Abstract
The transcription factor NFκB is a regulator of inflammatory and adaptive immune responses, yet only IκBα has been shown to limit NFκB activation and inflammatory responses. We investigated another negative feedback regulator, IκBε, in regulating B cell proliferation and survival. The loss of IκBε showed increased B cell proliferation and survival in response to both antigenic and innate stimulation. NFκB activity was elevated during late phase activation, but the dimer composition was stimulus-specific. In response to IgM, cRel dimers were elevated in IκBε-deficient cells, yet in response to LPS, RelA dimers were elevated also. The corresponding dimer-specific sequences were found in the promoters of hyper-activated genes. Using a mathematical model of the NFκB signaling system in B cells, we demonstrated that kinetic considerations of the IKK signaling input and IκBε’s interactions with RelA- and cRel-specific dimers could account for this stimulus-specificity. cRel is known to be the key regulator of B cell expansion. We found that RelA-specific phenotype in LPS-stimulated cells was physiologically relevant: unbiased transcriptome profiling identified the inflammatory cytokine, interleukin 6 (IL-6) to be hyper-activated in IκBε−/− B cells. When the IL-6 receptor was blocked, LPS-responsive IκBε−/− B cell proliferation was specifically reduced to near wild type levels. Our results provide novel evidence of a critical role of immune-response functions for IκBε in B cells; it regulates proliferative capacity via at least two mechanisms involving cRel and RelA-containing NFκB dimers. This study illustrates the importance of kinetic considerations in understanding the functional specificity of negative feedback regulators.
Introduction
The NFκB family of transcription factors controls expression of an extensive array of genes responsible for cell survival, proliferation, inflammation and immune regulation. This transcription factor family consists of a variety of dimers formed by combinations of five rel-homology-containing proteins, RelA, RelB, cRel, p50, p52. The activities of these dimers are regulated by members of the classical IκB protein family, namely IκBα, IκBβ, and IκBε. IκB proteins limit NFκB activity in the cellular basal state, but allow for NFκB activation when inflammatory stimuli result in their N-terminal specific serine phosphorylation by the NEMO-containing IκB kinase complex (IKK complex), specific lysine ubiquitination, and subsequent proteasome-dependent degradation (1). NFκB activity, however, is dynamic and transient. Both IκBα and IκBε are transcriptionally induced by NFκB, yet only IκBα has been shown to provide critical negative feedback functions (2,3).
While these insights have largely been derived from convenient cell line systems such as HeLa and mouse embryonic fibroblasts (MEFs), NFκB’s major physiological functions are in lymphocytes where it has a key role in regulating proliferation and survival during the adaptive immune response (4–14). Whereas in HeLa and MEF cells RelA:p50 is the predominant dimer, in B cells, upon activation with either antigenic stimulation through the B cell receptor by anti-IgM or pathogenic stimulation through the Toll-like receptor (TLR) by LPS, there is a significant increase in nuclear DNA binding activity of both RelA:p50 and cRel:p50 dimers (9,10,12,15). Interestingly, the majority of the evidence supports a critical role for cRel and p50 in controlling B-cell proliferation (16–19), but not RelA (20).
Although it is understood that the cRel:p50 dimer plays an essential role in B cell proliferation and survival, little is known about the mechanisms responsible for controlling cRel:p50 dimer activity. In fibroblasts, IκBα is known to be the primary regulator of the ubiquitous RelA:p50 dimer; IκBα-deficient fibroblasts show elevated basal levels, reduced activation, and prolonged duration of RelA:p50 activity in response to stimulation by the pro-inflammatory cytokine TNF-α (21). IκBε provides a secondary role, partially compensating for IκBα-deficiency, but IκBε-deficiency alone shows no discernible phenotype. Biochemical characterization suggests that IκBα preferentially binds RelA:p50 dimers, whereas IκBε associates not only with RelA- but also cRel-containing dimers (22–24). These differences suggest that IκBα and IκBε may have distinct physiological roles in controlling NFκB dimers. IκBε was reported to be a non-redundant regulator of cRel-dependent expression B-cell activating factor receptor (BAFFR) and CD40 (25), but how it controls cRel-containing dimers, or what other genes may be regulated remains unclear.
Here, we investigated the role of IκBε in controlling NFκB activity in B lymphocytes. Our results indicate that the ablation of IκBε allows for increased proliferation and survival in B cells stimulated with either IgM or LPS. In fact, we found that IκBε had a role in limiting not only cRel but also RelA-containing dimers, albeit in a stimulus-specific manner, as evidenced by both biochemical data and DNA motif signatures in hyper-regulated genes. Mathematical modeling was employed to show that a consideration of known kinetic differences between these proteins provides for a sufficient explanation. Further, we found that IκBε control of RelA in response to LPS was functionally relevant, as hyperinduction of IL6 in IκBε-deficient B-cells was shown to mediate hyper-expansion.
Materials and Methods
Cell isolation and culture
Spleens were harvested from C57Bl6 mice wild-type (Jackson Labs, Bar Harbor, MN), and C57Bl6 IκBε−/− mice (2). The collected spleens were homogenized using frosted glass slide grinding. For B cell isolation, homogenized splenocytes were incubated with anti-CD43 (Ly-48) microbeads for 15 minutes at room temperature. Following this incubation, the cells were washed with Hanks Buffered salt solution (HBSS) (Gibco 14170) containing 1% FCS, 10mM HEPES (Gibco 15630) and 1% FCS (Sigma F2442) and separated over a magnetic column (LS column MiltenyiBiotec 130-042-401). For B cells, purity was determined by flow cytometry using PE anti-B220 (eBioscience 12-0452-83), FITC anti-CD3 (eBioscience 11-0031-82), APC anti-CD4 (eBioscience 17-0041-83) and PerCP anti-CD8 (ebioscience 46-0081-82). Purity was consistently found to be between 92% and 95% (data not shown). For experiments where separation of marginal zone (MZ) B cell and follicular B cell was performed, anti-CD43 magnetically separated B cells were stained with APC anti-CD9 (eBioscience 17-0091-82) and B cell population separation was performed using a FACS-ARIA II cell sorter (BD Bioscience). Complete media consisting of RPMI-1640 (Gibco 11875), 10 mM HEPES (Gibco 15630), 1 mM Sodium Pyruvate (Gibco 11360), 1 mM non-essential amino acids (Gibco 11140), 0.055 mM β-mercaptoethanol (Gibco 21985), 100 units Penicillin/Streptomycin (Gibco 10378016) and 0.3 mg/ml glutamine was used to culture either B cells. B cells were stimulated with either 10 μg/ml anti-IgM (Jackson laboratories 115-006-020) or 10 μg/ml LPS (Sigma L2630).
Flow cytometry analysis of cell proliferation and survival
Purified B cells were stained with 5 μM CFSE (Invitrogen C1157) and culture in complete media with previously mentioned stimuli (see above). At various time points B cells were collected and stained with 7AAD (Invitrogen A1310). The B cells were analyzed for proliferation and survival using a C6 Accuri flow cytometer (BD biosciences). Gating of live B cells was determined by forward scatter and side scatter properties. Differences in cell proliferation was measured using FlowJo (Tree star inc.), and FlowMax (26), which determines maximum-likelihood cellular parameter sets for fraction of responding cells, times to division, and times to death for generations 0 to 10.
EMSA and Supershifts
Nuclear extracts were generated from B cells using high salt extraction. In brief, purified B cells were incubated with a low salt buffer (10mM HEPEs pH 7.9 (Gibco), 10 mM KCl (Thermo Fisher Scientific P217), 0.1 mM EGTA (Sigma E-4378), 0.1 mM EDTA (Thermo Fisher Scientific S312), 1mM DTT (Thermo Fisher Scientific BP172-5), 1 mM PMSF (Sigma P7626), 5 μg/ml apoprotein (Sigma A1153), 5 μg/ml leupeptin (Sigma L2884), 1 μM pepstatin A (Sigma P5318)) for 10 minutes on ice. Following this incubation, the cells were disrupted through the addition of NP-40 (US Biological N3500) to a final concentration of 0.5% and votexing for 15 seconds. Nuclei were pelleted away from the cytoplasmic fraction by centrifugation at 15,000 rpm for 1 minute and the cytoplasmic fraction was pipetted into a separate tube. The remaining nuclei were disrupted by a 20 minute incubation at 4°C in a high salt buffer (20 mM HEPES pH 7.9 (Gibco), 400 mM NaCl (Thermo Fisher Scientific S671), 1 mM EGTA (Thermo Fisher Scientific), 1 mM EDTA (Thermo Fisher Scientific), 20% Glycerol (Thermo Fisher Scientific), 1 mM DTT (Thermo Fisher Scientific), 1 mM PMSF(Sigma)). The nuclear fraction was collected following centrifugation at 15,000 rpm for 5 minutes. Equal amounts of nuclear extracts (1 μg) were pre-incubated for 20 minutes on ice in the presence or absence of antibodies specific for RelA (Santa Cruz Biotechnology sc-372), RelB (Santa Cruz Biotechnology sc-226), cRel (Santa Cruz Biotechnologies sc-71) or in combinations as follows; RelA/RelB, RelA/cRel and RelB/cRel. Following the pre-incubation with antibodies, [γ–32P]ATP (GE health) radio-labeled probe derived from HIV-κB sequence: 5′-GCTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGG-3′ was added and incubated at room temperature for an additional 15 minutes. The resulting DNA/protein/antibody complexes were resolved by electrophoresis on a 5% non-denaturing polyacrylamide gel and exposed to storage phosphor screen (GE healthcare) overnight before image development on at Typhoon 9200 Variable Mode Imager (GE healthcare). Images were analyzed and quantitated using ImageQuant™ (GE Health).
Western blot analysis
Whole cell lysates were prepared using RIPA buffer lysis of B cells. Cytoplasmic extracts and nuclear extracts were prepared as previously described (27,28). The resulting lysates and extracts were run on either 10% SDS-PAGE gels or 5%–14% Criterion Tris-HCl Gel (Bio Rad). The following antibodies were used to identify the protein of interest: p65 (Santa Cruz Biotechnology), IκBα (Santa Cruz Biotechnology), cRel (Santa Cruz Biotechnology) and actin (Santa Cruz Biotechnology). The resulting proteins were detected using the Bio-Rad ChemiDoc XRS System and SuperSignal West Femto Substrate Maximum Sensitivity Substrate (Thermo Scientific) to detect chemiluminescence released by HRP-labeled secondary antibodies.
Cytokine Neutralization and Receptor blocking
CFSE labeled B cells isolated from either wild type or IκBε−/− B cells were stimulated with either 10 μg/ml IgM or 10 μg/ml LPS in the presences of 2 μg/ml of the following antibodies: anti-mouse IL-1 alpha/IL-1F1 AF-400-NA (R&D systems), anti-mouse IL-1 beta/IL-1F2 AF-401-NA (R&D systems), anti- mouse IL-6 AF-406-NA (R&D systems), anti-mouse IL-1 receptor AF-480-NA (R&D systems), anti-mouse IL-6 receptor alpha AF1830 (R&D systems). Cell proliferation was measure using a C6 accuri flow cytometer (BD biosciences). Differences in cell proliferation was measured using FlowJo (Tree star inc.).
ELISA assays
Isolated B cells were plated at a concentration of 2 million cells per ml in the presences of 10 μg/ml IgM or 10 μg/ml LPS. At 2, 4, 8, 22, 34 and 48 hours cells were harvested, spun down and the supernatant was collected. The resulting supernatant was test for measured release of IL-1α (Mouse IL-1α ELISA MAX™ Deluxe, Biolegend) IL-1β (Mouse IL-1β ELISA MAX™, Biolegend) and Deluxe, IL-6 (Mouse IL-6 ELISA MAX™ Deluxe, Biolegend).
Transcriptome Analysis
Total RNA was isolated from IgM or LPS stimulated B cells isolated from wild type or IκBε−/− B cells over a four point time course. mRNA was extracted from 2 μg total RNA using oligo(dT) magnetic beads and fragmented at high temperature using divalent cations. Next cDNA library were generated using the IlluminaTruSeq kits and quantitation was performed using the Roche Light Cycler 480. Sequencing was performed on Illumina’s HiSeq 2000, according to the manufacturer’s recommendations by the BIOGEM core facility located at University of California, San Diego. Reads were aligned to the mouse mm10 genome and RefSeq genes (PMID 12045153, PMID 12466850) with Tophat (PMID 19289445). Cufflinks and CummRbund were used to ascertain differential expression of genes. Gene differential FPKMs were obtained from the cuffdiff program in the Tuxedo RNA-seq analysis suite. In R, we took the log base 2 of the FPKM values and used the wild type zero hour to normalize. The subset of genes that were induced by two fold in at least one of the wild type time points were identified and used to identify gene induced by IgM or LPS stimulation. Hyperinduced genes of the IκBε−/− samples were identified as being two fold greater than their corresponding wild type timepoint in two or more of the non-zero time points. Using R’s gplots package heat maps were created for fold change for all genes that made the above induction cut offs. Additionally, heat maps for latent genes induced no more than 0.3 log base 2 above their corresponding wild type time point were created. For both heat maps, the lists of genes mapped were recorded. The above process was done for both IgM and LPS stimulated conditions. Determination of NFκB motifs containing genes was performed using the NFκB PWMs previously identified by Bulyk ML et. al. (29) and converted into formats for HOMER motif discovery software (30). The promoter regions of genes found to be either hyper or latent induced were searched for the occurrence of these motifs. The percentage of genes from the identified gene lists that contained each motif versus the percent of promoters containing the NFκB motifs from genes identified as induced were graphed. Significance was determined using R’s stats package to do a Pearson’s Chi-squared test using the background counts as the expected values versus the percentage of hyper or latent induced genes containing the NFκB motifs.
Computational modeling of NFκB dimer activity
The computational model appends the previously published IκB models with the reactions that govern the generation of RelA- and cRel-containing NFκB dimers (Supplemental Figure 3). The model contains 36 species and 146 reactions (Supplemental Model Equations) governed by 74 parameters (Supplemental Table 1). The ODEs were solved numerically using MATLAB version R2013a (The MathWorks, Inc.) with subroutine ode15s, a variable order, multi-step solver. Prior to stimulation, the system was allowed to equilibrate from starting conditions to a steady state, defined as showing no concentration changes greater than 1% over a period of 4000 minutes. Stimulus-induced perturbation from the steady state was accomplished by direct modulation of IKK activity via a numerical input curve representing IgM or LPS stimulation (adapted from Werner 2008)(31). MATLAB model codes are available upon request.
Animal use
The animal protocols for this study were approved by the University of California, San Diego Animal Care and Use Committee.
Results
IκBε-deficiency in B cell subsets results in increased stimulus-responsive proliferation and survival
As NFκB controls B cell expansion, we sought to determine IκB regulators that limit NFκB activity in B-cells and thus B cell proliferation. Examining whole-cell extracts, we found that while IκBα protein levels rapidly decrease upon B-cell stimulation with IgM or LPS, IκBε protein levels decrease only slightly after stimulation and increase at late timepoints (Supplementary Figure 1A), suggesting a role in post-induction attenuation of NFκB.
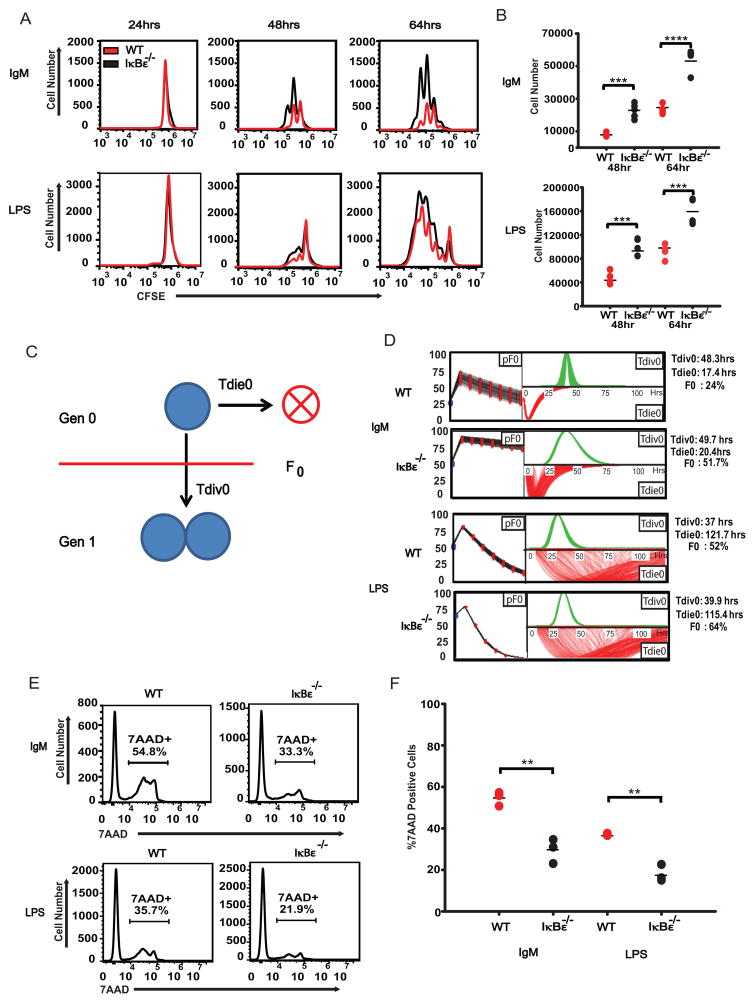
Using B cells magnetically purified from mixed splenocytes collected from wild type or IκBε−/− mice, we examined B cell expansion following ex vivo stimulation with 10 μg/ml IgM or 10 μg/ml LPS using CFSE dye dilution. We found that B cells lacking IκBε displayed increased expansion with either stimulus compared to wild type B cells (Figure 1A). Several repeats of the CFSE experiments yielded highly reproducible results (Figure 1B). To determine if these differences were the result of a proliferation or survival defect we used the computational phenotyping tool FlowMax (26), which parameterizes a modified cyton model to CFSE time courses and yields maximum likelihood non-redundant cellular parameters, such as the percentage of cells entering the proliferative program (pF), the time to the first division (Tdiv0), and the time to death (Tdie0) of cells not entering the proliferative program (Figure 1C). Using FlowMax, we found that the CFSE data indicates that IκBε−/− B cells are more likely to respond to the stimulus (pF0) than wild type cells under the same conditions (Figure 1D). In response to IgM, 51.7% of IκBε−/− B cells entered division compared to only 24% of wild type B cells. Following LPS stimulation 64% of the IκBε−/− B cells entered division, compared to 52% of their wild type counterparts. As only non-responding cells are susceptible to death (32–34), and the Tdie0 and Tdiv0 parameters showed little change in the knockout, this suggested that the death rates in IκBε−/− would be lower. Testing this prediction with 7AAD staining of responding cells at 24 hours, we indeed found lower percentages of dying cells in the IκBε−/− than wild type populations (Figure 1E and Figure 1F), 33.3% vs. 54.8% in response to IgM, and 21.9 vs. 37.7% in response to LPS).
Figure 1.
IκBε−/− B cells have increased proliferation and survival in response to both antigenic and inflammatory signals. B cells were isolated and purified from whole splenocytes of wild type and IκBε−/− using negative selection by CD43 magnetic beads. The separated B cells were stained with 1 nM CFSE and stimulated with either 10 μg/ml IgM (Jackson Labs) or 10 μg/ml LPS (Sigma). At designated time points, B cells were harvested and stained with 5 μg/ml 7AAD (Invitrogen) and analyzed for proliferation and death using flow cytometry. (A) B cells from the IκBε−/− mice displayed increased proliferation in response to both IgM and LPS at each time point. (B) The increased number of proliferating IκBε−/− B cells over that of wild type B cells was measured using Flowjo and the cell numbers were graphed. (C) Diagram depicting the fcyton model. In this model, stimulated cells undergo death over time (Tdie0) or enter division (pF0, fraction entering division and Tdiv0 time to division). (D) The CFSE proliferation profiles of the IκBε−/− and wild type B cells stimulated with either IgM or LPS where analyzed using FlowMax running the Pcyton model to predict the fraction of B cells responding to stimulation (pF0), the average time to division of undivided (Tdiv0) and average time to death of undivided (Tdie0) cells. The fraction of responding B cells (pF0) was greatly increased in both IgM and LPS stimulated IκBε−/− B cells when compared to wild type B cells. (E) 7AAD measurements of B cell death show an increased percentage of B cells in the wild type population undergo apoptotic death when compared to IκBε−/− B cells. (F) The percentage of 7AAD positive B cells from several experiments was measured using Flowjo and the percentages of 7AAD positive cells was graphed. Data shown in (A, B, D, E and F) are representative of at least four independent experiments. **P < 0.01, ***P < 0.005 and ****P <0.001 (unpaired t-test).
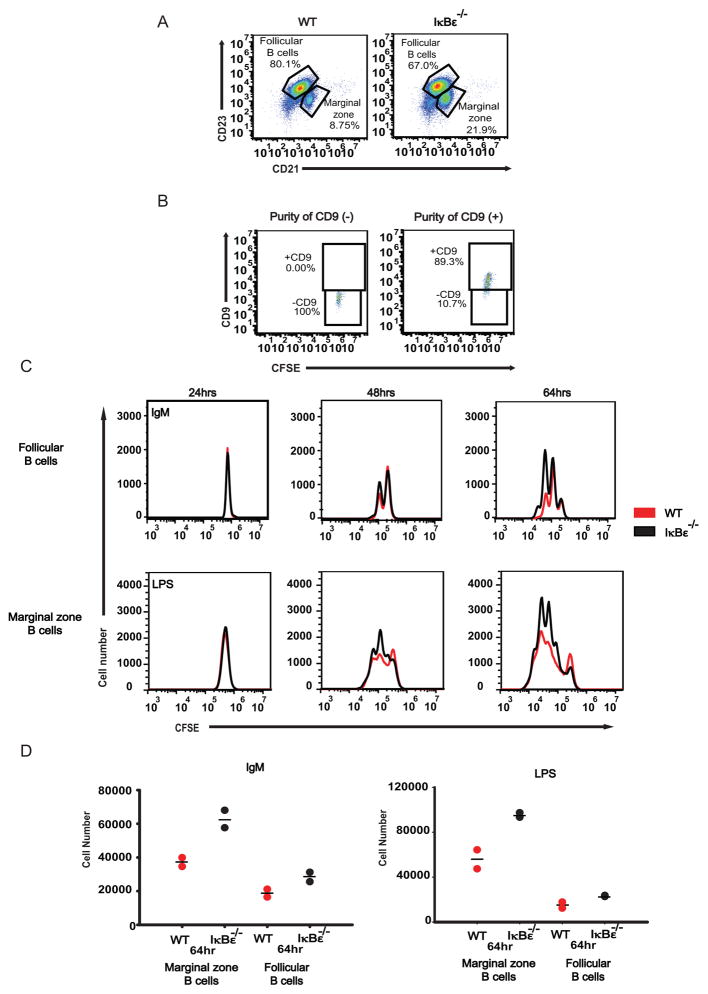
Examining the B-cell subsets in the spleen, we observed a higher percentage of marginal zone B cells as opposed to follicular B cell populations in IκBε−/− mice compared to wild type controls (Figure 2A). As marginal zone B cell maturation is more sensitive to NFκB activity than follicular B cell maturation, this observation is consistent with elevated NFκB activity following IκBε ablation. It also prompted us to question if the skewed distributions were responsible for the ex vivo expansion phenotype. To address this concern, we purified wild type and IκBε−/− marginal zone and follicular B cells away from each other, using APC anti-CD9 staining in conjunction with FACs cell sorting (35). The purity was determined to be consistently between 90% and 100% (Figure 2B). Purified follicular B cells whose physiological role is to respond to antigens, showed increased B-cell expansion in response to IgM stimulation when derived from IκBε−/− mice. Similarly, marginal zone B cells whose physiological role is to monitor for circulating endotoxin showed increased proliferation when derived from IκBε−/− mice (compared to wild type) and stimulated with LPS (Figure 2C and Figure 2D). These results indicate that the difference in proliferation and survival is a cell intrinsic B cell phenotype rather than a result of developmental changes that result in differences in the B cell subpopulation distributions.
Figure 2.
Follicular and Marginal zone IκBε−/− show increase proliferation. Follicular (FO) and Marginal zone (MZ) B cells population distributions were analyzed from whole splenocyte populations by flow cytometry using anti-B220, anti-CD21 and anti-CD23. (A) IκBε−/− B cells were found to have a distribution of 67% FO B cells and 21.9% MZ B cells. Wild type B cells were distributed as 80.1% FO B cells and 8.75% MZ B cells. (B) Marginal zone and Follicular B cells were separated from each other using FACS sorting of anti-CD9. Purity of the separated CD9+ and CD9- B cell populations was found to be between 90% and 100%. (C) IκBε−/− FO and MZ B cells showed increased proliferation over wild type FO and MZ B cells. (D) The increased number of proliferating IkBe−/− B cells over that of wild type B cells was measured using Flowjo and the cell numbers were graphed. Data shown in (A, B, C and D) are representative of two independent experiments.
IκBε provides negative feedback on both RelA- and cRel-containing dimers, albeit stimulus-specifically
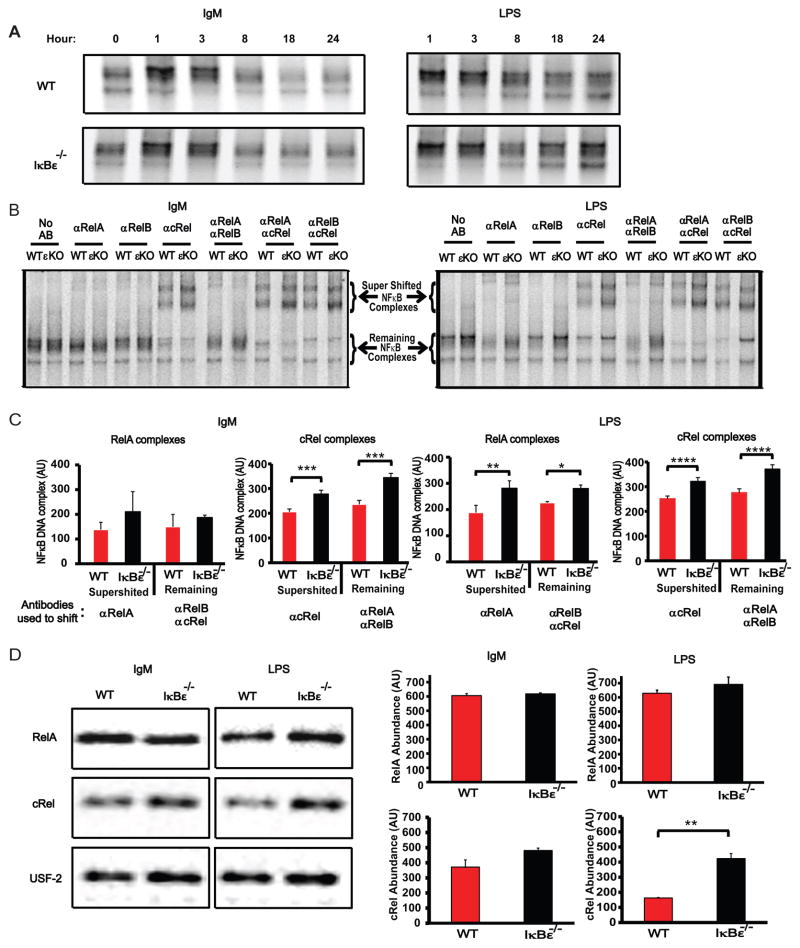
Given the B cell proliferation phenotype of IκBε deficiency, we sought to determine how the loss of IκBε affected NFκB dimer activation. In previous studies, IκBε had been shown to bind preferentially to cRel (22,24), suggesting that IκBε-deficiency may preferentially affect cRel-containing dimer activity. We prepared nuclear extracts from wild type and IκBε−/− B cells stimulated with IgM or LPS as previously described, and used them for electrophoretic mobility shift assays (EMSA) with a κB-site containing probe. NFκB heterodimers (all containing p50, but containing either RelA, cRel or RelB, see below) were found elevated in IκBε−/−B cells at later time points (18 and 24 hours) when compared to wild type B cells (Figure 3A), consistent with the high level of induction of this inhibitor seen in immunoblots of wild type cells (Supplementary Figure 1A). Supershift analysis of these B cells nuclear extracts using specific antibodies for the three activation domain-containing Rel proteins, RelA, cRel and RelB, were employed to quantitate both the supershifted complex observed with one antibody, as well as the remaining non-supershifted complex when two antibodies were used to ablate the activities of their cognate Rel proteins. We found greatly increased levels of the cRel:p50 dimer activity in IκBε−/−B cells extracts stimulated with either IgM or LPS (Figure 3B and 3C). Interestingly, examining RelA activity, we found that RelA nuclear activity was seen to be significantly increased in LPS-stimulated IκBε−/−B cells when compared to wild type B cells, but not in the IgM stimulation condition (Figure 3B and 3C). No difference was seen the basal levels of RelA and cRel (Supplementary Figure 1B), suggesting that this affect is induced during stimulation of the B cells. We employed immunoblots of nuclear extracts to examine the results further. Following three biological repeats we found statistically significant differences only for the hyperactivation of cRel in response to LPS, while other conditions showed the same trend as the EMSA results but did not pass the statistical test of significance (Figure 3D). Together, these biochemical data suggest that IκBε provides a key function in limiting cRel-containing dimer activity via a negative feedback loop, while it is critical for limiting RelA activity in response to only some stimuli but not others.
Figure 3.
NFκB activity is increased in IκBε−/− B cells. Purified B cells were collected and extracted into cytoplasmic and nuclear fraction at various time points. Nuclear extracts tested for total NFκB activity using EMSAs. (A) IκBε−/− B cells stimulated with either IgM or LPS exhibited increased NFκB activity at 18 and 24 hours following stimulation. (B) The 24 hours nuclear extracts were incubated with antibodies directed towards anti-RelA (αRelA), anti-RelB (αRelB), anti-cRel (αcRel) as well as combination of these antibodies (αRelA/αRelB, αRelA/αcRel, and αRelB/αcRel). Following a 20 minute incubation, 32P-labled probe was added and allowed to incubate for a further 15 minutes. The resulting samples were run on a 5% non-reducing acrylamide gel. (C) The resulting supershifts were quantitated using ImageJ and graphed below each shift. Both IgM and LPS stimulation had increases in the cRel/p50 activity in the IκBε−/− B cell extracts when compared to extracts of wild type B cells stimulated under the same conditions. LPS stimulated IκBε−/− B cell extracts had increased RelA activity. (D) Nuclear western blots for RelA and cRel were run and quantitated to determine if the increased cRel activity observed in the supershifts was the result of increased RelA and cRel levels in the IκBε−/− B cells. Increased cRel protein levels were observed in the nuclear extracts from IκBε−/− B cell when compared to wild type B cell extracts. Similar levels of RelA were found between wild type and IκBε−/− B cell nuclear extracts. Data shown in (A) are representative of two independent experiments. Data shown in (B, C and D) are representative of three independent experiments (n=3, error bars are represented as standard deviations). *P < 0.05, **P < 0.01, ***P < 0.005 and ****P <0.001 (unpaired t-test).
A mathematical model of RelA and cRel dynamics suggests a kinetic basis for IκBε’s stimulus-specific functions
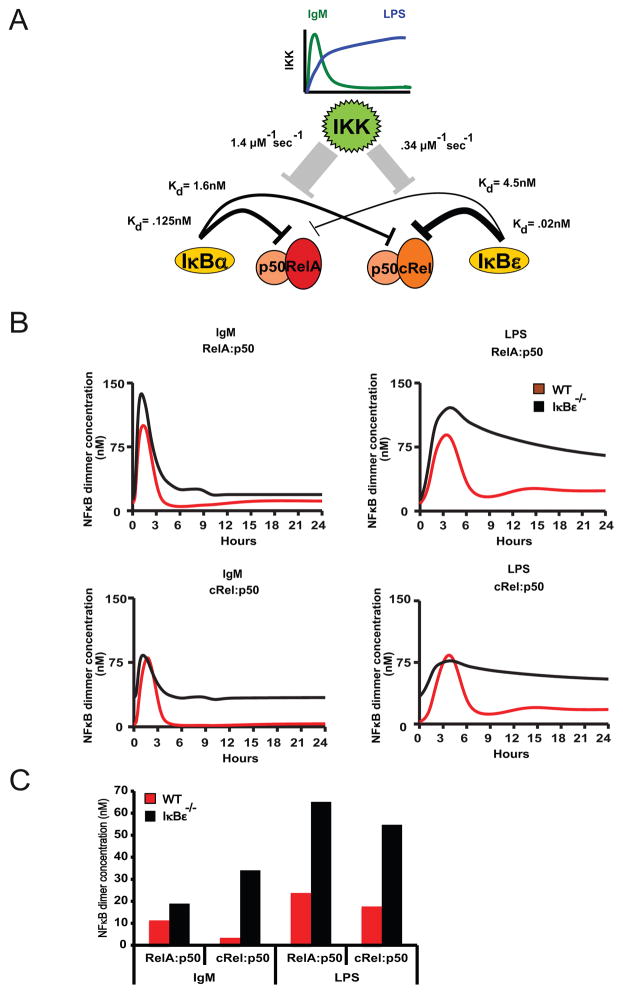
The observation that the signaling phenotypes are stimulus-specific may suggest that there are underlying stimulus-specific biochemical mechanisms such as a co-stimulatory signaling pathway that is activated by one stimulus but not another. An alternative, more parsimonious explanation may be that differential signaling kinetics may account for the stimulus-specific phenotypes. Using a mathematical modeling approach we sought to test the latter hypothesis. To begin, we summarized the known relative relationships between the two potential negative feedback regulators IκBα and IκBε in terms of their interactions with RelA and cRel-containing dimers, their differential responsiveness to IKK-induced degradation and the stimulus-specific dynamics of IKK activity (Figure 4A). Interestingly, we found that whereas NFκB-responsive IκBα gene expression required RelA, NFκB-responsive IκBε gene expression could be mediated by either RelA or cRel (Supplementary Figure 2). Next, we constructed a mathematical model with these parameters by adapting a previously established mathematical model (3) to include the cRel dimers and to recapitulate B cell-specific dynamic control of RelA- and cRel-containing dimers (see Supplementary Methods). Simulations of this model with the IgM-induced transient IKK activity showed that RelA:p50 dimer is barely affected by IκBε-deficiency, but that in response to LPS-induced long lasting IKK activity, RelA:p50 remains hyperactivated at late times (Figure 4B, top panels). In contrast, cRel:p50 was hyperactivated in either condition (Figure 4b, bottom panels). By quantitating the time course at 24hrs, the stimulus-agnostic effect on cRel and stimulus-specific effect on RelA is readily appreciated (Figure 4C); this graph in fact closely resembles the experimental results obtained biochemically (Figure 3C).
Figure 4.
Computation modeling of the IκBε−/− B cells’ NFκB activity identifies IκBε as the dominant regulator of cRel:p50 dimers. Hypothetical model of the IκBα and IκBε control of NFκB within B cells following IgM or LPS stimulation. (A) IgM and LPS stimulation results in two different IKK activity profiles; IgM stimulation has a transient IKK activity, while LPS results in lasting IKK activity. Activation of IKK results in the degradation of IκBs, and can release NFκB dimers into the nucleus. From previous data, we infer that IκBε has a very strong affinity for cRel:p50, while IκBα has a strong affinity for RelA:p50, but not as strong as IκBε-cRel:p50. IκBε has a very low affinity for RelA:p50, while IκBα has a weak affinity for cRel:p50, although the affinity between IκBα-cRel:p50 is stronger than IκBε-cRel:p50. (B) Data from our computation model illustrates the NFκB activity profiles for both wild type and IκBε-deficient B-cells under IgM and LPS stimulus. (C) The model is able to recapitulate the experimental late timepoints, in which IκBε-deficient B-cells have elevated cRel:p50 after both LPS and IgM stimulation. However, RelA:p50 is only elevated after LPS stimulation in IκBε-deficient B-cells.
These simulation results demonstrate that the kinetic argument is a sufficient explanation for the stimulus-specific phenotype seen in IκBε-deficient B-cells. We may summarize the kinetic argument as follows: In response to transient IKK signals, IκBα is capable of providing post-induction repression on its high affinity target RelA:p50, but less effectively on its low affinity target cRel:p50, which requires IκBε for complete suppression. However, in response to long lasting IKK signals, IκBα’s responsiveness to IKK renders it effectively neutralized; thus in this stimulation condition IκBε, which has lower responsiveness, plays an important role not only for its high affinity target cRel:p50 but also its low affinity target RelA:p50. Interestingly, the single specificity of the IκBα negative feedback loop for RelA:p50 and the dual specificity of the IκBε negative feedback loop for RelA;p50 and cRel:p50 are reflected in the dimer requirements for IκBα and IκBε inducible expression (Supplementary Figure S1). We note, that although reported kinetic relationships (Supplementary Methods) are consistent with this sufficiency argument, we cannot rule out that a stimulus-specific signaling pathway also plays a role in the described phenotype.
IκBε−/−B cells show increased expression of NFκB target genes
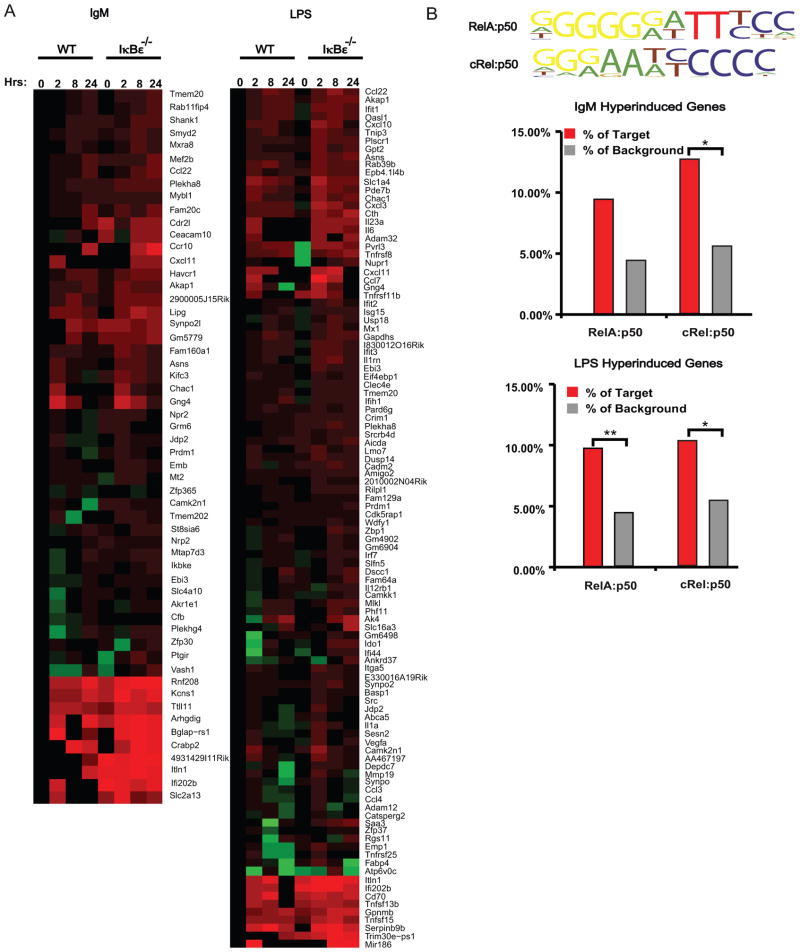
To further characterize the phenotype at the molecular level, we examined how the gene expression programs induced by IgM or LPS were affected by the loss of IκBε. To this end, we employed high-throughput sequencing of polyA RNA isolated from wild type and IκBε−/− B cells at 0, 2, 8 and 24 hours following stimulation with either IgM or LPS. Sequence data was converted into transcriptome levels using CummRbund (36), and these were normalized to the wild type 0 hour timepoint. We selected for genes induced in wild type B-cells by at least 2-fold by IgM and LPS in at least one timepoint. This resulted in 881 and 846 genes, respectively. We identified hyperinduced genes as those that showed at least a 2-fold change in the IκBε−/− B-cell data at two timepoints when compared to the corresponding wild type timepoints, resulting in 56 and 106 genes, respectively (Figure 5A).
Figure 5.
IκBε−/− B cells have an enrichment of NFκB dependent gene expression. Total RNA gene expression was obtained from RNAseq of both wild type and IκBε−/− B cells at 0, 2, 8 and 24 hours stimulated with IgM or LPS. Hyperinduced gene were identified as all genes having a 2-fold induction of at least one time point in wild type B cells and ≥ 2-fold induction at two timepoints in the IκBε−/− B cells were identified using fold change of the FPKM values. (A) The genes identified to be hyperinduced over wild type were displayed as heatmaps for the IgM and LPS stimulation. (B) Motifs for the NFκB dimers were loaded and used in Homer motif discovery software to search the promoter sequences of the identified induced and hyperinduced genes of the IκBε−/− B cells for occurrences of the listed NFκB motifs. The resulting genes containing one of these NFκB motifs were graphed as percentages over the total genes detected to be hyperinduced within the IκBε−/− samples. *P < 0.05 and **P < 0.01.
To determine if the identified hyperinduced genes are under the control of NFκB dimers, we utilized the HOMER motif discovery software adapted to perform searches of the known NFκB dimer motifs previously identified (29) and summarized here (Figure 5B top). We found enrichment for these κB-motifs within the promoter regions of genes hyperinduced in IκBε−/−B cells when compared to controls that were not hyperinduced (Figure 5B). Interestingly, we found that the enrichment of the cRel:p50 motif in hyperinduced genes was statistically significant in both the IgM and LPS condition; however, the RelA:p50 motif enrichment was statistically significant only when B-cells were activated with LPS, and not IgM. These results reflect the prior biochemical data that showed that whereas cRel:p50 is hyperactivated in IκBε−/− B-cells in both IgM and LPS conditions, RelA:p50 hyperactivation occurs primarily in response to LPS (Figure 3C).
Increased expression of IL-6 mediates the enhanced proliferation of IκBε-deficient B cells
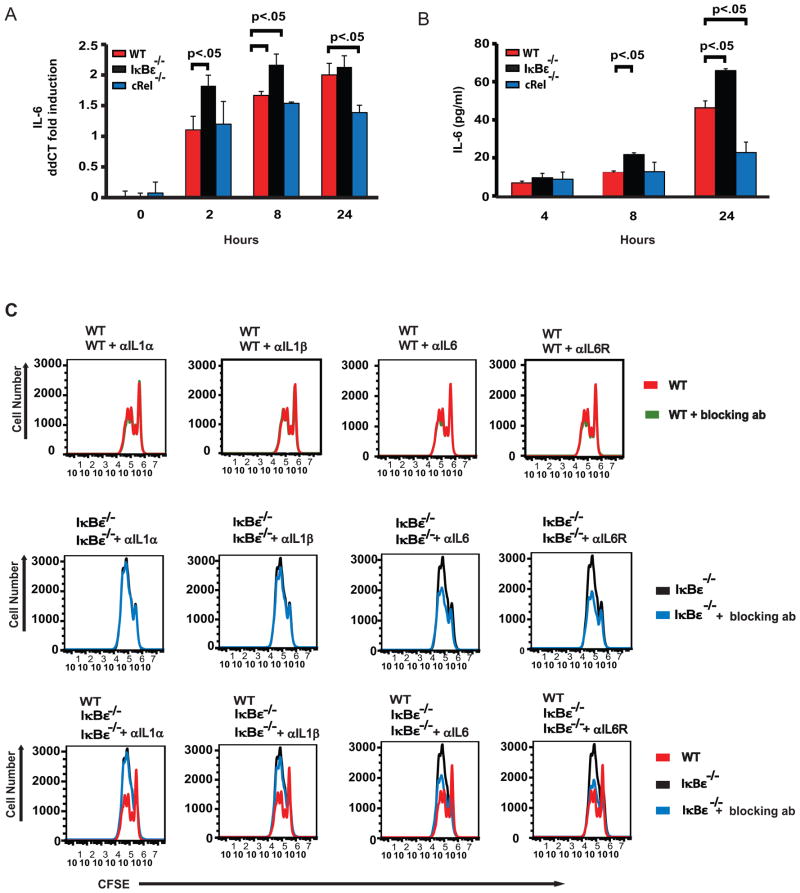
Among the NFκB target genes that were hyperinduced in LPS-stimulated IκBε−/− B-cells (Supplementary Table), we found inflammatory IL-6, which was initially discovered as a B cell stimulating and differentiation factor. Subsequent studies of IL-6’s effects on B cells have found that IL-6 allows for increased proliferation, enhanced differentiation and reduced apoptosis in B cells (37–40). The literature is unclear whether IL-6’s expression is cRel- or RelA-dependent (41–44). In our stimulation conditions, we found that IL-6 is robustly induced in response to LPS in wild-type cells both at the level of mRNA (Figure 6A) and secreted cytokine measured in supernatants (Figure 6B). As expected, IκBε−/− B cells show hyperinduction at both early and late times, with the strongest effect at the protein level being at the late timepoint of 24 hrs. Interestingly, cRel-deficient B-cells showed no mRNA reduction at early timepoints (2 and 8 hrs), though a significant deficiency was observed at 24 hrs. Further, IL-6 hyperinduction appeared to be LPS-specific, correlating with RelA-hyperactivity, and prior observations in other cell types that pointed to RelA-dependent expression (41–43). Together, these data suggest that IL-6 induction is first triggered by RelA and may then be enhanced by cRel-containing dimers.
Figure 6.
Release of IL-6 is enhanced in IκBε−/− B cells. B cells from wild type and IκBε−/− mice were isolated and stimulated with LPS. Supernatants and RNA extracts were collected at 0, 4, 8 and 24 hours. (A) Quantitative PCR results of wild type, IκBε−/− and cRel−/− B cells. IL-6 expression is enhanced by the loss of IκBε and is partially dependent on cRel. (B) ELISA assay for IL-6 in wild type, LPS stimulated IκBε−/− B cells have increased cytokine release of IL-6 when compared to wild type IκBε−/− and cRel−/− B cells. The loss of cRel reduced the amount of IL-6 released. (C) Neutralizing antibodies against the cytokines IL-1α, IL-1β and IL-6 or receptor blocking antibodies against the IL-1α, IL-1β and IL-6 receptor were used at a concentration of 2 μg/ml in B cell proliferation assays. Reduced proliferation of IκBε−/− B cells only occurred in the presences of antibodies directed against the individual cytokines or their receptors for the cytokines IL-6. Wild type B cell proliferation is only slightly reduced in the presence of the IL-6 antibody. Data shown in (A and B) are representative of at 3 independent experiments (n=3, error bars are represented as standard deviations). *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001 (unpaired t-test). Data shown in (C) are representative of 2 independent experiments.
We next asked if the enhanced proliferation of the LPS stimulated IκBε−/− B cells is potentially mediated by increased autocrine IL-6 co-stimulation of these cells. To test this hypothesis, we isolated B cells from wild type or IκBε−/− mice and stimulated them with LPS in the presence of either 2 μg/ml cytokine-neutralizing or 2 μg/ml receptor-blocking antibodies for IL-6. We used antibodies neutralizing IL-1ε and IL-1β as controls, as these pro-proliferative cytokines were not found to be hyperinduced in our transcriptomic profiling. Effects on B cell proliferation was measured using CFSE staining. We found that antibodies blocking IL-6 signaling had little effect on B cell expansion from wild type mice; however, using B cells from IκBε−/− mice we found a reduction in the B cell expansion to almost wild type levels (Figure 6C). Neither of the control antibodies had an effect on the LPS-triggered expansion of wild type or IκBε−/− B cells. These findings suggest that the increased production of IL-6 is at least in part, responsible for the increased proliferative capacity of IκBε−/− B cells.
Discussion
In this study, we identified IκBε as a key negative feedback regulator of cRel-containing NFκB dimers in B-cells, which thus limits B-cell proliferation in response to mitogenic stimulation. In contrast, to our understanding based on fibroblast studies, we found that IκBε also plays a non-redundant role in limiting the ubiquitous RelA-containing NFκB, albeit in a stimulus-specific manner. This result led to two insights, first from a physiological perspective we found that limiting RelA activation is relevant for controlling B-cell expansion, as neutralizing the expression of the RelA-target gene IL-6 mitigated the IκBε-deficient phenotype. Secondly, considering the NFκB system as a dynamical system, we conclude that the stimulus-specific functions of IκBε negative feedback are based on kinetics rather than the engagement of a stimulus-specific mechanism or pathway.
Members of the IκB protein family have been found to preferentially bind different NFκB members (22,23,45–47). Unlike IκBα, which is known to bind and regulate RelA-containing dimers, IκBε has been shown to bind with cRel homodimers and cRel:p50 heterodimers, although RelA-containing dimers may also be bound (22,23). Interestingly, we found that these binding specificities are also reflected in the specificity of their NFκB-responsive expression: whereas IκBα expression is highly NFκB inducible in a RelA-dependent manner, NFκB-inducible expression of IκBε may be mediated by RelA or cRel. Whereas IκBα is a negative feedback regulator dedicated to RelA-dimers, IκBε can be effective for both cRel- and RelA-dimers. As a regulator of two NFκB dimers, IκBε may thus also mediate cross-regulation between them, whose physiological relevance remains to be explored.
Whereas RelA is a critical regulator of the inflammatory response in tissue cells and in macrophages, cRel is critically required for B cell proliferation (16–19). Here we revealed that IκBε−/− B cells showed increased expansion in CFSE dye dilution studies. Using the computational phenotyping tool FlowMax revealed that IκBε−/− B cells stimulated with either IgM or LPS showed an increase in the percentage of B cells responding to stimulation (pF0) with little change in the time to division or death parameters. Because responding cells are protected from undergoing apoptosis, we also found an increase in survival by 7AAD staining. In a previous study, IκBε−/− B cells were found to show enhanced survival and enhanced expression of cRel (25) in unstimulated conditions, but the functions of IκBε within the dynamical context of B-cell expansion was not investigated. IκBα deficiency, however, was shown to result in increased B-cell proliferation (48). Though the underlying mechanism was not examined, it is likely that basal hyperactivity of RelA may result in hyper-expression of cRel in the IκBε knockout, mediating hyper-proliferation. Our study demonstrates the negative feedback function of IκBε plays a critical and non-redundant role in limiting B-cell expansion.
In examining the downstream mediators of this phenotype, the biochemical analysis identified the cRel:p50 dimer, which is a known activator of B-cell proliferation, as being misregulated in response to both BCR and TLR stimulation. Interestingly, we also identified the RelA:p50 dimer as being misregulated, albeit only in cells responding to LPS, not BCR stimulation, NFκB dimers may not only be distinguished biochemically and with immunological tools, but given their somewhat different binding sequence preferences, they may also be distinguished to some degree by the binding motifs present in downstream target genes. We employed this approach by identifying hyperinduced genes in the IκBε−/− B cells using RNAseq and screening their regulatory regions using Homer motif discovery software modified to search for NFκB dimer motifs identified by Bulyk ML et. al. (29). Intriguingly the cRel:p50 motif was found to be statistically overrepresented in hyperexpressed genes in response to both stimuli, but the RelA:p50 motif only rose to statistical significance in response to LPS. Thus, two lines evidence support the conclusion that the NFκB dimers under the control of IκBε are a function of the initiating stimulation.
Given the established role of cRel in B-cell expansion, we examined whether RelA-misregulation is functionally relevant. One LPS-specific misregulated target gene whose NFκB binding site conforms to the RelA-dimer motif is the cytokine IL-6. Remarkably, both the direct neutralization of IL-6 and the blocking of the IL-6 receptor reduced IκBε−/− B cells proliferation to near wild type levels, while only slightly affecting wild type B cell proliferation. These results suggest that the autocrine stimulation of IL-6 is at least partially responsible for the enhanced proliferation of IκBε−/− B cells in response to LPS, and that hyperactivation of RelA is indeed functionally relevant. Our investigation of the proliferative phenotype observed in IgM stimulated IkBe−/− B cells found IL-6 expression to be lacking, yet the IgM stimulated IkBe−/− B cells still displayed increased proliferation compared to wild type B cells. This difference between IgM and LPS stimulated IkBe−/− B cells could be explained by the differences in the downstream stimulation pathways of IgM and LPS. The RNA-seq data displayed in our study (Figure 5A) demonstrate that there is very little overlap of the genes upregulated by each pathway. We propose that stimulation with LPS leads to a proliferative phenotype which is dependent on IL-6 upregulation and stimulation, yet in the case of IgM stimulation, an upregulation of a different set of proliferative genes occurs.
How may the same negative feedback regulator target different signal transducers in response to different stimuli? By adapting an established mathematical model of the NFκB signaling module to B-cells, we showed here that a kinetic explanation is sufficient to account for the observations. Specifically, differences in the interaction parameters of IκBα and IκBε with RelA and cRel dimers, in conjunction with IκB’s differential responsiveness to IKK activities whose temporal profiles are in turn stimulus-specific, could reproduce the stimulus-specific control of the RelA dimer. The present study is thus an extension of previous work that demonstrated that kinetic differences between IκBα and IκBδ could impart them with stimulus-specific functions: whereas IκBα is critical for turning off NFκB activity following transient activation signals, IκBδ limits NFκB activity when the activation signals are long lasting (3). As our understanding of signaling systems improves and mathematical modeling is adopted more widely, we may expect to find an increasing number of examples in which considerations of the kinetics is critical for an understanding of the specificity of observed phenomena.
Supplementary Material
Acknowledgments
This study was supported in part by NIH R01AI083453 and P50G071573 (AH), NIH/NCI T32 CA009523 (BA), NSF-GTRF (MNS and RT).
Reference List
- 1.O’Dea E, Hoffmann A. The regulatory logic of the NF-kappaB signaling system. Cold Spring Harb Perspect Biol. 2010;2:a000216. doi: 10.1101/cshperspect.a000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 3.Shih VF, Kearns JD, Basak S, Savinova OV, Ghosh G, Hoffmann A. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci U S A. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerondakis S, Grumont R, Rourke I, Grossmann M. The regulation and roles of Rel/NF-kappa B transcription factors during lymphocyte activation. Curr Opin Immunol. 1998;10:353–359. doi: 10.1016/s0952-7915(98)80175-1. [DOI] [PubMed] [Google Scholar]
- 6.Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 7.Gerondakis S, Grumont RJ, Banerjee A. Regulating B-cell activation and survival in response to TLR signals. Immunol Cell Biol. 2007;85:471–475. doi: 10.1038/sj.icb.7100097. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 9.Grumont RJ, Gerondakis S. The subunit composition of NF-kappa B complexes changes during B-cell development. Cell Growth Differ. 1994;5:1321–1331. [PubMed] [Google Scholar]
- 10.Grumont RJ, I, Rourke J, O’Reilly LA, Strasser A, Miyake K, Sha W, Gerondakis S. B lymphocytes differentially use the Rel and nuclear factor kappaB1 (NF-kappaB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J Exp Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grumont RJ, Strasser A, Gerondakis S. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-kappaB regulated c-myc transcription. Mol Cell. 2002;10:1283–1294. doi: 10.1016/s1097-2765(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 12.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 13.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 14.Sriskantharajah S, Belich MP, Papoutsopoulou S, Janzen J, Tybulewicz V, Seddon B, Ley SC. Proteolysis of NF-kappaB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat Immunol. 2009;10:38–47. doi: 10.1038/ni.1685. [DOI] [PubMed] [Google Scholar]
- 15.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–60. doi: 10.1146/annurev.immunol.20.100301.064842. Epub;%2001 Oct 4.: 709–760. [DOI] [PubMed] [Google Scholar]
- 16.Pohl T, Gugasyan R, Grumont RJ, Strasser A, Metcalf D, Tarlinton D, Sha W, Baltimore D, Gerondakis S. The combined absence of NF-kappa B1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc Natl Acad Sci U S A. 2002;99:4514–4519. doi: 10.1073/pnas.072071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liou HC, Jin Z, Tumang J, Andjelic S, Smith KA, Liou ML. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int Immunol. 1999;11:361–371. doi: 10.1093/intimm/11.3.361. [DOI] [PubMed] [Google Scholar]
- 18.Hsia CY, Cheng S, Owyang AM, Dowdy SF, Liou HC. c-Rel regulation of the cell cycle in primary mouse B lymphocytes. Int Immunol. 2002;14:905–916. doi: 10.1093/intimm/dxf055. [DOI] [PubMed] [Google Scholar]
- 19.Gilmore TD, Kalaitzidis D, Liang MC, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23:2275–2286. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- 20.Horwitz BH, Zelazowski P, Shen Y, Wolcott KM, Scott ML, Baltimore D, Snapper CM. The p65 subunit of NF-kappa B is redundant with p50 during B cell proliferative responses, and is required for germline CH transcription and class switching to IgG3. J Immunol. 1999;162:1941–1946. [PubMed] [Google Scholar]
- 21.Klement JF, Rice NR, Car BD, Abbondanzo SJ, Powers GD, Bhatt PH, Chen CH, Rosen CA, Stewart CL. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whiteside ST, Epinat JC, Rice NR, Israel A. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simeonidis S, Liang S, Chen G, Thanos D. Cloning and functional characterization of mouse IkappaBepsilon. Proc Natl Acad Sci U S A. 1997;94:14372–14377. doi: 10.1073/pnas.94.26.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Nabel GJ. A new member of the I kappaB protein family, I kappaB epsilon, inhibits RelA (p65)-mediated NF-kappaB transcription. Mol Cell Biol. 1997;17:6184–6190. doi: 10.1128/mcb.17.10.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark JM, Aleksiyadis K, Martin A, McNamee K, Tharmalingam T, Williams RO, Memet S, Cope AP. Inhibitor of kappa B epsilon (IkappaBepsilon) is a non-redundant regulator of c-Rel-dependent gene expression in murine T and B cells. PLoS One. 2011;6:e24504. doi: 10.1371/journal.pone.0024504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shokhirev MN, Hoffmann A. FlowMax: A Computational Tool for Maximum Likelihood Deconvolution of CFSE Time Courses. PLoS One. 2013;8:e67620. doi: 10.1371/journal.pone.0067620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann A, Roeder RG. Cloning and characterization of human TAF20/15. Multiple interactions suggest a central role in TFIID complex formation. J Biol Chem. 1996;271:18194–18202. doi: 10.1074/jbc.271.30.18194. [DOI] [PubMed] [Google Scholar]
- 28.O’Dea EL, Kearns JD, Hoffmann A. UV as an amplifier rather than inducer of NF-kappaB activity. Mol Cell. 2008;30:632–641. doi: 10.1016/j.molcel.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-kappaB family DNA binding. Nat Immunol. 2012;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O’Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawkins ED, Turner ML, Dowling MR, van GC, Hodgkin PD. A model of immune regulation as a consequence of randomized lymphocyte division and death times. Proc Natl Acad Sci U S A. 2007;104:5032–5037. doi: 10.1073/pnas.0700026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawkins ED, Hommel M, Turner ML, Battye FL, Markham JF, Hodgkin PD. Measuring lymphocyte proliferation, survival and differentiation using CFSE time-series data. Nat Protoc. 2007;2:2057–2067. doi: 10.1038/nprot.2007.297. [DOI] [PubMed] [Google Scholar]
- 34.Hawkins ED, Markham JF, McGuinness LP, Hodgkin PD. A single-cell pedigree analysis of alternative stochastic lymphocyte fates. Proc Natl Acad Sci U S A. 2009;106:13457–13462. doi: 10.1073/pnas.0905629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Won WJ, Kearney JF. CD9 is a unique marker for marginal zone B cells, B1 cells, and plasma cells in mice. J Immunol. 2002;168:5605–5611. doi: 10.4049/jimmunol.168.11.5605. [DOI] [PubMed] [Google Scholar]
- 36.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burdin N, Van KC, Galibert L, Abrams JS, Wijdenes J, Banchereau J, Rousset F. Endogenous IL-6 and IL-10 contribute to the differentiation of CD40-activated human B lymphocytes. J Immunol. 1995;154:2533–2544. [PubMed] [Google Scholar]
- 38.Foussat A, Wijdenes J, Bouchet L, Gaidano G, Neipel F, Balabanian K, Galanaud P, Couderc J, Emilie D. Human interleukin-6 is in vivo an autocrine growth factor for human herpesvirus-8-infected malignant B lymphocytes. Eur Cytokine Netw. 1999;10:501–508. [PubMed] [Google Scholar]
- 39.Szczepek AJ, Belch AR, Pilarski LM. Expression of IL-6 and IL-6 receptors by circulating clonotypic B cells in multiple myeloma: potential for autocrine and paracrine networks. Exp Hematol. 2001;29:1076–1081. doi: 10.1016/s0301-472x(01)00682-8. [DOI] [PubMed] [Google Scholar]
- 40.Gado K, Domjan G, Hegyesi H, Falus A. Role of INTERLEUKIN-6 in the pathogenesis of multiple myeloma. Cell Biol Int. 2000;24:195–209. doi: 10.1006/cbir.2000.0497. [DOI] [PubMed] [Google Scholar]
- 41.Kawashima T, Murata K, Akira S, Tonozuka Y, Minoshima Y, Feng S, Kumagai H, Tsuruga H, Ikeda Y, Asano S, Nosaka T, Kitamura T. STAT5 induces macrophage differentiation of M1 leukemia cells through activation of IL-6 production mediated by NF-kappaB p65. J Immunol. 2001;167:3652–3660. doi: 10.4049/jimmunol.167.7.3652. [DOI] [PubMed] [Google Scholar]
- 42.Legrand-Poels S, Schoonbroodt S, Piette J. Regulation of interleukin-6 gene expression by pro-inflammatory cytokines in a colon cancer cell line. Biochem J. 2000;349(Pt 3):765–773. doi: 10.1042/bj3490765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouaaz F, Li M, Beg AA. A critical role for the RelA subunit of nuclear factor kappaB in regulation of multiple immune-response genes and in Fas-induced cell death. J Exp Med. 1999;189:999–1004. doi: 10.1084/jem.189.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumang JR, Hsia CY, Tian W, Bromberg JF, Liou HC. IL-6 rescues the hyporesponsiveness of c-Rel deficient B cells independent of Bcl-xL, Mcl-1, and Bcl-2. Cell Immunol. 2002;217:47–57. doi: 10.1016/s0008-8749(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 45.Tran K, Merika M, Thanos D. Distinct functional properties of IkappaB alpha and IkappaB beta. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam WF, Sen R. IkappaB family members function by different mechanisms. J Biol Chem. 2001;276:7701–7704. doi: 10.1074/jbc.C000916200. [DOI] [PubMed] [Google Scholar]
- 47.Malek S, Chen Y, Huxford T, Ghosh G. IkappaBbeta, but not IkappaBalpha, functions as a classical cytoplasmic inhibitor of NF-kappaB dimers by masking both NF-kappaB nuclear localization sequences in resting cells. J Biol Chem. 2001;276:45225–45235. doi: 10.1074/jbc.M105865200. [DOI] [PubMed] [Google Scholar]
- 48.Chen CL, Singh N, Yull FE, Strayhorn D, Van KL, Kerr LD. Lymphocytes lacking I kappa B-alpha develop normally, but have selective defects in proliferation and function. J Immunol. 2000;165:5418–5427. doi: 10.4049/jimmunol.165.10.5418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.