Abstract
The role of adaptation in the evolutionary process has been contentious for decades. At the heart of the century-old debate between neutralists and selectionists lies the distribution of fitness effects (DFE)—that is, the selective effect of all mutations. Attempts to describe the DFE have been varied, occupying theoreticians and experimentalists alike. New high-throughput techniques stand to make important contributions to empirical efforts to characterize the DFE, but the usefulness of such approaches depends on the availability of robust statistical methods for their interpretation. We here present and discuss a Bayesian MCMC approach to estimate fitness from deep sequencing data and use it to assess the DFE for the same 560 point mutations in a coding region of Hsp90 in Saccharomyces cerevisiae across six different environmental conditions. Using these estimates, we compare the differences in the DFEs resulting from mutations covering one-, two-, and three-nucleotide steps from the wild type—showing that multiple-step mutations harbor more potential for adaptation in challenging environments, but also tend to be more deleterious in the standard environment. All observations are discussed in the light of expectations arising from Fisher’s geometric model.
Keywords: adaptation, experimental evolution, Fisher’s geometric model (FGM), adaptive walk, distribution of fitness effects
CHARACTERIZING the distribution of fitness effects (DFE) of new mutations remains a central focus in both theoretical and empirical population genetics. Understanding the underlying DFE speaks to questions of fundamental importance to all of evolutionary biology: What is the relative role of drift vs. selection in driving evolution? What is the adaptive potential of a population upon facing a novel selective pressure? What is the relative importance of new vs. standing variation in dictating rates of positive selection?
Testable predictions describing the change in selective effects upon environmental change come from a classical model proposed by Fisher (1930). Fisher’s geometric model (FGM) provides an illustrative approach to understand adaptation to novel environments. In this model, the effect of a mutation is described by a random vector in an n-dimensional phenotype space, originating from the position of the current phenotype in that space (i.e., the wild-type state). Fitness is evaluated as the Euclidean distance gain toward a fitness optimum that the vector confers. Different environments are characterized by different fitness optima in phenotype space, which results in theoretical expectations concerning, for example, the proportion of beneficial mutations and the correlation of fitness effects between different environments. There are a number of hypotheses concerning the DFE that can be derived from FGM. For example, the number of beneficial mutations should increase upon environmental change (i.e., for a population that has become more distant from the optimum state), the effect sizes of beneficial mutations are proportional to the distance from the optimum, and the step size of mutations in adaptive walks should become progressively smaller as the optimum is neared (cf., e.g., Orr 1998, 2003; Martin and Lenormand 2006).
Efforts to quantify the DFE have come from three types of approaches: first, statistical inference from natural polymorphism and/or divergence data (e.g., Keightley and Eyre-Walker 2010; Schneider et al. 2011), in which sequence data of natural populations are used to gain information on the DFE; second, laboratory selection experiments (e.g., Lenski et al. 1991; Rozen et al. 2002; Melnyk and Kassen 2011; Sousa et al. 2012), in which populations of microbes or viruses are evolved in the laboratory and spontaneously occurring mutations are tracked; and finally, engineered-mutation-driven experimental evolution (e.g., Fowler et al. 2010; Hietpas et al. 2011), in which specific mutations are introduced and evaluated against each other and the wild type. Each approach has relative merits and weaknesses, and thus each has provided novel insight into evolutionary processes.
Within this third category, we have recently proposed a novel approach termed EMPIRIC (Hietpas et al. 2011), whereby fitness can be empirically estimated for a large number of mutations in a given region of a protein. In this method, mutant libraries are constructed consisting of every possible single-point mutation in the protein region to be interrogated, these mutants are competed in bulk culture for a number of generations, and individual growth rates are determined by deep sequencing over a number of time points. The advantages of the EMPIRIC approach compared with the above-mentioned approaches are the following:
By systematically studying all possible mutations in a chosen region, it allows one to infer the complete DFE of all new mutations (rather than simply the mutations observed in a laboratory selection experiment that is inherently unable to quantify the proportion of strongly deleterious mutations, for example), including the direct assessment of the relative proportions of deleterious, neutral, and beneficial mutations.
Due to the design of the experiment, the risk of secondary mutations in the course of the experiment is minimized—hence, the estimates are likely to represent the actual effect of single mutations.
As a consequence of the bulk competition, identical conditions for all mutations are certain. In particular, previous studies have shown that the EMPIRIC bulk competition produces quantitatively similar selection estimates to those of a binary competition—no frequency-dependent effects or other complications owing to the high number of mutants in the culture were observed (Hietpas et al. 2011, Figure S1B; Hietpas et al. 2013, Figure S5; and Roscoe et al. 2013, Figure 2B). However, this approach is limited in the sense of considering only the DFE of a single region (as opposed to laboratory selection experiments that inherently consider the whole genome), and (as with all such experimental approaches) the selective pressures chosen may not well represent the pressures faced by the organism in nature.
Figure 2.
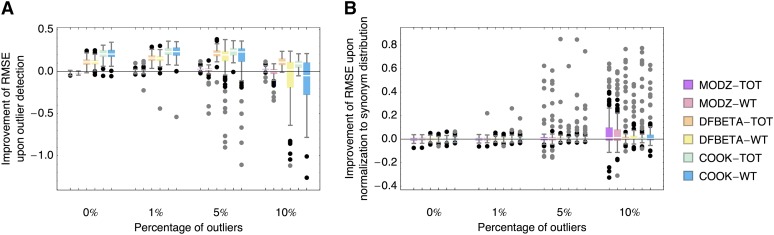
(A) Improvement of root mean square errors (RMSE) of linear regression estimates upon outlier detection, calculated as RMSE with outlier detection divided by RMSE without outlier detection. Improvement of up to 20% is possible under moderate amounts of outliers. With regression based on mutant-to-wild type (WT), outlier detection can be highly disadvantageous. (B) Improvement of RMSE upon normalization to wild-type-synonym distribution instead of single wild-type sequence, calculated as RMSE based on synonym distribution divided by RMSE based on single wild-type sequence. All analyses were performed on simulated data sets. Boxes represent the interquartile range, whiskers extend to the highest/lowest data point within the box ±1.5 times the interquartile range, and black and gray circles repesent close and far outliers, respectively.
Due to the growing availability—and decreasing cost—of such next-generation sequencing (NGS) and other high-throughput technologies, it is likely that sequencing and/or molecular genotyping will become the methods of choice for estimating fitness and for experimentally evaluating the fit of classic population-genetic models like the FGM. The estimation of fitness from NGS data presents unique challenges and sources of error in comparison to traditional methods, such as the counting of phenotypically marked genotypes, and computational tools to analyze such data are largely lacking (but for an example, see Fowler et al. 2011). Thus, we here introduce a Bayesian Monte Carlo Markov chain (MCMC) method to obtain statistically meaningful estimates of selection coefficients from EMPIRIC and related experiments. We use this method to analyze data sets containing 560 different codon changes from Hsp90 in Saccharomyces cerevisiae under six environmental conditions (including selective challenges of both temperature and salinity). We show that the experimental method results in highly accurate fitness measurements, even allowing for the detection of a deleterious synonymous codon, and further demonstrate the increased (and predicted) potential for beneficial mutations as the population is brought farther from the optimum phenotypic state. When comparing the DFEs consisting of one- vs. several-nucleotide steps, we observe a strong correspondence with what is predicted about the nature of adaptive walks by the FGM—namely, that there is a higher potential for beneficial mutations comprising multiple-nucleotide steps when the population is far from the phenotypic optimum, whereas such mutations are likely to have deleterious effects when the optimum is close.
Materials and Methods
EMPIRIC experimental procedure and environmental challenges
Library construction, bulk competitions, and sequencing were performed as previously described (Hietpas et al. 2013). Briefly, codon substitution libraries for the amino acid 582–590 region of yeast Hsp90 (Hsp82) were transformed into the DBY288 shutoff strain (can1-100 ade2-1 his3-11,15 leu2-3,12 trp1-1 ura3-1 hsp82::leu2 hsc82::leu2 ho::pgals-hsp82-his3) of S. cerevisiae by the lithium acetate method (Gietz and Woods 2002). The yeast library was amplified in nonselective galactose (Gal) medium with 100 μg/ml ampicillin (1.7 g yeast nitrogen base without amino acids, 5 g ammonium sulfate, 0.1 g aspartic acid, 0.02 g arginine, 0.03 g valine, 0.1 g glutamic acid, 0.4 g serine, 0.2 g threonine, 0.03 g isoleucine, 0.05 g phenylalanine, 0.03 g tyrosine, 0.04 g adenine hemisulfate, 0.02 g methionine, 0.1 g leucine, 0.03 g lysine, 0.01 g uracil per liter with 1% raffinose and 1% galactose) for 12 hr at 30° and transferred to selective dextrose medium (identical to Gal medium but raffinose and galactose replaced with 2% dextrose) at 30° for 8 hr to begin shutoff of the wild-type copy of Hsp90 . After 8 hr, the library culture was split into six distinct environmental conditions (25°, 30°, 36°, 25° + S, 30° + S, and 36° + S, where S represents the addition of 0.5 M sodium chloride), and the cultures were grown for 12–20 generations with samples taken at distinct time points and stored at −80°. Wild-type generation time was calculated by monitoring the increase in OD600 of a monoculture of DBY288 cells harboring the wild-type Hsp90 construct under each environmental condition. Yeast lysis, DNA isolation, and preparation for Illumina sequencing were performed as previously described (Hietpas et al. 2012). Sequencing was performed by the University of Massachusetts deep sequencing core facility and generated ≈30 million reads of 99% confidence at each read position as judged by PHRED scoring (Ewing and Green 1998; Ewing et al. 1998).
Timescaling and outlier detection
To account for the slower growth of S. cerevisiae in stressful environments, we scaled the data such that sampling times are specified in hours. In other words, we measure the growth rate per hour instead of per wild-type generation (as commonly done), which results in a less intuitive interpretation of the growth rates but at the same time allows for a more consistent comparison of growth rates across environments. Missing data points are replaced with zeros.
Similar to previous studies, 10 of the randomized codons that result in internal HpaII sites (endonuclease was used in processing samples) were removed from further analysis.
To correct for sequencing errors at single sampling points that can, for example, arise from amplification errors during the sequencing procedure (for some example trajectories, see Supporting Information, Figure S1), we performed an outlier-detection step on the basis of an individual mutant’s trajectory. We acknowledge that some fraction of these removed data points could reflect biologically interesting non-log-linear effects; however, it is likely that the majority are errors and so their removal is necessary. In Results and Discussion, we show that performance is optimized when using the log ratio of the mutant’s read number to the total number of reads at each individual time point. These ratios serve as the basis for an outlier detection using the DFBETA statistic, using a cutoff of 2 (Belsley et al. 1980).
Model underlying the MCMC and estimation procedure
Each mutant i ∈ {1, … , K} in the culture has an initial population size ci at the beginning of the experiment and is assumed to grow exponentially at a constant rate ri (cf. Figure S2). The true abundance of an exponentially growing mutant in the population, Ni(t), follows the equation where ci is the (true) population size of that mutant at time 0, ri is the growth rate of a particular mutant i, t ∈ {1, … , T} is the sampling time point in hours, and T is the number of sampling time points.
At each sampling point and for each mutant i, sequencing reads are drawn from a multinomial distribution with parameters nt (total number of reads at time t) and {p1, p2, pk}, where is the relative amount of mutant i in the population after t generations of exponential growth at rate ri. We assume that each sampling point t represents an independent drawing experiment of ni,t reads for each mutant i from the total amount of reads Hence, the overall likelihood is the product of the individual likelihoods, For each mutant, we estimate its initial population size ci and the corresponding growth rate ri. However, since we measure relative abundance of each mutation, only relative sizes and growth rates can be estimated, which reduces the effective number of estimated parameters by 2. Sampling points that were detected as outliers were excluded from the analysis for the respective mutant and time point. To preserve the overall read number for the multinomial sampling (and hence the multinomial probabilities), the sum of these dropped counts was added in an additional row for each time point at the end of the data set. This creates a set of at most T additional parameters that account for all outliers of the respective time point.
To allow for arbitrary distributions of growth rates and initial population sizes, we use improper priors for all estimated parameters that give equal prior probabilities to all attainable values of ri and ci (i.e., ri ∈ ℝ+ and ci ∈ ℕ), but the method allows for the implementation of any kind of prior assumptions. We implemented a Metropolis–Hastings algorithm in R to compute the stationary distribution of the MCMC. The variances of the proposal distributions were set such that the acceptance ratio was kept between 20 and 30%. A burn-in phase of 5,000,000 accepted parameter combinations and a subsequent estimation phase of 10,000,000 accepted values ensured convergence of the MCMC for the high number of parameters that are estimated simultaneously. Subsampling of every 1000th parameter combination resulted in the data sets used for further analysis. The R package “coda” was used to confirm convergence and mixing of the MCMC.
During the estimation phase, the wild-type sequence was normalized to growth rate r = 1 in every generation and its population size to 10,000—both values representing an arbitrary choice that does not affect the outcome of the estimation. A corrected mean of the growth rates of all mutations synonymous to the wild type is later used as a normalization constant, and all growth rates are rescaled accordingly. This approach improves the reproducibility of fitness estimates from full experimental replicates (cf. Results and Discussion). The original wild-type sequence is excluded from further analysis. The corrected mean is determined as the mean growth rate of those mutants that are synonymous to the wild type (excluding the original wild type itself) and that are within the mean plus or minus two standard deviations from the mean of the set of all mutants synonymous to the wild-type sequence. This accounts for potential outliers of the distribution of synonyms (cf. Results and Discussion). After this procedure, 560 single-nucleotide mutations (in all environments) remain for further analysis.
Test data sets
To evaluate experimental noise, the power of the MCMC, and the reliability of the outlier detection, we created 800 simulated data sets according to the above-described model with varying amounts of artificial noise. Distributions of fitness effects and initial population sizes were drawn from the empirical distributions of the respective parameters obtained from the 30° and 30° + S data sets to represent a fast- and a slow-growing sample data set. A distribution of artificial wild-type-synonym growth rates was drawn from a normal distribution with mean 1 and variance obtained from the empirical distribution of wild-type synonyms. Of note, we are not able to detect any sequencing errors that lie within the range of the distribution that arises from multinomial sampling. Therefore, we obtained the expected mean sampling number at a time point, mt, and its standard deviation σ. We then introduced outliers drawn as random values from a log-uniform distribution in the interval [1/10mt, 10mt], excluding the interval of the expected mean sampling number at a data point plus or minus four times its standard deviation (i.e., [mt − 4σ, mt + 4σ]). We used four different probabilities of outliers, f = 0, 0.01, 0.05, 0.1. Outliers were assigned independently for each data point. We then performed the outlier detection on all data sets, using the DFBETA statistic, and ran the MCMC for 50 simulated data sets for 30° + S both with and without outlier detection for f = 0, 0.01, 0.05.
Results and Discussion
Evaluation of the MCMC approach
We evaluated two different methods to perform a linear regression as a basis for outlier detection, utilizing the simulated data sets detailed in Materials and Methods. First, we based the regression on log ratios of mutant to wild-type reads (in the following, we abbreviate this approach to WT), which is the commonly used method (e.g., Hietpas et al. 2011)—however, we hypothesized that potential noise in the wild-type measurement that transfers to all measurements could cause deviations in the estimates. For comparison, we based the regression on log ratios of mutant reads to the sum of all reads for that time point (abbreviated by TOT). Compared with the former procedure, this introduces an error that is due to the nonexponentiality of a sum of exponentially growing individuals with different growth rates. Figure 1 and Figure 2 (as well as Figure S3 and Figure S4) show that this error is small compared with the potential noise that sampling variance and mismeasurement of the wild-type sequence introduce into each measurement—which suggests that for similar next-generation sequencing data sets containing many mutants with individually low read numbers, the log ratio of mutant-to-total read number (or to another, ideally large subset of read numbers) provides much better estimates than the commonly used mutant-to-wild-type ratio.
Figure 1.
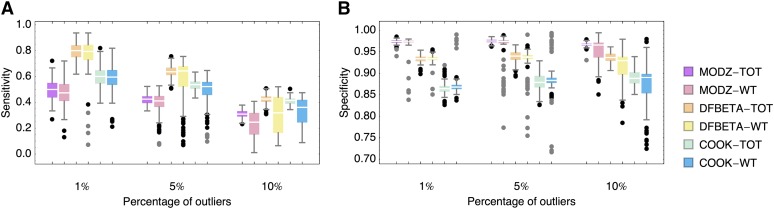
Sensitivity (A) and specificity (B) of outlier detection obtained from 100 simulated data sets each for three different percentages of outliers and six detection methods. Boxes represent the interquartile range (i.e., the 50% C.I.), whiskers extend to the highest/lowest data point within the box ±1.5 times the interquartile range, and black and gray circles represent close and far outliers, respectively. Clearly, DFBETA-TOT performs best in terms of sensitivity while maintaining a reasonable specificity. Note the increased amount of outliers when regression is based on mutant-to-wild-type ratio (WT) compared with mutant-to-total (TOT) regression.
Furthermore, we tested three commonly used outlier detection methods, based on the residuals of a linear regression of the above-described log ratios: a modified z-score, the DFBETA statistic, and Cook’s distances. These were chosen to represent different levels of complexity and differently conservative statistics. Whereas the modified z-score (MODZ) is solely based on the residuals of a simple linear regression, both DFBETA and Cook’s distances (COOK) are based on the change in parameters upon rejecting individual data points. Hereby, DFBETA calculates the effect on the estimated parameters (i.e., slope and intercept of the linear regression) separately, whereas COOK computes an aggregate measure of influence of a data point. As suggested in the literature (Belsley et al. 1980), we chose cutoffs of 3.5 for MODZ, 2 (as a conservative cutoff for small data sets) for DFBETA, and 4/n (where n is the number of sampling points) for COOK.
We classified the fraction of true positives (TP) (i.e., sampling points correctly classified as outliers), false positives (FP) (i.e., sampling points wrongly classified as outliers), true negatives (TN) (i.e., sampling points correctly classified as not being an outlier), and false negatives (FN) (i.e., true outliers that were not identified as such), as well as the sensitivity and the specificity for each of the regression methods combined with each of the detection methods. The results are shown in Figure 1 and Figure S3.
Next, we evaluated the root mean square error (RMSE) of growth rate estimates based on a linear regression with different outlier detection methods and under different normalization (cf. Figure 2 and Figure S4). Figure 2A shows that even if no outliers are present in the data, the outlier detection methods DFBETA and COOK improve the estimates. This owes to the fact that they filter a part of the sampling noise from the multinomial sampling. However, because not many real data points are discarded using DFBETA (cf. fraction of false positives in Figure S3), we consider this effect unproblematic. Regardless, the regression based on TOT demonstrates much better results than WT, likely owing to the inability to detect outliers in the wild-type sequence reliably. For the same reason, it is favorable to use a normalization of growth rates to the distribution of synonyms instead of a normalization to the wild-type sequence, as is shown in Figure 2B. Although the median improvement in the RMSE stays close to 0, the quality of estimates can, in some cases, increase by up to 80% if the normalization is based on the distribution of synonyms. Naturally, and apart from the above-described filtering of sampling noise, the knowledge of the true outliers is favorable in the case of many outliers (cf. Figure S4B).
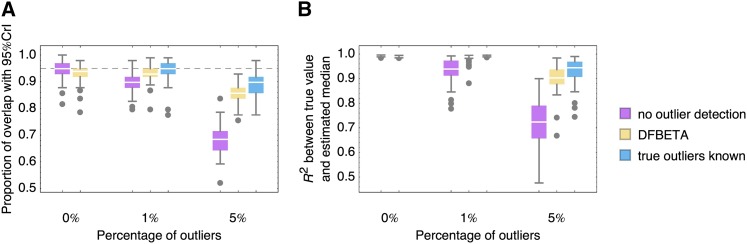
We also evaluated the fraction of mutations for which the 95% credibility interval of the MCMC estimate includes the true growth rate (Figure 3), comparing no outlier detection with the DFBETA detection method and the knowledge of the true outliers (notably a case that is unachievable for real data). Whereas the performance is similar if no outliers are present in the data set, there is a steep decline in the proportion of well-estimated growth rates if 1 or 5% of outliers are present and no outlier detection is conducted. At the same time, the performance with outlier detection is almost as good as if true outliers are known. However, we note that, even if the estimation of growth rates from EMPIRIC bulk competition experiments is on average very accurate, statements about single mutations should ideally be reinforced by experimental replicates or separate competition assays.
Figure 3.
Performance of MCMC on 50 simulated data sets for each percentage of outliers and each mode of outlier detection/knowledge. Boxes represent the interquartile range, whiskers extend to the highest/lowest data point within the box ±1.5 times the interquartile range, and gray circles represent outliers. (A) Proportion of mutations for which the 95% credibility interval of the MCMC estimate includes the true growth rate. Outliers are mostly due to cases of wrong normalization that yield systematic displacement of all posteriors, where the correlation between true and estimated values is not affected (see B). (B) Pearson’s R2 measure of correlation between the estimated median growth rates for each mutant in the data set and its true value. Correlation and 95% overlap are much improved for 1 and 5% outliers if outlier detection is performed.
In conclusion, all obtained results point toward DFBETA in combination with the regression approach TOT for outlier detection and a normalization based on the distribution of synonyms instead of the wild-type sequence only being the most reliable data processing steps that should be undertaken to filter sequencing errors.
Evaluation of the DFE in six environments
We next applied our novel MCMC approach to characterize changes in the DFE across six environmental conditions. Following Hietpas et al. (2011), deep sequencing of systematic mutant libraries was used to evaluate the fitness of 560 point mutants in a nine-amino-acid window of Hsp90. Hietpas et al. (2013) evaluated four environmental conditions and performed analyses only at the amino acid level based on log-linear regressions. Thus, we here add two additional environmental conditions, perform analyses based on the nucleotide level, and implement a Bayesian MCMC approach to incorporate uncertainty into our estimates based on an exponential growth model.
Hsp90 is a well-studied chaperone that aids in protein folding, especially at high temperatures (Borkovich et al. 1989). As such, the experimental treatments included three different temperatures (25°, 30°, and 36°), with the expected importance of Hsp90 increasing with temperature. Three additional treatments consisted of the addition of 0.5 M sodium chloride (NaCl) at each temperature (hereafter referred to as the 25S, 30S, and 36S treatments). Elevated NaCl does not induce increased Hsp90 levels (Gasch et al. 2000; Causton et al. 2001; Berry and Gasch 2008), although its basal activity is required for activation of the high-osmolarity glycerol pathway that provides for efficient growth in elevated salinity environments (Hohmann 2002; Yang et al. 2006, 2007; Hawle et al. 2007). Based on these observations and the absolute growth rates of wild-type yeast in the six environments (Table 1), we state the following (cf. also Hietpas et al. 2013):
Table 1. Relative growth rates of the standard wild type in the six environments.
| Temperature | Standard salinity | Elevated salinity |
|---|---|---|
| 25° | 0.63 | 0.3 |
| 30° | 1 | 0.45 |
| 36° | 0.83 | 0.33 |
Thirty degrees corresponds to the standard condition. Therefore, this environment represents the one that yeast is best adapted to. Every other environment represents some kind of “adaptive challenge”, which is reflected in lower absolute growth rates.
The constraint on the protein function of Hsp90 grows with increasing temperature. However, absolute growth rates imply that wild-type yeast is better able to cope with increased than with decreased temperature.
Absolute growth rates indicate that elevated salinity represents a greater environmental challenge than a change in temperature. However, elevated salinity does not result in higher constraint on Hsp90 protein function.
A description of our findings concerning the shape of the DFE in these six environments and the implications for the potential of adaptation to challenging environments follow.
The shape of the full DFE
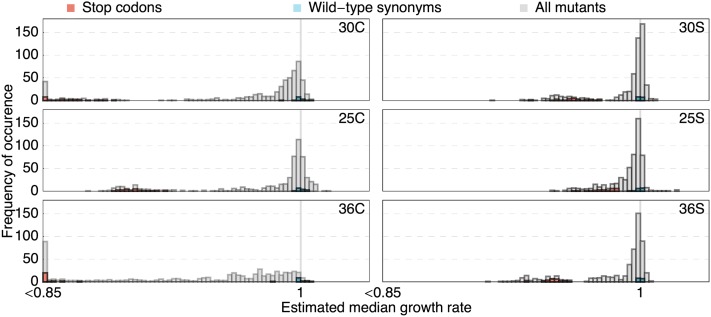
Figure 4 shows the histograms of the full DFEs obtained for all six environments. All DFEs but that for the 25S environment are bimodal, with one mode centered at wild-type-like fitness and the other mode centered at stop-codon-like fitness. It is likely that the bimodal shape is not seen for 25S because the absolute growth rate of the wild type is the lowest (cf. Table 1). Therefore, the difference between wild-type-like and stop-codon-like fitnesses is smaller. Second, we find the largest beneficials in the 25S environment, resulting in many low read numbers at later time points of the experiment that frequently induce overestimation of deleterious selection coefficients (cf. Figure S2). However, Figure 4 shows that we can clearly distinguish wild-type-like and strongly deleterious mutants although the relative location of the deleterious mode differs between environments dependent on the relative growth of the wild type between environments (generally slower growth at high salinity) and on the duration of the experiment.
Figure 4.
Histograms of the DFEs in all six environments, where the estimated median growth rate of each mutant relative to wild type (on the x-axis) is used as a proxy for the selection coefficient. All DFEs but the 25S environment show a bimodal shape. Subsets of wild-type synonyms and stop codons are depicted in blue and red, respectively.
The strong mutational constraint on the protein in the high-temperature environment (36°) in conjunction with a relatively high relative growth rate of the wild type results in the most clearly different DFE among the six environments, with a huge number of deleterious mutants, and almost no mutants better than wild-type average. In contrast, the 25°, 25S, and 30S environments, which present challenging environments, but wherein Hsp90 is potentially under less constraint, harbor adaptive potential reflected in many mutations that have higher growth rates than the wild-type average.
To compare the absolute numbers of beneficial and deleterious mutations, we chose a global cutoff of |r| < 0.004 that we define as the wild-type-like (“wt-like”) growth rate. This is a conservative cutoff based on the largest mean 95% credibility interval of wild-type-synonymous mutations across all six environments. Of note, the wt-like category is not necessarily the same as a neutral class of mutations. How many of our observed mutations are effectively neutral is highly dependent on the effective population size, which is probably very high in the bulk competition experiment (cf., e.g., Skelly et al. 2009, who estimate Ne ≈ 2.6 × 107). Since neutral mutations are expected to be those with s ≤ 1/Ne, the threshold for effective neutrality is probably very low and unlikely to be captured by any existing experimental method. In the following, we characterize and compare the DFEs in more detail, based on the above-described categorization.
Evaluating changes in selective pressure on the DFE—one-, two-, and three-step mutations
Because every possible codon was tested at every amino acid position, we can split the full data set into subsets containing mutations that are one-, two-, and three-nucleotide (nt) changes away from the wild-type sequence. These can be interpreted as representing different step numbers in a potential adaptive walk. Of note, only the one-step subset comprises all 81 possible single-nt mutations, whereas the other categories contain subsets of all possible two- and three-step walks in sequence space.
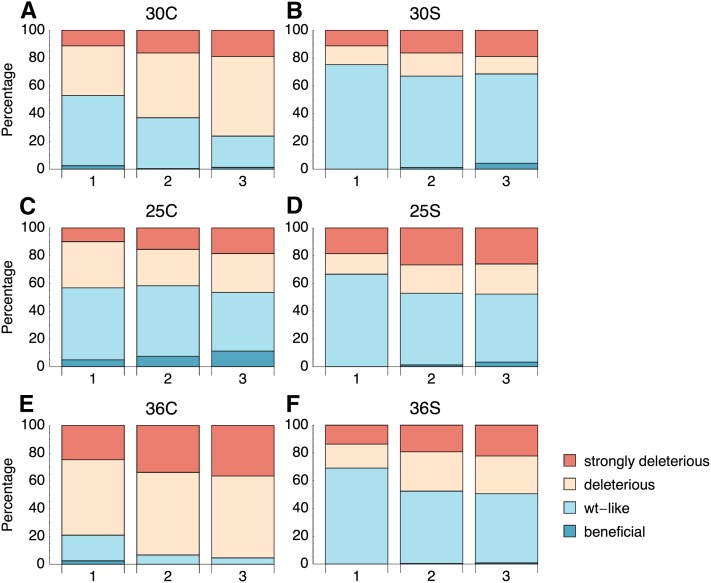
Figure 5 shows bar charts of the proportions of beneficial, wt-like, deleterious, and strongly deleterious mutants for each of the mutational step classes. We observe the following:
Figure 5.
Proportions of beneficial, wt-like, deleterious, and strongly deleterious mutations distinguished by mutational step size (1-, 2-, or 3-nt changes, as indicated on the x-axes of the bar charts) in the six environments. In general, the proportion of deleterious mutations decreases with the number of nt steps. In those environments that present a greater environmental challenge (B–D and F) the number of beneficial mutations increases with the mutational step size, whereas it tends to decrease for the standard environment and the high-temperature environment (A and E). An alternative representation of the mutational categories, based on the overlap of the posterior distribution with 1, can be found in Figure S5.
The proportion of wt-like mutations decreases with the number of nt changes, whereas the number of deleterious and strongly deleterious mutations increases. This is always true for the difference between one- and two-step mutations. This is consistent with a generally lower effect size of single-nt mutations compared with multiple-nt steps—but also with the population residing in a position in the sequence space that confers high mutational robustness (see also Figure 3D in Hietpas et al. 2011). This observation is consistent between all environments.
The proportion of beneficial mutations consistently increases with the number of nt steps in the 25°, 25S, 30S, and 36S environments (i.e., environments imposing a selective challenge that does not require higher Hsp90 expression). Conversely, the number of beneficials tends to decrease in the standard environment and the 36C environment. These observations show a stunning correspondence with predictions of the FGM, if one takes into account that we cannot categorize mutations of small fitness effect. In the standard environment, the optimum is close to the current phenotype, and each successive mutational step is likely to overshoot or move away from the optimum, resulting in fewer beneficial and wt-like mutations. On the other hand, if the optimum is shifted away from the current phenotype by presenting a new environmental challenge, there is more potential for mutations to have a beneficial effect, because they are more likely to leave the wt-like area. Hence more steps (or, alternatively, larger arrows) may more likely result in advantageous mutants.
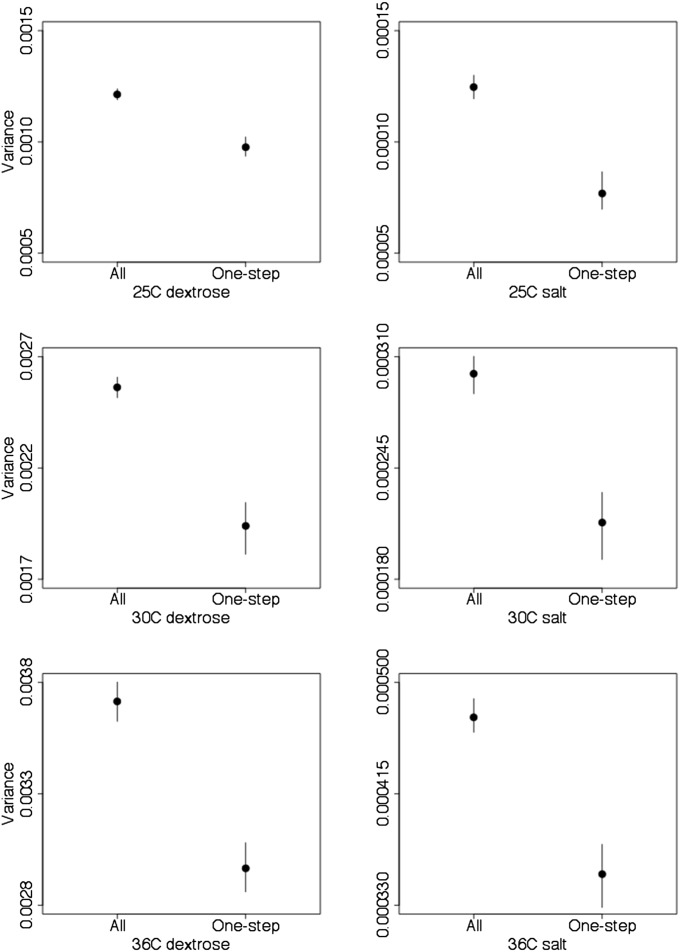
The variance of the DFE is higher at 30° and 36° than it is at 25°, as might be expected if Hsp90 is less important for fitness in the 25° environment. Variance is substantially lower for the one-step mutants in comparison to all mutants, again suggesting that the wild-type sequence is fairly robust to nearby changes in sequence space (Figure 6).
Figure 6.
Variance of the DFE for all mutants and the subset of single-nt mutants. Variance is substantially higher for the whole data compared with the single-step mutants. Median variance (solid circles) and 95% credibility intervals based on 10,000 samples from the posterior are shown.
We used a Kolmogorov–Smirnov (KS) test to evaluate whether the distributions of one-, two-, and three-step mutations are significantly different from each other and from the complete DFE. The resulting P-values are summarized in Table 2. We observe that the single-step DFE is significantly different from the other distributions, in particular for the 30° and 36° data sets, which are those that are suggestive of being fairly well adapted and close to the phenotypic optimum (cf. the following subsection). Hence, the difference in the distributions likely results not from differences in the beneficial tails of the distributions, but rather from a difference in the proportions of deleterious vs. wt-like mutations (cf. Figure 5). This implies that during adaptation, populations move not only to a local fitness peak but also to a location in the sequence space that confers mutational robustness, such that single-nt mutations are less likely to result in unfit progeny. This is in accordance with a recent study of an RNA virus, showing that mutational robustness evolves even on the level of codon usage (Lauring et al. 2012).
Table 2. Comparison of the one- (1), two- (2), and three (3)-step subsets with the full DFE, using a K-S test.
| Data set | 1 – all | 2 – all | 3 – all |
|---|---|---|---|
| 30° | 0.008 [0.001, 0.042] | 0.758 [0.479, 0.946] | 0.071 [0.022, 0.152] |
| 30S | 0.385 [0.141, 0.617] | 0.992 [0.830, 1.000] | 0.976 [0.801,0.998] |
| 25° | 0.268 [0.083, 0.393] | 0.983 [0.882, 0.999] | 0.510 [0.382, 0.637] |
| 25S | 0.104 [0.019, 0.280] | 0.851 [0.479, 0.996] | 0.887 [0.694, 0.986] |
| 36° | 0.013 [0.006, 0.022] | 0.994 [0.963, 0.999] | 0.150 [0.100, 0.202] |
| 36S | 0.034 [0.005, 0.112] | 0.999 [0.925, 1.000] | 0.549 [0.247, 0.843] |
Significant P-values are shown in boldface type. P-values are obtained as the median [95% C.I.] of 1000 samples from the posterior.
The distance to the optimum according to the FGM
Martin and Lenormand (2006) proposed that the distance to the phenotypic optimum according to the FGM can be estimated by fitting a so-called displaced-gamma distribution to the distribution of fitness effects. A displaced-gamma distribution has the shape of a negative gamma distribution, shifted by a parameter μ to accommodate beneficial mutations. To determine the respective distances to the phenotypic optimum in each environment, which is given by μ, we used a maximum-likelihood approach to fit displaced-gamma distributions to the wild-type-like mode of our DFEs for all six environments [the wild-type-like mode of the DFE for each environment was defined based on the cutoff determined during the initial data preparation (cf. Materials and Methods)]. The resulting distances to the optimum are summarized in Table 3 and a graphical representation of the fitted curves can be found in Figure S6. Largely consistent with what we have reported on the basis of amino acids (Hietpas et al. 2013), the distance to the optimum is lowest for the 36S and 36° environments that are under the highest Hsp constraint. The largest distances to the optimum are estimated for the 25S and 25° environments.
Table 3. Estimated distances [95% C.I.] to the optimum under the assumption of a displaced-gamma distribution, obtained from 100 samples from the posteriors.
| Standard salinity | Elevated salinity | |
|---|---|---|
| 25° | 0.0173 [0.0128, 0.0246] | 0.0235 [0.0219, 0.0249] |
| 30° | 0.0083 [0.0061, 0.0117] | 0.0106 [0.0090, 0.0114] |
| 36° | 0.0075 [0.0063, 0.0096] | 0.0064 [0.0051, 0.0085] |
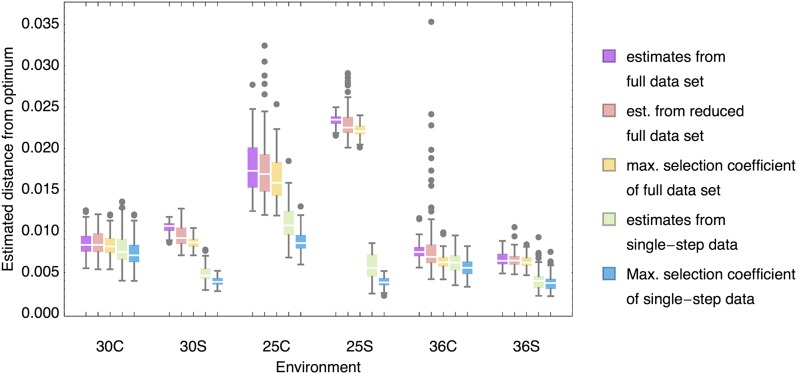
To test whether the displaced-gamma estimates from the single-nt subset differ significantly from those obtained from the full data set, we compared the posterior estimates of the distance to the optimum between data sets. To take into account the effect of the smaller single-step data sets, we repeated the fitting exercise with random subsets of the full data set of the same length as the single-step data. The results are summarized in Figure 7.
Figure 7.
Estimates of the distance to the optimum from full data sets, single-step data sets, and subsets of the full data with the same length as the single-step data set and maximum selection coefficients. The larger the estimated distance to the optimum from the full data set, the greater is the difference from the single-nt subset. Further, the estimated distance from the optimum appears largely determined by the maximum selection coefficient (equivalent to the estimated growth rate minus 1) identified in the data set. Estimates were obtained from 100 samples from the posterior for each data set and category. Boxes represent the interquartile range, whiskers extend to the highest/lowest data point within the box ±1.5 times the interquartile range, and gray circles represent outliers.
Clearly, the estimated distances to the optimum between the full and the single-step data set differ dramatically when the distance to the optimum is large (i.e., particularly in the 25° and 25S environments). Even though the estimation from subsets of the full data naturally results in a higher variance of estimates, this cannot account for the observed differences in the estimates. This has a number of implications:
The estimate of the distance to the optimum seems to be primarily determined by the maximum growth rate in the data set. Therefore, the estimates from the subsets of the full data are on average lower than those from the complete data. This implies that the estimate of μ is critically dependent on the successful sampling of beneficial mutants. This can be ensured only in large data sets or lengthy laboratory selection experiments.
In those environments that provide potential for adaptation (as suggested by a high number of beneficials), μ is generally underestimated when only the single-nt subset of the data is used—in fact, the maximum observed effect lies frequently above the proposed distance to the optimum estimated from the single-step data set. Hence, single-nt mutations are not sufficient to explore the adaptive landscape well enough to make a statement about the distance to the optimum. This corresponds well to the above observations of larger potential for adaptation to challenging environments in multiple-step mutations (cf. Figure 5 and discussion thereof).
The shape of the beneficial tail of the DFE
The shape of the beneficial tail of the DFE harbors important information about the course and the predictability of evolution. The FGM predicts bounded fitness tails if the optimum is close and an exponential distribution of beneficial fitness effects when the optimum is far away (Martin and Lenormand 2008). However, given that the distribution of beneficial effects indeed represents a tail distribution, all possible tail shapes can be summarized in the generalized Pareto distribution (GPD), and its shape parameter κ yields information about the domain of attraction of the underlying DFE (Beisel et al. 2007; Joyce et al. 2008). So far, most experimental studies have reported exponential tail distributions (e.g., Kassen and Bataillon 2006; MacLean and Buckling 2009), but there exist also reports of bounded beneficial tails (Rokyta et al. 2008; Bataillon et al. 2011), and a recent example of a potentially heavy-tailed distribution of beneficial effects that would not be accounted for by the FGM (Foll et al. 2014).
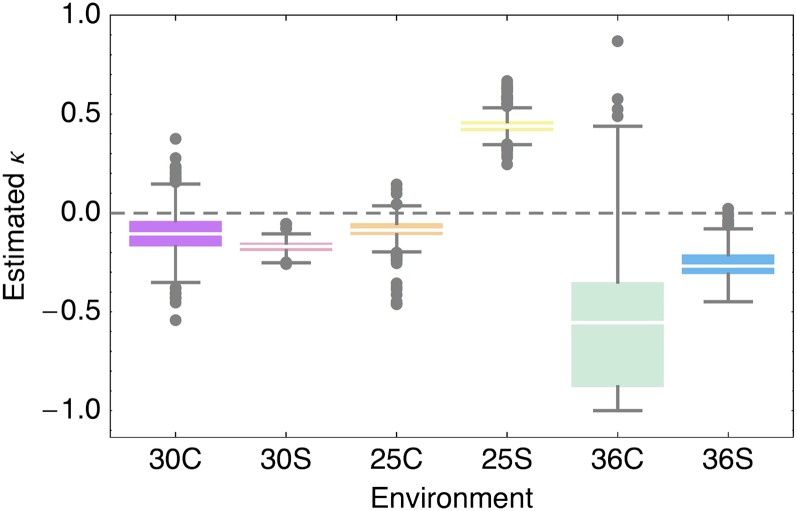
We fitted the GPD to the beneficial part of our data to compare the tail shapes across environments. The resulting shape parameter estimates are shown in Figure 8. For all but the 25S environment, the negative shape parameter estimates indicate that the DFE most likely belongs to the Weibull domain, i.e., that the DFE is bounded to the right. This would indicate that there is indeed a close optimum and that mutational effects cannot exceed a certain value. However, for the 25S environment, the shape estimates indicate that the DFE might belong to the Frechet domain, i.e., that it has a heavier-than-exponential tail. If the beneficial DFE is heavy-tailed, the size of beneficial fitness effects can be very large and the step size of adaptive evolution will be highly unpredictable.
Figure 8.
Estimated κ parameter under the generalized Pareto distribution (GPD) from 1000 samples from the posterior, taking into account all mutations with r > 1 in the sample. Negative κ estimates indicate a bounded beneficial DFE (i.e., Weibull domain of the DFE tail) for all but the 25S environment, suggesting a heavy-tailed distribution (i.e., DFE belonging to the Frechet domain of attraction). Because of the low number of beneficial mutations in the 36° environment, there is little power to estimate its tail shape.
Effects of synonymous mutations
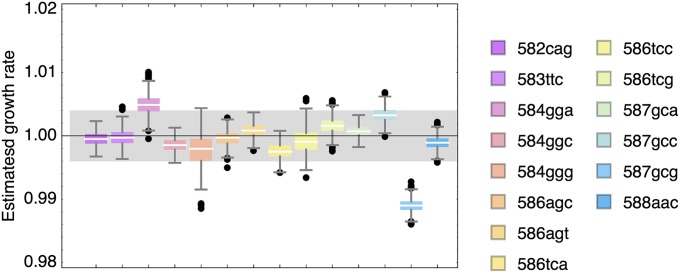
Many population-genetic statistics rely on the assumption that synonymous mutations in coding regions (i.e., mutations that do not change the amino acid and hence the protein sequence) have equal fitness [e.g., the McDonald–Kreitman test (McDonald and Kreitman 1991) and the HKA test (Hudson et al. 1987)]. However, an increasing number of studies have reported fitness differences between synonymous mutations (e.g., Singh et al. 2007). It is still unclear, however, how these fitness differences arise, although one common hypothesis suggests that, e.g., codon composition can affect RNA packaging (Sauna and Kimchi-Sarfaty 2011). The high precision of the EMPIRIC estimates enables us to explicitly study the distribution of synonymous fitness effects. Each of our data sets comprises 15 mutations that are synonymous to the wild-type sequence, and even within this small selection, we identify a deleterious outlier that persists across replicates and in three of the six different environments (cf. Figure 9 and Figure S7). The identified mutation 588aac represents an unpreferred change coding for asparagine.
Figure 9.
Box–whisker plot of the growth rates of the 15 mutations synonymous to the wild-type sequence in the standard environment (30°), based on 1000 samples from the posterior distribution. Boxes represent the interquartile range, whiskers extend to the highest/lowest data point within the box ±1.5 times the interquartile range, and black circles repesent outliers.
Genotype × environment interactions
While the overall shape of the DFE is similar between environments (Figure 4), different individual mutations may nonetheless have very different effects, depending on the environment. Such a scenario is referred to as a genotype × environment (G × E) interaction, wherein the fitness effect of a mutation depends on its environment. The consistency of evolutionary responses of populations to different environments will depend in large part on the extent of G × E, which can be captured as the covariance of fitness effects between environments. If covariance is high, then we expect populations to follow very similar trajectories, since the same mutations are beneficial or deleterious in each environment. By contrast, if covariance is low or negative, then different mutations would be expected to accumulate between populations. The extent and magnitude of G × E is of substantial current interest (e.g., Lazzaro et al. 2008; Gerke et al. 2010) and is typically inferred from studies of standing variation in natural populations. Thus, the current data set represents a unique opportunity to examine the extent to which fitness varies with environment for new mutations.
We find strong between-environment correlations for all mutants (cf. Table 4) and for the 81 single-step mutants (data not shown), with correlation coefficients ranging from 0.76 to 0.97 (all mutants) or from 0.80 to 0.97 (single-step mutants). This result suggests that, on the whole, the sign and magnitude of selection coefficients are consistent between environments. Further consideration, however, suggests that there is substantial variation in fitness effects between environments for particular classes of mutation. For example, among the 76 mutants that are beneficial in at least one environment, correlation coefficients range from −078 to 0.87 (cf. Table S1). Correlations between high salt and normal salt environments tend to be negative, indicating that mutations that are neutral or beneficial in salt tend to be deleterious in normal salinity and vice versa. Mutations that are deleterious in at least one environment (cf. Table S2) also have heterogeneous effects, albeit to a lesser extent than beneficial mutations, with correlation coefficients ranging from −0.12 to 0.86.
Table 4. Correlations and covariances between fitnesses in different environments for all mutants (n = 560).
| Environment | 25° | 25S | 30° | 30S | 36° | 36S |
|---|---|---|---|---|---|---|
| 25° | 0.0012 | 0.76 | 0.97 | 0.91 | 0.85 | 0.93 |
| (0.75–0.77) | (0.97–0.98) | (0.90–0.92) | (0.85–0.86) | (0.92–0.93) | ||
| 25S | 0.00029 | 0.00012 | 0.82 | 0.86 | 0.77 | 0.85 |
| (0.00029–0.00031) | (0.81–0.83) | (0.84–0.88) | (0.76–0.78) | (0.83–0.86) | ||
| 30° | 0.0017 | 0.00047 | 0.0026 | 0.95 | 0.89 | 0.97 |
| (0.00029–0.00031) | (0.00045–0.00048) | (0.94–0.96) | (0.88–0.89) | (0.96–0.97) | ||
| 30S | 0.00055 | 0.00017 | 0.00083 | 0.0003 | 0.84 | 0.95 |
| (0.00054–0.00056) | (0.00016–0.00017) | (0.00082–0.00085) | (0.83–0.84) | (0.95–0.96) | ||
| 36° | 0.0018 | 0.000528 | 0.0027 | 0.00088 | 0.0037 | 0.91 |
| (0.0018–0.0018) | (0.00051–0.00054) | (0.0027–0.0028) | (0.00086–0.00090) | (0.91–0.92) | ||
| 36S | 0.00070 | 0.00021 | 0.0011 | 0.00036 | 0.00128 | 0.00047 |
| (0.00069–0.00072) | (0.00020–0.00021) | (0.0010–0.0011) | (0.00035–0.00037) | (0.0012–0.0012) |
Correlation coefficients are given above the diagonal, covariances are below the diagonal, and within-environment variances are on the diagonal (boldface type). Values given are median and 95% credibility intervals from 10,000 samples of the posterior.
Conclusions
With a rich population-genetic framework that has been a century in the making describing the adaptive and stochastic forces governing the evolutionary process, and an impressively growing body of data sets from experimental evolution, the most important remaining piece for assembling this evolutionary puzzle is the development of a robust statistical framework for characterizing the underlying DFE. We here present a Bayesian MCMC approach shown to have a number of advantages over existing methodology. Using this developed framework, and utilizing a novel NGS experimental data set, we validate a number of predictions of Fisher’s geometric model: the DFE is dispersed as the population is brought farther from the optimum state, resulting in an increased number and magnitude of beneficial mutations; multiple-step mutations are beneficial only when the optimum is distant; and there is a strong genotype × environment interaction. Further, the precision of this estimation procedure has additionally provided insight into the nonneutrality of particular synonymous mutations. While these results are in themselves interesting within the context of furthering our understanding of the adaptive process, the statistical approach presented here additionally represents a robust methodology for characterizing the DFE in future NGS experimental data sets.
Supplementary Material
Acknowledgments
We thank Vital-IT and the Swiss Institute of Bioinformatics for computational resources and Adam Eyre-Walker, Daniel Wegmann, Matthieu Foll, Greg Ewing, Lisha Mathew, and an anonymous reviewer for helpful comments and discussion. This work is funded in part by grants from the Swiss National Science Foundation and a European Research Council starting grant (to J.D.J.), by grant R01-GM083038 from the National Institutes of Health (to D.N.B.), and by a National Sciences and Engineering Research Council Banting Fellowship (to A.W.).
Footnotes
Communicating editor: C. H. Langley
Literature Cited
- Bataillon T., Zhang T., Kassen R., 2011. Cost of adaptation and fitness effects of beneficial mutations in Pseudomonas fluorescens. Genetics 189: 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C. J., Rokyta D. R., Wichman H. A., Joyce P., 2007. Testing the extreme value domain of attraction for distributions of beneficial fitness effects. Genetics 176: 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsley, D. A., E. Kuh, and R. E. Welsch, 1980 Regression Diagnostics: Identifying Influential Data and Sources of Collinearity. John Wiley & Sons, New York/Hoboken, NJ. 8 [Google Scholar]
- Berry D. B., Gasch A. P., 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19: 4580–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S., 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9: 3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., et al. , 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M. C., Green P., 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Foll M., Poh Y.-P., Renzette N., Bank C., Shim H., et al. , 2014. Influenza virus drug resistance: a time-sampled population-genetics perspective. PLoS Genet. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Araya C. L., Fleishman S. J., Kellogg E. H., Stephany J. J., et al. , 2010. High-resolution mapping of protein sequence-function relationships. Nat. Methods 7: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Araya C. L., Gerard W., Fields S., 2011. Enrich: software for analysis of protein function by enrichment and depletion of variants. Bioinformatics 27: 3430–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Ramnarine S., Cohen B., 2010. Gene-environment interactions at nucleotide resolution. PLoS Genet. 6: e1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D. R., Woods R. A., 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Hawle P., Horst D., Bebelman J. P., Yang X. X., Siderius M., et al. , 2007. Cdc37p is required for stress-induced high-osmolarity glycerol and protein kinase C mitogen-activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p). Eukaryot. Cell 6: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas R., Roscoe B., Jiang L., Bolon D. N. A., 2012. Fitness analyses of all possible point mutations for regions of genes in yeast. Nat. Protoc. 7: 1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas R. T., Jensen J. D., Bolon D. N. A., 2011. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. USA 108: 7896–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas R. T., Bank C., Jensen J. D., Bolon D., 2013. Shifting fitness landscapes in response to altered environments. Evolution 67: 3512–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguadé M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce P., Rokyta D. R., Beisel C. J., Orr H. A., 2008. A general extreme value theory model for the adaptation of DNA sequences under strong selection and weak mutation. Genetics 180: 1627–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., Bataillon T., 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38: 484–488. [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2010. What can we learn about the distribution of fitness effects of new mutations from DNA sequence data? Philos. Trans. R. Soc. B 365: 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A. S., Acevedo A., Cooper S. B., Andino R., 2012. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 12: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B. P., Flores H. A., Lorigan J. G., Yourth C. P., 2008. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 4: e1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C., 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- MacLean R. C., Buckling A., 2009. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 5: e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution 60: 893–907. [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2008. The distribution of beneficial and fixed mutation fitness effects close to an optimum. Genetics 179: 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Melnyk A. H., Kassen R., 2011. Adaptive landscapes in evolving populations of Pseudomonas fluorescens. Evolution 65: 3048–3059. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D. R., Beisel C. J., Joyce P., Ferris M. T., Burch C. L., et al. , 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe B. P., Thayer K. M., Zeldovich K. B., Fushman D., Bolon D. N. A., 2013. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J. Mol. Biol. 425: 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A. G. M., Gerrish P. J., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Sauna Z. E., Kimchi-Sarfaty C., 2011. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12: 683–691. [DOI] [PubMed] [Google Scholar]
- Schneider A., Charlesworth B., Eyre-Walker A., Keightley P. D., 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189: 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Bauer DuMont V. L., Hubisz M. J., Nielsen R., Aquadro C. F., 2007. Patterns of mutation and selection at synonymous sites in Drosophila. Mol. Biol. Evol. 24: 2687–2697. [DOI] [PubMed] [Google Scholar]
- Skelly D. A., Ronald J., Connelly C. F., Akey J. M., 2009. Population genomics of intron splicing in 38 Saccharomyces cerevisiae genome sequences. Genome Biol. Evol. 1: 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A., Magalhães S., Gordo I., 2012. Cost of antibiotic resistance and the geometry of adaptation. Mol. Biol. Evol. 29: 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-X., Maurer K. C. T., Molanus M., Mager W. H., Siderius M., et al. , 2006. The molecular chaperone Hsp90 is required for high osmotic stress response in Saccharomyces cerevisiae. FEMS Yeast Res. 6: 195–204. [DOI] [PubMed] [Google Scholar]
- Yang X.-X., Hawle P., Bebelman J. P., Meenhuis A., Siderius M., et al. , 2007. Cdc37p is involved in osmoadaptation and controls high osmolarity-induced cross-talk via the MAP kinase Kss1p. FEMS Yeast Res. 7: 796–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.