Abstract
Although pregnancy-associated microchimerism is known to exist in humans, its clinical significance remains unclear. Fetal microchimerism has been documented in rhesus monkeys, but the trafficking and persistence of maternal cells in the monkey fetus and infant have not been fully explored. To investigate the frequency of maternal microchimerism in the rhesus monkey (Macaca mulatta), a real-time polymerase chain reaction (PCR) strategy was developed and validated to target polymorphic major histocompatibility complex (MHC) gene sequences. Informative PCR assays were identified for 19 of 25 dams and their respective offspring. Analyses were performed on tissues (thymus, liver, spleen, lymph nodes, and bone marrow) and peripheral blood mononuclear cells (PBMCs) collected prenatally and postnatally in a subset of animals. Seven of 19 monkeys had detectable maternal microchimerism in at least one compartment (range: 0.001–1.9% chimeric cells). In tissues, maternal microchimerism was found in 2 of 7 fetuses and 3 of 12 juveniles (1–1.5 years of age), and most of the animals that were positive had microchimeric cells in more than one tissue. Maternal microchimerism was detected in PBMCs from all (4 of 4) fetuses. These observations suggest that maternal microchimerism occurs in the rhesus monkey fetus and can be detected in tissues in a subset of offspring after birth.
Keywords: major histocompatibility complex, microchimerism, quantitative PCR, rhesus monkey, transplacental transfer
Introduction
During pregnancy, cells traffic bi-directionally by transplacental transport between the mother and the fetus.1 In humans, a small population of maternal cells can be detected in fetuses, representing maternal microchimerism.2-4 Conversely, the presence of fetal cells in the mother is known as fetal microchimerism.5,6 It has been suggested that trafficking of fetal antigens in the maternal circulation is important for the establishment of maternal-fetal tolerance, thus ensuring full term pregnancies without rejection of the fetus.7 Fetal T cells are specifically nonresponsive to non-inherited maternal alloantigens through the action of regulatory T cells.8 Central tolerance mechanisms may also be involved in the nonresponsiveness to non-inherited maternal alloantigens in chimeric subjects.9 The bi-directional exchange of cells during pregnancy can have a lasting impact on tolerance, which has been shown in transplantation studies.10 Fetal microchimerism in humans has been associated with the development of autoimmune diseases such as scleroderma.1 Thus, the ability to accurately detect and quantify microchimeric cells may be informative for investigations of the biological significance of microchimerism from a developmental and transplantation perspective.
Detection of microchimerism has typically relied on assays that distinguish between the genetic material of maternal and fetal cells, e.g., sex chromosome-specific assays to allow discrimination between mothers and male offspring.1,11,12 Alternatively, assays specific for polymorphic alleles in the major histocompatibility complex (MHC) or insertion-deletion (In-Del) polymorphisms have been used to detect microchimerism.13-15 Techniques such as fluorescence in situ hybridization, flow cytometry, or quantitative polymerase chain reaction (qPCR) allow for detection and measurement of microchimeric populations by cell counting and/or by comparison against known standards, with qPCR offering the advantage of higher throughput, better sensitivity, and reagent versatility.
Pregnancy-associated microchimerism has been extensively described in humans and in rhesus monkeys.16-18 Previous reports have clearly demonstrated the importance of rhesus monkeys for these studies because they directly parallel findings in humans and provide an important translational model.19 While the use of sensitive assays targeting the Y chromosome has proven highly informative for the assessment of fetal microchimerism and to confirm engraftment of cells post-transplantation,20,21 there is a need for additional assays that can target a larger population of animals regardless of gender. Given the association between microchimerism and immune tolerance as well as the importance of the rhesus monkey as a model of human health and disease, we sought to develop a technique capable of detecting and quantifying maternal microchimerism in the rhesus monkey. In this study, a panel of qPCR assays was designed and validated based on genetic polymorphisms in rhesus monkey MHC loci.
Results
Analysis of maternal microchimerism levels in rhesus monkeys using Mamu-MHC allele-specific qPCR assays
Maternal microchimerism testing was initially performed on 25 monkeys. Six offspring were excluded from analysis because maternal allele-specific qPCR assays could not be identified. Microchimerism analysis was performed on DNA extracted from tissues (thymus, liver, spleen, lymph nodes, and bone marrow) collected prenatally from seven fetuses and postnatally from 12 juveniles. In the latter group, testing was also done on DNA extracted from peripheral blood mononuclear cells (PBMCs) collected both prenatally and postnatally from four of 12 fetuses, whereas for the remaining eight neonates, testing was done on DNA extracted from PBMCs collected only at or after birth.
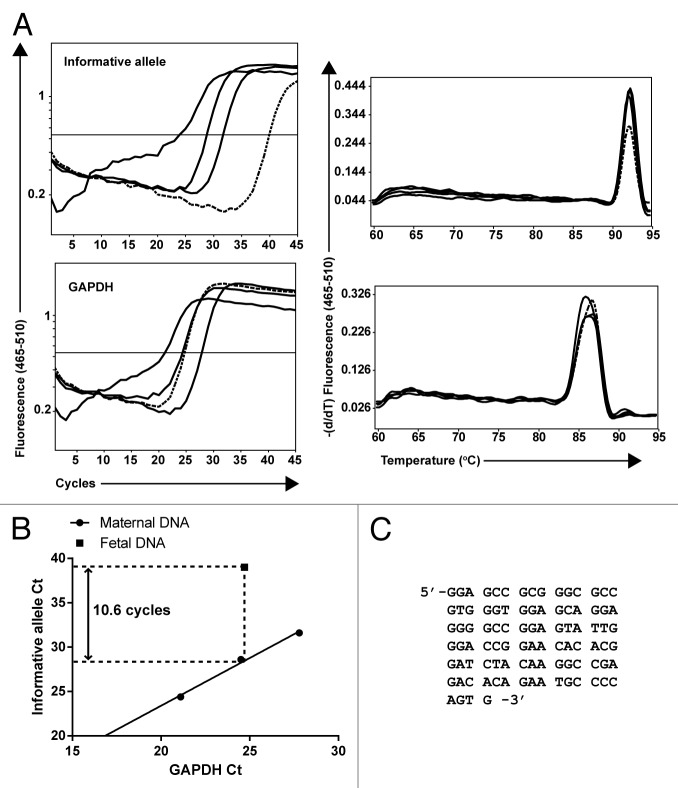
Each maternal-fetal pair was first genotyped using a panel of Mamu-MHC allele-specific qPCR assays (see Materials and Methods) to identify assays informative for investigating maternal microchimerism. Subsequently, the fetal DNA sample (containing at least 105 genomic equivalents of input DNA) and three 10-fold serial dilutions of maternal DNA were amplified in parallel using the assays for the informative MHC alleles and for GAPDH. Melting curve analysis and gel electrophoresis were used to verify that any signal detected was specific in both fetal and maternal DNA samples, and not due to nonspecific products or primer dimer artifacts. Results for a representative assay from one pair are shown in Figure 1A. The GAPDH assay Ct for each maternal sample dilution was plotted against the MHC informative allele assay Ct. A standard curve was generated and analyzed by performing linear regression. For the example shown in Figure 1B, the frequency of the maternal allele within the fetal sample, calculated using the ΔCt method, was 2-10.6 or 0.06%. To further verify specificity of the signal detected in the fetal sample, the PCR products amplified from the maternal and fetal DNA samples were purified and sequenced. The sequence obtained from the fetal sample was identical to that from the maternal sample (Figure 1C).
Figure 1. Analysis of maternal microchimerism in fetal rhesus monkey samples using informative Mamu-MHC allele assays. (A) Three 10-fold serial dilutions of maternal DNA (solid lines) were amplified with the informative allele assay identified during the initial screening and with the GAPDH assay to control for genomic input. In parallel, a concentration of fetal DNA equivalent to that used in the intermediate diluted maternal sample (dashed line) was amplified using the same assays. The specificity of the maternal microchimeric signal detected in the fetal sample was verified by dissociation analysis (right panels). (B) The Ct values from amplification of the maternal dilutions with the informative allele and GAPDH assays (circles) were plotted to create a standard curve used to calculate the level of maternal microchimerism in the fetal sample (square). In the example presented, there is a 10.6 cycle difference between the informative allele signal detected in the fetal sample and the extrapolated signal for an equivalent genomic input of maternal sample. The calculated microchimerism level in the fetal sample is 2-10.6 = 0.06%. (C) The products amplified from maternal and fetal DNA were purified and sequenced using the amplification primers. A full-length sequence was obtained from both the maternal and fetal sample as shown. The sequence obtained from the fetal sample was identical to the sequence obtained from the maternal sample.
Detection of maternal microchimerism in fetal rhesus monkey tissues
Ten maternal-fetal pairs were screened to identify informative alleles to quantify the level of maternal microchimerism in DNA extracted from fetal tissues, as described above. Three pairs were excluded from analysis because an informative allele assay could not be identified. The remaining seven fetuses were tested for maternal microchimerism in the thymus, spleen, liver, mesenteric lymph nodes, and bone marrow. In some cases, more than one informative allele was identified. In each case where maternal microchimerism was detected, direct sequencing of the PCR product (for informative allele assays yielding a product longer than 100 bp) or cloning followed by sequencing (for short amplicon assays) was used to confirm the 100% identity of the PCR product detected in the fetal sample relative to the product amplified from the maternal sample. Furthermore, in some cases, paternal DNA was also amplified and sequenced in parallel, using the informative allele assays to rule out the possibility that weak amplification of the inherited paternal allele in the fetal DNA was misinterpreted as maternal microchimerism. In those cases, paternal alleles were clearly distinguishable from maternal alleles by at least six nucleotide mismatches (data not shown).
A summary of maternal microchimerism levels in fetal tissues is presented in Table 1. Two of seven tested had detectable levels of microchimerism in at least one of the tissues analyzed (Table 1): in one (#2), microchimerism was detected in all five tissues with a range of 0.005% to 0.07%; in another (#6), a low-level positive result near the limit of detection was identified in the bone marrow in one of three independent experiments, each using two different assays, suggesting that a single copy or a small number of copies of the target sequence was present in the aliquots tested for that fetus. Within the limits of detection of the assays (ranging from 0.001% to 0.05%), no maternal microchimerism was detected in any tissue in five of the fetuses. Importantly, a similar number of cells was tested for each sample. Several of the informative allele-specific PCR assays demonstrated cross-amplification to a minor extent of MHC alleles in the fetal DNA other than the targeted non-inherited maternal alleles, despite amplification at relatively high annealing temperatures (Table 1). Sequencing of the amplified products allowed discrimination between maternal-specific DNA and weak cross-reactivity to fetal DNA.
Table 1. Maternal microchimerism detected in prenatal and postnatal tissues.
| Animal # | Sample | Informative allele assay | Maternal microchimerism, %* | ||||
|---|---|---|---|---|---|---|---|
| BM | LIV | THY | LN | SP | |||
| 1 | Prenatal | B24–540F + B28–600R† DQB*18:04‡ |
0 NS |
0 NS |
NS 0 |
0 0 |
0 0 |
| 2 | B42–206F + B30–255R B46–414F + B0–536R† |
0.0426 ± 0.0525 NS |
0.0384 ± 0.0228 NS |
0.0683 ± 0.0459 NS |
0.0078 ± 0.0056 NS |
0.0046 ± 0.0015 NS |
|
| 3 | For1 + rev51§ | 0 | 0 | 0 | 0 | 0 | |
| 4 | For1 + 1802rev1║ DQB*18:02¶ |
0 0 |
0 0 |
0 0 |
0 0 |
ND ND |
|
| 5 | A4*14F + A4*14R# | 0 | 0 | 0 | 0 | 0 | |
| 6 | B24–540F + B28–600R A03–113F + A03–249R† DQB*18:17F1+R2 |
+** NS +** |
0 NS 0 |
0 NS 0 |
0 0 0 |
0 NS 0 |
|
| 7 | B42–206F + B30–255R† DQB*06:01F154+R285†† |
NS 0 |
NS NS |
NS NS |
NS 0 |
NS 0 |
|
| 9 | Postnatal | DQB*18:11 | 0.0420 ± 0.0268 | 0 | 1.9177 ± 0.0977 | 0 | 0 |
| 11 | DPB1*06 | 0 | 0 | 0 | 0 | 0.0042 ± 0.0028 | |
| 17 | DQB*18:01 | 0 | 0.0084 ± 0.0037 | 0.0265 ± 0.0097 | 1.2593 ± 0.3005 | 0.1107 ± 0.1317 | |
BM = bone marrow; LIV = liver; THY = thymus; LN = mesenteric lymph nodes; SP = spleen; NS, non-specific; and ND, not determined. *At least two independent experiments were performed for each sample. Results are shown as mean ± standard deviation. †Non-specific signal detected, determined by sequencing PCR products from maternal and fetal DNA. ‡Limit of detection 0.05% calculated by spiking 10-fold serial dilutions of maternal DNA in background fetal DNA negative for the target allele. §Limit of detection 0.005%. ║Limit of detection 0.03%. ¶Limit of detection 0.003%. #Limit of detection 0.02%. **Detectable in 1 of 3 independent experiments (and shown by sequencing to be a true positive, +). ††Limit of detection 0.001%.
Transient maternal microchimerism levels in rhesus monkey PBMCs
Fifteen rhesus monkey maternal-infant/juvenile pairs were screened for informative alleles that could allow quantification of maternal microchimerism in DNA extracted from PBMCs. After exclusion of three pairs where an informative allele assay could not be identified, 12 pairs were tested for maternal microchimerism in PBMCs. Four offspring had both prenatal and postnatal samples available for microchimerism analyses whereas the remaining eight focused on postnatal samples because the fetal samples were used for other study-related assays. In the early third trimester, fetal blood was collected by cardiocentesis (left ventricle) under ultrasound guidance, an approach that minimizes the potential for maternal blood contamination. At this time point, maternal microchimerism was detected in fetal PBMCs from all four fetuses assessed (0.0236 ± 0.0056%, 0.0153 ± 0.0123%, 0.0011 ± 0.0008%, 0.0389 ± 0.0086% for #8–11, respectively). No microchimerism was detected in umbilical cord blood from these four animals at birth or in PBMCs postnatally (range 4 to 51 weeks). Eight other offspring were tested for maternal microchimerism in PBMCs at birth and subsequent postnatal time points. No microchimerism was detected in these eight infants/juveniles at any of the time points assessed (data not shown).
Persistence of maternal microchimerism in rhesus monkey tissues postnatally
Twelve juveniles assessed for microchimerism in PBMCs were also analyzed for maternal microchimerism in tissues (e.g., thymus, spleen, liver, mesenteric lymph nodes, and bone marrow) at ~1–1.5 years of age. Maternal microchimerism ranging from 0.004% to 1.9% was detected in at least one tissue in three out of 12 evaluated (Table 1). Microchimerism levels and tissue distribution varied widely between each of the animals.
Discussion
The results of this study suggest that it is possible to successfully quantify maternal microchimerism via an MHC allele-specific qPCR assay in rhesus monkeys. In many cases, multiple informative alleles were identified, increasing the likelihood of identifying primer pairs capable of detecting low levels of microchimerism with a high level of confidence.
In humans, microchimerism analysis based on HLA assays relies on initial genotyping of family members to identify non-inherited informative alleles, often using well-characterized sequence-specific oligonucleotide methods on commercial platforms.22 In rhesus monkeys, there are as many as 22 active MHC class I genes (compared with six in humans) and 10-fold higher sequence divergence in MHC relative to humans.23 This complexity renders MHC typing of rhesus monkeys difficult with current methodologies. A recent report described the use of massively parallel pyrosequencing for macaque MHC genotyping,24 although the widespread adoption of this technique may be limited because of the current cost. Despite the complexity of macaque MHC, this study has shown that a panel of 24 real-time allele-specific PCR assays targeting rhesus monkey MHC can be applied to analyze maternal microchimerism in this species.
In some cases, the qPCR assays used to analyze microchimerism cross-reacted with an inherited allele in the fetal sample at low efficiency, leading to misinterpretation of the level of microchimerism if the cross-reactive product had the same dissociation curve profile as the targeted non-inherited maternal allele. These cases could be ruled out as bona fide microchimerism by DNA sequencing. Each informative allele-specific qPCR assay was tested at several annealing temperatures to identify the optimal conditions for increased specificity at higher temperatures without a loss in sensitivity. While specificity could be further enhanced by introduction of an internal probe within the PCR product, SYBR Green chemistry was utilized in this study due to cost considerations.
Reports of chimerism in the setting of transfusion or transplantation rely on testing of a sample derived from the patient prior to treatment13 in order to demonstrate specificity through the absence of signal amplified using the informative assay. This type of control sample does not exist for analysis of maternal microchimerism. Rather, in some cases, paternal DNA was used as a control to demonstrate specificity of the assays for detecting maternal microchimerism. The use of this control was only informative for cases where the paternal DNA did not share the targeted non-inherited maternal allele. In other cases, where misinterpretation of maternal microchimerism could be due to weak cross-amplification of the inherited paternal or maternal allele, an appropriate control could not be identified. Furthermore, studies have failed to identify tissues that are consistently negative for microchimerism,25 precluding their use as a control for specificity. In fact, due to the persistence and variety of cell types identified as microchimeric in many studies,3,13,26 it is likely that cell trafficking between the mother and fetus includes stem or progenitor cells that can populate many different tissues. One strategy to obtain an animal-specific negative control sample for maternal microchimerism may be to clone a population of fetal cells in tissue culture by limiting dilution. This approach, however, is labor-intensive and exceeds the level of confidence described in most maternal microchimerism studies. Alternatively, droplet-based digital PCR technology27 may allow discrimination of maternal microchimeric DNA from background fetal DNA and accurate quantification of microchimerism, due to its ability to partition samples into a large number of nanodroplet reactions each potentially containing a single target DNA molecule.
A limitation of our method is that microchimerism can only be detected in the setting of maternal disparity at one or more target alleles. This method, by its very nature of employing non-inherited MHC polymorphisms, cannot detect microchimerism if the dam is homozygous at the targeted MHC locus or if the mother and fetus share a given allele. Since the MHC plays a critical role in regulating immune tolerance, it seems likely that the potential for microchimeric cells to engraft could be influenced by the context of the MHC relationship between mother and fetus. In fact, studies in mice have shown that the pattern of maternal-fetal histocompatibility is associated with differences in the microchimerism levels detected.28,29
Analysis of microchimerism in mice has been facilitated by the availability of transgenic strains whose cells can easily be tracked in a large background of wild-type cells by amplification of a transgene, such as green fluorescent protein (GFP),30 neomycin resistance,29 or firefly luciferase.25 This approach, however, is not currently feasible in the rhesus monkey model. Non-MHC polymorphisms have been used to identify chimerism in humans, such as the use of insertion-deletion31 and single-nucleotide polymorphisms,32,33 and Y-chromosome assays.26 The sequencing of the rhesus macaque genome and development of databases for genomic data analysis34,35 provide valuable resources for future design of non-MHC based PCR assays informative for detection of microchimerism in monkeys.
Previous work in humans and animal models has shown that pregnancy-associated microchimerism is a common event. In humans, maternal microchimerism was observed in 15 of 18 fetal lymph node samples tested8 while in mice, maternal microchimerism was noted in 51 of 60 animals in various organs within three weeks after birth.25 It is important to note that the mouse has a yolk sac placenta which differs from the placental structure of human and nonhuman primates.36 In our study, seven of 19 rhesus monkeys with informative alleles were positive for maternal microchimerism in at least one of the tested compartments, a considerably lower fraction. Human data also suggest that maternal cells may be found in a variety of fetal tissues.37 Of the two rhesus monkey fetuses that had detectable microchimerism, one showed the presence of maternal DNA in five of five tissues evaluated, indicating wide biodistribution.
It is possible that the assays employed in this study may be underestimating the frequency of maternal microchimerism in rhesus monkeys due to the difficulty of detecting microchimerism using MHC assays as a result of the relative complexity of the MHC locus in macaques compared with humans. Another possibility is that the number of cells used for screening limited the ability to detect more prevalent microchimerism in monkeys. As for the level of maternal microchimerism previously reported within a sample, it was shown to range from 0.0035–0.83% in human fetal lymph nodes8 and averaged around 0.16% in mice.25 Furthermore, in rhesus monkeys, fetal cells have been detected in the CD34+ fraction of maternal blood at an average level of 0.009%.18 The level of maternal microchimerism detected in the current study falls within the lower end of the range reported in these prior studies in regard to fetal monkey tissues, although results were higher in postnatal monkey tissues.
It is also possible that maternal or fetal administration of nonpathogenic SIVmac1A11 (described in Materials and Methods) could have skewed the measurement of microchimerism reported here by altering the relative abundance of immune cells in response to circulating virus. This possibility, however, is not likely because SIVmac1A11 is a nonpathogenic molecular clone that does not result in disease; maternal complete blood counts were all within normal limits (data not shown); and monkey #9 was a control (no exposure to SIVmac1A11) and clearly showed maternal microchimerism in peripheral blood within the range of the other animals tested (0.0153% vs. 0.0011–0.0389%, respectively). These examples suggest that exposure to nonpathogenic SIVmac1A11 did not impact the levels of microchimerism. While developmentally different than primates, a study focused on microchimerism in pigs, including those experimentally infected with porcine reproductive and respiratory syndrome virus, also suggested that infection did not alter the findings; it should be noted that the number of pigs in this study was small.38 Similarly, the relatively low number of monkeys in the current study with detectable microchimerism does not provide a definitive conclusion, although the results suggest that nonpathogenic SIV did not alter the outcomes.
It has been shown that maternal microchimeric cells engraft in fetal tissues and persist for decades in humans,3 similar to findings in rhesus monkeys.18 The study described herein suggests that maternal microchimerism in the peripheral blood of rhesus monkeys may be primarily detectable during gestation, comparable to fetal microchimerism. Since maternal microchimerism was detectable in PBMCs from all fetuses (4/4) evaluated but was only detectable in tissue samples from a fraction of animals (5/19) it is possible that tissue microchimerism may arise directly from maternal cells in the blood, but further studies would be required to explore this possibility.
In conclusion, these studies have demonstrated that a panel of MHC sequence-specific qPCR assays can be readily applied to the detection of pregnancy-associated maternal microchimerism in rhesus monkeys and that maternal microchimerism can be detected in postnatal tissues up to ~1.5 years of age. This method expands the current toolkit available for studies of fetal:maternal cell trafficking in this important translational model of human health and disease.
Materials and Methods
Animals
All animal procedures conformed to the requirements of the Animal Welfare Act and protocols were approved prior to implementation by the Institutional Animal Care and Use Committee at the University of California, Davis. Normally cycling, adult female rhesus monkeys (Macaca mulatta) (n = 25) with a history of prior pregnancy were selected based on negative testing for Mamu-A*01, A*02, B*08, and B*17 alleles, and bred with males that were positive for Mamu-A*01 and A*02 alleles and negative for B*08 and B*17 alleles. Pregnancies were established by timed mating and detected by ultrasound.39 Pregnancy in the rhesus monkey is divided into trimesters by 55-day increments, with 0–55 days of gestation representing the first trimester, 56–110 days of gestation representing the second trimester, and 111–165 days of gestation the third trimester (term 165 ± 10 days).40 Activities related to animal care (e.g., diet and housing) were performed according to California National Primate Research Center standard operating procedures (SOPs). Fetal growth and development were monitored by ultrasound, fetal blood was collected by cardiocentesis under ultrasound guidance in the early third trimester using established techniques,39 and maternal blood samples were collected during gestation and at pregnancy termination. Hysterotomies were performed (n = 10) to collect fetal tissues, using established protocols41 based on study assignment, and included controls (n = 4) or fetuses of dams that had been administered nonpathogenic SIVmac1A11 by the intraperitoneal (IP) route in the late first trimester (n = 6). Fetal body weights and measures were assessed after the collection of fetal blood, then all organs including the brain, lung, heart, thymus, spleen, liver, lymph nodes (axillary, inguinal, mesenteric), pancreas, adrenals, kidneys, reproductive tract including gonads, gastrointestinal tract (stomach, duodenum, jejunum, ileum, colon), skin, muscle, and bone marrow were removed and select organs weighed. A placental evaluation was also performed, and sections of the umbilical cord, membranes, decidua, and placenta obtained. Cell suspensions from thymus, liver, spleen, lymph nodes, and bone marrow were prepared according to established protocols.41
Cesarean sections were also performed at term (n = 15) using established methods and included collection of umbilical cord blood and a placental evaluation.42 Newborns were raised in the nursery according to established SOPs, with blood samples collected from a peripheral vessel at defined time points (weekly or monthly) under ketamine sedation (10 mg/kg).43 PBMCs were purified from whole blood (of the dam, fetus, or infant/juvenile) by density centrifugation over a Histopaque gradient (Sigma, Catalog #10771). Isolated cells were subsequently cryopreserved in fetal bovine serum (FBS) supplemented with 10% dimethyl sulfoxide (DMSO) prior to DNA extraction. Of these 15 animals, seven were fetal controls and received either pathogenic SIVmac239 or SIVmac251 postnatally (as part of a parallel series of studies). For five, either the dam (intravenous) or the fetus (IP) was administered nonpathogenic SIVmac1A11 in the late first trimester, and three were administered a lentiviral vector prenatally (IP) and SIVmac251 postnatally. Tissue harvests were performed according to established protocols at ~1 to 1.5 years postnatal age and five collected tissues were analyzed from each animal (thymus, liver, spleen, mesenteric lymph nodes, and bone marrow). Tissues were collected and weighed as described above, and cell suspensions prepared according to established protocols as noted.
DNA extraction for PCR genotyping
An estimated 5x106 cells were washed in 1 ml PBS and digested in 500 μl of PCR solution (100 mM KCl, 10 mM Tris pH 8.3, 2.5 mM MgCl2, 1% Tween-20, 1% NP40) and 75 μg of protease (Qiagen Inc., Cat#19155) at 60 °C for 1.5 h. Protease was inactivated by heating at 95 °C for 30 min. For typing reactions, genomic DNA was diluted 1:50 in PCR solution to a concentration of approximately 1000 genomic equivalents/5 μl.
Design and development of Mamu-MHC allele-specific real-time PCR assays
Four A locus primers were designed based on polymorphic regions in published Mamu-A gene sequences.44 Likewise, 10 B locus primers were designed based on polymorphic regions in Mamu-B gene sequences included within the Immuno Polymorphism Database.45,46 All A and B locus primers are located within the highly variable second and third exons. The A locus primers were combined to form two assays and the B locus primers were combined into six assays, each of which targets a single MHC class I allele. In addition, we also used techniques kindly provided by Dr. David Watkins, Department of Pathology, University of Miami Miller School of Medicine, as well as published assays,47-49 to form a panel consisting of 12 MHC allele-specific qPCR assays and one GAPDH reference assay used as a loading control. In addition to the MHC allele-specific assay panel, we performed sequence-based typing of Mamu-DQB for each animal in these studies and designed qPCR assays specific for the non-inherited DQB maternal allele for 12 maternal-fetal pairs. The real-time PCR primer sequences and amplicon characteristics (e.g., size and dissociation temperature) are shown in Table 2.
Table 2. Real-time PCR primer sequences and characteristics of amplicons used to identify informative alleles for microchimerism.
| Real-time PCR assay | Forward primer sequence (5′→3′) | Reverse primer sequence (5′→3′) | Amplicon size, bp | Dissociation temp, °C |
|---|---|---|---|---|
| MHC class I | ||||
| A03–113F + A03–249R | GCGACGCCGAGAGTCCGAGAGA | GCCGCGCAGGTTCCGCAGGGC | 137 | 92.7 |
| A04–271F + A02–487R | GGGTCTCACACCTACCAGGT | CGCCCTCCAGGTAGGTTCTGTGCTG | 217 | 93.0 |
| B42–206F + B30–255R | GACGCCGAGAGTCCGAGGAT | CCGTGTTCCGTGTCTGCTC | 88 | 91.2 |
| B0–206F + B0–278R | GACGCCGCGAGTCCGAGAGA | TCGGTCAGTCTGTGCCTGGG | 111 | 90.7 |
| B0–206F + B0–311R | GACGCCGCGAGTCCGAGAGA | GTTGTAGTAGCCGCGCAGGT | 144 | 92.1 |
| B46–414F + B24–536R | GCCTCCTCCGCGGGTACCGG | CTCCAGGTAGGCTCTGAACC | 161 | 92.2 |
| B46–414F + B44–558R | GCCTCCTCCGCGGGTACCGG | GGAGCCACTCCACGCACGTC | 183 | 93.0 |
| B24–540F + B28–600R | ACCGTTATGCGGAGCGGTTC | CTTCCCGTTCTCCAGGTGTC | 80 | 88.0 |
| A4*14F + A4*14R* | GGGACCCGACGGGCGCCTCCAA | GGCCCTCCAGGTAGACTCTGTC | 179 | 92.9 |
|
MHC class II |
||||
| DPB1*06F + DPB1*06R† | ATCTACAATCGGGAAGAGTACGT | CCTCGTCCAGTTCGTAGTTGTA | 172 | 90.8 |
| For1 + rev51‡ | CCTGTGCTACTTCACCAACG | ACCTCGTAGTTGTGCA | 252 | 92.4 |
| For1 + 1802rev1‡ | CCTGTGCTACTTCACCAACG | AGTCGTGCGGAGCTCTGA | 267 | 92.1 |
| DQB1*06:01 F154 + R285 | TGGACGGAGCGCGTGCGTTA | GCTGTTCCAGTACTCGGCGT | 132 | 90.9 |
| DQB1*06:01 F278 + R379 | GGAACAGCCAGAAGGACGTG | CTCTCCTCTGCAAGATCCC | 102 | 88.5 |
| DQB1*06:02 F241 + R374 | GCGGTGACGCCGCAGGGGCG | CTCTGCAAGATCCCGCGGTA | 134 | 91.1 |
| DQB1*15:01 F1 + R2 | GAGCGCGTGTGGAGTGTAGA | TGTTCCAGTACTCGGCACTG | 124 | 90.6 |
| DQB1*16:03 F41 + R99 | AGAGCGCGTGCGGTTAGTAG | CGAAGCGCGCGAACTCTTCG | 59 | 86.3 |
| DQB1*18:01 F2 + R2 | GCGGTGACGCCGCAGGGGCA | TGTATGCACACCGTGTCCAA | 98 | 89.1 |
| DQB1*18:02 F208 + R285 | GTGCGCTTCGACAGCGACTG | GCTGTTCCAGTACTCTGCAG | 78 | 92.1 |
| DQB1*18:04 F172 + R268 | CTTGTGACCAGACACATTTC | CGTCAGGTCGCCCCAGCGGA | 97 | 89.8 |
| DQB1*18:11 F248 + R335 | CGCCGCTGGGGCGGTCTTGG | CTGCACACCGTGTCCACCGA | 88 | 88.1 |
| DQB1*18:17 F1 + R2 | GTCTTGTGACCAGACACGTT | TTGCACACCGTGTCCACCGA | 166 | 92.9 |
| DQB1*18:17 F45 + R172 | GGGACGGAGCGCGTGCGTCT | TTCCAGTACTCGGCGCTAGG | 128 | 91.9 |
| DQB1*18:22 F192 + R270 | GAGAGCACCCGGGCGGAGGC | CTCGCCGCTGCAACGTCGT | 79 | 89.8 |
|
Reference |
||||
| GAPDHF + GAPDHR§ | GCACCACCAACTGCTTAGCAC | TCTTCTGGGTGGCAGTGATG | 105 | 86.0 |
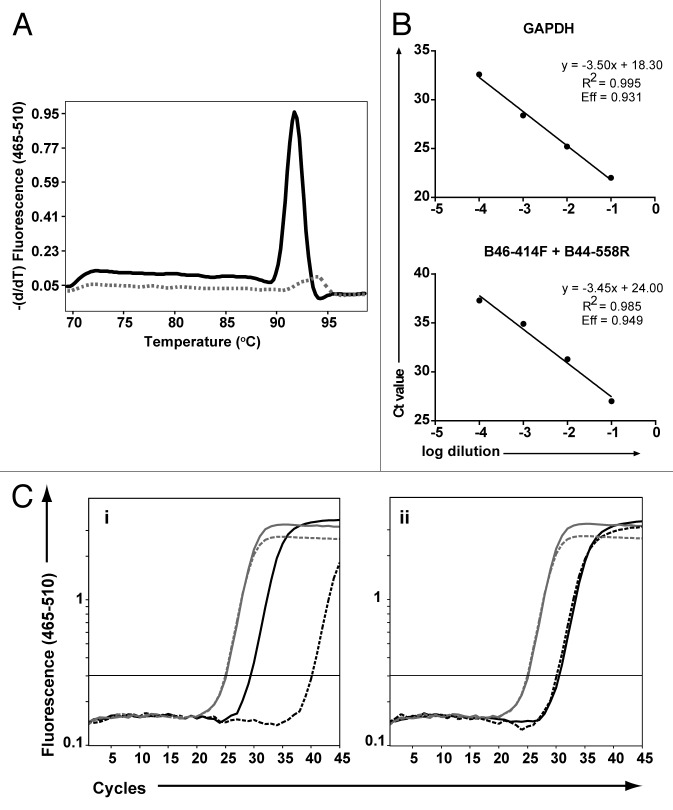
To validate the utility of the MHC allele-specific assay panel for detection of microchimerism, the assays were used to screen rhesus monkey DNA from at least five different animals. As shown by the melting curve analysis from one representative assay for Mamu-A1*003 (Figure 2A), the primer pairs were informative for distinguishing between rhesus monkey samples known to be positive or negative for this allele. The predicted sizes of the amplified products were verified on an ethidium-stained agarose gel (data not shown). Amplification of serial dilutions of DNA with the Mamu-MHC allele-specific assays showed linearity over at least four orders of magnitude. The linear dynamic range for the reference assay for GAPDH and a representative allele-specific MHC PCR assay (designed to target B*047) are shown in Figure 2B. To determine the limit of detection of maternal DNA in a fetal sample, serial dilutions of rhesus macaque DNA positive for the targeted MHC allele were spiked into a fixed amount of DNA negative for the targeted allele. The relative amount of DNA in each sample was determined by GAPDH qPCR using the 2-ΔCt method. The sensitivity of the Mamu-MHC allele-specific PCR assays, defined as the lowest amount of DNA positive for the targeted MHC allele detected in a background of DNA negative for the targeted allele, ranged from 0.001% to 0.05% (data not shown).
Figure 2. Performance of Mamu-MHC allele-specific real-time PCR assays used to identify microchimerism. (A) Samples from five different monkeys were amplified using A03–113F + A03–249R as a representative Mamu-MHC assay. Dissociation curves are shown for one animal positive for the target sequence (solid black curve) and one animal negative for the target sequence (dashed gray curve). (B) Four 10-fold serial dilutions of rhesus monkey DNA positive for the target sequence were spiked in PCR buffer (for GAPDH amplification) or in background rhesus monkey DNA negative for the target sequence (for MHC amplification). Standard curves are shown for GAPDH (used to control for total genomic input) and a representative Mamu-MHC assay. Amplification efficiency (Eff) was calculated based on the slope of the standard curve. (C) DNA from maternal-fetal monkey pairs was amplified with the assays shown in Table 2. Amplification curves are shown for GAPDH (in gray) to control for genomic input. Maternal samples are shown by solid lines and fetal samples are indicated by dashed lines. Examples of informative (Ci) and non-informative (Cii) allele assays for detection of maternal microchimerism are shown in black. The maternal sample is positive for the target sequence whereas the fetal sample may be positive for maternal microchimerism (Ci) or for the target sequence (Cii).
Identification of informative alleles for maternal microchimerism by real-time PCR
To determine which assays were informative for detection of maternal microchimerism, the MHC allele-specific qPCR panel was used to genotype samples from maternal-fetal rhesus monkey pairs. Five μl of the diluted DNA lysate were added to 10 μl of PCR buffer containing 5 mM MgCl2, 1 mM dNTPs (Bioline USA Inc., Catalog #39029), 1 μM of each primer (Integrated DNA Technologies), 0.25X SYBR Green I (Invitrogen, Catalog #S7585), and 0.7 U FastStart Taq (Roche Applied Science, Catalog #04738420001). Real-time PCR was performed in a 384-well format on a sequence detection system (LightCycler 480 II, Roche) with the following cycle conditions: 1 min at 95 °C followed by 45 cycles of 30 s at 95 °C, 30 s at 60 °C, and 45 s at 72 °C, with a melting curve analysis at the end of the reaction. Any weak maternal-specific signal detected in the fetus was considered as possibly derived from microchimeric cells that crossed the placenta from the dam into the fetus. An assay was defined as “informative” for maternal microchimerism if a Ct between 20 and 30 was detected in the maternal sample and if the Ct of the fetal sample was either undetectable or at least five cycles higher than that of the maternal sample for equivalent maternal and fetal DNA input. Examples of informative and non-informative alleles identified by qPCR genotyping are presented in Figure 2C, i and ii, respectively. Once informative alleles were identified, 10-fold serial dilutions of the DNA lysate were amplified in a 96-well format using different annealing temperatures ranging from 56–70 °C. Optimal annealing temperatures were chosen for each assay based on increased specificity at higher temperatures without a loss in sensitivity.
PCR analysis and quantification
To quantify the level of maternal microchimerism, fetal DNA and 10-fold serial dilutions of maternal DNA samples diluted in PCR solution were amplified with primers for informative allele(s) and GAPDH. Quantification of maternal microchimeric cells in fetal DNA was calculated using the ΔCt method with GAPDH as a reference gene. The specificity of the amplified products in the fetal samples was determined by melting curve analysis and gel electrophoresis in comparison to amplified products in the maternal samples.
Sequencing of PCR products
Thirty-five μl of the PCR product were electrophoresed in a 2% agarose gel containing ethidium bromide. The band was excised and purified using a QIAquick PCR purification kit (Qiagen, Catalog #28106). The purified PCR product was eluted into 30 μl of water and 20 μl was submitted with the 5′ and 3′ primers used during amplification for automated sequencing (MCLAB). Alternatively, the PCR product was directly purified using a QIAquick PCR purification kit prior to sequencing with the amplification primers. For amplicons shorter than 100 bp, the PCR product was cloned into pCRTM4-TOPO® using the TOPO® TA Cloning® kit for sequencing (Life Technologies, Catalog #K4575) and for transformation of One Shot® TOP10 chemically competent E. coli. Transformants were analyzed by PCR using M13 Forward(-20) and M13 Reverse primers and visualized by agarose gel electrophoresis. Amplicons of the correct size were purified using a QIAquick PCR purification kit prior to sequencing with the M13 primers. Sequence-based typing of Mamu-DQB1 was performed by amplifying exon 2 (270 bp) using the primers DQBF (5′-TCCCCGCAGA GGATTTCGTG-3′) and DQB*18R (5′-CGCTCACCTC GCCGCTGCAA-3′) and sequencing the PCR products using the amplification primers. Sequence analysis was performed using Sequencher® version 5.0 software (Gene Codes Corporation). Mamu-DQB sequences for each maternal-fetal pair were aligned and primers specific for the non-inherited maternal DQB allele were designed such that there was a 1–3 nucleotide mismatch at each primer 3′ end relative to the fetal DQB sequences.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors wish to thank Dr. David Watkins for providing PCR sequence-specific primers and Dr. Terry Ng for cloning assistance and expertise. These studies were supported by grants from the National Institutes of Health (#AI084109 and #AI090677) (to J.M.M.) and the California National Primate Research Center base-operating grant (#OD011107). J.M.M. is the recipient of the National Institutes of Health Director’s Pioneer Award as part of the National Institutes of Health Roadmap for Medical Research (#OD00329).
Glossary
Abbreviations:
- PCR
polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- MHC
major histocompatibility complex
- PBMCs
peripheral blood mononuclear cells
Footnotes
Previously published online: www.landesbioscience.com/journals/chimerism/article/27778
References
- 1.Gammill HS, Nelson JL. Naturally acquired microchimerism. Int J Dev Biol. 2010;54:531–43. doi: 10.1387/ijdb.082767hg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall JM, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Detection of maternal cells in human umbilical cord blood using fluorescence in situ hybridization. Blood. 1995;86:2829–32. [PubMed] [Google Scholar]
- 3.Maloney S, Smith A, Furst DE, Myerson D, Rupert K, Evans PC, Nelson JL. Microchimerism of maternal origin persists into adult life. J Clin Invest. 1999;104:41–7. doi: 10.1172/JCI6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saadai P, Lee TH, Bautista G, Gonzales KD, Nijagal A, Busch MP, Kim CJ, Romero R, Lee H, Hirose S, et al. Alterations in maternal-fetal cellular trafficking after fetal surgery. J Pediatr Surg. 2012;47:1089–94. doi: 10.1016/j.jpedsurg.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schröder J. Transplacental passage of blood cells. J Med Genet. 1975;12:230–42. doi: 10.1136/jmg.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzenberg LA, Bianchi DW, Schröder J, Cann HM, Iverson GM. Fetal cells in the blood of pregnant women: detection and enrichment by fluorescence-activated cell sorting. Proc Natl Acad Sci U S A. 1979;76:1453–5. doi: 10.1073/pnas.76.3.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlebacher A. Mechanisms of T cell tolerance towards the allogeneic fetus. Nat Rev Immunol. 2013;13:23–33. doi: 10.1038/nri3361. [DOI] [PubMed] [Google Scholar]
- 8.Mold JE, Michaëlsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, Lee TH, Nixon DF, McCune JM. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–5. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yunis EJ, Zuniga J, Romero V, Yunis EJ. Chimerism and tetragametic chimerism in humans: implications in autoimmunity, allorecognition and tolerance. Immunol Res. 2007;38:213–36. doi: 10.1007/s12026-007-0013-3. [DOI] [PubMed] [Google Scholar]
- 10.Dutta P, Burlingham WJ. Microchimerism: tolerance vs. sensitization. Curr Opin Organ Transplant. 2011;16:359–65. doi: 10.1097/MOT.0b013e3283484b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianchi DW, Flint AF, Pizzimenti MF, Knoll JH, Latt SA. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci U S A. 1990;87:3279–83. doi: 10.1073/pnas.87.9.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, Lee TH. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion. 2001;41:1524–30. doi: 10.1046/j.1537-2995.2001.41121524.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee TH, Paglieroni T, Ohto H, Holland PV, Busch MP. Survival of donor leukocyte subpopulations in immunocompetent transfusion recipients: frequent long-term microchimerism in severe trauma patients. Blood. 1999;93:3127–39. [PubMed] [Google Scholar]
- 14.Lambert NC, Erickson TD, Yan Z, Pang JM, Guthrie KA, Furst DE, Nelson JL. Quantification of maternal microchimerism by HLA-specific real-time polymerase chain reaction: studies of healthy women and women with scleroderma. Arthritis Rheum. 2004;50:906–14. doi: 10.1002/art.20200. [DOI] [PubMed] [Google Scholar]
- 15.Lee TH, Chafets DM, Reed W, Wen L, Yang Y, Chen J, Utter GH, Owings JT, Busch MP. Enhanced ascertainment of microchimerism with real-time quantitative polymerase chain reaction amplification of insertion-deletion polymorphisms. Transfusion. 2006;46:1870–8. doi: 10.1111/j.1537-2995.2006.00992.x. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez DF, Tarantal AF. Quantitative analysis of male fetal DNA in maternal serum of gravid rhesus monkeys (Macaca mulatta) Pediatr Res. 2003;53:18–23. doi: 10.1203/00006450-200301000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez DF, Tarantal AF. Fetal gender determination in early first trimester pregnancies of rhesus monkeys (Macaca mulatta) by fluorescent PCR analysis of maternal serum. J Med Primatol. 2003;32:315–9. doi: 10.1046/j.1600-0684.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez DF, Leapley AC, Lee CI, Ultsch MN, Tarantal AF. Fetal CD34+ cells in the maternal circulation and long-term microchimerism in rhesus monkeys (Macaca mulatta) Transplantation. 2005;79:142–6. doi: 10.1097/01.TP.0000144468.71962.AA. [DOI] [PubMed] [Google Scholar]
- 19.Lo YM. Fetal DNA in maternal plasma/serum: the first 5 years. Pediatr Res. 2003;53:16–7. doi: 10.1203/00006450-200301000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Reitsma MJ, Harrison MR, Pallavicini MG. Detection of a male-specific sequence in nonhuman primates through use of the polymerase chain reaction. Cytogenet Cell Genet. 1993;64:213–6. doi: 10.1159/000133579. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Lan F, Gan J, Yao X, Reisner Y. Donor-type chimerism determination by competitive polymerase chain reaction (PCR) in a primate model for bone marrow transplantation. Transplantation. 1999;68:1573–7. doi: 10.1097/00007890-199911270-00023. [DOI] [PubMed] [Google Scholar]
- 22.Chan WF, Atkins CJ, Naysmith D, van der Westhuizen N, Woo J, Nelson JL. Microchimerism in the rheumatoid nodules of patients with rheumatoid arthritis. Arthritis Rheum. 2012;64:380–8. doi: 10.1002/art.33358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–15. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, et al. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–6. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su EC, Johnson KL, Tighiouart H, Bianchi DW. Murine maternal cell microchimerism: analysis using real-time PCR and in vivo imaging. Biol Reprod. 2008;78:883–7. doi: 10.1095/biolreprod.107.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996;93:705–8. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George D, Czech J, John B, Yu M, Jennings LJ. Detection and quantification of chimerism by droplet digital PCR. Chimerism. 2013;4:102–8. doi: 10.4161/chim.25400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonney EA, Matzinger P. The maternal immune system’s interaction with circulating fetal cells. J Immunol. 1997;158:40–7. [PubMed] [Google Scholar]
- 29.Kaplan J, Land S. Influence of maternal-fetal histocompatibility and MHC zygosity on maternal microchimerism. J Immunol. 2005;174:7123–8. doi: 10.4049/jimmunol.174.11.7123. [DOI] [PubMed] [Google Scholar]
- 30.Khosrotehrani K, Johnson KL, Guégan S, Stroh H, Bianchi DW. Natural history of fetal cell microchimerism during and following murine pregnancy. J Reprod Immunol. 2005;66:1–12. doi: 10.1016/j.jri.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Alizadeh M, Bernard M, Danic B, Dauriac C, Birebent B, Lapart C, Lamy T, Le Prisé PY, Beauplet A, Bories D, et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood. 2002;99:4618–25. doi: 10.1182/blood.V99.12.4618. [DOI] [PubMed] [Google Scholar]
- 32.Lo YM, Lo ES, Watson N, Noakes L, Sargent IL, Thilaganathan B, Wainscoat JS. Two-way cell traffic between mother and fetus: biologic and clinical implications. Blood. 1996;88:4390–5. [PubMed] [Google Scholar]
- 33.Maas F, Schaap N, Kolen S, Zoetbrood A, Buño I, Dolstra H, de Witte T, Schattenberg A, van de Wiel-van Kemenade E. Quantification of donor and recipient hemopoietic cells by real-time PCR of single nucleotide polymorphisms. Leukemia. 2003;17:621–9. doi: 10.1038/sj.leu.2402856. [DOI] [PubMed] [Google Scholar]
- 34.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 35.Malhi RS, Sickler B, Lin D, Satkoski J, Tito RY, George D, Kanthaswamy S, Smith DG. MamuSNP: a resource for Rhesus Macaque (Macaca mulatta) genomics. PLoS One. 2007;2:e438. doi: 10.1371/journal.pone.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsey EM, Houston ML, Harris JW. Interactions of the trophoblast and maternal tissues in three closely related primate species. Am J Obstet Gynecol. 1976;124:647–52. doi: 10.1016/0002-9378(76)90068-5. [DOI] [PubMed] [Google Scholar]
- 37.Jonsson AM, Uzunel M, Götherström C, Papadogiannakis N, Westgren M. Maternal microchimerism in human fetal tissues. Am J Obstet Gynecol. 2008;198:e1–6. doi: 10.1016/j.ajog.2007.09.047. [DOI] [PubMed] [Google Scholar]
- 38.Karniychuk UU, Van Breedam W, Van Roy N, Rogel-Gaillard C, Nauwynck HJ. Demonstration of microchimerism in pregnant sows and effects of congenital PRRSV infection. Vet Res. 2012;43:19. doi: 10.1186/1297-9716-43-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarantal AF. Ultrasound imaging in rhesus (Macaca mulatta) and long-tailed (Macaca fascicularis) macaques: reproductive and research applications. In: Wolfe-Coote S, ed. The Laboratory Primate. London: Academic Press, 2005:317-52. [Google Scholar]
- 40.Tarantal AF, Gargosky SE. Characterization of the insulin-like growth factor (IGF) axis in the serum of maternal and fetal macaques (Macaca mulatta and Macaca fascicularis) Growth Regul. 1995;5:190–8. [PubMed] [Google Scholar]
- 41.Jimenez DF, Lee CI, O’Shea CE, Kohn DB, Tarantal AF. HIV-1-derived lentiviral vectors and fetal route of administration on transgene biodistribution and expression in rhesus monkeys. Gene Ther. 2005;12:821–30. doi: 10.1038/sj.gt.3302464. [DOI] [PubMed] [Google Scholar]
- 42.Tarantal AF, Goldstein O, Barley F, Cowan MJ. Transplantation of human peripheral blood stem cells into fetal rhesus monkeys (Macaca mulatta) Transplantation. 2000;69:1818–23. doi: 10.1097/00007890-200005150-00015. [DOI] [PubMed] [Google Scholar]
- 43.Tarantal AF, McDonald RJ, Jimenez DF, Lee CC, O’Shea CE, Leapley AC, Won RH, Plopper CG, Lutzko C, Kohn DB. Intrapulmonary and intramyocardial gene transfer in rhesus monkeys (Macaca mulatta): safety and efficiency of HIV-1-derived lentiviral vectors for fetal gene delivery. Mol Ther. 2005;12:87–98. doi: 10.1016/j.ymthe.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Lobashevsky AL, Thomas JM. Six mamu-A locus alleles defined by a polymerase chain reaction sequence specific primer method. Hum Immunol. 2000;61:1013–20. doi: 10.1016/S0198-8859(00)00177-4. [DOI] [PubMed] [Google Scholar]
- 45.Non-Human Primates IPD-MHC. (NHP): The Immuno Polymorphism Database [Internet]. Cambridge: European Bioinformatics Institute. 2003 - [cited 2010 Nov 30]. Available from: http://www.ebi.ac.uk/ipd/mhc/nhp/
- 46.Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2013;41:D1234–40. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vigon N, Sauermann U. Sequence-based typing techniques for rhesus macaque MhcMamu-DQB1 allow the identification of more than 35 alleles. Tissue Antigens. 2002;59:88–94. doi: 10.1034/j.1399-0039.2002.590203.x. [DOI] [PubMed] [Google Scholar]
- 48.Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A. 2005;102:1626–31. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcondes MC, Burdo TH, Sopper S, Huitron-Resendiz S, Lanigan C, Watry D, Flynn C, Zandonatti M, Fox HS. Enrichment and persistence of virus-specific CTL in the brain of simian immunodeficiency virus-infected monkeys is associated with a unique cytokine environment. J Immunol. 2007;178:5812–9. doi: 10.4049/jimmunol.178.9.5812. [DOI] [PubMed] [Google Scholar]