Abstract
Background
Tumor recurrence remains the major clinical complication of meningiomas, the majority of recurrences occurring among WHO grade I/benign tumors. In the present study, we propose a new scoring system for the prognostic stratification of meningioma patients based on analysis of a large series of meningiomas followed for a median of >5 years.
Methods
Tumor cytogenetics were systematically investigated by interphase fluorescence in situ hybridization in 302 meningioma samples, and the proposed classification was further validated in an independent series of cases (n = 132) analyzed by high-density (500K) single-nucleotide polymorphism (SNP) arrays.
Results
Overall, we found an adverse impact on patient relapse-free survival (RFS) for males, presence of brain edema, younger patients (<55 years), tumor size >50 mm, tumor localization at intraventricular and anterior cranial base areas, WHO grade II/III meningiomas, and complex karyotypes; the latter 5 variables showed an independent predictive value in multivariate analysis. Based on these parameters, a prognostic score was established for each individual case, and patients were stratified into 4 risk categories with significantly different (P < .001) outcomes. These included a good prognosis group, consisting of approximately 20% of cases, that showed a RFS of 100% ± 0% at 10 years and a very poor-prognosis group with a RFS rate of 0% ± 0% at 10 years. The prognostic impact of the scoring system proposed here was also retained when WHO grade I cases were considered separately (P < .001).
Conclusions
Based on this risk-stratification classification, different strategies may be adopted for follow-up, and eventually also for treatment, of meningioma patients at different risks for relapse.
Keywords: iFISH, meningioma, recurrence, risk stratification, SNP arrays
Meningiomas are usually considered to be slowly growing, clinically benign tumors that can be cured by conventional surgical procedures.1–3 However, between 10% and 30% of patients who undergo complete tumor resection and around 60% of patients who undergo subtotal tumor resection show tumor recurrence at 10 years in association with a significantly poorer overall survival.4,5 So far, multiple different independent prognostic factors have been identified in meningiomas, and some prognostic scoring systems have been proposed6–10 to predict the outcome of individual patients at the time of diagnosis.2 Among such prognostic factors, tumor cytogenetics, together with WHO tumor grade, extent of tumor resection, patient age, and tumor localization have proven to be particularly informative.4,11–14
Although risk stratification, based on individual prognostic factors and some combinations of prognostic factors,6,15,16 has been proven effective for predicting patient outcome, it has not been fully adopted in routine clinical practice. Among other reasons is the fact that both the individual risk factors and prognostic scores proposed so far have still failed to predict the outcome for a significant proportion of patients included in both the low- and the high-risk categories. For example, atypical and anaplastic tumors show a greater recurrence rate than WHO grade I meningiomas, but the majority of meningioma relapses (around 80% of all recurrences) still occur among WHO grade I cases.14 Similarly, when using the Maillo et al15 score, which was developed on the basis on chromosome 14 abnormalities, patient age, and tumor histopathology, a significant number of recurrences are still observed among the good-prognosis category, particularly in WHO grade I/benign meningiomas (eg, approximately 12% of all relapses). At the same time, a significant fraction of the high-risk cases (eg, approximately 50% of high-risk WHO grade I tumors) are relapse-free long-term survivors. Based on all of the above, there is an urgent need for a more reliable risk stratification classification for meningiomas to provide adequate definition of the most efficient follow-up strategies and the potential adoption of different treatment approaches on an individual patient basis.
Here we analyzed a series of 302 meningioma cases with a median follow-up of more than 5 years for the most relevant clinical and biological features of the disease, including tumor cytogenetics as assessed by interphase fluorescence in situ hybridization (iFISH). Our major goal was to identify a combination of prognostic factors that could be used to stratify meningioma patients according to their risk of recurrence. Such risk stratification should therefore allow identification of both a good-prognosis group of patients who do not require follow-up and a poor-prognosis group for whom closer monitoring, including potential adoption of additional treatment measures, would be required for early diagnosis and/or prevention of tumor recurrence, particularly among histologically benign/grade I meningiomas. An additional group of 132 cases, studied by single nucleotide polymorphism (SNP) arrays, was included in this study for validation purposes.
Materials and Methods
Patients and Samples
Overall, 302 patients (91 males, 211 females; mean age of 60 ± 15y; range, 6y–87y), who had been diagnosed with meningioma at the Neurosurgery Service of the University Hospital of Salamanca, were prospectively included in this study. Participants were enrolled in the study after informed consent had been given in accordance with the guidelines of the local Ethics Committee and the Declaration of Helsinki. The great majority of participants (283/302; 94%) underwent complete tumor resection; this included 35 of 38 grade II/III tumors (n = 30 and 8 cases, respectively). Adjuvant radiotherapy was given, in addition to surgery, to 21 participants with WHO grade II/III tumors. One participant with anaplastic meningioma also received systemic chemotherapy in addition to radiotherapy.
Histological tumor diagnosis was performed according to the WHO criteria.17 At the moment of closing this study, 42 of 302 participants (14%) had relapsed after a median follow-up of 65 months. From the whole series, 41 cases were excluded from survival analyses because follow-up data were not available (n = 30 cases) or the participant had died shortly after surgery (n = 11 cases). Follow-up studies were performed according to a standard clinicobiological protocol that included MRI techniques performed 3 months after surgery and every 12 months thereafter. Additional MRI studies were performed whenever clinical signs and/or symptoms were noted and/or a relapse was suspected. Immediately after surgical removal, part of the fresh tumor tissue was frozen in liquid nitrogen and stored at −80°C until used for iFISH and/or SNP array studies. In addition, EDTA-anticoagulated peripheral blood (PB) samples were also obtained from each participant at diagnosis and processed in parallel with the tumor.
iFISH Studies
For all freshly frozen tumor samples obtained after surgery, iFISH analyses were performed to identify the numerical alterations of 11 distinct chromosomes by using a panel of probes specific for those chromosomes and chromosomal regions more frequently altered in meningiomas (1p36/1q25, 7, 9p34, 10, 14q32.3, 15q22, 17q21, 18q21, 22q11.2, X and Y). All probes were obtained from Vysis Inc. and were used in double stainings, as previously described in detail:18 LSI BCR/ABL dual-color probe for chromosomes 9 and 22; LSI PML/RAR-α dual-color probe for chromosomes 15 and 17; LSI IgH/BCL2 dual-color probe for chromosomes 14 and 18; 1p36/1q25 dual-color probe for chromosome 1; CEP 7 and 10 DNA probes conjugated with Spectrum Orange and Spectrum Green, respectively; and CEP X (Spectrum Orange) and CEP Y (Spectrum Green) for chromosomes X and Y, respectively.
Copy Number Alterations by SNP Arrays
Copy number (CN) alterations were analyzed by SNP arrays in a subset of 50 samples previously reported in the literature using the GeneChip Human Mapping 250K Nsp and 250K Sty arrays (Affymetrix) according to the manufacturer's instructions, as previously described in detail.19 DNA from paired (frozen) tumor tissue and normal PB samples was purified using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions. DNA purity and integrity were determined with a NanoDrop-1000 spectrophotometer (Nano-Drop Technologies) and by conventional electrophoretic procedures in 1% agarose gel, respectively. The SNP call rate per array was always ≥92% (range, 92%–99.8%). Overall, 200 .CEL files were obtained. In addition, another external series of 82 meningiomas20 analyzed by 100K Affymetrix SNP arrays, which also had data about relevant tumor characteristics and patient survival available at the GEO public database (access code: GSE16583), was also included in this part of the study. Follow-up data were available for 108 of 132 of these cases, and 17 of them (13%) had relapsed after a median follow-up of 59 months (range, 1–115 months).
For the analysis of SNP array data, the GCOS version 1.3 (Affymetrix), the Copy Number Analysis Tool (CNAT v4.0, Affymetrix), dChip 2007 (http//www.dchip.org; Dana Farber Institute), and the GeneChip Genotyping Analysis (GTYPE 4.1; Affymetrix) software programs were used. CN values were calculated for each SNP array and plotted according to chromosomal localization. Genotypes were generated using the BRLMM algorithm included in the Genotyping Console software version 3.0.2 (Affymetrix). Based on the results obtained from normal PB samples, cutoff values of ≤1.30 and ≥2.50 (arbitrary units) were used to establish CN losses and gains, respectively.
Statistical Methods
Standard deviation values, range, median and mean were calculated for all continuous variables, and frequencies were used for categorical variables. To establish the statistical significance of differences observed between groups, the Student t test and the Mann–Whitney U test were used for continuous variables, whereas the chi-square test was used for qualitative variables. Relapse free-survival (RFS) curves were plotted according to the method of Kaplan–Meier, and the (1-sided) log-rank test was used to establish the statistical significance of differences observed between survival curves. Multivariate analysis of prognostic factors for RFS was performed using the Cox stepwise regression model. In this part of the study, only those variables showing a significant association with RFS in the univariate analysis were included. For all statistical analyses, the SPSS version 15.0 software package was used (SPSS Inc). P values <.05 were considered to be associated with statistical significance.
Results
Histopathological Features and Cytogenetic Profile of Meningiomas
Two-hundred sixty-four meningiomas (87%) were WHO grade I tumors, while only 30 cases (10%) were grade II and 8 (3%) were grade III meningiomas. Around half of the tumors corresponded to meningothelial meningiomas (n = 147; 49%). The rest of the tumors were 57 (19%) transitional, 34 (11%) psammomatous, 24 (8%) atypical, 17 (5%) fibroblastic, 7 (2%) angioblastic, 5 (2%) anaplastic, 5 (2%) secretory, 3 (1%) chordoid, and 2 (1%) rhabdoid meningiomas; the remaining case was a papillary tumor. According to tumor localization, most meningiomas (n = 276; 91%) corresponded to intracranial tumors (35% cranial base, 21% convexity, 18% parasagittal, 12% falcine, 3% tentorial, and 2% intraventricular; only 26 (9%) were spinal meningiomas. Brain edema was found in 173 of 302 (57%) cases and evaluated according to its extension as light (smaller or equal to the volume of the tumor) in 72 cases (24%), moderate (double the volume of the tumor) in 65 cases (21%), and severe edema (more than twice the volume of the tumor) in 36 cases (12%).
From the 302 meningiomas analyzed, 90 (30%) displayed no cytogenetic alterations by iFISH for the 11 chromosomes analyzed. In contrast, the other 212 (70%) cases showed numerical alterations for ≥1 chromosome. As expected, chromosome 22 was the most frequently altered chromosome (173/302 cases; 57%), its alteration mainly consisted of monosomy 22/22q deletions (166/173 altered cases). Other recurrently altered chromosomes included chromosome Y in males (26/91 cases; 28%), chromosome 1p (67/302 cases; 22%), chromosome 14 (47/302 cases; 16%), chromosome X in females (28/211 cases; 13%), chromosome 1q (37/302 cases; 12%), and chromosome 10 (31/302 cases; 10%). Overall, chromosomal losses were more frequently observed than gains (55% vs 2% for chromosome 22, 26% vs 2% for chromosome Y, 21% vs 1% for chromosome 1p, 13% vs 3% for chromosome 14, 12% vs 1% for chromosome X, and 6% vs 4% for chromosome 10), except for chromosome 1q, which was more frequently gained (10%) than lost (2%). All other chromosomes analyzed were found to be altered at lower (≤8%) frequencies (Supplementary Table 1).
Around half of all cytogenetically altered cases (n = 106; 35%) showed isolated alterations of a single chromosome, whereas the other half (n = 106; 35%) displayed cytogenetic alterations (losses and/or gains) involving ≥2 chromosomes (Table 1). Among those cases carrying isolated chromosomal alterations, the most frequent pattern consisted of loss of one chromosome 22 or del(22q) (83/106 cases; 78%), while isolated involvement of other chromosomes was restricted to a few cases. Isolated loss of chromosome 1p was found in 9 tumors, loss of chromosome Y in 4, and loss of chromosome 10 in 2. Losses of chromosomes X and 9 were found in one case each, and; isolated gains of chromosomes 1q and 14 were detected in 5 and one cases, respectively.
Table 1.
Clinical and biological characteristics of the meningioma cases included in this study (n = 302) and their association with disease outcome in those 261 cases with available follow-up data
| Variables | Patient Distribution (%) | No. of Recurrences (%) | P value | % Patients Relapse-free |
75% RFS (months) | Univariate Analysis (P value) | Multivariate Analysis # |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| 5y-RFS* | 10y-RFS * | 15y-RFS * | P value | HR (95% CI) | ||||||
| Age | ||||||||||
| <55 years | 97 (32%) | 23 (24%) | .001 | 78% ± 5% | 70% ± 7% | 59% ± 9% | 85 | .005 | .01 | 3 (1–5) |
| ≥55 years | 205 (68%) | 19 (9%) | 92% ± 2% | 83% ± 4% | 77% ± 6% | NR | ||||
| Sex | ||||||||||
| Male | 91 (30%) | 18 (20%) | .04 | 80% ± 5% | 68% ± 8% | 63% ± 9% | 98 | .02 | ||
| Female | 211 (70%) | 24 (11%) | 90% ± 3% | 83% ± 4% | 73% ± 6% | 172 | ||||
| Cytogenetic profile | ||||||||||
| Diploid karyotype | 90 (30%) | 5 (6%) | <.001 | 94% ± 3% | 90% ± 6% | 78% ± 12% | NR | .001 | .002 | |
| One altered chromosome | 106 (35%) | 10 (9%) | 91% ± 3% | 88% ± 4% | 80% ± 7% | NR | ||||
| Monosomy22/del(22q) | 83 (27%) | 7 (8%) | 92% ± 4% | 92% ± 4% | 82% ± 8% | NR | 7 (1–33) | |||
| Other | 23 (8%) | 3 (13%) | 89% ± 8% | 74% ± 15% | 74% ± 15% | 82 | 15 (4–64) | |||
| Complex karyotype | 106 (35%) | 27 (25%) | 79% ± 5% | 63% ± 7% | 55% ± 8% | 78 | 17 (3–113) | |||
| WHO grade | ||||||||||
| Grade I | 264 (87%) | 28 (11%) | <.001 | 90% ± 2% | 82% ± 4% | 74% ± 5% | 172 | <.001 | ||
| Grade II | 30 (10%) | 12 (40%) | 64% ± 10% | 56% ± 11% | 37% ± 17% | 22 | <.001 | 7 (3–17) | ||
| Grade III | 8 (3%) | 2 (25%) | 86% ± 13% | – | – | 85 | 6 (1–30) | |||
| Tumor localization | ||||||||||
| Convexity | 63 (21%) | 10 (16%) | N.S. | 90% ± 5% | 76% ± 8% | 64% ± 11% | 122 | <.001 | <.001 | |
| Parasagittal | 56 (18%) | 7 (13%) | 91% ± 5% | 85% ± 7% | 78% ± 9% | 237 | ||||
| Falcine | 35 (12%) | 3 (9%) | 89% ± 6% | 89% ± 6% | 89% ± 6% | NR | ||||
| Cranial base (anterior) | 38 (13%) | 6 (16%) | 78% ± 9% | 78% ± 9% | 52% ± 22% | 172 | 17 (4–75) | |||
| Cranial base (middle) | 47 (15%) | 6 (13%) | 93% ± 5% | 76% ± 10% | 68% ± 12% | 127 | ||||
| Cranial base (posterior) | 22 (7%) | 3 (14%) | 82% ± 10% | 82% ± 10% | 82% ± 10% | NR | ||||
| Tentorial | 11 (3%) | 2 (18%) | 78% ± 14% | 78% ± 14% | – | NR | ||||
| Intraventricular | 4 (2%) | 3 (75%) | 0% | – | – | 9 | 17 (4–84) | |||
| Spinal | 26 (9%) | 2 (8%) | 96% ± 4% | 77% ± 18% | 77% ± 18% | NR | ||||
| Tumor histology | ||||||||||
| Meningothelial | 147 (49%) | 18 (12%) | <.001 | 90% ± 3% | 83% ± 4% | 75% ± 6% | 172 | <.001 | ||
| Fibroblastic | 17 (5%) | 2 (12%) | 92% ± 8% | 0% | 0% | 82 | ||||
| Transitional | 57 (19%) | 6 (11%) | 89% ± 6% | 74% ± 14% | 59% ± 17% | 109 | ||||
| Psammomatous | 34 (11%) | 1 (3%) | 95% ± 5% | 95% ± 5% | 95% ± 5% | NR | ||||
| Angioblastic | 7 (2%) | 0 (0) | 100% | 100% | – | NR | ||||
| Secretory | 5 (2%) | 1 (20%) | 67% ± 27% | – | – | 19 | ||||
| Atypical | 24 (8%) | 12 (50%) | 57% ± 11% | 49% ± 12% | 25% ± 18% | 21 | ||||
| Chordoid | 3 (1%) | 0 (0) | – | – | – | NR | ||||
| Anaplastic | 5 (2%) | 2 (40%) | 80% ± 18% | 40% ± 30% | – | 85 | ||||
| Rhabdoid | 2 (1%) | 0 (0) | – | – | – | NR | ||||
| Papillary | 1 (0) | 0 (0) | – | – | – | NR | ||||
| Edema | ||||||||||
| No | 128 (43%) | 11 (9%) | .03 | 92% ± 3% | 85% ± 5% | 82% ± 6% | NR | .01 | ||
| Yes | 173 (57%) | 31 (18%) | 83% ± 3% | 73% ± 5% | 59% ± 8% | 109 | ||||
| Tumor size | ||||||||||
| <30 mm | 66 (22%) | 3 (5%) | 96% ± 3% | 87% ± 9% | 87% ± 9% | NR | <.001 | .001 | ||
| 30–50 mm | 139 (47%) | 15 (11%) | <.001 | 93% ± 3% | 79% ± 6% | 76% ± 6% | 237 | |||
| >50 mm | 92 (31%) | 24 (26%) | 72% ± 6% | 69% ± 6% | 49% ± 10% | 51 | 5 (1–21) | |||
Results expressed as number of cases and percentage between brackets or as * percentage of cases ± SE (standard error);
# The category with the best prognosis was selected as reference group.
Abbreviations: CI, confidence interval; HR, hazard ratio; NR, 75% relapse-free survival not reached; RFS, relapse-free survival.
Except for chromosome 10 in the whole series and chromosomes Y and X in males, alterations of all other individual chromosomes were significantly more frequent among WHO grade II/III versus grade I tumors (P ≤ .001; Supplementary Table 1). In line with these findings, diploid tumors and cases with single chromosomal alterations were more frequently (P ≤ .001) observed among grade I meningiomas. Eighty-two of 90 (91%) diploid cases and 101 of 106 (95%) tumors with only one altered chromosome corresponded to WHO grade I tumors, while more complex cytogenetic patterns predominated among grade II/III meningiomas; 25 of 38 (66%) grade II/III tumors showed a complex karyotype. Of note, tumors with complex karyotypes represented more than half of the male cases (51/91 cases; 56%), whereas they only accounted for 26% of female cases (55/211 cases; P ≤ .001).
Prognostic Impact of Tumor Cytogenetics and Other Relevant Clinical and Histopathological Features of the Disease
Tumor cytogenetics, as assessed by iFISH, showed a significant association with the incidence of relapses and participants' RFS (Supplementary Table 1). Accordingly, alterations of chromosomes 1p, 1q, 7, 9, 10, 14, 18, and 22 in the whole series and chromosome X in females were associated with both a higher incidence of relapses (P < .05; Supplementary Table 1) and/or a significantly shorter RFS (P < .03; Supplementary Table 1 and Supplementary Fig. 1). Multivariate analysis, including all individual chromosomes with a significant impact on RFS in the univariate study, showed that only the alteration of chromosome 14 (P = .001), gains of chromosome 7 (P = .047) and losses of chromosome 18 (P < .001) had an independent predictive value for a poorer outcome (Supplementary Table 1). In turn, when the 3 major iFISH cytogenetic subgroups of meningiomas were considered, participants with tumors carrying a complex karyotype showed a significantly shorter RFS than those showing a diploid karyotype and presence of isolated chromosomal alterations (75% RFS of 78 months vs not reached, respectively; P = .001) (Table 1; Fig. 1H). Of note, individual chromosomes lost their independent prognostic value once these 3 cytogenetic profiles were included in the analysis (data not shown). Participants carrying tumors with complex karyotypes showed lower 5-, 10-, and 15-year RFS rates than those with diploid karyotypes or isolated chromosomal alterations (79% ± 5% vs 94% ± 3% and 91% ± 3%; 63% ± 7% vs 90% ± 6% and 88% ± 4%; and 55% ± 8% vs 78% ± 12% and 80% ± 7%, respectively) (Table 1). Of note, within those cases with an isolated chromosomal alteration, participants who had isolated monosomy 22/del(22q) showed a slightly better RFS than those with isolated alterations of other chromosomes: 75% RFS not reached versus 82 months (P > .05); 5-, 10- and 15-years RFS rates of 92% ± 4% versus 89% ± 8%; 92% ± 4% versus 74% ± 15%; and 82% ± 8% versus 74% ± 15%, respectively (Table 1). Of note, combined assessment of chromosomes 1p, 7, 14, 18, and 22 represented the minimal chromosomal panel that could be applied for identification of the 3 most common subtypes of genetic patterns used in the prognostic scoring system.
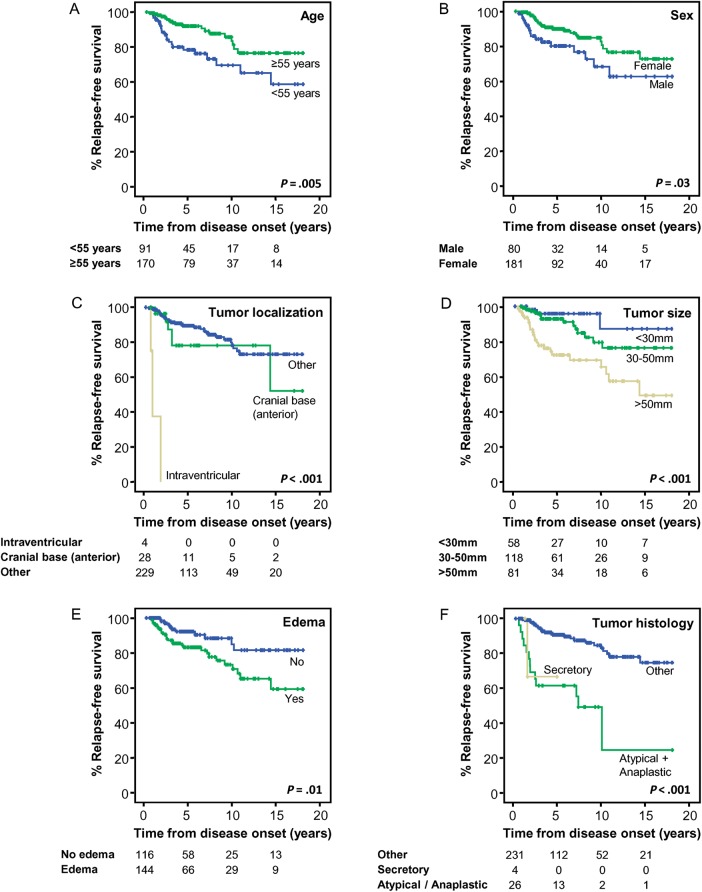
Fig. 1.
Clinical, biological, and genetic features of meningiomas that showed a significant impact on patient relapse-free survival (RFS; n = 261). Relapse-free survival curves of meningioma cases according to patient age (A), sex (B), tumor localization (C), and size (D), presence versus absence of edema (E), tumor histology (F), WHO grade (G), and interphase fluorescence in situ hybridization (iFISH) cytogenetic profile (H). The last 2 panels (I and J) show patients' RFS curves according to the new prognostic scoring system here proposed (I) and that previously reported by Maillo et al15 (J).
In addition to tumor cytogenetics, several other clinical and histopathological features of the disease also showed an adverse impact on participants' RFS, namely younger age (<55 years; P = .005), male sex (P = .02), WHO grade II/III tumor (P < .001), atypical or anaplastic tumor histopathology (P < .001), intraventricular or anterior cranial base tumor localization (P < .001), tumor size >50 mm (P < .001), and presence of brain edema (P = .01) (Table 1 and Fig. 1A–G). Multivariate analysis of prognostic factors showed that tumor cytogenetics (P = .002), together with the WHO grade (P < .001), localization (P < .001), size (P = .001), and participant age (P = .01), represented the best combination of independent prognostic factors for predicting RFS of meningioma cases (Table 1). Of note, the Simpson grade of resection did not show a significant (P > .05) impact on participant RFS.
Based on the above 5 variables (patient age, tumor cytogenetics, WHO grade, localization, and size), a prognostic score was established for each individual participant using the criteria described in Table 2. Once this score was applied, participants were divided into 4 risk groups: (i) low-risk cases with a score ≤1 (n = 62), (ii) intermediate-1 (int-1) cases with a score between 2 and 4 (n = 170), (iii) intermediate-2 (int-2) cases with a score between 5 and 6 (n = 59), and (iv) high-risk cases with a score ≥7 (n = 11). Participants included in these 4 risk categories showed progressively shorter 75% RFS rates (P < .001) from low (not reached) to int-1 (not reached), int-2 (38 months), and high-risk cases (15 months). Similarly, progressively lower 5-, 10- and 15-year RFS rates were also found for low (100% ± 0%); int-1 (93% ± 2%, 85% ± 5%, and 75% ± 7%, respectively); int-2 (70% ± 7%, 59% ± 16%, and 45% ± 11%, respectively), and high-risk cases (50% ± 16%, 0% ± 0%, and 0% ± 0%, respectively) (Table 2; Fig. 1I). For comparison purposes, RFS curves for the same series of cases, as defined according to the scoring system previously proposed by Maillo et al,15 are shown in Fig. 1J.
Table 2.
Scoring criteria used for those 5 variables included in the new prognostic scoring system proposed in this study
| Score | Age (years) | WHO grade | Cytogenetic Profile |
Tumor Size (mm) | Tumor Localization |
|||
|---|---|---|---|---|---|---|---|---|
| 0 | ≥55 | I | Diploid karyotype | <30 | Other | |||
| 1 | <55 | II | Monosomy 22/del(22q) | 30–50 | Cranial base (anterior) | |||
| 2 | III | One altered chromosome (other than Chr22) | >50 | Intraventricular | ||||
| 3 | Complex karyotype | |||||||
| Risk Group (overall score) | Patient Distribution (%) | No. of Recurrences (%) | P value | % of Patient Relapse-free | 75% RFS (months) | P value | ||
| 5y-RFS* | 10y-RFS* | 15y-RFS* | ||||||
| All WHO grade tumors | ||||||||
| Low (0–1) | 62 (21%) | 0 (0%) | <.001 | 100% ± 0% | 100% ± 0% | 100% ± 0% | NR | <.001 |
| Intermediate-1 (2–4) | 170 (56%) | 15 (9%) | 93% ± 2% | 85% ± 5% | 75% ± 7% | NR | ||
| Intermediate-2 (5–6) | 59 (20%) | 21 (36%) | 70% ± 7% | 59% ± 8% | 45% ± 11% | 38 | ||
| High (≥7) | 11 (4%) | 6 (55%) | 50% ± 16% | 0% ± 0% | 0% ± 0% | 15 | ||
| WHO grade I tumors | ||||||||
| Low (0–1) | 59 (22%) | 0 (0%) | <.001 | 100% ± 0% | 100% ± 0% | 100% ± 0% | NR | <.001 |
| Intermediate-1 (2–4) | 163 (62%) | 14 (9%) | 94% ± 2% | 85% ± 5% | 76% ± 7% | NR | ||
| Intermediate-2 (5–6) | 40 (15%) | 12 (30%) | 72% ± 8% | 61% ± 10% | 49% ± 15% | 51 | ||
| High (≥7) | 2 (1%) | 2 (100%) | 0% ± 0% | 0% ± 0% | 0% ± 0% | 15 | ||
Results expressed as number of cases and percentage between brackets or as *percentage of cases ± SE (standard error).
Abbreviations: NR, 75% relapse-free survival not reached; RFS, relapse-free survival.
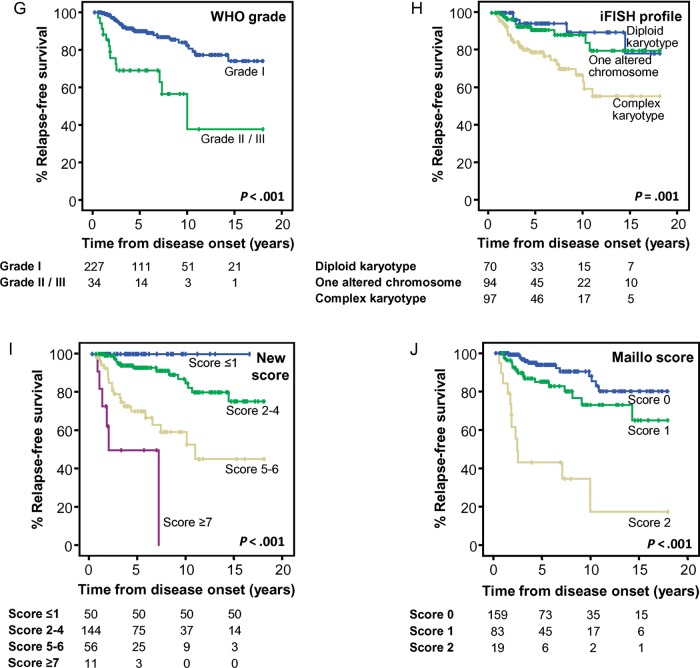
Most interestingly, when WHO grade I tumors were exclusively considered, tumor cytogenetics retained their prognostic significance (P = .01; Table 3; Fig. 2D) together with patient age, tumor localization, and size. Thus, complex karyotypes, age < 55 years, intraventricular and anterior cranial base localization, and a tumor size ≥50 mm were all associated with shorter 75% RFS rates both in the univariate (P = .01, P = .02, P < .001, and P = .002, respectively; Table 3; Fig. 2A–C) and in the multivariate analyses (P < .001, P = .007, P = .002 and P = .03, respectively; Table 3). When the proposed score was specifically adjusted for WHO grade I tumors (Table 2), the significantly different outcome of low-, int-1, int-2, and high-risk cases was retained, with 75% RFS rates of not reached, not reached, 51 months, and 15 months, respectively (P < .001; Table 2; Fig. 2E). This clearly improved the prediction obtained with the previously proposed Maillo et al score, particularly for the low-risk group (Fig. 2F).
Table 3.
Clinical and biological characteristics of WHO grade I meningiomas (n = 264) and their association with disease outcome in 227 cases with available follow-up data
| Variables | Patient Distribution (%) | No. of Recurrences (%) | P value | % Patients Relapse-free |
75% RFS (months) | Univariate Analysis (P value) | Multivariate Analysis # |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| 5y-RFS* | 10y-RFS* | 15y-RFS* | P value | HR (95% CI) | ||||||
| Age | ||||||||||
| <55 years | 81 (31%) | 15 (19%) | .009 | 83% ± 5% | 76% ± 7% | 63% ± 10% | 130 | 0.02 | .007 | 3 (1–8) |
| ≥55 years | 183 (69%) | 13 (7%) | 94% ± 2% | 86% ± 4% | 81% ± 5% | NR | ||||
| Sex | ||||||||||
| Male | 72 (27%) | 10 (14%) | NS | 87% ± 5% | 73% ± 9% | 66% ± 10% | 109 | NS | ||
| Female | 192 (73%) | 18 (9%) | 91% ± 3% | 86% ± 4% | 77% ± 6% | NR | ||||
| Cytogenetic profile | ||||||||||
| Diploid karyotype | 82 (31%) | 4 (5%) | .01 | 96% ± 3% | 91% ± 6% | 79% ± 12% | NR | .01 | <.001 | |
| One altered chromosome | 101 (38%) | 8 (8%) | 93% ± 3% | 90% ± 4% | 82% ± 7% | NR | ||||
| Monosomy22/del(22q) | 81 (31%) | 6 (7%) | 93% ± 3% | 93% ± 3% | 83% ± 8% | NR | 12 (2–68) | |||
| Other | 20 (7%) | 2 (10%) | 93% ± 6% | 78% ± 15% | 78% ± 15% | NR | 31 (6–160) | |||
| Complex Karyotype | 81 (31%) | 16 (20%) | 82% ± 5% | 67% ± 8% | 62% ± 9% | 109 | 32 (3–296) | |||
| Tumor localization | ||||||||||
| Convexity | 55 (21%) | 7 (13%) | NS | 90% ± 5% | 85% ± 7% | 70% ± 11% | 131 | <.001 | .002 | |
| Parasagittal | 44 (17%) | 3 (7%) | 94% ± 4% | 86% ± 9% | 86% ± 9% | NR | ||||
| Falcine | 29 (11%) | 1 (3%) | 95% ± 5% | 95% ± 5% | 95% ± 5% | NR | ||||
| Cranial base (anterior) | 37 (14%) | 6 (16%) | 78% ± 9% | 78% ± 9% | 52% ± 22% | 172 | 21 (5–99) | |||
| Cranial base (middle) | 43 (16%) | 5 (12%) | 96% ± 4% | 78% ± 10% | 70% ± 12% | 127 | ||||
| Cranial base (posterior) | 21 (8%) | 2 (10%) | 87% ± 9% | 87% ± 9% | 87% ± 9% | NR | ||||
| Tentorial | 11 (4%) | 2 (18%) | 78% ± 14% | 78% ± 14% | 78% ± 14% | NR | ||||
| Intraventricular | 2 (1%) | 1 (50%) | 0% | – | – | 23 | 11 (1–113) | |||
| Spinal | 22 (8%) | 1 (5%) | 80% ± 18% | 80% ± 18% | 80% ± 18% | NR | ||||
| Tumor histology | ||||||||||
| Meningothelial | 147 (56%) | 18 (12%) | NS | 90% ± 3% | 82% ± 4% | 75% ± 6% | 172 | NS | ||
| Fibroblastic | 17 (6%) | 2 (12%) | 92% ± 8% | – | – | 82 | ||||
| Transitional | 57 (22%) | 6 (11%) | 89% ± 6% | 74% ± 14% | 59% ± 17% | 109 | ||||
| Psammomatous | 34 (13%) | 1 (3%) | 95% ± 5% | 95% ± 5% | 95% ± 5% | NR | ||||
| Angioblastic | 4 (1%) | 0 (0%) | – | – | – | NR | ||||
| Secretory | 5 (2%) | 1 (20%) | 67% ± 27% | – | – | 19 | ||||
| Edema | ||||||||||
| No | 120 (46%) | 9 (8%) | NS | 94% ± 3% | 86% ± 5% | 83% ± 6% | NR | NS | ||
| Yes | 143 (54%) | 19 (13%) | 86% ± 4% | 79% ± 5% | 65% ± 9% | 131 | ||||
| Tumor size | ||||||||||
| <30 mm | 63 (24%) | 3 (5%) | .001 | 96% ± 3% | 87% ± 9% | 87% ± 9% | NR | .002 | .03 | |
| 30–50 mm | 117 (45%) | 8 (7%) | 96% ± 2% | 85% ± 6% | 81% ± 7% | NR | ||||
| >50 mm | 79 (31%) | 17 (22%) | 77% ± 6% | 73% ± 6% | 55% ± 11% | 78 | 4 (1–20) | |||
Results expressed as number of cases and percentage between brackets or as *percentage of cases ± SE (standard error);
# the category with the best prognosis was selected as reference group.
Abbreviations: CI,confidence interval; HR, hazard ratio; NR, 75% RFS not reached; NS, not statistically significant (P > .05); RFS, relapse-free survival.
Fig. 2.
Clinical, biological, and genetic features of WHO grade I meningiomas with a significant impact on patient relapse-free survival (RFS; n = 227). RFS curves of WHO grade I meningioma cases grouped according to patient age (A), tumor localization (B), size (C), and interphase fluorescence in situ hybridization (iFISH) cytogenetic profile (D). RFS curves are shown for the same cases grouped according to the new prognostic scoring system proposed here (E) and that previously reported by Maillo et al15 (F).
Validation of iFISH Profiles by High-density Copy Number Arrays
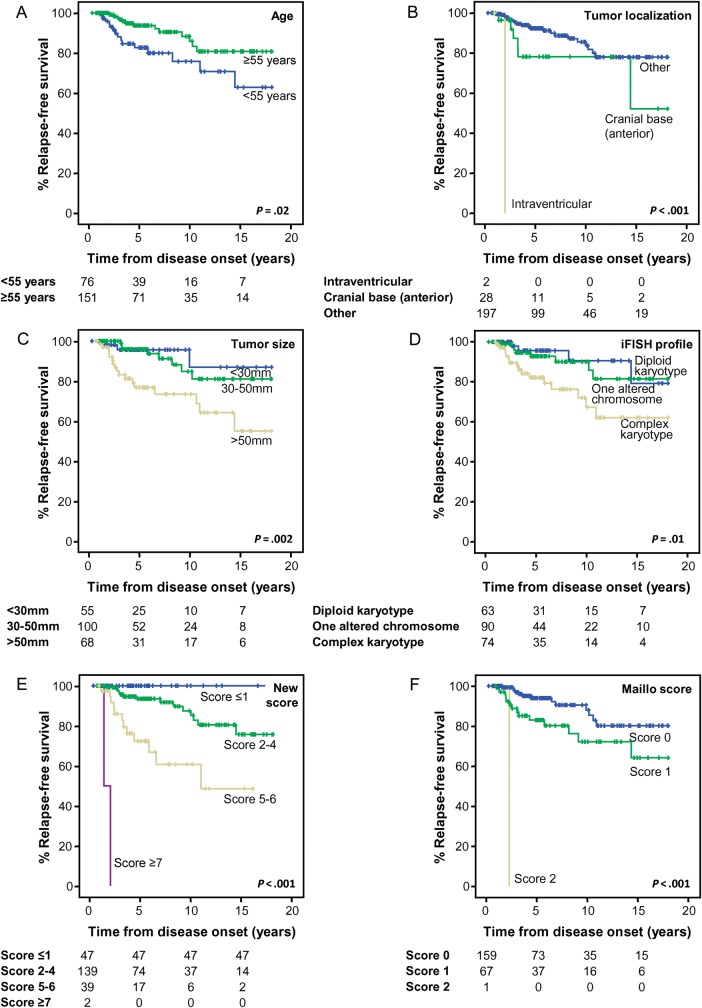
To validate the iFISH cytogenetic profiles, the prognostic value of CN profiles obtained for the 24 human chromosomes by SNP arrays was evaluated in an additional group of 132 cases pooled from our institution (n = 50)19 and another series from the literature (n = 82)20 with publicly available data. Based on SNP arrays, 41 (31%) of these 132 cases showed no CN alterations for any of the 24 chromosomes evaluated, 40 cases (30%) showed isolated alterations of a single chromosome (including monosomy 22/del[22q] in 35/40 cases), and 51 participants (39%) displayed complex karyotypes. Once again, the majority of WHO grade II/III tumors showed a CN pattern compatible with a complex karyotype (27/34 cases; 79%), whereas low grade meningiomas most frequently had a diploid karyotype or isolated alterations of a single chromosome (74/98 cases; 76%). In addition, meningioma cases carrying complex karyotypes associated with CN alterations of ≥2 chromosomes also showed a poorer outcome than cases with a diploid karyotype or isolated chromosomal alterations (P = .005; Fig. 3A). Similarly, we could also confirm in this validation series the improved predictive value of the new prognostic scoring system defined above (Fig. 3B) over the Maillo et al score (Fig. 3C), particularly for the low-risk category.
Fig. 3.
Impact of tumor karyotype, defined by the chromosomal copy number profile, as analyzed by SNP arrays on relapse-free survival (RFS) of meningioma cases. (A) RFS curves of meningioma cases classified according to the copy number patterns (diploid, isolated alteration of a single chromosome, and complex karyotypes) for the 2 series of cases analyzed by single-nucleotide polymorphism (SNP) arrays (n = 132) published so far in the literature. Only cases with follow-up data (n = 108) were considered for RFS analysis. (B) RFS curves of the same cases grouped according to the new prognostic scoring system proposed here. (C) RFS curves of the same cases grouped according to the prognostic scoring system previously reported by Maillo et al.15 In both panels B and C the cases from the Lee series were adjusted for the information available (data on tumor localization and size were not available).
Discussion
Tumor recurrence remains the major clinical complication of meningioma.2,4 Although multiple prognostic factors have long been identified2,4,5,21,22 and some prognostic classifications including tumor cytogenetics have been proposed,8,9,23 predicting tumor recurrence on an individual patient basis still remains a challenge.2,4 Because of this, a relatively uniform, close follow-up of every newly diagnosed patient has been adopted in most centers. In fact, a significant percentage (12% to >50%) of all recurrences still occur among the good-prognosis case categories; at the same time, a substantial fraction of all high-risk cases are relapse-free long-term survivors.2,4,14 Altogether, these illustrate the need for improved prognostic stratification systems based on long-term analysis of large numbers of cases and assessment of all potentially relevant prognostic parameters at diagnosis.
In the present study, we analyzed a large series of meningioma cases with a uniform median follow-up of >5 years, from which the most relevant clinicobiological, histopathological, and cytogenetic features of the disease had been systematically investigated at diagnosis. Our major goal was to identify a combination of prognostic factors that could stratify meningioma patients into different risk categories. Such risk categories should consider both very low-risk patients who do not require follow-up and a poor-prognosis category in which additional/alternative therapeutic measures might be required and considered in the future.
Based on our results, 4 groups of meningioma cases with distinct prognoses were defined by a combination of 5 features that emerged as independent prognostic factors. Thus, approximately 20% of cases were classified as having a very good prognosis with no relapses at 10 years. In contrast, intermediate-2 and high-risk cases displayed a high frequency of relapses with RFS rates at 10 years of 59% ± 8% and 0% ± 0%, respectively. To the best of our knowledge, this is one of the largest series of meningioma cases reported so far in which tumor cytogenetics were systematically investigated together with other relevant clinical and histopathological features of the disease. In line with previous observations by our group and other groups,2,3,5,6,21,22,24 WHO grade combined with tumor cytogenetics, patient age, tumor size, and localization emerged as the most relevant prognostic factors. Of note, once WHO grade I meningiomas were separately considered, the other 4 parameters retained their independent prognostic value. Consequently, the newly proposed scoring system, based on these 4 prognostic factors, was also retained at the expense of a decreased frequency of cases included in the intermediate-2 and poor-prognosis risk categories. Altogether, these results indicate that the new classification proposed here adds valuable prognostic information to that of the WHO grade alone and leads to a more refined risk stratification of meningiomas.
Apart from the WHO grade, tumor cytogenetics have emerged as being particularly informative. For decades now, the cytogenetic profile of tumor cells has been recurrently associated with the clinical outcome of meningioma patients.8,9,12,13,16,23,25,26 In fact, many different individual chromosomal alterations have been associated with the outcome of meningioma patients, particularly the loss of chromosomes 1p, 10, and 14.4,16,27,28 Of note, monosomy 1412,14,15,28–30 has even emerged as an independent prognostic factor for RFS,14,15 especially when associated with other specific chromosomal alterations (eg, del[1p36]).14 These latter findings may help explain why the presence of monosomy 14 in our series did not retain its independent prognostic value, as all cases showing monosomy 14 also showed complex karyotypes that frequently included del(1p36) (data not shown). Although a higher frequency of complex karyotypes was found among grade II/III versus grade I meningiomas, a significant fraction of these cases showed coexistence of multiple chromosomal alterations in association with a worse outcome, which helps explain the independent prognostic value of tumor cytogenetics over and above the WHO grade. Implementation of cytogenetic studies in routine clinical practice could be easily achieved by using either inexpensive CN oligonucleotide arrays (FullChromaArray, patent number 201231829) containing probes for the analysis of copy number alteration of the 24 human chromosomes or FISH technique based on a relatively limited number of probes for a few chromosomes (eg, chromosomes 1p, 7, 14, 18, and 22).
The prognostic impact of tumor cytogenetics was further confirmed in another independent series of meningioma cases (GEO 42624)19 in which SNP arrays were used to assess the CN alteration profiles, therefore confirming our iFISH results.
Other adverse prognostic factors that retained their independent prognostic value in our series included younger age (<55 years), tumor size >50 mm, and tumor localization at intraventricular and anterior cranial base areas; these features of meningiomas have been previously described by our group and other groups as being relevant prognostic factors.2,3,5,6,21 While several studies have shown a worse outcome for younger patients,5,31,32 other studies were not able to confirm the prognostic value of patient age.33,34 The mechanisms still remain to be elucidated for the adverse impact of tumor size and both intraventricular and anterior cranial base localization on RFS. Despite this, it could be hypothesized that larger tumor size and specific tumor localizations are associated with patterns of local tissue tumor infiltration and behavior that may lead to lower probability of successful complete tumor resection. In this regard, tumors localized in the convexity are typically considered to be curable by surgical resection,32,35,36 while intraventricular and skull-based meningiomas (especially those localized in the petroclival region and those that show involvement of the cavernous sinus and the orbit) are associated with a less favorable outcome because of the more complex resection procedures necessary to prevent neurological sequelae.2,3 Despite this, parasagittal/falcine37,38 and non-skull base tumors10,21,24 have also been associated with a worse prognosis in other series. Therefore, the specific reasons for the association between tumor localization and participant prognosis deserve further investigation. Only a small percentage of participants in our series did not undergo complete tumor resection, and no significant correlation was found between tumor localization and recurrence. In fact, this small group showed an apparently similar distribution by localization to that observed for the other tumors.
Most interestingly, our results also indicated that the new prognostic scoring system proposed here clearly improved the predictive value of tumor grade as well as other previously proposed prognostic scoring systems (eg, the Maillo et al score15), particularly for identifying the subgroup of meningioma cases with a very good prognosis that remained relapse-free at 10 years and would probably not have required close monitoring. This new prognostic scoring system also allowed identification of a small group of cases, mainly those with grade II/III tumors together with a few grade I meningiomas, that showed a dismal outcome with a recurrence rate at 10 years of 100% ± 0% and would require closer monitoring and/or alternative treatment strategies.
In summary, here we propose a new prognostic classification for meningioma patients based on tumor size, localization, and cytogenetics as well as patient age and the WHO grade. This classification allows stratification of meningioma patients at diagnosis into 4 risk categories associated with significantly different relapse rates. These patients may potentially benefit from different follow-up strategies as well as distinct treatment approaches.
Supplementary Material
Funding
This work was partially supported by grants from the Fundação para a Ciência e Tecnologia (PIC/IC/83108/2007, FCT, Portugal); Fondo de Investigaciones Sanitarias (FIS/FEDER 06/0312 and RETICC RD06/0020/0035, RD06/0020/0059 and RD12/0036/0048, Instituto de Salud Carlos III (ISCIII/FEDER), Ministerio de Sanidad y Consumo, Madrid, Spain), Caja Burgos (Spain), Fundación MMA (exp 75312010 and 87692011, Madrid, Spain), and Consejeria Sanidad Junta de Castilla y León, Gerencia Regional de Salud: GRS689/A/11. Patrícia Domingues is supported by grant (SFRH/BD/64799/2009) from FCT. Maria Dolores Tabernero is supported by IECSCYL (Fundación Instituto de Estudios Ciencias de la Salud de Castilla y León, Soria, Spain).
GEO accession no. GSE 42624.
Supplementary Material
Acknowledgments
Patrícia Domingues performed experiments, analyzed results, and wrote the manuscript. Pablo Sousa, Alvaro Otero, Jesus Maria Gonçalves, and Laura Ruiz provided the samples and clinical follow-up of the patients. Maria Celeste Lopes and Catarina de Oliveira supervised the study. Alberto Orfao designed the research, supervised the study, and wrote/reviewed the manuscript. Maria Dolores Tabernero designed the research, supervised the study, analyzed/interpreted data, and reviewed the manuscript.
Conflict of interest statement. None declared.
References
- 1.Mawrin C, Perry A. Pathological classification and molecular genetics of meningiomas. J Neurooncol. 2010;99:379–391. doi: 10.1007/s11060-010-0342-2. [DOI] [PubMed] [Google Scholar]
- 2.Saraf S, McCarthy BJ, Villano JL. Update on meningiomas. Oncologist. 2011;16:1604–1613. doi: 10.1634/theoncologist.2011-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marosi C, Hassler M, Roessler K, et al. Meningioma. Crit Rev Oncol Hematol. 2008;67:153–171. doi: 10.1016/j.critrevonc.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Yew A, Trang A, Nagasawa DT, et al. Chromosomal alterations, prognostic factors, and targeted molecular therapies for malignant meningiomas. J Clin Neurosci. 2013;20:17–22. doi: 10.1016/j.jocn.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Stafford SL, Perry A, Suman VJ, et al. Primarily resected meningiomas: outcome and prognostic factors in 581 Mayo Clinic patients, 1978 through 1988. Mayo Clin Proc. 1998;73:936–942. doi: 10.4065/73.10.936. [DOI] [PubMed] [Google Scholar]
- 6.Perry A, Stafford SL, Scheithauer BW, et al. Meningioma grading: an analysis of histologic parameters. Am J Surg Pathol. 1997;21:1455–1465. doi: 10.1097/00000478-199712000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Espinosa AB, Tabernero MD, Maillo A, et al. The cytogenetic relationship between primary and recurrent meningiomas points to the need for new treatment strategies in cases at high risk of relapse. Clin Cancer Res. 2006;12:772–780. doi: 10.1158/1078-0432.CCR-05-1480. [DOI] [PubMed] [Google Scholar]
- 8.Urbschat S, Rahnenfuhrer J, Henn W, et al. Clonal cytogenetic progression within intratumorally heterogeneous meningiomas predicts tumor recurrence. Int J Oncol. 2011;39:1601–1608. doi: 10.3892/ijo.2011.1199. [DOI] [PubMed] [Google Scholar]
- 9.Ketter R, Urbschat S, Henn W, et al. Application of oncogenetic trees mixtures as a biostatistical model of the clonal cytogenetic evolution of meningiomas. Int J Cancer. 2007;121:1473–1480. doi: 10.1002/ijc.22855. [DOI] [PubMed] [Google Scholar]
- 10.Ketter R, Rahnenfuhrer J, Henn W, et al. Correspondence of tumor localization with tumor recurrence and cytogenetic progression in meningiomas. Neurosurgery. 2008;62:61–69. doi: 10.1227/01.NEU.0000311062.72626.D6. [DOI] [PubMed] [Google Scholar]
- 11.Pham MH, Zada G, Mosich GM, et al. Molecular genetics of meningiomas: a systematic review of the current literature and potential basis for future treatment paradigms. Neurosurg Focus. 2011;30:E7. doi: 10.3171/2011.2.FOCUS1117. [DOI] [PubMed] [Google Scholar]
- 12.Pfisterer WK, Coons SW, Aboul-Enein F, et al. Implicating chromosomal aberrations with meningioma growth and recurrence: results from FISH and MIB-I analysis of grades I and II meningioma tissue. J Neurooncol. 2008;87:43–50. doi: 10.1007/s11060-007-9498-9. [DOI] [PubMed] [Google Scholar]
- 13.Pfisterer WK, Hank NC, Preul MC, et al. Diagnostic and prognostic significance of genetic regional heterogeneity in meningiomas. Neuro Oncol. 2004;6:290–299. doi: 10.1215/S1152851704000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maillo A, Orfao A, Espinosa AB, et al. Early recurrences in histologically benign/grade I meningiomas are associated with large tumors and coexistence of monosomy 14 and del(1p36) in the ancestral tumor cell clone. Neuro Oncol. 2007;9:438–446. doi: 10.1215/15228517-2007-026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maillo A, Orfao A, Sayagues JM, et al. New classification scheme for the prognostic stratification of meningioma on the basis of chromosome 14 abnormalities, patient age, and tumor histopathology. J Clin Oncol. 2003;21:3285–3295. doi: 10.1200/JCO.2003.07.156. [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Ketter R, Henn W, et al. Histopathologic indicators of recurrence in meningiomas: correlation with clinical and genetic parameters. Virchows Arch. 2006;449:529–538. doi: 10.1007/s00428-006-0285-3. [DOI] [PubMed] [Google Scholar]
- 17.Louis DN, Ohgaki H, Wiestler OT, et al. WHO Classification of Tumors of the Central Nervous System. 4th ed. Lyon, France: IARC press; 2007. pp. 163–172. [Google Scholar]
- 18.Sayagues JM, Tabernero MD, Maillo A, et al. Incidence of numerical chromosome aberrations in meningioma tumors as revealed by fluorescence in situ hybridization using 10 chromosome-specific probes. Cytometry. 2002;50:153–159. doi: 10.1002/cyto.10075. [DOI] [PubMed] [Google Scholar]
- 19.Tabernero MD, Maillo A, Nieto AB, et al. Delineation of commonly deleted chromosomal regions in meningiomas by high-density single nucleotide polymorphism genotyping arrays. Genes Chromosomes Cancer. 2012;51:606–617. doi: 10.1002/gcc.21948. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Liu J, Patel S, et al. Genomic landscape of meningiomas. Brain Pathol. 2010;20:751–762. doi: 10.1111/j.1750-3639.2009.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P, Ma W, Yin S, et al. Three risk factors for WHO grade II and III meningiomas: A study of 1737 cases from a single center. Neurol India. 2013;61:40–44. doi: 10.4103/0028-3886.107928. [DOI] [PubMed] [Google Scholar]
- 22.Cahill KS, Claus EB. Treatment and survival of patients with nonmalignant intracranial meningioma: results from the Surveillance, Epidemiology, and End Results Program of the National Cancer Institute. Clinical article. J Neurosurg. 2011;115:259–267. doi: 10.3171/2011.3.JNS101748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ketter R, Henn W, Niedermayer I, et al. Predictive value of progression-associated chromosomal aberrations for the prognosis of meningiomas: a retrospective study of 198 cases. J Neurosurg. 2001;95:601–607. doi: 10.3171/jns.2001.95.4.0601. [DOI] [PubMed] [Google Scholar]
- 24.Kane AJ, Sughrue ME, Rutkowski MJ, et al. Anatomic location is a risk factor for atypical and malignant meningiomas. Cancer. 2011;117:1272–1278. doi: 10.1002/cncr.25591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zang KD. Meningioma: a cytogenetic model of a complex benign human tumor, including data on 394 karyotyped cases. Cytogenet Cell Genet. 2001;93:207–220. doi: 10.1159/000056986. [DOI] [PubMed] [Google Scholar]
- 26.Pfisterer WK, Hendricks WP, Scheck AC, et al. Fluorescent in situ hybridization and ex vivo 1H magnetic resonance spectroscopic examinations of meningioma tumor tissue: is it possible to identify a clinically-aggressive subset of benign meningiomas? Neurosurgery. 2007;61:1048–1059. doi: 10.1227/01.neu.0000303201.62123.5c. [DOI] [PubMed] [Google Scholar]
- 27.Mihaila D, Gutierrez JA, Rosenblum ML, et al. Meningiomas: analysis of loss of heterozygosity on chromosome 10 in tumor progression and the delineation of four regions of chromosomal deletion in common with other cancers. Clin Cancer Res. 2003;9:4435–4442. [PubMed] [Google Scholar]
- 28.Cai DX, Banerjee R, Scheithauer BW, et al. Chromosome 1p and 14q FISH analysis in clinicopathologic subsets of meningioma: diagnostic and prognostic implications. J Neuropathol Exp Neurol. 2001;60:628–636. doi: 10.1093/jnen/60.6.628. [DOI] [PubMed] [Google Scholar]
- 29.Barbera S, San Miguel T, Gil-Benso R, et al. Genetic changes with prognostic value in histologically benign meningiomas. Clin Neuropathol. 2013;32:311–317. doi: 10.5414/NP300580. [DOI] [PubMed] [Google Scholar]
- 30.Tabernero MD, Espinosa AB, Maillo A, et al. Characterization of chromosome 14 abnormalities by interphase in situ hybridization and comparative genomic hybridization in 124 meningiomas: correlation with clinical, histopathologic, and prognostic features. Am J Clin Pathol. 2005;123:744–751. [PubMed] [Google Scholar]
- 31.van Alkemade H, de Leau M, Dieleman EM, et al. Impaired survival and long-term neurological problems in benign meningioma. Neuro Oncol. 2012;14:658–666. doi: 10.1093/neuonc/nos013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz J, Martinez A, Hernandez S, et al. Clinicopathological variables, immunophenotype, chromosome 1p36 loss and tumour recurrence of 247 meningiomas grade I and II. Histol Histopathol. 2010;25:341–349. doi: 10.14670/HH-25.341. [DOI] [PubMed] [Google Scholar]
- 33.Ildan F, Erman T, Gocer AI, et al. Predicting the probability of meningioma recurrence in the preoperative and early postoperative period: a multivariate analysis in the midterm follow-up. Skull Base. 2007;17:157–171. doi: 10.1055/s-2007-970554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasuya H, Kubo O, Tanaka M, et al. Clinical and radiological features related to the growth potential of meningioma. Neurosurg Rev. 2006;29:293–296. doi: 10.1007/s10143-006-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko KW, Nam DH, Kong DS, et al. Relationship between malignant subtypes of meningioma and clinical outcome. J Clin Neurosci. 2007;14:747–753. doi: 10.1016/j.jocn.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Palma L, Celli P, Franco C, et al. Long-term prognosis for atypical and malignant meningiomas: a study of 71 surgical cases. J Neurosurg. 1997;86:793–800. doi: 10.3171/jns.1997.86.5.0793. [DOI] [PubMed] [Google Scholar]
- 37.Ayerbe J, Lobato RD, de la Cruz J, et al. Risk factors predicting recurrence in patients operated on for intracranial meningioma. A multivariate analysis. Acta Neurochir (Wien) 1999;141:921–932. doi: 10.1007/s007010050398. [DOI] [PubMed] [Google Scholar]
- 38.Vranic A, Popovic M, Cor A, et al. Mitotic count, brain invasion, and location are independent predictors of recurrence-free survival in primary atypical and malignant meningiomas: a study of 86 patients. Neurosurgery. 2010;67:1124–1132. doi: 10.1227/NEU.0b013e3181eb95b7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.