cargo binding of kinesin-3. (A) Localization of an EE cluster, labeled with GFP-Rab5a (arrowhead) in a cell expressing the peptide pHk361–385 for 2 h. Note that clusters initially appear subapically but later shift to the cell end (tip). (B) Motility of GFP-Rab5a–labeled EEs in a cell expressing the peptide pHk361–385 for 2 h. A subapical cluster is indicated by an asterisk. Cell end is indicated by tip and the dotted line. See also Video 7. (C) Mean run length of EEs in control cells and cells expressing pHk361–385 for 2 h. Bars represent data from two experiments and are means ± SE; sample size is indicated. Significant difference is indicated: ***, P < 0.0001. (D) Contrast-inverted kymographs showing anterograde motility of kinesin-3–GFP in control cells, after expressing the peptide pHk361–385, and in Δhok1 and Δhok1 cells expressing Hok1Δ361–385. Kinesin-3 signals are strong in the control but weak in all mutants. (E) Kinesin-3-GFP numbers in control cells, after expressing the peptide pHk361–385, and in Δhok1– and hok1–null cells expressing HokΔ361–385. Note that native kinesin-3 levels are shown. All numbers represent two experiments and are given as means ± SD; sample size is indicated. Motor number estimation is based on comparison with an internal calibration standard (Schuster et al., 2011c) and assuming that kinesin-3 is a dimer (Hammond et al., 2009). Red dotted lines indicate medians. Images in A, B, and D were adjusted in brightness, contrast, and γ settings. Horizontal bars are in micrometers, and vertical bars are in seconds.
Kinesin-3 numbers on EEs drop before dynein binding
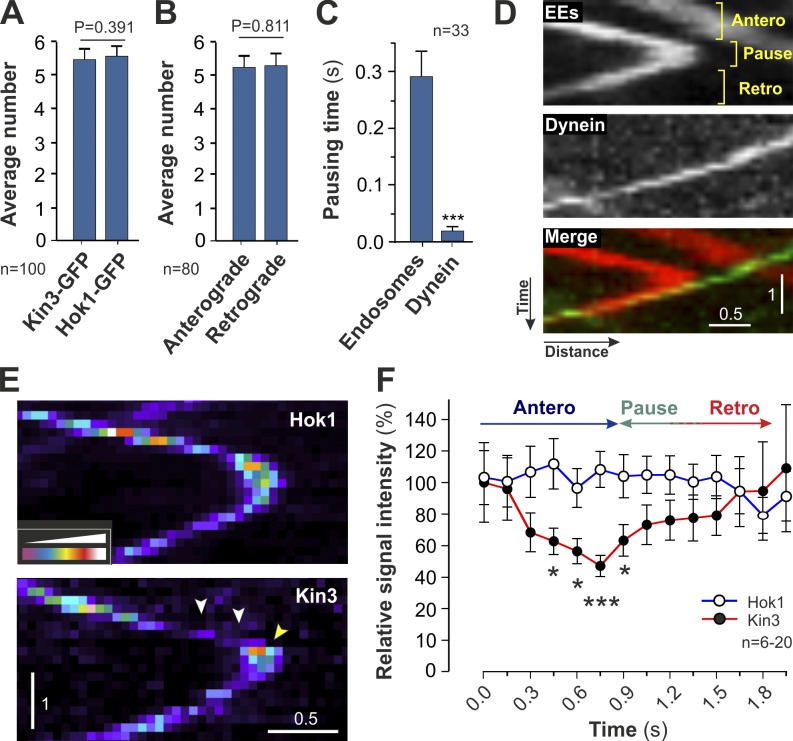
Our results indicate that Hok1 mediates attachment of both kinesin-3 and dynein to EEs. In U. maydis, retrograde EE motility is driven by single dynein motors, whereas anterograde motility is based on several kinesin-3 motors (Schuster et al., 2011c). We confirmed this by co-observation of dynein and EEs. We found that 93.6% of all anterograde to retrograde turns within the apical 10 µm of the cell are associated with a retrograde-moving dynein signal (n = 44 signals). As these signals represent single dyneins (Fig. 3 B; Schuster et al., 2011c; this study), individual dyneins can overcome a “team” of kinesin-3 motors. We reasoned that Hok1 could influence the outcome of a potential tug-of-war by reducing the number of kinesin-3 molecules during dynein attachment and retrograde EE motility. First, we tested whether this is a random process by determining the location where EEs turn from anterograde kinesin-3–driven to retrograde dynein-mediated motility. We focused our analysis on the apical 10 µm of the cell, where MTs are largely unipolar (Fig. S1 B; Schuster et al., 2011b). We found that most EEs turn at the end of MTs, where dynein is concentrated and interaction between dynein and EEs is increased (Fig. S5 D, bar labeled MT ends; Lenz et al., 2006; Schuster et al., 2011a). However, no preferential turning point was found in these subapical regions (Kruskal–Wallis test, P = 0.6601). This suggests that anterograde to retrograde turning of EEs is a stochastic process. We determined the number of kinesin-3 motors and associated Hok1 proteins on moving EEs and found that on average five kinesin-3 and five Hok1 proteins bind to a single organelle (Fig. 6 A). This suggests that Hok1 forms a 1:1 complex with kinesin-3, and we considered whether dynein and kinesin-3 compete for an interaction with Hok1. If so, less kinesin-3 would be attached to the EEs during retrograde dynein-driven motility. We did not find such a difference (Fig. 6 B). This makes a simple competition between dynein and kinesin-3 for binding to Hok1 or its associated adapters unlikely. Alternatively, a Hok1-related mechanism may transiently lower kinesin-3 attachment before dynein binding, thereby modulating the tug-of-war. We investigated this by tracing the binding of dynein to EEs in cells expressing mCherry-Rab5a and GFP fluorescent dynein heavy chain. We observed that anterograde to retrograde turning of EEs is accompanied by a pause in motility that lasted for ∼300 ms (Fig. 6, C and D). This suggested a “battle” between motors in a presumed tug-of-war. However, dynein did not pause when binding to an EE (Fig. 6 C) but transported the organelles immediately toward the minus ends of MTs (Fig. 6 D). This suggests that anterograde kinesin-3 activity is reduced before dynein binds to EEs. We therefore investigated whether release of Hok1-GFP and/or kinesin-3–GFP induces organelle pausing. We found that Hok1-GFP signals remained equal in intensity during anterograde motility, during pausing, and during subsequent retrograde motion (Fig. 6, E and F, Hok1). In contrast, kinesin-3 GFP fluorescence intensity reduced significantly before pausing of EEs but recovered during retrograde dynein-driven motility (Fig. 6, E [Kin3, drop shown by arrowheads] and F). This suggests that the Hok1 complex releases kinesin-3 before dynein binding, thereby favoring dynein-driven retrograde motion over kinesin-3 in a tug-of-war.
Figure 6.
Kinesin-3 numbers drop before anterograde to retrograde turning of EEs. (A) Numbers of Hok1-GFP and kinesin-3–GFP in signals moving bidirectionally. Bars are means ± SE of two experiments; sample size is indicated. No significant difference is found, P = 0.391. Numbers were estimated using an internal calibration standard (Schuster et al., 2011c) and assume proteins are dimers (Xu et al., 2008; Hammond et al., 2009). (B) Numbers of kinesin-3–GFP in anterograde and retrograde signals. Bars are means ± SE of two experiments; sample size is indicated. No significant difference is found, P = 0.811. Numbers were estimated using an internal calibration standard (Schuster et al., 2011c) and assume proteins are dimers (Hammond et al., 2009). (C) Pausing time of EEs and dynein before retrograde motility. Bars are means ± SE of two experiments; sample size is indicated. (D) Kymographs showing the binding of GFP3-dynein (green) to a mCherry-Rab5a–labeled EE (red). An anterograde (antero)-moving organelle pauses (pause) before it binds dynein and turns to retrograde motility (retro). (E) False-colored kymographs showing Hok1-GFP and kinesin-3-GFP during anterograde to retrograde turning. Kinesin-3 signals drop just before the organelle pauses (arrowheads). Recovery of the fluorescent signal begins during pause (yellow arrowhead). Note signal variations caused by changes in focal plane and that stationary signals are brighter as they are more focused. (F) Mean intensities of Hok1-GFP and kinesin-3-GFP during anterograde to retrograde turning of EEs. Hok1 numbers remain stable, whereas kinesin-3 signals drop before the pausing phase. Data points are means ± SE from a representative experiment, and sample sizes are indicated. Significant difference is indicated: *, P < 0.05; ***, P < 0.0001. Images in D and E were adjusted in brightness, contrast, and γ settings. Horizontal bars are in micrometers, and vertical bars are in seconds.
Controlling motor to cargo attachment by Hook proteins may be conserved
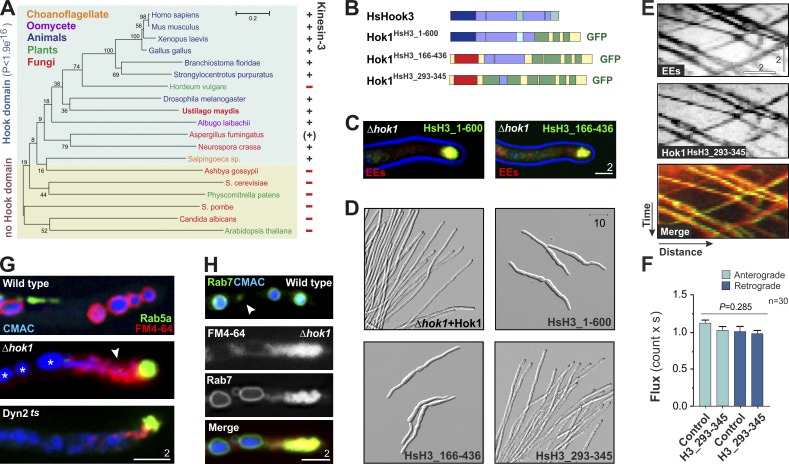
The central part of the CC domain shows high amino acid sequence identity with Hook3 (Fig. 4 D), suggesting that its role in controlling kinesin-3 recruitment may be evolutionarily ancient. We searched publicly available genome data for Hook and kinesin-3. We found Hook proteins, characterized by a Hook domain in the Pfam server (P < 1.9 × 10−16), and kinesin-3 motors, characterized by a Kif1A-like motor domain and a C-terminal DUF3694 and/or pleckstrin homology (PH) domain, in all opisthokonts (Fig. 7 A), which include animals, fungi, and choanoflagellate protozoa (Steenkamp et al., 2006). Hook appears absent from plants, except barley, and is present in the oomycete Albugo laibachii, which is closely related to brown algae (Beakes et al., 2012). However, plants do not carry kinesin-3, suggesting that barley may have acquired Hook by horizontal gene transfer. Interestingly, both Hook and kinesin-3 are absent from the Saccharomycotina (Candida albicans, Saccharomyces cerevisiae, and Ashbya gossypii) and Schizosaccharomyces pombe, arguing that this group has lost the machinery for long-range MT-dependent motility during evolution. Collectively, these results suggest that Hook proteins are the universal cargo adapter of kinesin-3.
Figure 7.
The function of Hok1 is conserved. (A) Phylogenetic tree of Hook and closest non-Hook proteins and the correlation with the presence of kinesin-3 motors. No Kif1A-like sequence was found at NCBI for A. fumingatus, and no Hook was listed for A. nidulans. The tree is based on Hook domains, identified in Pfam, and calculated in MEGA 5.10 (Tamura et al., 2011). (B) Domain organization of human Hook3 and the chimeric proteins Hok1HsH3_1–600, Hok1HsH3_166–436, and Hok1HsH3_293–345. (C) Colocalization of Hok1HsH3_1–600 and Hok1HsH3_166–436 with mCherry-Rab5a on apical EEs. (D) Morphology of Δhok1 expressing Hok1, Hok1HsH3_1–600, Hok1HsH3_166–436, and Hok1HsH3_293–345. The conserved region of human Hook3 restores the mutant phenotype in U. maydis. (E) Co-motility of Hok1HsH3_293–345 and mCherry-Rab5a–labeled EEs. See also Video 8. (F) Bidirectional flux of EEs in wild type and Δhok1 expressing Hok1HsH3_293–345. Bars represent two experiments and are means ± SE; sample size is indicated. No significant difference was found (P = 0.285, Kruskal–Wallis test). (G) Endocytic sorting defect in Δhok1 and dynein (Dyn2ts) mutants treated with the dye FM4-64. In wild type, the dye appears in the vacuoles (stained with Cell Tracker Blue CMAC), whereas in the mutants, it accumulates next to GFP-Rab5a–carrying EEs and does not reach the vacuoles (asterisks). The arrowhead indicates the FM4-64–stained “cloud.” (H) The late endocytic compartment in Δhok1. In wild-type cells, the late endosomes marker Rab7 localizes to vacuoles and motile structures (arrowhead). In Δhok1, Rab7 colocalizes with the apical FM4-64–positive cloud, whereas the localization on vacuoles is not affected. Images in C, D, E, G, and H were adjusted in brightness, contrast, and γ settings. Horizontal bars are in micrometers, and the vertical bar is in seconds.
As Hook proteins are present in both fungi and humans, we tested for functional conservation. Hook3 shows high sequence similarities within the functionally important conserved CC stretch, and we therefore set out to restore EE motility in Δhok1 mutants by expressing human Hook3 mutant proteins. In humans, Hook3 localizes to Golgi (Walenta et al., 2001). To target proteins to EEs, we fused the N-terminal 600 aa of Hook3 to the C-terminal part of Hok1 (Hok1HsH3_1–600; Fig. 7 B) and expressed it in Δhok1 mutants. The chimeric protein was correctly targeted to EEs (Fig. 7 C) but did not rescue the Δhok1 phenotype (Fig. 7 D). We therefore integrated the entire CC region (Hok1HsH3_166–436) and the central conserved stretch (aa 293–345; Hok1HsH3_293–345) of human Hook3 into Hok1 (Fig. 7 B). Both proteins targeted correctly (Fig. 7 C), but only Hok1HsH3_293–345 was able to restore EE motility and hyphal growth of Δhok1 mutants (Fig. 7, D–F; and Video 8). We conclude that the conserved region within the N-terminal CC is most likely involved in regulation of motor–cargo interactions, which may be a conserved function of Hook proteins from fungi to humans.
In D. melanogaster, Hook is implied in late endocytic sorting to lysosomes (Krämer and Phistry, 1996; Sunio et al., 1999). To test whether Δhok1 mutants show a similar phenotype, we used the lipophilic styryl dye FM4-64 (Vida and Emr, 1995), which enters the fungal vacuole/lysosome via endocytic sorting (Fischer-Parton et al., 2000; Wedlich-Söldner et al., 2000). Indeed, FM4-64 appeared in the vacuoles of wild-type cells within 30 min, but no dye was found in EEs or other compartments (Fig. 7 G, wild type). In contrast, in Δhok1, FM4-64 did not reach the vacuole (Fig. 7 G, Δhok1, vacuoles indicated by asterisks) and accumulated near the apical cluster of EEs (Fig. 7 G, Δhok1, arrowhead). To test whether FM4-64–positive “clouds” are part of the late endosomal compartment, we introduced the late endosomal marker GFP-Rab7 (Higuchi et al., 2014) into Δhok1. In wild type, GFP-Rab7 localized to vacuoles and associated structures that are most likely late endosomes (Fig. 7 H, wild type). In Δhok1 mutants, GFP-Rab7 colocalized with the FM4-64 cloud (Fig. 7 H, Δhok1), suggesting that Hok1 is required for organization of the late endosomal compartment. Interestingly, a similar FM4-64 distribution defect was found in EE motility–defective dynein mutants (Fig. 7 G, Dyn2ts). These results suggest that Hook-mediated EE motility is essential for endocytic sorting to the vacuole/lysosome compartment. This is consistent with the described role of Hook in late endocytic sorting in D. melanogaster (Sunio et al., 1999).
Discussion
Here, we report identification of the fungal Hook protein Hok1 as part of an adapter complex that coordinates attachment of dynein and kinesin-3. Hook proteins are known to be involved in endocytic protein trafficking, Golgi organization, and cilium formation (Krämer and Phistry, 1996, 1999; Sunio et al., 1999; Walenta et al., 2001; Ge et al., 2010; Baron Gaillard et al., 2011; Maldonado-Báez et al., 2013), suggesting that they play fundamental roles in eukaryotic cells. Hook proteins bind simultaneously to MTs and organelles and were therefore implicated in the linkage of cargo to cytoskeleton (Walenta et al., 2001; Linstedt, 2004; Baron Gaillard et al., 2011). We demonstrate that Hok1 localizes on rapidly moving organelles, where it mediates dynein and kinesin-3 attachment to EEs. This suggests a more dynamic role for Hook proteins in eukaryotic cells. It was previously noted that the loss of Hook resembles a defect in dynein function (Walenta et al., 2001; Szebenyi et al., 2007). Furthermore, Hook from worms links the nucleus to the dynein complex (Malone et al., 2003). Thus, it is possible that Hook proteins are conserved adapters for kinesin-3 and dynein on organelles. This notion is supported by our findings that Hook and kinesin-3 are paired and conserved across the opisthokonts and that Hook mutants in U. maydis and D. melanogaster share a late endocytic sorting defect (Sunio et al., 1999; this study). We also found that a chimeric protein of Hok1 and human Hook3 is functional in U. maydis, suggesting that the core function in motor coordination is conserved. However, only the conserved CC stretch of human Hook3 was functional when integrated into Hok1, whereas several other Hok1-Hook3 chimeric proteins failed to rescue the Δhok1 phenotype. Therefore, we consider it likely that the detailed mechanism of Hook function is different between fungi and humans.
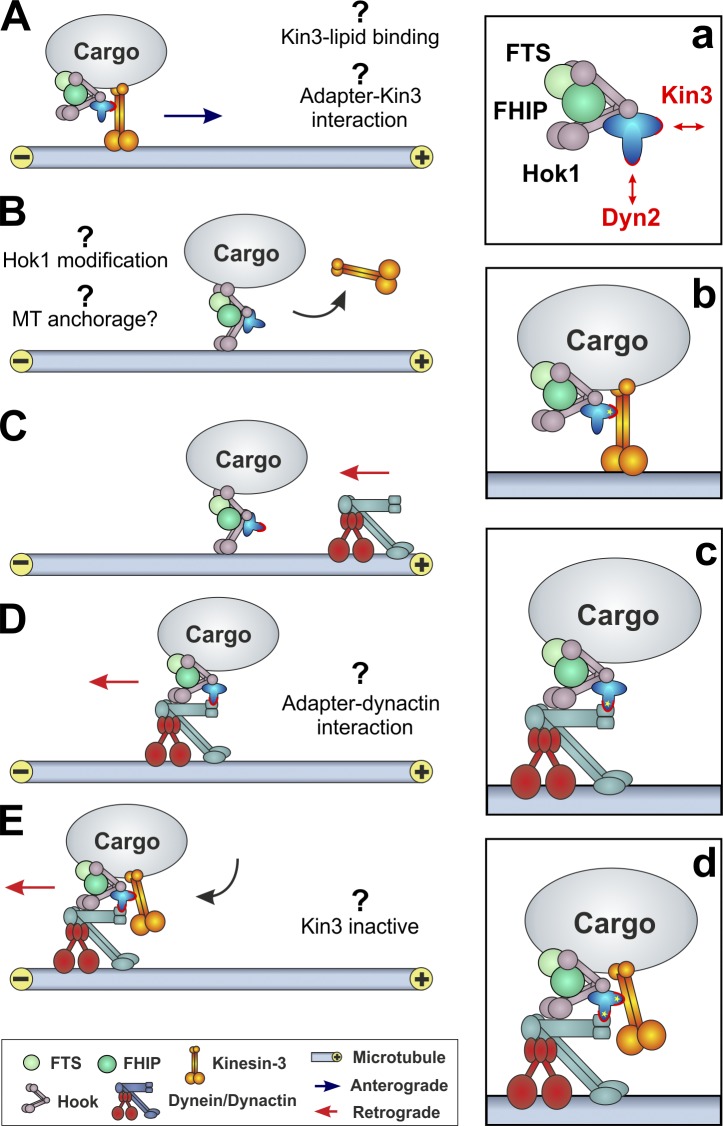
A key question is how Hok1 performs its role in regulating EE motility. Our data suggest that the role of Hook proteins in motor coordination may be conserved. We therefore set out to interpret our results in the context of a published study on the function of animal Hook proteins (Xu et al., 2008). We report that Hok1 is a central part of a larger protein complex that includes FTS and FHIP homologues Fts1 and Fhp1. In humans, both proteins interact with a conserved part of the C-terminal domain of Hook proteins (Xu et al., 2008). Fts1 and Fhp1 bind more strongly to Hok225–930 but only weakly to Hok1625–930. Thus, binding to Hok1 requires both the C-terminal region and the CC domain. This suggests that FHIP and FTS bridge between both domains (Fig. 8, inset a). The function of both proteins is currently not known, but their ability to bind to the EE-targeting C terminus of Hok1 raises the possibility that Fts1 and Fhp1 participate in anchoring the Hok1 adapter complex to EEs.
Figure 8.
Model of Hok1 in controlling EE motility. (A) Fts1, Fhp1, and Hok1 form a complex that may contain additional, yet unknown, adapter proteins that link dynein and kinesin-3 to this Hok1 complex (b and c). Kinesin-3 binds to Hok1 and, to a lesser extent, directly to EE membranes. (B) Before dynein binding, the Hok1 complex releases kinesin-3. The cargo stops moving but remains attached to the MT. This may reflect the ability of Hok1 to bind MTs, as described for human Hook proteins (Walenta et al., 2001). (C) Dynein leaves the MT plus ends and travels toward the pausing EE, where it interacts with the Hok1 complex. This could involve the dynactin complex, as shown in the fungus A. nidulans (c; Zhang et al., 2011). (D and E) During pausing and while dynein moves the cargo to minus ends of MTs, kinesin-3 rebinds to the Hok1 complex. Speculative parts of the model are indicated by question marks.
Surprisingly, we did not find an interaction of Hok1 with dynein or kinesin-3 when precipitating Hok1 from cell extracts. This suggests that transient interaction only occurs on EE membranes. Additional adapter proteins may bridge between the N-terminal region of Hok1, which we show here is essential for motor binding and kinesin-3 and dynein (Fig. 8, inset a). In humans, the HAP1 binds to kinesin-1 and the dynactin subunit p150Glued (Engelender et al., 1997; Li et al., 1998; McGuire et al., 2006) and links them to the disease protein huntingtin to enable coordination of intracellular membrane trafficking (Caviston and Holzbaur, 2009). Our results suggest that a similar adapter could link Hok1 and kinesin-3/dynein. The DENN/MADD protein binds to the stalk of the mammalian kinesin-3 motors Kif1a and Kif1B and links them to synaptic vesicles via the small GTPase Rab3 (Niwa et al., 2008). However, we did not find a homologue of DENN/MADD in the genome of U. maydis. Therefore, a similar link to Rab-GTPases and Hok1 is unlikely. We conject that an unknown Hok1-interacting adapter needs to be identified (Fig. 8, inset a). Our immunoprecipitation experiments only identified Fts1 and Fhp1 as Hok1 interactors, raising the possibility that these proteins participate in the binding of Hok1 to motors. Further studies are needed to address this question.
We found that ∼20% of kinesin-3 remained bound to EEs in the absence of Hok1. This suggests an additional, Hok1-independent way of interacting with the cargo. It was shown that the C-terminal PH domain of ameba kinesin-3 directly binds phosphatidylinositol (4,5) bisphosphate (Klopfenstein et al., 2002). U. maydis Kin3 contains a conserved PH domain (Wedlich-Söldner et al., 2002b), suggesting that the motor can bind to EE membranes independently of the Hok1 complex. As this membrane-bound kinesin-3 is active, it ultimately drives EEs to plus ends in Δhok1 mutants. However, the reduced number of kinesin-3 on EE results in a decrease in run length. This corresponds well with findings that extended run length of cargo depends on multiple kinesin motors (Klumpp and Lipowsky, 2005; Beeg et al., 2008; Derr et al., 2012; Reis et al., 2012). We therefore conclude that the Hok1 adapter complex increases the motor number on organelles, thereby enabling extended runs of up to 90 µm (Schuster et al., 2011b). We reported recently that this extended motility is required to distribute ribosomes and mRNA efficiently in the elongated hyphal cells (Higuchi et al., 2014).
Our results suggest that the same number of kinesin-3 and Hok1 resides on anterograde-moving EEs (Fig. 8 B). A change in transport direction is initiated by the release of kinesin-3 from the Hok1 adapter, which itself stays on the EEs. The trigger for this event is unknown, but we find no preference for turning of the transport direction along the apical MTs. Thus, we consider it likely that the release of kinesin-3 is a stochastic event. This does not exclude the possibility that higher order control via phosphorylation/dephosphorylation could participate in the release of kinesin-3. Indeed, phosphorylation of huntingtin recruits kinesin-1 to cargo vesicles, which results in a longer anterograde run length of vesicles (Colin et al., 2008). A similar mechanism was reported for coordinated bidirectional transport of amyloid precursor protein in axons. Here, phosphorylation of the kinesin-1– and dynactin-binding scaffold protein JIP1 enhances the association of kinesin-1, thereby supporting anterograde motility. In contrast, dephosphorylation releases kinesin-1 and most likely fosters binding of dynein–dynactin (Fu and Holzbaur, 2013). Although it is tempting to speculate that kinesin-3 is released from the EEs upon dephosphorylation of a Hok1-interacting adapter, experimental evidence for such a mechanism is currently missing.
The release of kinesin-3 reduces anterograde forces and results in a pause of EE motility. Surprisingly, we found that these pausing EEs do not diffuse away from the MT. This suggests that they are anchored to the track. Whether Hok1 is involved in this anchorage is unknown, but human Hook is able to bind MTs (Walenta et al., 2001). Although we did not find strong association of Hok1 with MTs, the protein may mediate transient interaction of EEs with MTs (Fig. 8 B). Subsequently, dynein binds to the pausing organelles (Fig. 8, C and D). This situation is most likely specific for U. maydis, in which dynein is released from the MT plus ends and binds to EEs during retrograde movements, whereas anterograde EEs do not carry dynein (Schuster et al., 2011c). We suggest that a conformational change in the Hok1 adapter complex releases Kin3. This may allow efficient interaction of dynein–dynactin with the adapter complex. Currently, we do not know which part of the dynein–dynactin complex binds to the Hok1 adapter. In Aspergillus nidulans, the dynactin complex component p25 is required for EE motility (Fig. 8, D and E, insets c and d; Zhang et al., 2011). However, in mammalian cells, the intermediate chains of dynein interact with an unknown factor on signaling endosomes (Mitchell et al., 2012), which might resemble a Hook adapter. It remains to be shown whether Hook proteins control mammalian endosome motility.
Finally, we show that kinesin-3 returns to the EEs, whereas dynein takes the cargo toward MT minus ends. We found five kinesin-3 motors on retrograde-moving EEs, but these are not taking over. How this is achieved remains obscure, but one possibility is that during retrograde motility, kinesin-3 is bound in an inactive state. Indeed, a retrograde to anterograde turn in transport is always associated with the release of dynein, but in most cases, these organelles pause before returning to anterograde motility (Schuster et al., 2011c). This suggests that kinesin-3 does not force the turning. In other words, the release of dynein and the switch to anterograde motility are most likely not simple consequences of a tug-of-war. Instead, kinesin-3 appears to need the pause time to take over, which could reflect activation after dynein release. The mechanism by which dynein releases and kinesin-3 is activated may involve Hok1-associated proteins and will be subject to future investigations.
Conclusion
In this study, we show that Hok1 mediates both dynein and kinesin-3 recruitment to EEs and thus regulates bidirectional EE motility. We provide evidence that the core role in controlling motor attachment is conserved and show that the pairing of Hook and kinesin-3 is conserved across the opisthokonts. Thus, we propose that Hook proteins are most likely general regulatory adapters for kinesin-3 and dynein in cargo transport in eukaryotes. Although our work establishes this novel role of Hook proteins, it also opens up numerous new questions. Among these are (a) what are the adapters that bridge between Hook and the motors, (b) how does the Hok1 complex “know” when to release kinesin-3, (c) what is the role of FTS and FHIP in the adapter complex, and (d) how is the activity status of kinesin-3 regulated? More research on the Hok1-interacting proteins might shed light on these questions. However, the identification of Hook as a coordinator of kinesin-3 and dynein activity opens new avenues to better understanding membrane trafficking in eukaryotic cells.
Materials and methods
Growth conditions
All cultures of U. maydis were grown for 8–15 h at 28°C in a complete medium (Holliday, 1974) containing 1% (wt/vol) glucose, shaking at 200 rpm. Hyphal growth was induced overnight by shifting to nitrate minimal (NM) medium supplemented with 1% (wt/vol) glucose (Brachmann et al., 2001). To induce expression of the pHk361–385 peptide under the crg promoter (Bottin et al., 1996), cells were grown in NM liquid medium containing 1% (wt/vol) arabinose for 2 h (Brachmann et al., 2001). The temperature-sensitive mutant strain AB5Dyn2ts_GRab5a was grown at 22°C and was shifted to restrictive conditions (32°C) for 2 h to perform the FM4-64 experiment or overnight to assess EE distribution and motility.
Strains and plasmids
U. maydis strains and plasmids used in this study are summarized in Table 1, and their usage throughout the study is summarized in Table S1. Recombinant DNA was introduced into U. maydis by homologous recombination. All plasmids were generated using standard techniques and in vivo recombination in S. cerevisiae using strains DS94 (MATα ura3-52 trp1-1 leu2-3 his3-111 lys2-801; Tang et al., 1996) and FY834 (MATα his3 Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63; Winston et al., 1995) and following published protocols (Knop et al., 1999; Raymond et al., 1999).
Table 1.
Strains and plasmids used in this study
| Strain name | Genotype | Source |
| AB33 | a2 PnarbW2 PnarbE1, bleR | Brachmann et al., 2001 |
| EMD5 (AB33_GRab5a) | a2 PnarbW2 PnarbE1, bleR/pCoGRab5a | This study |
| AB33ΔKin3 | a2 PnarbW2 PnarbE1, bleR, Δkin3, natR | Higuchi et al., 2014 |
| AB33ΔKin3_GRab5a | a2 PnarbW2 PnarbE1, bleR, Δkin3, natR/pCoGRab5a | Schuster et al., 2011b |
| AB5Dyn2ts_GRab5a | a2 PnarbW2 PnarbE1, bleR, Pdyn2-dyn2ts, hygR/pCoGRab5a | Schuster et al., 2011b |
| AB33GRab5a | a2 PnarbW2 PnarbE1, bleR/poNGRab5a | Schuster et al., 2011a |
| AB33ΔHok1 | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR | This study |
| AB33ΔHok1_Hok1 | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pCHok1G | This study |
| EMD5_Hok1 | a2 PnarbW2 PnarbE1, bleR/pCoGRab5a/pHok1_SE | This study |
| AB33GRab5a_ΔHok1 | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/poNGRab5a | This study |
| AB33_mChRab5a_Hok1G | a2 PnarbW2 PnarbE1, bleR, Phok1-hok1-egfp, hygR/pomChRab5a | This study |
| AB33ΔHok1_mChRab5a | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a | This study |
| AB33ΔHok1_mChRab5a_Hok1625–930G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1625–930G | This study |
| AB33_mChTub1_Hok11–224G | a2 PnarbW2 PnarbE1, bleR/pHomChTub1/pHok11–224G | This study |
| AB33ΔHok1_mChRab5a_Hok1225–930G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1225–930G | This study |
| AB33ΔHok1_mChRab5a_Hok11–224G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok11–224G | This study |
| AB33ΔHok1_mChRab5a_Hok1225–624G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1225–624G | This study |
| AB33ΔHok1_mChRab5a_Hok11–624G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok11–624G | This study |
| AB33ΔHok1_mChRab5a_Hok11–624PXG | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok11–624PXG | This study |
| AB33G3Dyn2 | a2 PnarbW2 PnarbE1, bleR, Pdyn2-3gfp-dyn2, natR | This study |
| AB33G3Dyn2_ΔHok1 | a2 PnarbW2 PnarbE1, bleR, Pdyn2-3gfp-dyn2, natR, Δhok1, hygR | This study |
| AB33G3Dyn2_ΔHok1_mChRab5a | a2 PnarbW2 PnarbE1, bleR, Pdyn2-3gfp-dyn2, natR, Δhok1, hygR/poCmChRab5a | This study |
| AB33mCh3Dyn2_ΔHok1_Hok11–624G | a2 PnarbW2 PnarbE1, bleR, Pdyn2-3mcherry-dyn2, natR, Δhok1, hygR/pHok11–624G | This study |
| AB33ΔHok1_mChRab5a_Hok1Δ225–624G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1Δ225–624G | This study |
| AB33ΔHok1_mChRab5a_Hok11–224PXG | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok11–224PXG | This study |
| AB33_mChRab5a_Hok1G_↑pHk361–385 | a2 PnarbW2 PnarbE1, bleR, Phok1-hok1-egfp, hygR/pomChRab5a/pHk361–385 | This study |
| AB33ΔHok1_mChRab5a_Hok1Δ361–385G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1Δ361–385G | This study |
| AB33ΔHok1_mChRab5a_Hok1Δ333–355G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1Δ333–355G | This study |
| AB33GRab5a_↑pHk361–385 | a2 PnarbW2 PnarbE1, bleR/poNGRab5a/pHk361–385 | This study |
| AB33Kin3G | a2 PnarbW2 PnarbE1, bleR, Pkin3-kin3-egfp, hygR | Schuster et al., 2011b |
| AB33Kin3G_ΔHok1 | a2 PnarbW2 PnarbE1, bleR, Pkin3-kin3-egfp, hygR, Δhok1, natR | This study |
| AB33Kin3G_↑pHk361–385 | a2 PnarbW2 PnarbE1, bleR, Pkin3-kin3-egfp, hygR/pHk361–385 | This study |
| AB33Kin3G_ΔHok1_Hok1Δ361–385 | a2 PnarbW2 PnarbE1, bleR, Pkin3-kin3-egfp, hygR, Δhok1, natR/pHok1Δ361–385HA | This study |
| AB33G3Dyn2_mChRab5a | a2 PnarbW2 PnarbE1, bleR, Pdyn2-3gfp-dyn2, natR/pomChRab5a | Schuster et al., 2011c |
| AB33ΔHok1_mChRab5a_pHok1H3_1–600G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1H3_1–600G | This study |
| AB33ΔHok1_mChRab5a_pHok1H3_166–436G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1H3_166–436G | This study |
| AB33ΔHok1_mChRab5a_pHok1H3_293–345G | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/pomChRab5a/pHok1H3_293–345G | This study |
| AB33_mChRab5a_Fts1G | a2 PnarbW2 PnarbE1, bleR, Pfts1-fts1-egfp, hygR/pomChRab5a | This study |
| AB33_mChRab5a_Fhp1G | a2 PnarbW2 PnarbE1, bleR, Pfhp1-fhp1-egfp, hygR/pomChRab5a | This study |
| AB33_mChRab5a_Hok1G_ΔKin3 | a2 PnarbW2 PnarbE1, bleR, Phok1-hok1-egfp, hygR, Δkin3, cbxR/pomChRab5a | This study |
| AB33GRab7 | a2 PnarbW2 PnarbE1, bleR/poGRab7 | Higuchi et al., 2014 |
| AB33ΔHok1_GRab7 | a2 PnarbW2 PnarbE1, bleR, Δhok1, hygR/poGRab7 | This study |
| AB33nRFP | a2 PnarbW2 PnarbE1, bleR/poNLS3RFP | Schuster et al., 2011a |
| FB2N107G | a2b2 Pnup107-nup107-egfp, bleR | Steinberg et al., 2012 |
| pCoGRab5a | Potef-egfp-rab5a, cbxR | Higuchi et al., 2014 |
| pΔKin3 | Δkin3, natR | Schuster et al., 2011c |
| poNGRab5a | Potef-egfp-rab5a, natR | Schuster et al., 2011a |
| pHΔHok1 | Δhok1, hygR | This study |
| pHok1_SE | Phok1-hok1, SE, hygR | This study |
| pCHok1G | Phok1-hok1-egfp, cbxR | This study |
| pomChRab5a | Potef-mcherry-rab5a, natR | Schuster et al., 2012 |
| pHok1G | Phok1-hok1-3egfp, hygR | This study |
| pHok1625–930G | Phok1-hok1625–930-egfp, cbxR | This study |
| pHok1225–930G | Phok1-hok1225–930-egfp, cbxR | This study |
| pHok11–224G | Phok1-hok11–224-egfp, cbxR | This study |
| pHok1225–624G | Phok1-hok1225–624-egfp, cbxR | This study |
| pHok11–624G | Phok1-hok11–624-egfp, cbxR | This study |
| pHok11–624PXG | Phok1-hok11–624-yup14–148-egfp, cbxR | This study |
| pHomChTub1 | Potef-mCherry-tub1, hygR | Schuster et al., 2012 |
| pNG3Dyn2 | Pdyn2-3gfp-dyn2, natR | This study |
| poCmChRab5a | Potef-mcherry-rab5a, cbxR | This study |
| pHok1Δ225–624G | Phok1-hok1Δ225–624-egfp, cbxR | This study |
| pHok11–224PXG | Phok1-hok11–224-yup14–148-egfp, cbxR | This study |
| pHk361–385 | Pcrg-hok1361–385, cbxR | This study |
| pHok1Δ361–385G | Phok1-hok1Δ361–385-egfp, cbxR | This study |
| pHok1Δ333–355G | Phok1-hok1Δ333–355-egfp, cbxR | This study |
| pHok1Δ361–385HA | Phok1-hok1Δ361–385-HA, cbxR | This study |
| pHok1H3_1–600G | Phok1-hshook31–600-hok1625–930-egfp, cbxR | This study |
| pHok1H3_166–436G | Phok1-hok11–224-hshook3166–436-hok1625–930-egfp, cbxR | This study |
| pHok1H3_293–345G | Phok1-hok11–332-hshook3293–345-hok1386–930-egfp, cbxR | This study |
| pKin3G_H | Pkin3-kin3-egfp, hygR | Schuster et al., 2011c |
| pNΔHok1 | Δhok1, natR | This study |
| pFts1G | Pfts1-fts1-egfp, hygR | This study |
| pFhp1G | Pfhp1-fhp1-egfp, hygR | This study |
| pCΔKin3 | Δkin3, cbxR | This study |
| poGRab7 | Potef-egfp-rab7, cbxR | Higuchi et al., 2014 |
| pN107G | Pnup107-nup107-egfp, bleR | Theisen et al., 2008 |
a and b, mating type loci; P, promoter; a hyphen indicates fusion; Δ, deletion; a slash indicates ectopically integrated; hygR, hygromycin resistance; bleR, phleomycin resistance; natR, nourseothricin resistance; cbxR, carboxin resistance; ts, temperature-sensitive allele; crg, conditional arabinose-induced promoter; otef, constitutive promoter; nar, conditional nitrate reductase promoter; E1 and W2, genes of the b mating type locus; SE, self-replicating plasmid; mCherry, monomeric Cherry; kin3, kinesin-3; dyn2, C-terminal half of the dynein heavy chain; tub1, tubulin α; nup107, nucleoporin; PX, PX domain from the putative endosomal t-SNARE Yup1; rab5a and rab7, small endosomal GTPases; HA, human influenza HA tag; hok1, Hook protein in U. maydis; hshook3, Hook3 from human; fts1, FTS homologue; fhp1, FHIP homologue.
The plasmid pCoGRab5a (Higuchi et al., 2014), which contains the carboxin resistance cassette and rab5a gene fused to EGFP under the potef promoter (Spellig et al., 1996), was linearized and integrated ectopically into strain AB33 (Brachmann et al., 2001), resulting in AB33_GRab5a, which was used for the UV mutagenesis. To obtain AB33ΔKin3 (Higuchi et al., 2014), plasmid pΔKin3 (Schuster et al., 2011c), containing a nourseothricin resistance cassette flanked by the kin3 promoter and the sequence downstream of the kin3 gene carried by the pTZ19R vector (Fermentas), was digested with PvuI and integrated into the kin3 locus of strain AB33 (Brachmann et al., 2001). Correct integration was confirmed by PCR and by Southern blotting. The strain AB33_mChTub1 was made by ectopic integration of plasmid pHomChTub1 (Schuster et al., 2012) into AB33 (Brachmann et al., 2001), resulting in AB33_mChTub1. The plasmid pHomChTub1 contains the hygromycin resistance cassette and tubulin-α gene (tub1) fused to mCherry under the potef promoter (Spellig et al., 1996). To obtain the AB33_mChRab5a strain, plasmid pomChRab5a (Schuster et al., 2011c), which contains the nourseothricin resistance cassette and rab5a gene fused to mCherry under the potef promoter, was linearized and integrated ectopically into strain AB33. To obtain strain AB33mCh3Dyn2, plasmid pDyn2_3mCh (Schuster et al., 2011c), containing a triple mCherry tag C-terminally fused to a dynein heavy chain gene (dyn2), was digested with DraI and HindIII and homologously integrated into AB33 (Brachmann et al., 2001). Correct integration was confirmed by Southern blotting.
pNG3Dyn2.
This plasmid contains the native promoter of the dyn2 gene followed by a triple GFP and the first 1,762 bp of the open reading frame of dyn2. To integrate into the native locus, a second fragment, containing a part of the promoter and the nourseothricin resistance cassette, was fused in front of the full-length promoter. The plasmid pGFP3Dyn2 (Lenz et al., 2006) was digested with NotI to remove the hygromycin resistance cassette, which was replaced by the nourseothricin resistance cassette. This plasmid was transformed into strain AB33 (Brachmann et al., 2001), resulting in strain AB33G3Dyn2. Integration of triple GFP into the dyn2 locus was confirmed by Southern blotting.
pHΔHok1.
In this plasmid, the endogenous hok1 gene is replaced by the hygromycin resistance gene; the promoter sequence and a downstream sequence allow this plasmid to integrate into the hok1 locus, which results in a deletion of the hok1 open reading frame. To obtain the plasmid pHΔHok1, a 1,077-bp fragment, containing the hok1 promoter, and a 1,055-bp fragment of genomic DNA, containing the downstream sequence of the hok1 gene, were amplified by PCR using sets of primers fEB382-rEB383 and fEB385-rEB384, respectively (primers sequences are summarized in Table S2). Obtained fragments were cloned by in vivo recombination in the yeast S. cerevisiae DS94, flanked by the hygromycin resistance cassette, into a 5,088-bp region of the pNEBcbx-yeast plasmid (Schuster et al., 2011a), which was digested by EcoRI and HindIII. For transformation into the U. maydis strains AB33 (Brachmann et al., 2001), AB33GRab5a (Schuster et al., 2011a), AB33G3Dyn2, and AB33mCh3Dyn2, the plasmid pHΔHok1 was digested with AlwNI and integrated into the hok1 locus, resulting in AB33ΔHok1, AB33GRab5a_ΔHok1, AB33G3Dyn2_ΔHok1, and AB33mCh3Dyn2_ΔHok1, respectively. Deletion of the hok1 gene was confirmed by PCR and Southern blotting. Plasmids pomChRab5a (Schuster et al., 2011c) and poGRab7 (Higuchi et al., 2014) were integrated ectopically into the strain AB33ΔHok1, resulting in AB33ΔHok1_mChRab5a and AB33ΔHok1_GRab7, respectively.
pNΔHok1.
In this plasmid, the endogenous hok1 gene is replaced by the nourseothricin resistance gene, which is flanked by a native promoter of hok1 and a downstream sequence of the hok1 gene region. To obtain the plasmid pNΔHok1, the plasmid pHΔHok1 was digested by HindIII and AflII to remove the hygromycin resistance cassette, and a 1,524-bp region encoding the nourseothricin resistance cassette was cloned into the plasmid using in vivo recombination in S. cerevisiae DS94 and primers fEB395-rEB396. For transformation into the U. maydis AB33Kin3G strain (Schuster et al., 2011b), the plasmid pNΔHok1 was digested with AlwNI, resulting in AB33Kin3G_ΔHok1. Deletion of the hok1 gene was confirmed by PCR and Southern blotting.
pHok1_SE.
This plasmid is able to self-replicate in U. maydis and contains the hok1 promoter (−1,002 bp), the hok1 gene, and the hygromycin resistance cassette. To generate this plasmid, the self-replicating plasmid pNEBUC-yeast (provided by S. Kilaru, University of Exeter, Exeter, England, UK) was constructed through in vivo recombination in the yeast S. cerevisiae DS94. A 2,680-bp fragment, containing the yeast URA3 marker and 2µ ori, was amplified from plasmid pEYA2 (Invitrogen) and was cloned into the plasmid pNEBUC, which was linearized by SacI. The resulting plasmid pNEBUC-yeast contained the ampicillin resistance cassette, an Escherichia coli origin of replication, a U. maydis autonomously replicating sequence, and the carboxin resistance cassette. Next, a 4,859-bp fragment containing the hok1 promoter and gene was amplified from the genomic DNA of the U. maydis strain 521 using sets of primers fAB1-rAB8 and cloned into the plasmid pNEBUC-yeast, digested with PvuII and XbaI, using a yeast recombination technique and S. cerevisiae strain FY834. Finally, the carboxin resistance cassette was replaced by the hygromycin resistance cassette, obtained as a 2,896-bp fragment from plasmid pGFP3Dyn2 (Lenz et al., 2006), by digestion of the obtained plasmid pNEBUC-Hok1 with NotI. The plasmid was transformed into EMD5 strain, resulting in EMD5_Hok1 (all strains are listed in Table 1).
pCHok1G.
This plasmid contains a hok1 gene fused to EGFP. The hok1 gene is located behind the hok1 promoter (−1,002 bp), and the plasmid contains the carboxin resistance cassette. To obtain the plasmid, first a yeast bacterial shuttle vector that contained a single SspI restriction site within the carboxin resistance cassette was generated in the yeast S. cerevisiae DS94. The pNEBcbx-yeast-SspI plasmid was derived from the plasmid pNEBcbx-yeast (Schuster et al., 2011a), which was linearized by ZraI and modified by replacement of a fragment containing the ZraI site flanked by two SspI sites with a fragment containing one NdeI site amplified from the pKin3G plasmid (Wedlich-Söldner et al., 2002b), using primers fEB39 and rEB40. This plasmid was next used to obtain the pHok1G plasmid, in which a 1,002-bp region encoding the hok1 promoter (−1,002 bp) and the hok1 gene was amplified from genomic DNA of U. maydis strain 521, using sets of primers fAB85-rAB86 and fAB87-rAB88, respectively. A 1,045-bp region encoding EGFP and the Tnos terminator was amplified from the plasmid pKin3G (Wedlich-Söldner et al., 2002b), using primers fAB33-rEB14 and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834. The plasmid pHok1G was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1, resulting in AB33ΔHok1_Hok1G.
pHok1G3.
The plasmid consists of a 1,003-bp fragment near the 3′ end of the hok1 gene followed by triple egfp and the Tnos terminator, the hygromycin resistance cassette, and a 999-bp fragment downstream of the hok1 gene. The plasmid was generated through in vivo recombination in the yeast S. cerevisiae DS94. Fragments 1,003 and 999 bp were amplified with 30-bp overhangs from genomic DNA of the U. maydis strain 521 using sets of primers fEB378-rEB379 and fEB376-rEB377, respectively, and cloned with a 5,271-bp fragment encoding triple EGFP, the Tnos terminator, and the hygromycin cassette and a 4,968-bp fragment encoding an ampicillin resistance cassette, an E. coli replication origin, the yeast URA3 marker, and 2µ ori. Both fragments were derived from the plasmid pGrc1-3G (Schuster et al., 2011b) by digestion with SacI and MluI (a 5,271-bp fragment) and with PsiI and AgeI (a 4,968-bp fragment). The plasmid pHok1G3 was digested with PsiI and BamHI and homologously integrated into the hok1 locus of strain AB33_mChRab5a, resulting in AB33_mChRab5a_Hok1G. Integration of a single EGFP into the hok1 locus was confirmed by PCR and Southern blotting.
pHok1225–930G.
This plasmid contains a hok1 gene, truncated in the N-terminal region (Δ1–224 aa) and fused to EGFP. The hok1 gene is located behind the hok1 promoter (−1,002 bp), and the plasmid contains the carboxin resistance cassette. A 1,002-bp region encoding the hok1 promoter (−1,002 bp) and a region of 225–930 aa were amplified from genomic DNA of U. maydis strain 521, using sets of primers fAB85-rAB86 and fAB87-rAB88, respectively. A 1,045-bp region encoding EGFP and the Tnos terminator was amplified from the plasmid pKin3G (Wedlich-Söldner et al., 2002b), using primers fAB33-rEB14, and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834. The plasmid pHok1225–930G was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1225–930G.
pHok1625–930G.
This plasmid contains a hok1 gene truncated in the N-terminal region and the middle part (Δ1–624 aa), fused to egfp. The construct was placed under the hok1 promoter (−1,002 bp) and contains the carboxin resistance cassette. The plasmid was generated through in vivo recombination in the yeast S. cerevisiae FY834. To obtain a backbone with the carboxin resistance cassette and a C-terminal egfp gene followed by the Tnos terminator, the plasmid pNEB-cbx-yeast-pkin3_kin31–369PX-GFP was digested with NaeI and SexAI producing a fragment of 7,990 bp. The fragments encoding the promoter (−1,002 bp) and a region of 625–930 aa were amplified from the genomic DNA of the U. maydis strain 521 using sets of primers fAB85-rAB86 and fAB133-rAB88, respectively. The plasmid pHok1625–930G was digested with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1625–930G.
pHok11–224G.
This plasmid contains a fragment of the hok1 gene encoding only the N-terminal region (aa 1–224), fused to EGFP. To obtain the plasmid pHok11–224G, the promoter (−1,002 bp), followed by the first 224 aa of the hok1 gene, was amplified by PCR, thereby introducing the restrictions sites of KpnI and NcoI. The PCR product was digested with KpnI and NcoI, producing a fragment of 1,669 bp. To obtain the backbone with the carboxin resistance cassette and a C-terminal egfp gene, followed by the Tnos terminator, the plasmid p123 (Aichinger et al., 2003) was digested with KpnI and NcoI, producing a fragment of 5,396 bp. After a two fragment ligation, the plasmid pHok11–224G carried the carboxin and ampicillin resistance cassettes, an E. coli replication origin, the native promoter of the hok1 gene, the first 224 aa of the hok1 gene, and a C-terminal EGFP plus the Tnos terminator. The plasmid pHok11–224G was linearized with SspI and integrated in the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a and AB33_mChTub1, resulting in AB33ΔHok1_mChRab5a_Hok11–224G and AB33_mChTub1_Hok11–224G, respectively.
pHok11–624G.
This plasmid contains a region encoding the first 624 aa of Hok1, fused to EGFP. To obtain the plasmid, a 2,934-bp region, encoding the hok1 promoter (−1,002 bp) and the first 624 aa of Hok1, was amplified from genomic DNA of U. maydis strain 521, using primers fEB408-rEB405. A region encoding EGFP, the Tnos terminator, and a fragment of the carboxin resistance cassette was amplified from the plasmid pKin3G (Wedlich-Söldner et al., 2002b), using primers fEB407-rEB14, and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834. The plasmid pHok11–624G was linearized with AgeI and integrated into the succinate dehydrogenase locus of strains AB33ΔHok1_mChRab5a and AB33mCh3Dyn2_ΔHok1, resulting in AB33ΔHok1_mChRab5a_Hok11–624G and AB33mCh3Dyn2_ΔHok1_Hok11–624G, respectively.
pHok1225–624G.
This plasmid contains a region encoding a middle domain of Hok1, fused to EGFP. To obtain the plasmid, a 1,035-bp region, encoding hok1 promoter (−1,002 bp) and a region of 225–624 aa of Hok1, was amplified from genomic DNA of U. maydis strain 521, using primers fEB408-rEB418. A region encoding EGFP, the Tnos terminator, and a fragment of the carboxin resistance cassette was amplified from the pKin3G plasmid (Wedlich-Söldner et al., 2002b), using primers fEB419-rEB14, and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834. The plasmid pHok1225–624G was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1225–624G.
pHok11–624PXG.
This plasmid contains a region encoding first 624 aa from Hok1 and the PX domain from Yup1 (Wedlich-Söldner et al., 2000), fused to EGFP. To obtain the plasmid pHok11–624PXG, a 2,935-bp region, encoding the hok1 promoter (−1,002 bp) and the first 624 aa of Hok1, was amplified from the genomic DNA of U. maydis strain 521, using primers fEB408-rEB402. A 438-bp region, encoding the PX domain from the endosomal t-SNARE Yup1 (aa 4–148) and flanking overhangs, was amplified from the pSl-Yup-RFP-hygromycin plasmid (provided by U. Fuchs) using primers fEB403-rEB294. A region encoding EGFP, the Tnos terminator, and a fragment of the carboxin resistance cassette was amplified from the pKin3G plasmid (Wedlich-Söldner et al., 2002b), using primers fEB111-rEB14, and cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the S. cerevisiae strain FY834. The plasmid pHok11–624PXG was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok11–624PXG.
poCmChRab5a.
This plasmid contains mCherry fused to rab5a. To obtain it, plasmid popaGRab5a (Schuster et al., 2011a) was digested with NdeI and AgeI, which provided the rab5a gene, its terminator, and half of the plasmid backbone, containing half of the carboxin and the ampicillin resistance cassettes and the E. coli replication origin (a 4,337-bp fragment). The same plasmid was digested with AgeI and NcoI to receive a 2,338-bp fragment, containing the second half of the backbone with the second half of the carboxin resistance cassette and the potef promoter. To replace the photoactivatable GFP with a single mCherry, the plasmid pomChRab5a (Schuster et al., 2011c) was digested with NcoI and NdeI (a 732-bp fragment). After a three-fragment ligation, the resulting plasmid carried the constitutive potef promoter followed by mCherry, the open reading frame, and the terminator of the rab5a gene. The plasmid was linearized with SspI and ectopically integrated into AB33G3Dyn2_ΔHok1, resulting in AB33G3Dyn2_ΔHok1_mChRab5a.
pHok1Δ225–624G.
This plasmid contains the hok1 gene, truncated in the middle region (Δ225–624 aa), fused to EGFP. To obtain the plasmid, a 1,704-bp region encoding the hok1 promoter (−1,002 bp) and a region of 1–224 aa of Hok1 was amplified from genomic DNA of U. maydis strain 521, using primers fEB408-rEB413. A remaining 978-bp region, encoding aa 625–930, was amplified from genomic DNA of U. maydis strain 521, using primers fEB428-rEB375. A region containing EGFP, the Tnos terminator, and a fragment of the carboxin resistance cassette was amplified from the pKin3G plasmid (Wedlich-Söldner et al., 2002b), using primers fEB389-rEB14, and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the S. cerevisiae strain FY834. The plasmid pHok1Δ225–624G was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1Δ225–624G.
pHok11–224PXG.
This plasmid contains a region encoding the first 224 aa from Hok1 and the lipid-binding PX domain from Yup1 (Wedlich-Söldner et al., 2000), fused to EGFP. To obtain this plasmid, pHok11–224PX-HA was digested with AflII and BclI, providing a fragment containing the carboxin and ampicillin resistance cassettes, an E. coli replication origin, and the native promoter of the hok1 gene (a 7,990-bp fragment). The same plasmid was digested with BclI and NcoI to obtain a region encoding the first 224 aa of the hok1 gene and a region encoding 4–148 aa of the yup1 gene (a 1,291-bp fragment). The EGFP gene was obtained by digestion of the plasmid p123 (Aichinger et al., 2003) with NcoI and AflII (a 769-bp fragment). After a three-fragment ligation, the plasmid pHok11–224PXG was digested with SspI and integrated ectopically into strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok11–224PXG.
pHok1Δ361–385G.
This plasmid contains the hok1 gene, truncated in the conserved amino acid stretch (Δ361–385) and fused to EGFP. To obtain the plasmid, a 2,112-bp region encoding the hok1 promoter (−1,002 bp) and 1–360 aa of Hok1 was amplified from genomic DNA of U. maydis strain 521 using primers fEB408-rEB426. The remaining 1,695 bp, encoding a region of 386–930 aa, was amplified from genomic DNA of U. maydis strain 521 using primers fEB427-rEB375. A region containing EGFP, the Tnos terminator, and a fragment of the carboxin resistance cassette was amplified from the pKin3G plasmid (Wedlich-Söldner et al., 2002b) using primers fEB389-rEB14 and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834. The plasmid pHok1Δ361–385G was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1Δ361–385G.
pHok1Δ333–355G.
This plasmid contains the hok1 gene truncated in the conserved amino acid stretch (Δ333–355 aa), fused to EGFP. To obtain the plasmid, a 2,028-bp region, encoding hok1 promoter (−1,002 bp) and 1–332 aa of Hok1, was amplified from genomic DNA of U. maydis strain 521 using primers fEB408-rEB424. The remaining 1,786 bp, encoding a region of aa 356–930, was amplified from genomic DNA of U. maydis strain 521 using primers fEB425-rEB375. A region containing EGFP, the Tnos terminator, and a fragment of the carboxin resistance cassette was amplified from the pKin3G plasmid (Wedlich-Söldner et al., 2002b) using primers fEB389-rEB14 and was cloned into the pNEBcbx-yeast-SspI plasmid, digested with EcoRI and SacI, by in vivo recombination in the yeast S. cerevisiae FY834. The plasmid pHok1Δ333–355G was linearized with SspI and integrated into the succinate dehydrogenase locus of strain AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1Δ333–355G.
pHk361–385.
This plasmid contains a sequence encoding a short and conserved region of Hok1, fused behind the inducible crg promoter (Bottin et al., 1996). The construct was obtained by amplification of a 134-bp fragment, encoding aa 361–385 of Hok1 and containing 30 bp of the crg promoter and the Tnos terminator, from genomic DNA of U. maydis strain 521 using primers fEB422-rEB423. The fragment was cloned into the yeast vector pcrgPeb1211–268 (Schuster et al., 2011a) by using in vivo recombination in the yeast S. cerevisiae. The plasmid pHk361–385 was digested with AgeI and integrated ectopically into strains AB33_mChRab5a_Hok1G, AB33GRab5a (Schuster et al., 2011a), and AB33Kin3G (Schuster et al., 2011b), resulting in AB33_mChRab5a_Hok1G_↑pHk361–385, AB33GRab5a_↑pHk361–385, and AB33Kin3G_↑pHk361–385, respectively. The integration of the peptide construct was confirmed by PCR.
pHok1Δ361–385HA.
This plasmid contains the hok1 gene, truncated in the conserved stretch of aa 361–385, fused to human influenza HA tag. To replace EGFP and integrate the HA tag, the plasmid pHok1Δ361–385G was digested with AgeI and cloned with a 462-bp fragment (containing the AgeI restriction site) near the 3′ end of the gene, with a 96-bp fragment encoding HA tag and the overhangs and with a 1,470-bp region encoding the Tnos terminator and a part of the carboxin resistance cassette. All three fragments were amplified from the plasmid pHok1Δ361–385G using sets of primers fEB388-rEB469, fAB90-rAB97, and fEB370-rSK49, respectively. The plasmid pHok1Δ361–385HA was linearized with SspI and integrated into the succinate dehydrogenase locus of AB33Kin3G_ΔHok1, resulting in AB33Kin3G_ΔHok1_Hok1Δ361–385.
pHok1H3_1–600G.
This plasmid contains the carboxin resistance cassette, the C terminus of the hok1 (aa 625–930), and a 600-aa region amplified from human hook3 (aa 1–600) followed by egfp. To obtain the plasmid, pHok1Δ361–385G was digested with MscI and BclI and fused to a 222-bp fragment, amplified from the plasmid pHok1Δ361–385G, a 1,861-bp fragment encoding the first 600 aa amplified from human HeLa cDNA, and a 772-bp region encoding a part of the hok1 C terminus, amplified from the plasmid pHok1Δ361–385G using sets of primers fEB416-rEB418, fEB463-rEB464, and fEB452-rEB453, respectively. The plasmid pHok1H3_1–600G was linearized with SspI and integrated into the succinate dehydrogenase locus of AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1H3_1–600G.
pHok1H3_166–436G.
This plasmid contains the carboxin resistance cassette and the hok1 gene truncated in the CC region (Δ225–624 aa), which was replaced with a 271-aa region amplified from human hook3 (aa 166–436) followed by EGFP. To obtain the plasmid, pHok1Δ361–385G was digested with MscI and PflMI and fused to a 400-bp fragment encoding a part of the N terminus of hok1, amplified from the plasmid pHok1Δ361–385G, a 873-bp fragment encoding the region of aa 166–436, amplified from human HeLa cDNA, and a 772-bp region encoding a part of the hok1 C terminus, amplified from the plasmid pHok1Δ361–385G using sets of primers fEB451-rEB413, fEB449-rEB450, and fEB452-rEB453, respectively. The plasmid pHok1H3_166–436G was linearized with SspI and integrated into the succinate dehydrogenase locus of AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1H3_166–436G.
pHok1H3_293–345G.
This plasmid contains the carboxin resistance cassette and the hok1 gene truncated in the conserved amino acid stretch (Δ333–385 aa), which was replaced with a 53-aa region amplified from human hook3 (aa 293–345) followed by EGFP. To obtain the plasmid, pHok1Δ361–385G was digested with MscI and PspXI and fused to a 724-bp fragment, amplified from the plasmid pHok1Δ361–385G, a 221-bp fragment encoding the conserved stretch amplified from human HeLa cDNA, and a 1,489-bp region encoding EGFP, the Tnos terminator, and a part of the carboxin resistance cassette, amplified from the plasmid pHok1Δ361–385G using sets of primers fEB451-rEB424, fEB455-rEB456, and fEB454-rEB453, respectively. The plasmid pHok1H3_293–345G was linearized with SspI and integrated into the succinate dehydrogenase locus of AB33ΔHok1_mChRab5a, resulting in AB33ΔHok1_mChRab5a_Hok1H3_293–345G.
pFts1G.
This plasmid contains a 1,176-bp fragment near the 3′ end of the fts1 gene followed by EGFP, the Tnos terminator, the hygromycin resistance cassette, and a 1,077-bp downstream sequence of the fts1 gene. The plasmid was generated through in vivo recombination in the yeast S. cerevisiae FY834. Fragments 1,176 and 1,077 bp were amplified with 30-bp overhangs from genomic DNA of the U. maydis strain 521 using sets of primers fEB490-rEB491 and fEB483-rEB484, respectively, and cloned with a 4,101-bp fragment encoding EGFP, the Tnos terminator, and the hygromycin cassette and a 4,899-bp fragment encoding an ampicillin resistance cassette, an E. coli replication origin, the yeast URA3 marker, and 2µ ori. Both fragments were derived from the plasmid pKin3G_H (Schuster et al., 2011b) by digestion with SacI and PmlI (a 4,101-bp fragment) and with PsiI and SacI (a 4,899-bp fragment). The plasmid pFts1G was digested with PsiI and BamHI and homologously integrated into the fts1 locus of strain AB33_mChRab5a, resulting in AB33_mChRab5a_Fts1G. Integration into the fts1 locus was confirmed by PCR and Southern blotting.
pFhp1G.
This plasmid contains a 1,082-bp fragment near the 3′ end of the fhp1 gene followed by EGFP, the Tnos terminator, the hygromycin resistance cassette, and a 1,074-bp downstream sequence of the fhp1 gene. The plasmid was generated through in vivo recombination in the yeast S. cerevisiae FY834. Fragments 1,082 and 1,074 bp were amplified with 30-bp overhangs from genomic DNA of the U. maydis strain 521 using sets of primers fEB492-rEB493 and fEB494-rEB495, respectively, and cloned with a 4,101-bp fragment encoding EGFP, the Tnos terminator, and the hygromycin cassette and a 4,899-bp fragment encoding an ampicillin resistance cassette, an E. coli replication origin, the yeast URA3 marker, and 2µ ori. Both fragments were derived from the plasmid pKin3G_H (Schuster et al., 2011b) by digestion with SacI and PmlI (a 4,101-bp fragment) and with PsiI and SacI (a 4,899-bp fragment). The plasmid pFhp1G was digested with PsiI and BamHI and homologously integrated into the fhp1 locus of strain AB33_mChRab5a, resulting in AB33_mChRab5a_Fhp1G. Integration into the fhp1 locus was confirmed by PCR and Southern blotting.
pCΔKin3.
To replace the nourseothricin resistance cassette with carboxin, plasmid pΔKin3 (Schuster et al., 2011c) was digested with NotI. The obtained plasmid pCΔKin3 was digested with KpnI and SphI and integrated into the strain AB33_mChRab5a_Hok1G, resulting in AB33_mChRab5a_Hok1G_ΔKin3. Integration into the kin3 locus was confirmed by PCR and Southern blotting.
UV mutagenesis
The U. maydis strain AB33_GRab5a, expressing ectopically integrated GFP-Rab5a, was plated onto 120 × 120–mm complete medium agar plates containing 1% (wt/vol) glucose, at ∼6,000 cells per plate. To obtain a survival curve, cells were UV radiated using an ultraviolet cross-linker (wavelength, 254 nm; CL 508S; UVITEC) at 20–55 mJ/cm2 and grown in the dark. 32 mJ/cm2 results in 3% survival and was used to mutagenize strain AB33_GRab5a. Colonies were grown in darkness for 5–6 d and, subsequently, replica plated onto NM plates supplemented with glucose. After 3–4 d at 28°C, colonies showing the short hyphal growth phenotype were selected and microscopically investigated.
Sequencing
Whole genome sequencing was performed using genomic DNA, which was isolated as previously described (Hoffman and Winston, 1987). 2 µg DNA was diluted in TE buffer (Tris-EDTA; 200-µl total volume) and fragmented using a Bioruptor (Diagenode) at a 30-min cycle of medium power of 30 s on and 30 s off on ice. DNA was concentrated and purified using a QIAquick column (QIAGEN), and fragmentation of the DNA was confirmed using the DNA chip (Bioanalyzer 7500; Agilent Technologies). A sequencing library was prepared using SPRIworks (Beckman Coulter) with a 300–600-bp size selection. The library was amplified for 15 cycles of PCR using Phusion DNA polymerase (New England Biolabs, Inc.) and purified using 0.8 volumes AMPure XP beads (Beckman Coulter). The mean (mode) insert size of the library was 279 bp (ranging between 200 and 600 bp) and was determined using a Bioanalyzer 7500 DNA chip. The library was pooled with other bar-coded libraries, denatured, and diluted to 5.5 pmol clustered on a cBot (Illumina, Inc.). 50-bp paired-end sequencing, with a custom bar-coding read, was completed on a HiSeq 2000 (Illumina, Inc.) using TruSeq v3 reagents (Illumina, Inc.). Reads were demultiplexed, using the CASAVA 1.8 software pipeline (Illumina, Inc.) into FASTQ files. This allowed zero mismatches in the bar codes with 29,853,306 numbers of reads, which were filtered using the fastq-mcf program from the ea-utils package, applying –x 0.01, –q 20, –p 10, and –u. Reads were subsequently remapped to the U. maydis reference genome from the Munich Information Center for Protein Sequences Ustilago maydis Genome Database using the BWA (Burrows–Wheeler Aligner; Li and Durbin, 2009), using the settings bwa aln –t 8 –q 20 and bwa sampe –a 600. Conversion from SAM to BAM files, duplicate PCR read removal, and mpileup files were generated using the SAMtools software package (Li et al., 2009). Bespoke perl scripts were used to identify SNPs using mpileup files (based on minimum read depth of 10 and minimum base identity of 95%) and identify which genes they occurred in. Positions that showed a base difference between the mutant strain (EMD5) and strain AB33G_GRab5a were considered as candidates for the mutations that produced observed phenotypic differences.
Identification of mutations in the EMD5 mutant
Genomic DNA were extracted from mutant EMD5 and U. maydis strain AB33_GRab5a and sequenced using a HiSeq 2000. Short reads were aligned against the published genome, and SNPs were discovered, based on minimum read depth of 10 and minimum base identity of 95%.
Laser-based fluorescence microscopy
Fluorescence microscopy was performed as previously described (Schuster et al., 2011a). In brief, the fungal cells were placed onto a 2% agar cushion and directly observed using a motorized inverted microscope (IX81; Olympus), equipped with a Plan Apochromat 100×, 1.45 NA oil total internal reflection fluorescence objective or U Plan S Apochromat 60×, 1.35 NA oil objective (Olympus). The fluorescent tags were exited using a VS-LMS4 Laser Merge System with solid-state lasers (488 nm/50 mW or 75 mW and 561 nm/50 mW or 75 mW; Visitron Systems). Photobleaching experiments were performed using a 405-nm/60-mW diode laser, which was decreased by a neutral density 0.6 filter, resulting in 15-mW output power, coupled into the light path by an adaptor (OSI-IX 71; Visitron Systems). The 405-nm laser was controlled by a controller (UGA-40; Rapp OptoElectronic) and VisiFRAP 2D FRAP control software for Meta Series 7.5.x (Visitron Systems). Synchronized observation of RFP and GFP fluorescence was performed using an imager (Dual-View Micro; Photometrics) equipped with a dual-line beam splitter (z491/561; Chroma Technology Corp.) with an emission beam splitter (565 DCXR; Chroma Technology Corp.), an ET-Band pass 525/50 (Chroma Technology Corp.), and a single band pass filter (BrightLine HC 617/73; Semrock). Images were captured using a camera (CoolSNAP HQ2; Photometrics/Roper Scientific). All parts of the system were under the control of the software package MetaMorph (Molecular Devices). Unless otherwise noted, all statistical analysis was performed using Mann–Whitney testing, and all values given in the text are means ± SD of at least two experiments. All statistical analysis was performed using Prism 4.03 (GraphPad Software).
Protein colocalization experiments
Colocalizations of EEs and Hok1, the truncated protein Hok11–624G, or the chimeras of Hok11–624-PXG and Hok1H3_293–345G cells were performed using the strains AB33_mChRab5a_Hok1G, AB33ΔHok1_mChRab5a_Hok11–624G, AB33ΔHok1_mChRab5a_Hok11–624-PXG, and AB33ΔHok1_mChRab5a_pHok1H3_293–345G. The strains were placed onto a 2% agar cushion and transferred to the microscope. A region of 10 µm in length 5 µm behind the hyphal tip was photobleached by a 100-ms light pulse of the 405-nm laser (60 mW) at 80% laser power. Subsequently, 100 frames were taken using a Dual-View Micro imager, with the 488-nm laser (90% output) and the 561-nm laser (25% output power) at a 150-ms exposure time, binning of 2. Colocalization kymographs were generated from the acquired image series using the MetaMorph software.
To colocalize dynein with the truncated protein Hok11–624G, cells of strain AB33mCh3Dyn2_ΔHok1_Hok11–624G were placed onto a 2% agar cushion and transferred to the microscope. A region of 30–40 µm in length 3 µm behind the hyphal tip was photobleached by a 100-ms light pulse of the 405-nm laser (80% laser power). Subsequently, 50 frames were taken using the Dual-View Micro imager, with the 488-nm laser (90% output power) and the 561-nm laser (80% output power) at a 150-ms exposure time, binning of 2. Kymographs were generated from the acquired image series using the MetaMorph software.
Colocalization between Fts1 and EEs and Fhp1 and EEs was analyzed in strains AB33_mChRab5a_Fts1G and AB33_mChRab5a_Fhp1G, respectively, by photobleaching of a region of hyphae behind the hyphal tip, using a 100-ms laser pulse (405-nm laser and 100% output power) followed by acquisition of 150 frames, using the Dual-View Micro imager, the 488-nm laser at 100% output power, a 561-nm laser at 25% output power, an exposure time of 150 ms, and binning of 2.
Colocalization kymographs were generated using the MetaMorph software. To determine the colocalization of apical dynein and EEs in Δhok1 mutants, MTs were disrupted during microscopic observation by incubating hyphal cells of the AB33G3Dyn2_ΔHok1_mChRab5a strain for 5–7 min in liquid media containing 30 µM benomyl (Sigma-Aldrich) and placed on a 2% agar cushions, supplemented with benomyl, for microscopic observation.
Analysis of motor and EE motility
Retrograde flux of dynein was analyzed in control strain AB33G3Dyn2 and the Δhok1 mutant AB33G3Dyn2_ΔHok1. Cells were placed onto a 2% agar cushion and transferred to the microscope. A region of 15 µm in length 5 µm behind the hyphal tip was photobleached by a 100-ms light pulse of the 405-nm laser (80% laser power) followed by immediate observation using the 488-nm laser (100% output power), at 150-ms exposure time and binning of 1. Flux rates were determined in kymographs, generated using the MetaMorph software.
To analyze retrograde and anterograde flux in strains AB33_mChRab5a_Hok1G and AB33ΔHok1_mChRab5a_Hok11–624G, cells were observed on 2% agar cushions. A region of 10 µm in length 5 µm behind the hyphal tip was photobleached, using a 100-ms light pulse of the 405-nm laser at 80% laser power followed by acquisition of 100 frames, using the 488-nm laser at 100% output power at 150-ms exposure time and binning of 1. Flux rates were determined in kymographs, generated using the MetaMorph software.
The flux of EEs was analyzed in strains AB33_mChRab5a_Hok1G, AB33ΔHok1_mChRab5a_Hok1Δ333–355G, AB33ΔHok1_mChRab5a_Hok1Δ361–385G, and AB33_mChRab5a_Hok1G_↑pHk361–385. Cells were grown in their respective media, placed onto a 2% agar cushion, and transferred to the microscope. A region of 10 µm in length 5 µm behind the hyphal tip was photobleached, using a 100-ms, 405-nm laser pulse (80% output power) followed by observation, using the 561-nm laser at 50% output power at an exposure time of 150 ms and binning of 1. Flux rates were determined in kymographs, generated using the MetaMorph software.
EE run length was analyzed in strains AB33GRab5a (Schuster et al., 2011a) and AB33GRab5a_↑pHk361–385. Control cells were placed onto a 2% agar cushion and transferred to the microscope. To clear the cell from interfering signals, a region of 40–50 µm in length, starting ∼5 µm behind the hyphal tip, was photobleached using a 100-ms light pulse at 405 nm (80% laser output power). Subsequently, 150 frames were taken using the 488-nm laser at 25% output power at 150-ms exposure time and binning of 1. For strain AB33GRab5a_↑pHk361–385, the expression of pHk361–385 was induced by shifting the cells for 2 h in 1% arabinose-containing media. After this, the cells were directly observed using the aforementioned conditions. Run length of the organelles was determined in kymographs, generated using the MetaMorph software.
Pausing time of dynein and EEs during anterograde to retrograde turning was analyzed in strain AB33G3Dyn2_mChRab5a. Cells were placed on a thin layer of 2% agarose, and regions of ∼10 µm at ∼5 µm behind the hyphal tip were photobleached, using a 100-ms laser pulse (405-nm laser and 80% output power) followed by acquisition of 75 frames, using the Dual-View Micro imager, a 488-nm laser at 80% output power, a 561-nm laser at 50% output power, an exposure time of 150 ms, and binning of 1. Pausing time of the organelles and motors was determined in kymographs, generated using the MetaMorph software.
EE turning was analyzed in the strain AB33GRab5a (Schuster et al., 2011a) and measured in the first 10 µm of each hypha after photobleaching the region of the first 15 µm from the tip using a 100-ms laser pulse (405-nm laser and 100% output power) followed by acquisition of 150 frames, using the 488-nm laser at 50% output power at an exposure time of 150 ms and binning of 1. Antero- to retrograde turning of EEs was determined in kymographs, generated using the MetaMorph software.
Hyphal cell length was measured in strains AB33ΔHok1_Hok1G, AB33ΔHok1_mChRab5a_Hok11–624PXG, and AB33ΔHok1_mChRab5a, which were grown up overnight in NM liquid medium containing 1% glucose (wt/vol). In the case of bipolar hyphae of the mutant strains, only longer hypha was measured using MetaMorph software.
Analysis of motor and Hok1 numbers
Quantitative analysis of fluorescent intensities of kinesin-3, Hok1, and dynein numbers of Kin3-GFP, Hok1-GFP, and GFP3Dyn2 was performed using the nucleoporin Nup107-GFP as an internal calibration standard, following published procedures (Schuster et al., 2011a). Strain FB2NupG was placed on a 2% agar cushion, and images of Nup107-GFP at the upper level of the nucleus were acquired with a 150-ms exposure time. The integrated intensity of single weak Nup107-GFP signals, representing 16 Nup107-GFP in a single nuclear pore, was corrected for the adjacent background. Corrected values were plotted, and the mean integrated intensity for a single Nup107-GFP was estimated. The fluorescent intensities of moving signals of Kin3-GFP, Hok1-GFP, and GFP3Dyn2 were determined in strains AB33Kin3G (Schuster et al., 2011b), AB33Kin3G_↑pHk361–385, AB33Kin3G_Hok1Δ361–385, AB33Kin3G_ΔHok1, AB33_mChRab5a_Hok1G, AB33G3Dyn2_mChRab5a (Schuster et al., 2011c), and AB33G3Dyn2_ΔHok1_mChRab5a. Cells were grown in their respective media, placed onto a 2% agar cushion, and transferred to the microscope. The first 10 µm of the hypha was photobleached by a 100-ms light pulse of the 405-nm laser (80% laser output power). After 5 s, an image series of 15 frames was taken, using the 488-nm laser at 100% output power and an exposure time of 150 ms, with binning of 1. Signals that were in focus in the first plane and that moved toward the hyphal tip were considered. For retrograde moving of Kin3-GFP and GFP3Dyn2 signals, a region of 10 µm, 5 µm behind the hyphal tip, was photobleached by a 100-ms light pulse of the 405-nm laser (80% laser output power). After 5 s, an image series of 15 frames was taken, using the 488-nm laser at 100% output power and an exposure time of 150 ms, with binning of 1. Signals that were in focus in the first plane and that moved toward the hyphal tip were considered. The integrated intensity value for these signals was measured in the first frame and corrected by the adjacent background. The numbers of Kin3-GFP and Hok1-GFP were estimated by comparing their fluorescent intensities to the mean intensity of a single Nup107-GFP. This also took into account that Kin3 and Hok1 are most likely dimers (Xu et al., 2008; Hammond et al., 2009).
Fluctuation of Kin3-GFP and Hok1-GFP fluorescence during anterograde to retrograde turning was performed using kymographs. The integrated intensity in each plane was measured and corrected for the adjacent background and for frame-to-frame bleaching, which was determined in each kymograph.
Visualizing the endocytic pathway
Endocytic sorting was analyzed in strains AB33GRab5a (Schuster et al., 2011a), AB33GRab5a_ΔHok1, AB5Dyn2ts_GRab5a, AB33GRab7 (Higuchi et al., 2014), and AB33ΔHok1_GRab7. Hyphal growth was induced by adding a1/a2 pheromone at a final concentration of 2.5 µg/ml (Fuchs et al., 2006) and incubated for 3–4 h at 22°C. After hyphae formation, the dye FM4-64 (Invitrogen/Molecular Probes) was added to 500-µl cultures at a final concentration of 1 µg/ml (Wedlich-Söldner et al., 2000). Cells were incubated for 10 min and sedimented at 3,000 rpm for 1 min. Subsequently, they were washed with 500 µl of fresh media followed by 20-min incubation in dye-free media. The vacuoles were labeled with CellTracker blue CMAC (7-amino-4-chloromethylcoumarin; Invitrogen/Molecular Probes), which specifically stains the acidic vacuolar compartment in U. maydis (Steinberg et al., 1998). The dye was added to the culture pretreated with FM4-64 with a final concentration of 20.9 µg/ml. Cells were incubated for 5 min and sedimented, washed, and placed onto a 2% agar cushion for microscopic observation.
Protein extraction, native gel electrophoresis, and Western blotting
Expression of Hok1 proteins was confirmed by Western Blotting. To this end, hyphal cell extracts were generated from 100-ml overnight cultures, grown to OD600 0.5–1.8, by disruption of liquid nitrogen–frozen U. maydis cells in a mixer mill (MM400; Retsch) at 30/s for 2 × 2.5 min. Thawed cell extract, organelles, and cell debris were resuspended in 0.1–0.5 ml of 50 mM Hepes, 50 mM KCl, 1 mM EGTA, and 1 mM MgCl2, pH 7.0, complemented with protease inhibitor (cOmplete Mini; Roche). Debris was removed by centrifugation in a Heraeus Biofuge Stratos (Thermo Fisher Scientific), using rotor 3331 at 23,300 rpm for 30 min at 4°C. The protein concentration of the cell extracts was determined using protein assay reagent (500-0006; Bio-Rad Laboratories, Inc.), and ∼50 µg of each sample was loaded on 8% SDS-polyacrylamide gels. This was followed by transfer to nitrocellulose membrane (GE Healthcare) for 45 min at 190 mA in a semidry blot chamber (Fastblot; Analytik Jena). Each blot was blocked for 1 h with 5% nonfat milk in TBS–1% Tween 20 and incubated overnight at 4°C with anti-GFP mouse IgG monoclonal antibodies in a 1:5,000 dilution (11814460001; Roche) or anti-HA high affinity rat monoclonal antibodies in a 1:1,000 dilution (11867423001; Roche) followed by incubation with HRP-conjugated anti–mouse IgG in a 1:4,000 dilution (W402B; Promega) or with HRP-conjugated goat anti–rat IgG in a 1:5,000 dilution (62-9520; Invitrogen) for 1–2 h at room temperature. The loading of even amounts of cell extract was confirmed, using of mouse anti–α-tubulin antibodies in a 1:4,000 dilution (Oncogene Science), followed by HRP-conjugated anti–mouse IgG in a 1:4,000 dilution. All blots were developed using ECL Plus Western Blotting Detection system, following the manufacturer’s instructions (GE Healthcare).
To compare the migration of Hok1G, Hok1Δ333–355G, and Hok1Δ361–385G in 8% native gels, cell extract was prepared from strains AB33_mChRab5a_Hok1G, AB33ΔHok1_mChRab5a_Hok1Δ361–385G, and AB33ΔHok1_mChRab5a_Hok1Δ361–385G using BRB80 buffer (80 mM Pipes, 1 mM EGTA, and 1 mM MgCl2, pH 6.9), complemented with protease inhibitor (cOmplete Mini). ∼15 µg of each sample was loaded on 8% polyacrylamide gels lacking SDS. Proteins were detected by Western blotting.
Immunoprecipitation and mass spectrometry
Interacting proteins were identified using Hok1-GFP and the GFP-Trap method (ChromoTek) followed by mass spectrometry. Cells were sedimented and disrupted using a mixer mill (MM400) twice for 2.5 min at a frequency of 30/s in the presence of liquid nitrogen. Extraction buffer (10 mM Hepes, 50 mM KCl, 1 mM EGTA, and 1 mM MgCl2, pH 7.0, containing protease inhibitors) was added, and the mixture was centrifuged for 30 min at 50,000 g at 4°C. The supernatant was collected, and the protein concentration was determined using a Bradford assay. Appropriate sample volumes were incubated with ChromoTek beads for 2 h at 4°C followed by three washes in extraction buffer. The beads were then analyzed by mass spectrometry. For this, proteins were subjected to an in-solution trypsin digest and analyzed using a quadrupole time of flight system (Q-TOF 6520; Agilent Technologies) coupled to a 1200 Series HPLC-Chip interface system. 1 µl was loaded onto a micro-C18 reverse-phase analytical column (75 µm × 150 mm; Protein ID Chip; Agilent Technologies). A hit was regarded as a probable identification when a minimum of two unique peptides achieved a score of >7 with matching forward–reverse values and percentage of score peak intensities >60%. For further details see Dagdas et al. (2012). The U. maydis protein database was obtained from the Munich Information Center for Protein Sequences Ustilago maydis Genome Database.
Bioinformatics
All sequences were obtained from the NCBI Protein Database with the following accession numbers: (a) Hook protein sequences: U. maydis Hok1 (um05551.1), XP_761698; D. melanogaster Hook and human Hook1, AAC09298; human Hook3, NP_115786; Salpingoeca sp., XP_004988159; A. laibachii, CCA23185; Mus musculus Hook3, NP_997542; Gallus gallus, XP_003643087; Xenopus laevis, NP_001085515; Strongylocentrotus purpuratus, XP_796541; Aspergillus fumingatus, XP_750495; Neurospora crassa, XP_956252; Hordeum vulgare, BAK00177; and Branchiostoma floridae, XP_002602613. (b) Non-Hook proteins that are first hits in BLAST when using human Hook1: Physcomitrella patens, XP_001758626; Arabidopsis thaliana, XP_002887245; C. albicans, XP_710120; S. cerevisiae Btn2p, EEU04378; A. gossypii FDAG1, AEY95210; and S. pombe, Q09684. (c) Kinesin-3 protein sequences: Salpingoeca sp., XP_004997839; human Kif1A, NP_001230937; M. musculus Kif1A, NP_032466; G. gallus Kif1A-like, XP_003641781; D. melanogaster Unc104-like, NP_611155; X. laevis, NP_001138546; U. maydis Kin3, XP_762398; N. crassa Kin2, ESA42610; A. nidulans, XP_680816; S. purpuratus, XP_003728932; B. floridae, XP_002602366; and A. laibachii, CCA17471. (d) Kinesin motors with the highest e-values when compared with the human Kif1A motor domain: H. vulgare, BAK04987; C. albicans, XP_713761; A. gossypii, NP_984241; A. thaliana, CAB88133; P. patens, XP_001754280; and S. pombe, NP_594686. (e) Hook-interacting proteins: U. maydis UmFhp1, XP_757710; human HsFHIP_1, NP_115503; U. maydis UmFts1, XP_756598; and human HsFTS_b, EAW82809.
The degree of sequence identity and similarity between proteins was determined by using EMBOSS Needle. The protein domain predictions were performed in InterProScan and Pfam (http://pfam.sanger.ac.uk/search), CCs were predicted by the COILS server, sequence alignments were performed using ClustalW, and the isoelectric point of the conserved region in the CC of Ustilago Hok1 was estimated using the EXPASY server (http://web.expasy.org/compute_pi/). The phylogenetic tree was calculated in MEGA 5.10 (Tamura et al., 2011). Sequence searches were performed in in BLAST (basic local alignment search tool; NCBI).
Online supplemental material
Fig. S1 shows a schematic overview of kinesin-3 and dynein in mediating bidirectional EE motility in U. maydis. Fig. S2 provides additional information on the genetic screen, including the phenotype of kinesin-3 and dynein mutants, the appearance of the EMD5 mutant on plates, and the rescue of EMD5 and Δhok1 by wild-type Hok1. Fig. S3 shows Western blots of cell extracts from Δhok1 mutants expressing various Hok1 mutant proteins. Fig. S4 shows the colocalization of the N-terminal domain of Hok1 and MTs, additional kymographs of retrograde motility of dynein in control and Δhok1, and the domain organization and EE localization of the Hok1-interacting proteins Fts1 and Fhp1. Fig. S5 shows the phenotype of a Δhok1 mutant expressing Hok1Δ333–355, the position of Hok1, Hok1Δ333–355, and Hok1Δ361–385 in a native polyacrylamide gel, the localization of Hok1 on Rab5a-positive EEs in a kinesin-3–null mutant, and the position of anterograde to retrograde EE turnings within the apical 10 µm, at which MTs are unipolar. Table S1 shows the experimental usage of all strains, and Table S2 summarizes the primers used for cloning. Videos show bidirectional motility of EEs in wild-type, EMD5, and Δhok1 mutants (Video 1), localization of Hok1 on EEs (Video 2), localization of the chimeric protein Hok11–624PX on moving EEs (Video 3), covisualization of cytoplasmic dynein and EEs in Δhok1 (Video 4), covisualization of Hok11–624GFP and EE (Video 5) and Hok11–624GFP and dynein in Δhok1 mutants (Video 6), EE motility in the presence of the peptide pHk1361–385 (Video 7), and covisualization of Hok1HsH3_293–345 and EEs in Δhok1 mutants (Video 8). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201309022/DC1.
Supplementary Material
Acknowledgments
This work is dedicated to Prof. Manfred Schliwa, who introduced G. Steinberg to the fascinating world of molecular motors.
We thank Dr. C. Hemetsberger for genetic screening, Dr. K. Paszkiewicz and Dr. K. Moore from the Exeter Sequencing Facility for sequencing, Dr. Hannah Florance from the Exeter Mass Spectrometry Facility for mass spectrometry experiments, Dr. H. Dawe for human cDNA, Drs. U. Fuchs and S. Kilaru for plasmids, Dr. S. Gurr for comments on the manuscript, and Dr. X. Xiang for sharing unpublished results.
This work was supported by Wellcome Trust (097835/Z/11/Z) and the Biotechnology and Biological Sciences Research Council (BB/J009903/1).
The authors declare no competing financial interests.
Author contributions: E. Bielska and M. Schuster performed experiments, generated strains, and analyzed data. Y. Roger and A. Berepiki generated strains. D.M. Soanes identified the mutations in EMD5. N.J. Talbot discussed data. G. Steinberg conceived the project, analyzed data, and wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- CC
- coiled coil
- EE
- early endosome
- FHIP
- FTS- and Hook-interacting protein
- FTS
- fused toes
- MT
- microtubule
- NM
- nitrate minimal
- PH
- pleckstrin homology
- PX
- Phox
References
- Aichinger C., Hansson K., Eichhorn H., Lessing F., Mannhaupt G., Mewes W., Kahmann R. 2003. Identification of plant-regulated genes in Ustilago maydis by enhancer-trapping mutagenesis. Mol. Genet. Genomics. 270:303–314 10.1007/s00438-003-0926-z [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hammer J.A., III 2010. Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22:479–487 10.1016/j.ceb.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron Gaillard C.L., Pallesi-Pocachard E., Massey-Harroche D., Richard F., Arsanto J.P., Chauvin J.P., Lecine P., Krämer H., Borg J.P., Le Bivic A. 2011. Hook2 is involved in the morphogenesis of the primary cilium. Mol. Biol. Cell. 22:4549–4562 10.1091/mbc.E11-05-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beakes G.W., Glockling S.L., Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi”. Protoplasma. 249:3–19 10.1007/s00709-011-0269-2 [DOI] [PubMed] [Google Scholar]
- Beeg J., Klumpp S., Dimova R., Gracià R.S., Unger E., Lipowsky R. 2008. Transport of beads by several kinesin motors. Biophys. J. 94:532–541 10.1529/biophysj.106.097881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottin A., Kämper J., Kahmann R. 1996. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253:342–352 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl G., Kämper J., Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063 10.1046/j.1365-2958.2001.02699.x [DOI] [PubMed] [Google Scholar]
- Brady S.T. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 317:73–75 10.1038/317073a0 [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Holzbaur E.L. 2009. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 19:147–155 10.1016/j.tcb.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. 2008. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27:2124–2134 10.1038/emboj.2008.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas Y.F., Yoshino K., Dagdas G., Ryder L.S., Bielska E., Steinberg G., Talbot N.J. 2012. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science. 336:1590–1595 10.1126/science.1222934 [DOI] [PubMed] [Google Scholar]
- Davidse L.C., Flach W. 1977. Differential binding of methyl benzimidazol-2-yl carbamate to fungal tubulin as a mechanism of resistance to this antimitotic agent in mutant strains of Aspergillus nidulans. J. Cell Biol. 72:174–193 10.1083/jcb.72.1.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon S.W., Nascimento A., Serpinskaya A.S., Gelfand V.I. 2005. Regulation of bidirectional melanosome transport by organelle bound MAP kinase. Curr. Biol. 15:459–463 10.1016/j.cub.2004.12.074 [DOI] [PubMed] [Google Scholar]
- DeGiorgis J.A., Petukhova T.A., Evans T.A., Reese T.S. 2008. Kinesin-3 is an organelle motor in the squid giant axon. Traffic. 9:1867–1877 10.1111/j.1600-0854.2008.00809.x [DOI] [PubMed] [Google Scholar]
- Derr N.D., Goodman B.S., Jungmann R., Leschziner A.E., Shih W.M., Reck-Peterson S.L. 2012. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 338:662–665 10.1126/science.1226734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S., Sharp A.H., Colomer V., Tokito M.K., Lanahan A., Worley P., Holzbaur E.L., Ross C.A. 1997. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 6:2205–2212 10.1093/hmg/6.13.2205 [DOI] [PubMed] [Google Scholar]
- Fischer-Parton S., Parton R.M., Hickey P.C., Dijksterhuis J., Atkinson H.A., Read N.D. 2000. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198:246–259 10.1046/j.1365-2818.2000.00708.x [DOI] [PubMed] [Google Scholar]
- Fridolfsson H.N., Ly N., Meyerzon M., Starr D.A. 2010. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol. 338:237–250 10.1016/j.ydbio.2009.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M.M., Holzbaur E.L. 2013. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol. 202:495–508 10.1083/jcb.201302078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Manns I., Steinberg G. 2005. Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell. 16:2746–2758 10.1091/mbc.E05-03-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Hause G., Schuchardt I., Steinberg G. 2006. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell. 18:2066–2081 10.1105/tpc.105.039388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Frank C.L., Calderon de Anda F., Tsai L.H. 2010. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 65:191–203 10.1016/j.neuron.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.P. 2004. Hither and yon: a review of bi-directional microtubule-based transport. Phys. Biol. 1:R1–R11 10.1088/1478-3967/1/2/R01 [DOI] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Blasius T.L., Li Z., Jiang Y., Jih G.T., Meyhofer E., Verhey K.J. 2009. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7:e72 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks A.G., Perlson E., Ross J.L., Schroeder H.W., III, Tokito M., Holzbaur E.L. 2010. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20:697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Ashwin P., Roger Y., Steinberg G. 2014. Early endosome motility spatially organizes polysome distribution. J. Cell Biol. 204:343–357 10.1083/jcb.201307164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., Tanaka Y. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68:610–638 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Hoepfner S., Severin F., Cabezas A., Habermann B., Runge A., Gillooly D., Stenmark H., Zerial M. 2005. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 121:437–450 10.1016/j.cell.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Hoffman C.S., Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 57:267–272 10.1016/0378-1119(87)90131-4 [DOI] [PubMed] [Google Scholar]
- Holliday R. 1974. Ustilago maydis. In Handbook of Genetics. Vol. 1 King R.C., editor Plenum Press, New York: 575–595 [Google Scholar]
- Jolly A.L., Gelfand V.I. 2011. Bidirectional intracellular transport: utility and mechanism. Biochem. Soc. Trans. 39:1126–1130 10.1042/BST0391126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M.K., Wilder I.B., Oakley B.R. 1992. Amino acid alterations in the benA (β-tubulin) gene of Aspergillus nidulans that confer benomyl resistance. Cell Motil. Cytoskeleton. 22:170–174 10.1002/cm.970220304 [DOI] [PubMed] [Google Scholar]
- Kämper J., Kahmann R., Bölker M., Ma L.J., Brefort T., Saville B.J., Banuett F., Kronstad J.W., Gold S.E., Müller O., et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 444:97–101 10.1038/nature05248 [DOI] [PubMed] [Google Scholar]
- Klopfenstein D.R., Tomishige M., Stuurman N., Vale R.D. 2002. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 109:347–358 10.1016/S0092-8674(02)00708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S., Lipowsky R. 2005. Cooperative cargo transport by several molecular motors. Proc. Natl. Acad. Sci. USA. 102:17284–17289 10.1073/pnas.0507363102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15(10B):963–972 [DOI] [PubMed] [Google Scholar]
- Krämer H., Phistry M. 1996. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J. Cell Biol. 133:1205–1215 10.1083/jcb.133.6.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer H., Phistry M. 1999. Genetic analysis of hook, a gene required for endocytic trafficking in Drosophila. Genetics. 151:675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar A., Tripathy S.K., Xu J., Mattson M.K., Anand P., Sigua R., Vershinin M., McKenney R.J., Yu C.C., Mogilner A., Gross S.P. 2011. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. Proc. Natl. Acad. Sci. USA. 108:18960–18965 10.1073/pnas.1107841108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J.H., Schuchardt I., Straube A., Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 10.1038/sj.emboj.7601119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesche R., Peetz A., van der Hoeven F., Rüther U. 1997. Ft1, a novel gene related to ubiquitin-conjugating enzymes, is deleted in the Fused toes mouse mutation. Mamm. Genome. 8:879–883 10.1007/s003359900604 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.H., Gutekunst C.A., Hersch S.M., Li X.J. 1998. Interaction of huntingtin-associated protein with dynactin P150Glued. J. Neurosci. 18:1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A.D. 2004. Positioning the Golgi apparatus. Cell. 118:271–272 10.1016/j.cell.2004.07.015 [DOI] [PubMed] [Google Scholar]
- Maldonado-Báez L., Cole N.B., Krämer H., Donaldson J.G. 2013. Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J. Cell Biol. 201:233–247 10.1083/jcb.201208172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- McGuire J.R., Rong J., Li S.H., Li X.J. 2006. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J. Biol. Chem. 281:3552–3559 10.1074/jbc.M509806200 [DOI] [PubMed] [Google Scholar]
- Miki H., Setou M., Kaneshiro K., Hirokawa N. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA. 98:7004–7011 10.1073/pnas.111145398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.J., Blasier K.R., Jeffery E.D., Ross M.W., Pullikuth A.K., Suo D., Park J., Smiley W.R., Lo K.W., Shabanowitz J., et al. 2012. Trk activation of the ERK1/2 kinase pathway stimulates intermediate chain phosphorylation and recruits cytoplasmic dynein to signaling endosomes for retrograde axonal transport. J. Neurosci. 32:15495–15510 10.1523/JNEUROSCI.5599-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.J., Klumpp S., Lipowsky R. 2008. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. USA. 105:4609–4614 10.1073/pnas.0706825105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S., Tanaka Y., Hirokawa N. 2008. KIF1Bβ- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat. Cell Biol. 10:1269–1279 10.1038/ncb1785 [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Pownder T.A., Sexson S.L. 1999. General method for plasmid construction using homologous recombination. Biotechniques. 26:134–141 [DOI] [PubMed] [Google Scholar]
- Reis G.F., Yang G., Szpankowski L., Weaver C., Shah S.B., Robinson J.T., Hays T.S., Danuser G., Goldstein L.S. 2012. Molecular motor function in axonal transport in vivo probed by genetic and computational analysis in Drosophila. Mol. Biol. Cell. 23:1700–1714 10.1091/mbc.E11-11-0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Ashwin P., Lin C., Severs N.J., Steinberg G. 2011a. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 30:652–664 10.1038/emboj.2010.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Fink G., Collemare J., Roger Y., Steinberg G. 2011b. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell. 22:3645–3657 10.1091/mbc.E11-03-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lipowsky R., Assmann M.A., Lenz P., Steinberg G. 2011c. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. USA. 108:3618–3623 10.1073/pnas.1015839108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Treitschke S., Kilaru S., Molloy J., Harmer N.J., Steinberg G. 2012. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J. 31:214–227 10.1038/emboj.2011.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V., Rai A.K., Ramaiya A.J., Barak P., Mallik R. 2009. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. USA. 106:19381–19386 10.1073/pnas.0906524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T., Bottin A., Kahmann R. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503–509 [DOI] [PubMed] [Google Scholar]
- Splinter D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8:e1000350 10.1371/journal.pbio.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp E.T., Wright J., Baldauf S.L. 2006. The protistan origins of animals and fungi. Mol. Biol. Evol. 23:93–106 10.1093/molbev/msj011 [DOI] [PubMed] [Google Scholar]
- Steinberg G., Perez-Martin J. 2008. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 18:61–67 10.1016/j.tcb.2007.11.008 [DOI] [PubMed] [Google Scholar]
- Steinberg G., Schliwa M., Lehmler C., Bölker M., Kahmann R., McIntosh J.R. 1998. Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111:2235–2246 [DOI] [PubMed] [Google Scholar]
- Steinberg G., Schuster M., Theisen U., Kilaru S., Forge A., Martin-Urdiroz M. 2012. Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J. Cell Biol. 198:343–355 10.1083/jcb.201201087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunio A., Metcalf A.B., Krämer H. 1999. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol. Biol. Cell. 10:847–859 10.1091/mbc.10.4.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G., Hall B., Yu R., Hashim A.I., Krämer H. 2007. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 8:32–46 10.1111/j.1600-0854.2006.00511.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Halleck M.S., Schlegel R.A., Williamson P. 1996. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 272:1495–1497 10.1126/science.272.5267.1495 [DOI] [PubMed] [Google Scholar]
- Theisen U., Straube A., Steinberg G. 2008. Dynamic rearrangement of nucleoporins during fungal “open” mitosis. Mol. Biol. Cell. 19:1230–1240 10.1091/mbc.E07-02-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480 10.1016/S0092-8674(03)00111-9 [DOI] [PubMed] [Google Scholar]
- Vale R.D., Reese T.S., Sheetz M.P. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 42:39–50 10.1016/S0092-8674(85)80099-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Williams J.C., Varma D., Barnhart L.E. 2004. Dynein: An ancient motor protein involved in multiple modes of transport. J. Neurobiol. 58:189–200 10.1002/neu.10314 [DOI] [PubMed] [Google Scholar]
- van Spronsen M., Mikhaylova M., Lipka J., Schlager M.A., van den Heuvel D.J., Kuijpers M., Wulf P.S., Keijzer N., Demmers J., Kapitein L.C., et al. 2013. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 77:485–502 10.1016/j.neuron.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Vida T.A., Emr S.D. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta J.H., Didier A.J., Liu X., Krämer H. 2001. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152:923–934 10.1083/jcb.152.5.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Bölker M., Kahmann R., Steinberg G. 2000. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19:1974–1986 10.1093/emboj/19.9.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Schulz I., Straube A., Steinberg G. 2002a. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell. 13:965–977 10.1091/mbc.01-10-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Straube A., Friedrich M.W., Steinberg G. 2002b. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21:2946–2957 10.1093/emboj/cdf296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M.A. 2004. Bidirectional transport along microtubules. Curr. Biol. 14:R525–R537 10.1016/j.cub.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S.L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 11:53–55 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- Xu L., Sowa M.E., Chen J., Li X., Gygi S.P., Harper J.W. 2008. An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol. Biol. Cell. 19:5059–5071 10.1091/mbc.E08-05-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yao X., Fischer L., Abenza J.F., Peñalva M.A., Xiang X. 2011. The p25 subunit of the dynactin complex is required for dynein–early endosome interaction. J. Cell Biol. 193:1245–1255 10.1083/jcb.201011022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.