Abstract
Ubiquitin is a post-translational modifier with proteolytic and non-proteolytic roles in many biological processes. At mitochondria, it performs regulatory homeostatic functions and contributes to mitochondrial quality control. Ubiquitin is essential for mitochondrial fusion, regulates mitochondria-ER contacts, and participates in maternal mtDNA inheritance. Under stress, mitochondrial dysfunction induces ubiquitin-dependent responses that involve mitochondrial proteome remodeling and culminate in organelle removal by mitophagy. In addition, many ubiquitin-dependent mechanisms have been shown to regulate innate immune responses and xenophagy. Here, we review the emerging roles of ubiquitin at mitochondria.
Keywords: biogenesis, dynamics, mitochondria, mitophagy, ubiquitin
Introduction
The 76-amino acid protein ubiquitin is the founding member of the ubiquitin-like (UBL) protein family that is known for its regulatory functions in a large variety of different cellular pathways in the cytoplasm and nucleus [1, 2]. Ubiquitin can be covalently attached to target proteins as a single moiety, but can also form chains through internal lysine residues [3]. As several lysines in ubiquitin can be used, many different chain types are possible, which have distinct cellular functions [4]. Ubiquitin chains linked by its lysine 48 (K48) are best known as a signal for degradation by the ubiquitin proteasome system (UPS), whereas regulatory functions have been attributed to other types of chains [5]. Deubiquitylases (DUBs), which remove ubiquitin chains, render ubiquitylation reversible and offer possibilities for regulation [6]. Ubiquitylation begins with the activation of the modifier by E1 enzymes, followed by its transfer to E2-conjugating enzymes, and its ligation to the target substrate by E3 ubiquitin ligases [3]. Three major types of E3 ligases have been described, the HECT, the RING, and the RING-between-RING (RBR) ligases [4, 7]. HECT ligases covalently accept ubiquitin before transferring it to the substrate, whereas RING ligases promote substrate ubiquitylation by bringing together the E2 and the substrate and RBR ligases have a RING/HECT hybrid mechanism of action.
More recent studies have demonstrated that ubiquitin orchestrates mitochondrial functions [8, 9]. Mitochondria are unique organelles, bound by two membranes, and harboring their own DNA (mtDNA), which encodes essential respiratory chain subunits and therefore enables energy production. Several subcompartments can be distinguished within mitochondria, the outer membrane (OM), intermembrane space (IMS), inner membrane (IM), and matrix. The electron flow through the mitochondrial electron transport chain in the IM builds up an electric potential difference between the matrix and the IMS sides. This difference in proton concentration is then converted into ATP production by the ATP synthase. Besides being the ATP powerhouse, mitochondria are essential for a number of other metabolic pathways, including the synthesis of iron-sulfur clusters and phospholipids. Moreover, mitochondria participate in many cellular processes, such as apoptosis, calcium buffering, aging, cellular differentiation, and antiviral immune responses [10–12].
Here, we review our current knowledge on how ubiquitylation influences mitochondrial activities. On the one hand, ubiquitylation maintains mitochondrial homeostasis and regulates interorganelle communications and developmental programs. On the other hand, ubiquitylation is essential for pathogen defense and, under stress, for mitochondrial quality control (QC) and mitophagy.
Ubiquitin is a master regulator of mitochondrial dynamics
Mitochondria are highly dynamic organelles, the morphology of which is dictated by balanced fusion and fission events [10–14] (Fig 1A). The predominance of one over the other leads to a range of morphologies, from an interconnected mitochondrial network to the presence of a multitude of small, dispersed mitochondria. Mitochondrial fission is driven by Drp1 (Dnm1 in yeast), whereas fusion requires mitofusins—Mfn1 and Mfn2 in mammals and Fzo1 in yeast—in the OM and OPA1 (Mgm1 in yeast) in the IM. All three belong to a special class of GTPases, the dynamin-related proteins (DRPs), which provide the mechanical forces necessary for membrane remodeling [15, 16]. Drp1/Dnm1 is a cytosolic protein that is recruited to mitochondria when fission is initiated and forms spirals around mitochondrial tubules (Fig 1B). Interestingly, endoplasmic reticulum (ER) tubules were found to wrap around mitochondria, marking the sites for mitochondrial division by Drp1/Dnm1 [17]. Mitofusins are exposed to the cytoplasm but anchored to the OM by two transmembrane domains [18] (Fig 1B). In contrast, Opa1/Mgm1 is present in the IM and IMS and therefore does not contact the cytoplasm [18].
Figure 1.
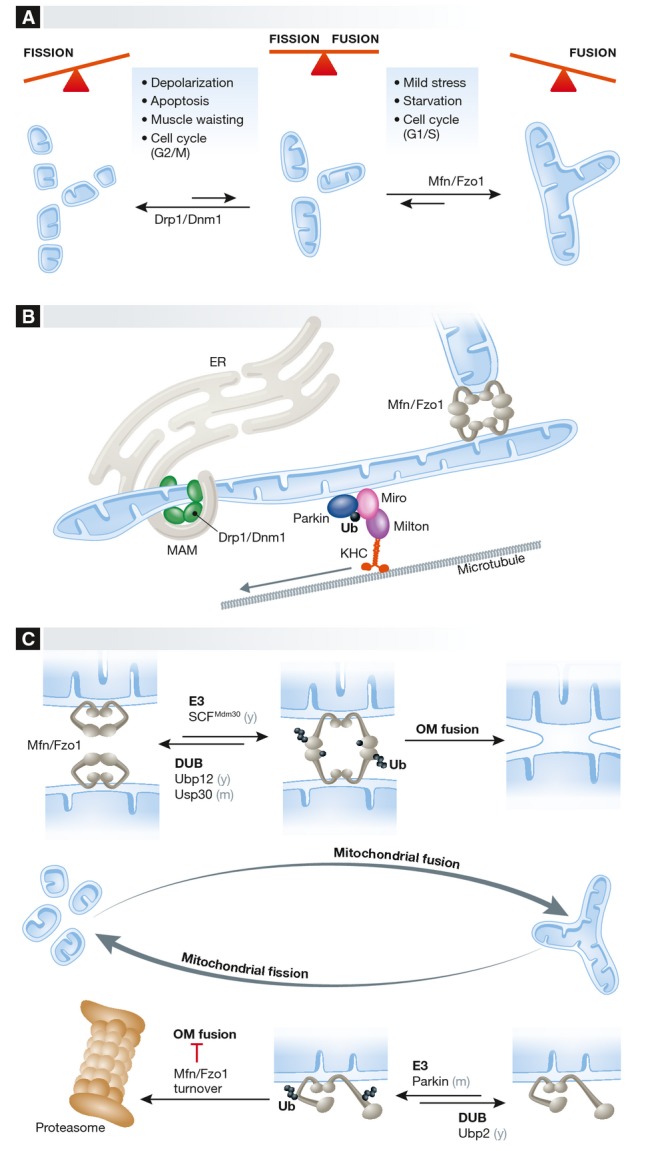
(A) Balanced fusion and fission events establish a large variety of mitochondrial network topologies in different cells and tissues and in response to various stimuli. (B) Mitochondria are physically tethered: to each other via mitofusins during fusion, to the mitochondrial transport machinery via Miro proteins, and to the ER via various protein complexes to allow the exchange of metabolites and signaling molecules and to mark sites for mitochondrial fission by Drp1. (C) Ubiquitylation of mitofusins has a dual function in yeast and mammals. Constitutive ubiquitylation, depending on the E3 ligase SCFMdm30 and the DUB Ubp12 in yeast and USP30 in mammals, promotes mitochondrial fusion. In contrast, ubiquitylation of mitofusins inhibits mitochondrial fusion upon growth arrest in yeast or in response to mitochondrial depolarization in mammals. The E3 ubiquitin ligase Parkin and other ligases ubiquitylate Mfn1 and Mfn2, marking them for proteasomal turnover. The DUB Ubp2 was identified in yeast to reverse Fzo1 ubiquitylation resulting in its stabilization. Ub, ubiquitin. Mfns, mitofusins. See Glossary for the other definitions and text for details.
A number of regulatory mechanisms that fine-tune opposing fusion and fission events have been described. Phosphorylation and ubiquitylation of mitofusins modulate the fusion of mitochondrial membranes [13]. In turn, mitochondrial fission is regulated by various post-translational modifications of Drp1, including phosphorylation, S-nitrosylation, SUMOylation, glycosylation, and ubiquitylation [14]. Recent groundbreaking discoveries identified proteins and protein complexes that link mitochondria to the cytoskeleton and to both the cell cortex and the ER, also contributing to the proper segregation of mtDNA [19–23] (Fig 1B). Interestingly, increasing evidence suggests that ubiquitylation by the same E3 ubiquitin ligases regulates fusion, fission, motility, and ER contacts of mitochondria. This interdependent regulation of dynamic morphological changes allows an efficient response to different physiological challenges.
Mitochondrial fusion
Mitofusins show a distinct ubiquitylation pattern that is conserved throughout evolution from yeast to mammals [24–26]. Mutations in Mfn2 cause the Charcot–Marie–Tooth type 2A peripheral neuropathy, and ubiquitylation and degradation of mitofusin was suggested to be implicated in the disease [27]. Once it was recognized that mitofusins are ubiquitylated and degraded [24–26, 28–34], several E3 ligases and DUBs acting on yeast, fly, and mammalian mitofusins were identified [13]. The analysis of these players revealed that ubiquitin is a double-faced regulator of mitochondrial fusion (Fig 1C). On the one hand, ubiquitylation triggers the conserved, proteasome-dependent degradation of mitofusins [24–26, 28–37] (Fig 1C, lower panel), leading to the inhibition of mitochondrial fusion, which fragment due to ongoing fission events. On the other hand, the ubiquitylation of mitofusins plays a positive, critical role for mitochondrial fusion [38–41] (Fig 1C, upper panel) as it can also activate them [40–44] (Sidebar A).
The most detailed mechanistic insight into how ubiquitylation of mitofusins promotes fusion is currently available for yeast. Fzo1 is ubiquitylated by the E3 ligase SCFMdm30 and deubiquitylated by Ubp12, which are soluble proteins that also associate with mitochondria [24, 40, 45, 46] (Fig 1C, upper panel). The positive role of Fzo1 ubiquitylation in fusing mitochondria appears to be linked to Fzo1 oligomerization [40] (Fig 1C, upper panel). In analogy to other DRP-mediated membrane remodeling events [15, 16], mitochondrial fusion requires the self-assembly of yeast and mammalian mitofusins [38, 39, 47]. In addition, OM fusion depends on the intermolecular cross-talk between Fzo1 monomers: The conserved lysine 464 is initially ubiquitylated and K48-linked ubiquitin chains are subsequently formed on lysine 398 of a different Fzo1 monomer [40, 43] (Fig 1C, upper panel). In mammals, ubiquitylated forms of Mfn1 and Mfn2 that promote mitochondrial fusion have been recently shown to be deubiquitylated by USP30 [41]. USP30 was known to localize to mitochondrial OM and regulate mitochondrial elongation [48]. Now, inhibition of USP30 was shown to efficiently revert the mitochondrial fusion defects of the single Mfn1-and Mfn2-knockout cell lines [41]. This strongly suggests that in mammals, both mitofusins are ubiquitylated in order to allow for fusion, in line with the absolute requirement of Fzo1 ubiquitylation for mitochondrial fusion in yeast [40]. Importantly, oxidized glutathione promotes disulfide-bond-mediated oligomerization of mammalian Mfn1 and Mfn2 and thereby directly activates mitochondrial fusion [49]. This suggests that cysteine-and lysine-mediated covalent modifications of mitofusin together orchestrate mitochondrial fusion (Sidebar A).
The pathway that triggers mitochondrial fragmentation by ubiquitylating and marking mitofusin for degradation by the proteasome has been longer known (Fig 1C, lower panel). In yeast, proliferation arrest by cell cycle blockage at the G1 phase induces Fzo1 proteolytic breakdown and organelle fragmentation, independently of the SCFMdm30 [28, 29]. In mammals, ubiquitylation and turnover of Mfn1 and/or Mfn2 occurs under various stress conditions—such as apoptosis, depolarization of mammalian mitochondria, and muscle atrophy—but also during progression through the cell cycle [32, 34, 36, 50]. Parkin and Gp78—an ER membrane-anchored ubiquitin E3 ligase involved in the ERAD pathway—ubiquitylates both Mfn1 and Mfn2 after mitochondrial depolarization or upon apoptosis induction [26, 31, 32, 51] (Fig 1C, lower panel). Parkin is encoded by Park2, whose loss-of-function mutations are associated with early onset of Parkinson's disease (PD), the most common neurodegenerative movement disorder [52]. Therefore, Parkin—a cytosolic RBR E3 ubiquitin ligase [53–55]—has been subject to extensive research. Notably, once ubiquitylated by Parkin, mammalian and fly mitofusins were shown to be recognized and extracted from the membrane by the p97/VCP/Cdc48 AAA-ATPase, and subsequently degraded by the proteasome [32, 48, 56]. Cdc48 and its co-factor Vms1 play a role in the QC of OM proteins in worms and yeast, which lack Parkin [57]. Upon oxidative stress, Vms1 translocates to mitochondria and appears to contribute to the degradation of mitochondrial OM proteins [57]. However, the involvement of Vms1 in the turnover of Fzo1 is controversial [57, 58] (Sidebar A).
Specific ubiquitylation and turnover of one of the two mammalian mitofusins has also been described [34, 36, 50] (Sidebar A). Genotoxic and other stresses were shown to induce an apoptotic response that requires JNK-dependent phosphorylation and subsequent ubiquitylation of Mfn2 by Huwe1—a HECT family ubiquitin ligase also termed Mule/ARF-BP1/HectH9/E3Histone/Lasu1. This was the first demonstration of the relevance of mitofusin phosphorylation [34]. MAPL/Mul1 also leads to the specific ubiquitylation of Mfn2 in response to muscle-wasting stimuli, targeting it to the proteasome [50]. MAPL is a SUMO E3 ligase integral to the OM, with two TM domains and a carboxyl terminal RING domain facing the cytosol [59, 60]. MAPL sumoylates Drp1 and regulates mitochondrial fission [60]. Although suggested to exert ubiquitin ligase activities as well [50, 59, 61], in vitro studies demonstrated a clear preference of MAPL for SUMO instead of ubiquitin [60]. Equally unknown is why the E3 ligase March5/MITOL specifically ubiquitylates Mfn1 but not Mfn2, in this case during the G2/M phase of the cell cycle [36, 62]. March5, a RING-type E3 ubiquitin ligase, harbors four transmembrane domains embedded in the OM and an amino terminal RING domain facing the cytosol [63, 64]. March5-mediated ubiquitylation and degradation of Mfn1 leads to mitochondrial fragmentation, perhaps to facilitate equal partitioning of cellular material to the two daughter cells, overcoming the hyperfused giant network formed during G1/S phase [36]. The control of mitochondrial morphology by the degradation of mitofusins could thus play an important role in the regulation of cell proliferation and differentiation [65].
Mitochondrial fission
Drp1/Dnm1 is a major hub for the regulation of mitochondrial fission. Several physiological and pathological stimuli lead to post-translational modifications of Drp1, which allow the coupling of mitochondrial division to mitosis and orchestrate the response to hypoxia, apoptosis, and mitophagy [14, 66]. The best-studied Drp1 modifications are SUMOylation and multiple phosphorylation events mediated by several kinases [14], but Drp1 ubiquitylation has also been observed. Interestingly, the hyperfused giant mitochondrial network formed during G1/S phase discussed above also seems to involve Drp1 ubiquitylation by the APC/CCdh1 E3 ubiquitin ligase complex, a central regulator of the M to G1 phase transition [67]. In addition to APC/CCdh1, the ubiquitin E3 ligases Parkin and March5/MITOL have been shown to modify Drp1 and one of its membrane anchors, Fis1.
Parkin induces the proteasomal degradation of Fis1 and Drp1 [68, 69], which can be co-immunoprecipitated with ubiquitin [68], suggesting that Parkin ubiquitylates Drp1 and Fis1. These observations, combined with the inverse correlation between Parkin expression levels and Drp1-dependent mitochondrial fragmentation [70], indicated that ubiquitin and Parkin are inhibitory for mitochondrial fission. Interestingly, a similar role was attributed to March5/MITOL, which interacts with Drp1 and human Fis1, leading to their ubiquitylation and proteasomal-dependent turnover [63, 64]. These observations, combined with an elongation of mitochondrial tubules upon March5 overexpression and a fragmentation of mitochondria upon March5 inactivation, led to the initial proposal that March5 is an inhibitor of fission [63, 64]. However, the model that March5 ubiquitylates Drp1 to induce its proteolytic turnover, thus elongating mitochondria due to unopposed fusion events, is controversial. In fact, the opposite mitochondrial phenotypes were observed in a latter study, including the formation of long and interconnected mitochondria upon March5 inactivation [71]. In this study, mutations in the RING domain of March5 decreased the cellular mobility of Drp1, suggesting that March5 promotes the ubiquitin-dependent recruitment of Drp1 to mitochondria [71]. Consistently, the expression of a March5 variant with a mutant RING domain restored tubular mitochondria in Mfn1−/− and Mfn2−/− cells, as expected from the simultaneous inhibition of mitochondrial fusion and fission [71]. In conclusion, although mitochondrial fission is clearly regulated by ubiquitylation of Drp1 and Fis1, further studies are required to dissect the roles and mechanisms involved.
Mitochondrial transport
Mitochondrial dynamics are particularly important in highly polarized cells, such as neurons, which depend on the long-range transport of mitochondria to ensure energy supply [72–74]. Mitochondria must be transported from the cell body to neurites, properly sustained, and removed if damaged. In fact, more than one quarter of the total mitochondria are actively moving, leading to constant changes in mitochondrial density in different synapses [72–74].
Mitochondrial transport occurs along microtubule tracks that are linked to mitochondria by the Miro, Milton, and kinesin heavy chain motor complex [72–74]. Miro1/RHOT1 and Miro2/RHOT2 are integral Rho-like GTPase proteins in the OM, each containing two GTPase motifs and a pair of EF hands involved in calcium binding [75, 76]. Milton is an adaptor protein that binds to both Miro and the kinesin heavy chain motor complex, and thereby loads mitochondria onto microtubules for anterograde axonal transport [75, 77]. Mitochondrial motility is regulated by the stability of the adaptor protein Miro. Mitochondrial depolarization results in the ubiquitylation of Miro by Parkin, which triggers its proteolytic breakdown [78–80]. Loss of Miro releases the mitochondria–motor complex bridge, arresting mitochondrial movement. Mfn2 was found to interact with both Miro and Milton proteins [81], possibly explaining the correlation of mitochondrial movement with fusion frequency [82]. Consistently, Mfn2 knockdown in neuronal culture cells impairs mitochondrial mobility. Similarly, mutations in Mfn2 associated with CMT2A impair the axonal transport of mitochondria [83] (Sidebar A).
Mitochondria-ER interactions
The existence of a special domain of the ER that is in contact with mitochondria—termed mitochondria-associated membranes (MAM)—is long known [84]. MAMs are enriched in enzymes of the lipid metabolism, as well as in proteins involved in calcium buffering, but also Mfn2 and Drp1 were found at the mitochondrial-ER contact sites [85]. MAMs physically associate with mitochondria via protein tethers, such as the yeast ER-mitochondria encounter structure (ERMES), which is composed of the ER protein Mmm1 and the mitochondrial OM proteins Mdm34, Mdm10 and Mdm12 [19]. Mdm34 is ubiquitylated by the SCFMdm30 [86]. Both MDM30 and MDM34 were originally found in a screen for genes implicated in mitochondrial morphology defects [45, 87]. However, how ubiquitylation affects the function of Mdm34 is not yet known. Interestingly, in addition to SCFMdm30, mitochondrial morphology is also regulated by the E3 ubiquitin ligase SCFMfb1 by an unknown mechanism [46, 88].
In addition to the ERMES complex, a number of other proteins have been proposed to tether ER to mitochondria in mammalian cells, including Mfn2 [89–92]. Moreover, March5/MITOL was shown to interact with the mitofusins Mfn1 and Mfn2 [62, 64]. A potential link between these findings stems from the observation that Mfn2 ubiquitylation by March5/MITOL contributes to interdependent functions of MAM and mitochondria [93]. March5/MITOL specifically catalyzes the formation of K63-linked ubiquitin chains on Mfn2 K129, which is not conserved in Mfn1 [93]. Notably, the ubiquitylation-deficient mutant Mfn2K129R still interacts with March5/MITOL [93], suggesting that there might be other roles for March5/MITOL in Mfn2 regulation. Mfn2 ubiquitylation by March5/MITOL does not affect its turnover, but rather affects ER-mitochondrial contacts and calcium exchange. Interestingly, the most frequent mutation in Mfn2 found in CMT2A patients, R94Q, is incompetent in promoting ER-mitochondria interaction [92]. These results suggest that March5/MITOL could be involved in CMT2A pathogenesis. The relevance of Mfn2 for ER-mitochondrial interactions is highlighted by the finding that the HIV-1 Vpr protein participates in the ubiquitylation and turnover of Mfn2 by the CUL4 E3 ligase, consequently leading to reduced ER-mitochondrial contacts [94]. These studies demonstrate that ubiquitin regulates mitochondria-ER contacts in different ways and that the E3 ubiquitin ligases that participate in cell cycle progression also modulate organelle contacts (Sidebar A).
Ubiquitin and mitochondrial quality control
Ubiquitin contributes to mitochondrial QC in various ways. The UPS mediates the proteolytic breakdown of nuclear-encoded pre-proteins before their import into mitochondria [95, 96], whereas mitochondrial proteases degrade damaged proteins present within the organelle [97] (Fig 2A). Severe damage or depolarization of mitochondria triggers the recruitment of Parkin to the mitochondrial surface [98], where it ubiquitylates OM proteins initiating their proteasomal degradation [99–101] and culminating in mitophagy, the selective autophagic removal of the whole organelle [102–104] (Fig 2B). Finally, ubiquitin participates in apoptosis via the regulation of the integral OM protein Mcl1, as reviewed recently [105].
Figure 2.
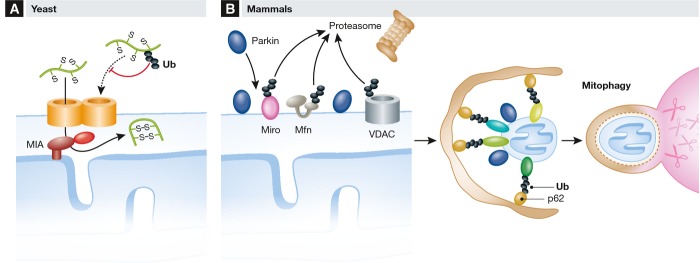
(A) Regulation of mitochondrial proteostasis by the UPS in yeast. Nuclear-encoded mitochondrial precursor proteins, such as substrates of the MIA pathway, are ubiquitylated and degraded by the UPS in the cytosol, limiting their accumulation in mitochondria. (B) Ubiquitin-dependent mitophagy of dysfunctional mitochondria in mammals. The recruitment of the E3 ubiquitin ligase Parkin to the OM of depolarized organelles leads to the ubiquitylation and proteasomal degradation of OM proteins, such as Miro, Mfns, and VDAC, inhibiting various processes, including mitochondrial transport or fusion. It also triggers mitophagy through the recruitment of adaptor factors for the autophagy machinery, such as p62. Ub, ubiquitin. Mfns, mitofusins. See Glossary for the other definitions and text for details.
Mitochondrial biogenesis
The vast majority of mitochondrial proteins are encoded by the nucleus and synthesized in the cytosol prior to their import into mitochondria. Dedicated machineries are present in each mitochondrial subcompartment that recognize, import, and sort newly imported proteins to their final destination within the organelle [106]. Several cytosolic kinases phosphorylate the protein import machinery in the OM, demonstrating that post-translational modifications regulate mitochondrial biogenesis, at least in yeast [107]. Ubiquitylation and proteasomal degradation, on the other hand, appear to control the influx of precursor proteins from the cytosol to mitochondria, as has been recently demonstrated for IMS proteins in yeast [96]. IMS proteins are imported by the MIA pathway through a folding-trap mechanism. The UPS regulates the mitochondrial proteome by constitutively ubiquitylating and degrading MIA substrates, suggesting that it can act as a negative regulator of mitochondrial biogenesis (Fig 2A). This is also consistent with the ubiquitylation of many nuclear-encoded mitochondrial proteins observed in proteome-wide studies [108–111] (Sidebar A).
Removal of damaged mitochondria through mitophagy
Pioneering work from the Youle laboratory has established a central role for the Ser/Thr kinase Pink1 and Parkin in the selective removal of damaged mitochondria [98]. Pink1 is an unstable mitochondrial IM protein that is rapidly degraded by the rhomboid protease PARL after import and maturation in healthy mitochondria [112]. A C-terminal proteolytic fragment is released from the IMS to the cytoplasm and degraded by the UPS through the N-end rule pathway [113]. Loss of membrane potential prevents the import of Pink1 into mitochondria and results in its accumulation at the OM and Parkin recruitment [98]. Pink1 activates Parkin, promoting the formation of a Parkin-ubiquitin thioester intermediate [114], perhaps by directly phosphorylating Parkin [115]. Parkin then ubiquitylates OM proteins, triggering their degradation by the UPS and thereby reshaping the protein composition of the mitochondria [99–101] (Fig 2B). Notably, if stabilized by proteasomal inhibitors, mature Pink1 can directly regulate cytosolic Parkin by binding to its RING1 domain, inhibiting Parkin activity [116, 117]. This suggests a new regulatory layer of the Pink1–Parkin complex that awaits the elucidation of whether at least some processed Pink1 is spared from degradation in healthy cells.
A recent genome-wide siRNA screen for factors affecting Parkin recruitment to mitochondria revealed an essential role of TOMM7, a subunit of the TOM complex, the protein translocase in the OM [118]. Binding of Pink1 to the TOM complex stabilizes it at the OM and promotes the recruitment of Parkin to mitochondria [118, 119]. Other factors involved in Parkin recruitment are HSPA1L, a widely distributed but low abundant member of the HSP70 family, and BAG4, a putative nucleotide-exchange factor of HSPA1L [118]. Both proteins interact physically with Parkin but have opposing roles in Parkin translocation to the OM [118].
Parkin accumulation at the surface of depolarized mitochondria allows the ubiquitylation of a variety of OM proteins with K48-and K63-linked chains, triggering their proteasomal degradation [99, 100] (Fig 2B). A systematic approach—using antibody capture of the diglypeptide combined with SILAC to monitor depolarization-dependent changes in abundance of the targets—recently expanded the knowledge of how Parkin acts on its substrates [101]. Parkin specificity was shown to be driven primarily by substrate recruitment or proximity, rather than by binding to specific target sequences [101]. In addition, Parkin dramatically altered the ubiquitylation status of the cytosolic domains of OM proteins, dependent on its active site residue C431, which is mutated in some PD patients.
The currently favored view is that general ubiquitylation at the mitochondrial surface of the damaged organelles recruits ubiquitin-binding adaptors, such as p62, which interact with the autophagic machinery [98, 99, 120, 121] (Fig 2B). However, a subset of OM proteins—such as VDAC—have also been suggested to have a specific role in mitophagy [99, 122]. An analysis of Parkin-mediated autophagy substrates in flies highlights how specific autophagy can be for individual proteins/complexes [123]. Selective mitophagy was also observed in yeast, which lack Parkin [124], but a role for ubiquitin has not yet been shown. Moreover, different pathways for selective mitophagy exist in mammalian cells [102], and two mitophagy-inducing conditions—oxygen or iron depletion—have been recently described. Hypoxia-induced mitophagy is regulated by FUNDC1 and phosphorylation and occurs in a Parkin-and seemingly ubiquitin-independent manner [125]. In turn, the Parkin-independent iron depletion-induced mitophagy pathway involves a metabolic switch from oxidative phosphorylation to glycolysis without depolarization of the engulfed mitochondria [117, 126] (Sidebar A).
Mitochondrial dynamics and mitophagy
The processes of fusion, fission, and movement of mitochondria are intimately linked to mitophagy. Starvation and mild oxidative stress induce hyperfusion of mitochondria and spare them from mitophagy [35, 49, 127, 128] (Fig 1A). In contrast, mitochondrial depolarization triggers mitochondrial fragmentation and mitophagy of the damaged organelles (Fig 1A). It is noteworthy that fission events, independently of stress insults, generate a mitochondrial population with uneven membrane potential, which affects the probability for subsequent fusion events [129, 130]. The impaired fusion of depolarized mitochondria allows their segregation from the mitochondrial network as fragments and therefore facilitates their removal by mitophagy. Similarly, damage leads to an arrest of mitochondrial movement in highly differentiated cells, such as neurons (see above) [30–32, 79] (Fig 2B). Interestingly, Parkin overexpression led to an increase in life span in flies and this could reflect the beneficial effects of an increased rate of mitophagy [37]. Consistently, it led to a decrease in Mfn2 levels that contributed to clearance of damaged mitochondria and increased mitochondrial activity [37].
Gp78 also activates mitophagy upon mitochondrial depolarization [51]. Although recognizing both Mfn1 and Mfn2 as substrates, Gp78-induced mitophagy depends only on ubiquitylation of Mfn1 [51]. In addition to Parkin and Gp78, the E3 ligase MAPL/Mul1 was recently shown to participate in mitophagy [50]. MAPL-dependent UPS turnover of Mfn2 was shown to facilitate mitophagy during skeletal muscle wasting, a process that is essential for recycling amino acids from proteins of the skeletal muscle [50, 131].
Recent evidence suggests that Mfn2 is involved in the QC surveillance of cardiac mitochondria [132]. In the heart, damaged-induced phosphorylation of Mfn2 at Thr111 and Ser442 was found to recruit Parkin to mitochondria. Consistently, mitochondrial dysfunction over time contributes to age-related heart failure, and heart-specific Mfn2−/− mice developed cardiomyopathies. However, earlier studies had demonstrated that Parkin can still induce mitophagy in Mfn1−/− Mfn2−/− mouse embryonic fibroblasts [98]. Although it is well established that activation of Parkin triggers mitophagy, whether the turnover of specific proteins such as Mfn2 is necessary remains to be clarified (Sidebar A).
Parkin and Parkinson's disease
PD is an important neurodegenerative disease that affects 1% of the population over 55 years old [133]. Genetic mutations are responsible for about 10% of all PD cases [134]. Mutations in Park2 cause autosomal recessive forms of PD and account for about 50% of the familial cases and 20% of the early-onset cases of PD [52]. Therefore, impaired mitophagy was suggested to contribute to neurodegeneration [135]. However, it should be noted that the majority of the findings indicating a role of Parkin in mitophagy were obtained in tumor cells upon depolarization of mitochondria and overexpression of Parkin. Parkin translocation to mitochondria was not observed in primary neurons or in mouse models accumulating mitochondria marked with ubiquitin, suggesting that alternative pathways do exist [103, 136]. Moreover, even in cases where Parkin recruitment to mitochondria was observed, it did not induce mitophagy [137]. Neither the analysis of PD mice models nor of park2 KO mice provided supportive evidence for Parkin-mediated mitophagy in vivo [138–140]. However, a progressive degeneration of dopaminergic neurons was observed after ablating park2 in adult mice [141], suggesting that the lack of PD phenotypes in PD mouse models might be explained by developmental compensation [141]. Similarly, the differences in Parkin recruitment and mitophagy induction observed between immortalized cells and neuronal cultures could arise from variations in protocols [142]. For example, Parkin was robustly recruited to neuronal cells cultured in the absence of antioxidants, which may counteract the action of chemical uncouplers; whether this can cause mitophagy requires further investigation [143].
Many Parkin substrates have already been identified, affecting a wide range of signaling and stress metabolic pathways, which is consistent with the broad neuroprotective capacity of Parkin. For instance, Parkin ubiquitylates and triggers proteolytic breakdown of PARIS, thus releasing the repression of the transcription factor PGC1-α that induces mitochondrial biogenesis [141]. Cellular stress recruits Parkin to the LUBAC complex [144], leading to the formation of linear ubiquitin chains on NEMO. Consequently, OPA1 transcription is upregulated, inhibiting apoptosis [144]. Thus, the relative contribution of various pathways modulated by Parkin to the pathogenesis of PD remains to be clarified (Sidebar A).
Maternal inheritance of mtDNA
In contrast to the nuclear DNA, mtDNA is maternally inherited [145, 146]. Several species-dependent mechanisms ensuring selective removal of sperm-derived mtDNA have been described. Sperm-derived mammalian mitochondria are marked with ubiquitin during spermatogenesis, and this was proposed to constitute the specific sorting signal for proteasomal-dependent elimination of male-derived mammalian mitochondria [147]. However, the lysosome has also been suggested to play a role [148]. Indeed, paternal mitochondria are actively disposed of by mitophagy after fertilization at early stages of C. elegans embryogenesis [149, 150]. Nevertheless, the relevance of these findings for mammals was recently challenged, because elimination of sperm mitochondria in mice was found not to be dependent on autophagy [151]. Although ubiquitylation of the sperm tail was observed, murine sperm mitochondria were not degraded post-fertilization. Rather, the most motile sperm that reached the oviduct were already depleted of mtDNA. Therefore, although a role of ubiquitin has been established, further studies are required to understand whether mitophagy is generally required to ensure maternal inheritance of mtDNA.
Mitochondria, ubiquitin, and antiviral defense
The innate immune system responds to pathogens such as bacteria and viruses using receptors from two different families, culminating in the activation of the NF-κB and type I interferon signaling pathways [152] (Fig 3). Double-stranded viral RNA is recognized in the cytoplasm by the RNA helicase RIG-1 [153]. Upon binding to viral RNA, RIG-1 is activated and exposes its CARD signaling domains, which associate with unanchored K63-linked ubiquitin chains. This leads to the formation of heterotetramers that interact with and activate the mitochondrial OM protein MAVS, a central player of the defense response (Fig 3). MAVS dimers or higher-order multimers recruit several downstream signaling effectors, such as the TRAF family members, which trigger the production of type I interferons and other cytokines [154–156]. MAVS polymers were recently shown to bind the E3 ubiquitin ligases TRAF2, TRAF5, TRAF6, and LUBAC, which collectively promote ubiquitylation reactions that recruit NEMO to the MAVS signaling complex, leading to the activation of the transcription factors IRF3 and NF-κB [157] (Fig 3).
Figure 3.
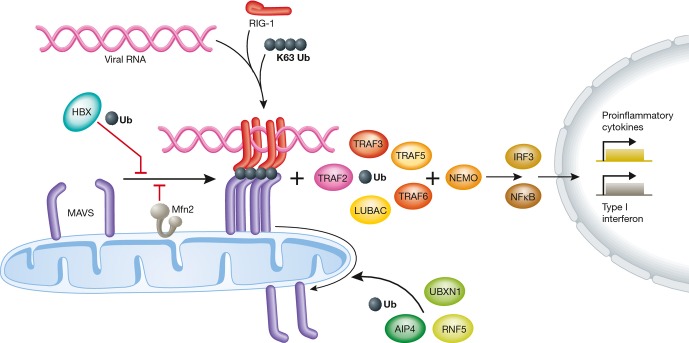
K63-linked ubiquitin chains, viral RNA, and the helicase RIG-1 participate in the assembly of a signal platform on mitochondria that contains MAVS. This allows several E3 ubiquitin ligases to activate LUBAC and TRAF transcription factors, which induce innate immunity genes. Moreover, ubiquitin is required for disassembly of the MAVS complex, terminating signal transduction. On the other hand, ubiquitin and Mfn2 impair MAVS assembly before signaling initiation. Ub, ubiquitin. See Glossary for the other definitions and text for details.
The majority of the mechanisms that terminate the antiviral response involve the proteolytic breakdown of MAVS. For example, the RNA-binding protein PCBP2 negatively regulates the immune response by recruiting the E3 HECT ligase AIP4 to MAVS, leading to its ubiquitylation and UPS-dependent turnover [158]. Similarly, the E3 RING ligase RNF5 attaches K48-linked ubiquitin chains at K362 and K461 of MAVS, targeting it for degradation after viral infection [159]. Interestingly, several viruses suppress the innate immune response, for example, by expressing specific proteases or—in the case of hepatitis B virus—by interaction of its protein X (HBX) with MAVS, thus targeting it for UPS-dependent degradation [160] (Fig 3).
The disruption of MAVS self-interactions precludes MAVS interaction with the TRAF proteins and represents another mechanism of curtailing MAVS activation [161]. The UBXN1 protein, a member of the ubiquitin-binding UBX protein family, binds to MAVS via its ubiquitin-associated domain and impairs antiviral responses [161]. Mfn2 has also been proposed to inhibit MAVS oligomerization by binding to it via its HR1 region [156, 162] which is in line with the fact that Mfn2−/− mouse embryonic fibroblasts have an increased antiviral response [162] (Fig 3).
Interestingly, most of the genes involved in the immune response to Sindbis virus are required for Parkin-mediated mitophagy, pointing to an intimate relationship between mitophagy and xenophagy, which is the autophagic degradation of incoming pathogens [163]. The E3 ubiquitin ligase SMURF1, for example, was identified as a mediator in both xenophagy and mitophagy [163]. Mitophagy is also related to the autophagic eradication of bacteria, because the innate immune response to bacterial infection involves Parkin [164]. Thus, multiple functions of ubiquitylation at the surface of mitochondria are emerging, acting upstream and downstream of MAVS during the immune response (Sidebar A).
Sidebar A:In need of answers
How are ubiquitylated mitofusins diverted from proteasomal turnover to promote mitochondrial fusion? Is ubiquitylation of Mfn1 and Mfn2 critical for embryonic viability?
How are ubiquitylated OM proteins degraded by 26S proteasomes? Which additional components are involved and how are they regulated?
Which properties distinguish Mfn1 from Mfn2, making them specific targets for selective ubiquitylation in apoptosis, muscle wasting, and cell cycle progression?
How are mitochondrial fusion and fission coordinated by ubiquitin and other post-translational modifications, for example, during the cell cycle? How does ubiquitin regulate cell proliferation and differentiation?
Is mitochondrial fusion controlled by the range of post-translational modifications that regulate mitochondrial fission? Conversely, how does ubiquitylation regulate mitochondrial fission?
How does ubiquitin regulate interorganellar contacts of mitochondria?
To which extent does the UPS-dependent turnover of mitochondrial precursor proteins in the cytosol contribute to the regulation of mitochondrial biogenesis?
What is the precise role of ubiquitylation and OM protein turnover for mitophagy? How does it affect aging and neurodegeneration?
What is the contribution of Pink1/Parkin-mediated mitophagy to mitochondrial quality control in vivo? How is Parkin activity regulated? What are the pathophysiological implications of the modification of its different targets?
How critical is ubiquitylation of mitochondrial proteins for the cellular immune response?
Conclusions
Ubiquitin serves as an important regulator of mitochondrial dynamics, surveys mitochondrial damage, and regulates innate immune responses in many ways. The ubiquitylation and turnover of several OM proteins contributes to maintaining mitochondrial homeostasis. We are only beginning to understand the emerging regulatory functions of ubiquitin, such as its essential role in promoting mitochondrial fusion or its wide participation in the initiation or termination of the innate immune response. The considerable research efforts in this active field hold the promise for more surprises to come, completing our picture of this master regulator of mitochondria. Future studies will shed light on the pathophysiological mechanisms of mitochondria-related diseases and thus define the contribution of the different processes regulated by ubiquitin, possibly identifying new therapeutic targets.
“Ubiquitylation: mechanism and functions” Review series
Previous issues of EMBO reports include:
Building and remodeling Cullin-RING E3 ubiquitin ligases, by Wade Harper et al
Ubiquitin in the immune system, by Henning Walczak et al
RBR E3 ligases at work, by Judith Smit and Titia Sixma
Other reviews in this series, which will be published in consecutive issues of EMBO reports, will cover:
Regulation of stem cell function by protein ubiquitylation, by Iannis Aifantis et al
Understanding ubiquitylation one structure at a time, by Ronald Hay et al
Acknowledgments
We are grateful to J. Dohmen for critical reading of the manuscript and to J. Dohmen, F. Anton, and M. Baker for stimulating discussions. This work was supported by grants of the Deutsche Forschungsgemeinschaft and the European Research Council (AdG 233078) to T.L.
Glossary
- AIP4
atrophin-1-interacting protein 4
- APC/C
Anaphase-promoting complex/cyclosome
- ATP
adenosine triphosphate
- BAG4
BCL2-associated athanogene 4
- CARD
caspase activation and recruitment domains
- Cdc48
cell division cycle 48
- Cdh1
cadherin 1
- CMTA
Charcot–Marie–Tooth type 2A
- CUL4
cullin 4
- Dnm1
yeast dynamin-related protein 1
- DRP
dynamin-related proteins
- Drp1
mammalian dynamin-related protein 1
- DUB
deubiquitylating enzyme
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- ERMES
ER-mitochondria encounter structure
- Fis1
mitochondrial fission 1
- Fzo1
yeast mitofusin, fuzzy onions homolog 1
- Gp78
glycoprotein 78
- GTPase
guanosine triphosphate hydrolase
- HBX
hepatitis B virus protein X
- HECT
homologous to the E6-AP carboxyl terminus
- HIV-1
human immunodeficiency virus 1
- HR1
heptad repeat 1
- HSP70
heat shock 70-kDa protein
- HSPA1L
heat shock 70-kDa protein 1-like
- Huwe1
HECT, UBA, and WWE domain containing 1
- IRF3
interferon regulatory transcription factor 3
- JNK
Jun N-terminal kinase
- LUBAC
linear ubiquitin chain assembly complex
- MAPL
mitochondrial-anchored protein ligase
- March5
membrane-associated ring finger (C3HC4) 5
- MAVS
mitochondrial antiviral signaling protein
- Mcl1
myeloid cell leukemia sequence 1
- Mdm
mitochondrial distribution and morphology (10,12,30,34)
- Mfb1
mitochondria-associated F-box protein 1
- Mfn
mammalian mitofusin
- Mgm1
mitochondrial genome maintenance 1
- MIA
mitochondrial intermembrane space import and assembly
- Miro1,2
mitochondrial Rho GTPase 1,2
- MITOL
mitochondrial ubiquitin ligase
- Mmm1
maintenance of mitochondrial morphology
- mtDNA
mitochondrial DNA
- NEMO
NF-κB essential modulator
- Opa1
optic atrophy 1
- PARIS
Parkin interacting substrate
- Park2
autosomal recessive juvenile Parkinson disease-2
- PARL
presenilin-associated rhomboid-like
- PCBP2
poly(rC) binding protein 2
- PGC1-α
peroxisome proliferator-activated receptor γ coactivator 1-α
- Pink1
PTEN-induced putative kinase 1
- RHOT1,2
ras homolog family member T1,2
- RIG-1
retinoic acid-inducible gene 1
- RING
really interesting new gene
- RNF5
RING finger protein 5
- SCF
Skp1/Cullin/F-box protein
- siRNA
small-interfering RNA
- SMURF1
SMAD-specific E3 ubiquitin protein ligase 1
- SUMO
small ubiquitin-related modifier
- TOM
translocase of the outer mitochondrial membrane
- TRAF
TNF receptor-associated factor
- UBL
ubiquitin-like
- Ubp
ubiquitin-specific protease
- UBX
ubiquitin regulatory X domain
- USP30
ubiquitin-specific protease 30
- VCP
valosin-containing peptide
- Vms1
VCP/Cdc48-associated mitochondrial stress-responsive
- Vpr
viral protein R
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Varshavsky A. The early history of the ubiquitin field. Protein Sci. 2006;15:647–654. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28:321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 6.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 7.Smit J, Sixma T. RBR E3 ligases at work. EMBO Rep. 2014;15:142–154. doi: 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 9.Livnat-Levanon N, Glickman MH. Ubiquitin-proteasome system and mitochondria-reciprocity. Biochim Biophys Acta. 2011;1809:80–87. doi: 10.1016/j.bbagrm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 11.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escobar-Henriques M, Anton F. Mechanistic perspective of mitochondrial fusion: tubulation vs. fragmentation. Biochim Biophys Acta. 2013;1833:162–175. doi: 10.1016/j.bbamcr.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochim Biophys Acta. 2013;1833:150–161. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 16.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 17.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5:1–16. doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swayne TC, Zhou C, Boldogh IR, Charalel JK, McFaline-Figueroa JR, Thoms S, Yang C, Leung G, McInnes J, Erdmann R, Pon LA. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr Biol. 2011;21:1994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci USA. 2013;110:E458–E467. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klecker T, Scholz D, Fortsch J, Westermann B. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J Cell Sci. 2013;126:2924–2930. doi: 10.1242/jcs.126045. [DOI] [PubMed] [Google Scholar]
- 23.Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin-Proteasome-dependent Degradation of a Mitofusin, a Critical Regulator of Mitochondrial Fusion. Mol Biol Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS ONE. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amiott EA, Cohen MM, Saint-Georges Y, Weissman AM, Shaw JM. A mutation associated with CMT2A neuropathy causes defects in Fzo1 GTP hydrolysis, ubiquitylation, and protein turnover. Mol Biol Cell. 2009;20:5026–5035. doi: 10.1091/mbc.E09-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neutzner A, Youle RJ. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- 29.Escobar-Henriques M, Westermann B, Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J Cell Biol. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- 34.Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell. 2012;47:547–557. doi: 10.1016/j.molcel.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci USA. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park YY, Cho H. Mitofusin 1 is degraded at G2/M phase through ubiquitylation by MARCH5. Cell Div. 2012;7:25. doi: 10.1186/1747-1028-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci USA. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anton F, Fres JM, Schauss A, Pinson B, Praefcke GJ, Langer T, Escobar-Henriques M. Ugo1 and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer membrane fusion. J Cell Sci. 2011;124:1126–1135. doi: 10.1242/jcs.073080. [DOI] [PubMed] [Google Scholar]
- 39.Cohen MM, Amiott EA, Day AR, Leboucher GP, Pryce EN, Glickman MH, McCaffery JM, Shaw JM, Weissman AM. Sequential requirements for the GTPase domain of the mitofusin Fzo1 and the ubiquitin ligase SCFMdm30 in mitochondrial outer membrane fusion. J Cell Sci. 2011;124:1403–1410. doi: 10.1242/jcs.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anton F, Dittmar G, Langer T, Escobar-Henriques M. Two deubiquitylases act on mitofusin and regulate mitochondrial fusion along independent pathways. Mol Cell. 2013;49:487–498. doi: 10.1016/j.molcel.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Yue W, Chen Z, Liu H, Yan C, Chen M, Feng D, Yan C, Wu H, Du L, Wang Y, Liu J, Huang X, Xia L, Liu L, Wang X, Jin H, Wang J, Song Z, Hao X, Chen Q. Small natural molecule uncovers the regulatory mechanism for mitochondrial fusion through inhibition of a deubiquitinase. Cell Res. 2013 doi: 10.1038/cr.2014.20. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiedemann N, Stiller SB, Pfanner N. Activation and degradation of mitofusins: two pathways regulate mitochondrial fusion by reversible ubiquitylation. Mol Cell. 2013;49:423–425. doi: 10.1016/j.molcel.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 43.Wrighton KH. Organelle dynamics: deubiquitylating mitofusin. Nat Rev Mol Cell Biol. 2013;14:130–131. doi: 10.1038/nrm3524. [DOI] [PubMed] [Google Scholar]
- 44.Escobar-Henriques M. Ubiquitylation of mammalian mitofusins promotes mitochondrial fusion. Cell Res. 2014 doi: 10.1038/cr.2014.23. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fritz S, Weinbach N, Westermann B. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol Biol Cell. 2003;14:2303–2313. doi: 10.1091/mbc.E02-12-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dürr M, Escobar-Henriques M, Merz S, Geimer S, Langer T, Westermann B. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol Biol Cell. 2006;17:3745–3755. doi: 10.1091/mbc.E06-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol Biol Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shutt T, Geoffrion M, Milne R, McBride HM. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 2012;13:909–915. doi: 10.1038/embor.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lokireddy S, Wijesoma IW, Teng S, Bonala S, Gluckman PD, McFarlane C, Sharma M, Kambadur R. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, Nabi IR. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013;24:1153–1162. doi: 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 53.Trempe JF, Sauvé V, Grenier K, Seirafi M, Tang MY, Ménade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 54.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, Ashrafi K, Glickman MH, Rutter J. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esaki M, Ogura T. Cdc48p/p97-mediated regulation of mitochondrial morphology is Vms1p-independent. J Struct Biol. 2012;179:112–120. doi: 10.1016/j.jsb.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 59.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B, Huang J, Li HL, Liu T, Wang YY, Waterman P, Mao AP, Xu LG, Zhai Z, Liu D, Marrack P, Shu HB. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–910. doi: 10.1038/cr.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park YY, Lee S, Karbowski M, Neutzner A, Youle RJ, Cho H. Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J Cell Sci. 2010;123:619–626. doi: 10.1242/jcs.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2-and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitra K. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. BioEssays. 2013;35:955–964. doi: 10.1002/bies.201300011. [DOI] [PubMed] [Google Scholar]
- 66.Otera H, Ishihara N, Mihara K. New insights into the function and regulation of mitochondrial fission. Biochim Biophys Acta. 2013;1833:1256–1268. doi: 10.1016/j.bbamcr.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Horn SR, Thomenius MJ, Johnson ES, Freel CD, Wu JQ, Coloff JL, Yang CS, Tang W, An J, Ilkayeva OR, Rathmell JC, Newgard CB, Kornbluth S. Regulation of mitochondrial morphology by APC/CCdh1-mediated control of Drp1 stability. Mol Biol Cell. 2011;22:1207–1216. doi: 10.1091/mbc.E10-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui M, Tang X, Christian WV, Yoon Y, Tieu K. Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J Biol Chem. 2010;285:11740–11752. doi: 10.1074/jbc.M109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, Zhou J, Chen Q. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lämmermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, Bouman L, Vogt-Weisenhorn D, Wurst W, Tatzelt J, Haass C, Winklhofer KF. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarz TL. Mitochondrial trafficking in neurons. Cold Spring Harb Perspect Biol. 2013;5:1–15. doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lovas JR, Wang X. The meaning of mitochondrial movement to a neuron's life. Biochim Biophys Acta. 2013;1833:184–194. doi: 10.1016/j.bbamcr.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 76.Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- 77.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weihofen A, Thomas KJ, Ostaszewski BL, Cookson MR, Selkoe DJ. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry. 2009;48:2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, Takahashi R, Hattori N, Imai Y, Lu B. Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Twig G, Liu X, Liesa M, Wikstrom JD, Molina AJ, Las G, Yaniv G, Hajnoczky G, Shirihai OS. Biophysical properties of mitochondrial fusion events in pancreatic beta-cells and cardiac cells unravel potential control mechanisms of its selectivity. Am J Physiol Cell Physiol. 2010;299:C477–C487. doi: 10.1152/ajpcell.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 85.Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM) Biochim Biophys Acta. 2013;1833:213–224. doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 86.Ota K, Kito K, Okada S, Ito T. A proteomic screen reveals the mitochondrial outer membrane protein Mdm34p as an essential target of the F-box protein Mdm30p. Genes Cells. 2008;13:1075–1085. doi: 10.1111/j.1365-2443.2008.01228.x. [DOI] [PubMed] [Google Scholar]
- 87.Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kondo-Okamoto N, Ohkuni K, Kitagawa K, McCaffery JM, Shaw JM, Okamoto K. The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol Biol Cell. 2006;17:3756–3767. doi: 10.1091/mbc.E06-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005;24:717–729. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 92.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 93.Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, Inatome R, Yanagi S. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 94.Huang CY, Chiang SF, Lin TY, Chiou SH, Chow KC. HIV-1 Vpr triggers mitochondrial destruction by impairing Mfn2-mediated ER-mitochondria interaction. PLoS ONE. 2012;7:e33657. doi: 10.1371/journal.pone.0033657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr Opin Cell Biol. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bragoszewski P, Gornicka A, Sztolsztener ME, Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol Cell Biol. 2013;33:2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anand R, Langer T, Baker MJ. Proteolytic control of mitochondrial function and morphogenesis. Biochim Biophys Acta. 2013;1833:195–204. doi: 10.1016/j.bbamcr.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 98.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 100.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ertel F, Nguyen M, Roulston A, Shore GC. Programming cancer cells for high expression levels of Mcl1. EMBO Rep. 2013;14:328–336. doi: 10.1038/embor.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 107.Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 108.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 109.Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc Natl Acad Sci USA. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 116.Fedorowicz MA, de Vries-Schneider RL, Rüb C, Becker D, Huang Y, Zhou C, Alessi Wolken DM, Voos W, Liu Y, Przedborski S. Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy. EMBO Rep. 2014;15:86–93. doi: 10.1002/embr.201337294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ivatt RM, Whitworth AJ. The many faces of mitophagy. EMBO Rep. 2014;15:5–6. doi: 10.1002/embr.201338224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hasson SA, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, Wang C, Heman-Ackah SM, Hessa T, Guha R, Martin SE, Youle RJ. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, II, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun Y, Vashisht AA, Tchieu J, Wohlschlegel JA, Dreier L. Voltage-dependent anion channels (VDACs) recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J Biol Chem. 2012;287:40652–40660. doi: 10.1074/jbc.M112.419721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abeliovich H, Zarei M, Rigbolt KT, Youle RJ, Dengjel J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun. 2013;4:2789. doi: 10.1038/ncomms3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 126.Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, Martinou JC. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu X, Weaver D, Shirihai O, Hajnoczky G. Mitochondrial “kiss-and-run”: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Attaix D, Taillandier D. The missing link: mul1 signals mitophagy and muscle wasting. Cell Metab. 2012;16:551–552. doi: 10.1016/j.cmet.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 132.Chen Y, Dorn GW., II PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lees AJ, Hardy J, Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 134.Blesa J, Phani S, Jackson-Lewis V, Przedborski S. Classic and new animal models of Parkinson's disease. J Biomed Biotechnol. 2012;2012:845618. doi: 10.1155/2012/845618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Exner N, Lutz AK, Haass C, Winklhofer KF. Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31:3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rakovic A, Shurkewitsch K, Seibler P, Grunewald A, Zanon A, Hagenah J, Krainc D, Klein C. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J Biol Chem. 2013;288:2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frank-Cannon TC, Tran T, Ruhn KA, Martinez TN, Hong J, Marvin M, Hartley M, Treviño I, O'Brien DE, Casey B, Goldberg MS, Tansey MG. Parkin deficiency increases vulnerability to inflammation-related nigral degeneration. J Neurosci. 2008;28:10825–10834. doi: 10.1523/JNEUROSCI.3001-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc Natl Acad Sci USA. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Grenier K, McLelland GL, Fon EA. Parkin-and PINK1-dependent mitophagy in neurons: will the real pathway please stand up? Front Neurol. 2013;4:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joselin AP, Hewitt SJ, Callaghan SM, Kim RH, Chung YH, Mak TW, Shen J, Slack RS, Park DS. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- 144.Müller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, Lichtenthaler SF, Motori E, Hrelia S, Wurst W, Trümbach D, Langer T, Krappmann D, Dittmar G, Tatzelt J, Winklhofer KF. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 145.Sato M, Sato K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim Biophys Acta. 2013;1833:1979–1984. doi: 10.1016/j.bbamcr.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 146.Wolff JN, Gemmell NJ. Mitochondria, maternal inheritance, and asymmetric fitness: why males die younger. BioEssays. 2013;35:93–99. doi: 10.1002/bies.201200141. [DOI] [PubMed] [Google Scholar]
- 147.Sutovsky P, McCauley TC, Sutovsky M, Day BN. Early degradation of paternal mitochondria in domestic pig (Sus scrofa) is prevented by selective proteasomal inhibitors lactacystin and MG132. Biol Reprod. 2003;68:1793–1800. doi: 10.1095/biolreprod.102.012799. [DOI] [PubMed] [Google Scholar]
- 148.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod. 2000;63:582–590. doi: 10.1095/biolreprod63.2.582. [DOI] [PubMed] [Google Scholar]
- 149.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 150.Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 151.Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, Ouyang YC, Hou Y, Schatten H, Sun QY. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci USA. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Koshiba T. Mitochondrial-mediated antiviral immunity. Biochim Biophys Acta. 2013;1833:225–232. doi: 10.1016/j.bbamcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 153.Berke IC, Li Y, Modis Y. Structural basis of innate immune recognition of viral RNA. Cell Microbiol. 2013;15:386–394. doi: 10.1111/cmi.12061. [DOI] [PubMed] [Google Scholar]
- 154.Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hou FJ, Sun LJ, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:841–841. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sasaki O, Yoshizumi T, Kuboyama M, Ishihara T, Suzuki E, Kawabata S, Koshiba T. A structural perspective of the MAVS-regulatory mechanism on the mitochondrial outer membrane using bioluminescence resonance energy transfer. Biochim Biophys Acta. 2013;1833:1017–1027. doi: 10.1016/j.bbamcr.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 157.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 159.Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol. 2010;184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 160.Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, Cheng X, Zhang Y, Yang X, Wang Y, Wen C, Wu Q, Shi W, Zhong H. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol. 2010;185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 161.Wang P, Yang L, Cheng G, Yang G, Xu Z, You F, Sun Q, Lin R, Fikrig E, Sutton RE. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 2013;3:1057–1070. doi: 10.1016/j.celrep.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 163.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]


