Background: Dnmt1 faithfully propagates DNA methylation patterns to the next generation.
Results: The DNA methylation activity of Dnmt1 was stimulated by the direct interaction of the SRA domain of Uhrf1 and Dnmt1.
Conclusion: The SRA facilitates DNA accession to the catalytic center.
Significance: The RFTS and SRA interaction contributes to the correct feeding of the hemi-methylated DNA to the catalytic center of Dnmt1.
Keywords: DNA Methylation, DNA Methyltransferase, Epigenetics, Gene Regulation, Gene Silencing
Abstract
Dnmt1 is responsible for the maintenance DNA methylation during replication to propagate methylation patterns to the next generation. The replication foci targeting sequence (RFTS), which plugs the catalytic pocket, is necessary for recruitment of Dnmt1 to the replication site. In the present study we found that the DNA methylation activity of Dnmt1 was DNA length-dependent and scarcely methylated 12-bp short hemi-methylated DNA. Contrarily, the RFTS-deleted Dnmt1 and Dnmt1 mutants that destroyed the hydrogen bonds between the RFTS and catalytic domain showed significant DNA methylation activity even toward 12-bp hemi-methylated DNA. The DNA methylation activity of the RFTS-deleted Dnmt1 toward 12-bp hemi-methylated DNA was strongly inhibited on the addition of RFTS, but to a lesser extent by Dnmt1 harboring the mutations that impair the hydrogen bond formation. The SRA domain of Uhrf1, which is a prerequisite factor for maintenance methylation and selectively binds to hemi-methylated DNA, stimulated the DNA methylation activity of Dnmt1. The SRA to Dnmt1 concentration ratio was the determinant for the maximum stimulation. In addition, a mutant SRA, which had lost the DNA binding activity but was able to bind to Dnmt1, stimulated the DNA methylation activity of Dnmt1. The results indicate that the DNA methylation activity of Dnmt1 was stimulated on the direct interaction of the SRA and Dnmt1. The SRA facilitated acceptance of the 12-bp fluorocytosine-containing DNA by the catalytic center. We propose that the SRA removes the RFTS plug from the catalytic pocket to facilitate DNA acceptance by the catalytic center.
Introduction
DNA methylation of cytosine is a key epigenetic modification and crucial for normal development of mammals (1–3). The global methylation patterns in the genome are established at an early stage of embryogenesis and in germ cells and then propagated to the next generation thereafter in a cell lineage-dependent manner. DNA methyltransferases, e.g. Dnmt3a and Dnmt3b, are responsible mainly for the establishment of DNA methylation patterns, and another DNA methyltransferase, Dnmt1, for the maintenance of the patterns.
Mouse Dnmt1 is a large molecule comprising 1620 amino acid residues. The N-terminal 243-amino acid sequence undergoes independent folding (4), this domain playing a role as a platform for it to interact with many factors such as transcription repressor DMAP1 (5), Dnmt3a and Dnmt3b (6), Rb2 (7), proliferating cell nuclear antigen (PCNA) (8), DNA (4), protein kinase CDKL5 (9), and casein kinase Ck1 δ/ϵ (10). It was reported that PCNA, which is known to slide on replicating DNA with DNA polymerase, recruits Dnmt1 to facilitate maintenance methylation (8). However, it was also reported that a Dnmt1 lacking this independent N-terminal domain containing the PCNA binding sequence exhibits processive DNA methylation activity toward hemi-methylated DNA, which represents maintenance DNA methylation (11). Following the independent N-terminal domain, there are multiple domains (see Fig. 1A): the replication foci targeting sequence (RFTS),3 a Zn-finger like CXXC motif (CXXC), two bromo-adjacent homology domains, and the catalytic domain. The catalytic domain is highly homologous to those of the bacterial DNA (cytosine-5) methyltransferase family (12). The RFTS is necessary for Dnmt1 to target a replication site (13). Recently, we determined the three-dimensional structure of mouse Dnmt1 lacking the N-terminal platform domain by x-ray crystallography (14). The RFTS, CXXC motif, two bromo-adjacent homology domains, and catalytic domain show almost independent folding. However, a striking feature is that the RFTS plugs the catalytic pocket. Substrate DNA cannot gain access to the catalytic center unless the RFTS is removed from the catalytic pocket. Recently, Song et al. reported the co-crystal structure of the Dnmt1 catalytic domain with fluorocytosine-containing DNA (15). Comparison of the two three-dimensional structures clearly indicates that the RFTS domain must be removed from the catalytic pocket for maintenance DNA methylation.
FIGURE 1.
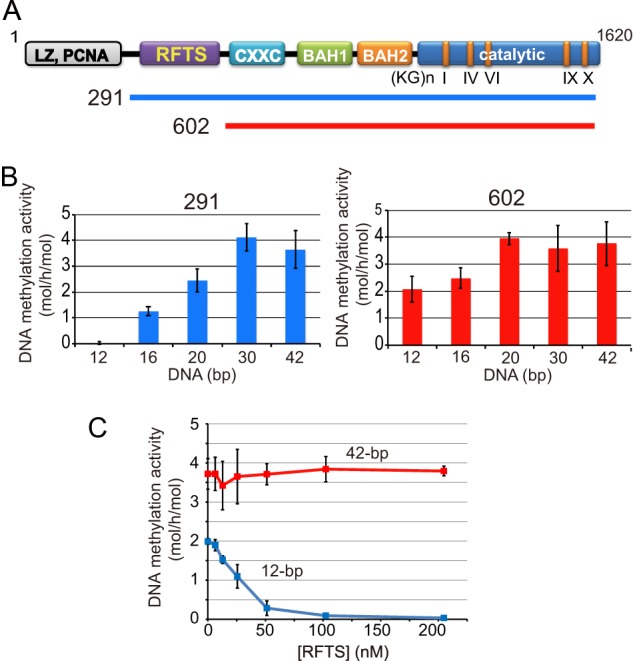
DNA length-dependent DNA methylation activities of Dnmt1(291–1620) and Dnmt1(602–1620). A, schematic illustrates Dnmt1. Dnmt1 has a multidomain structure (4, 14). The Dnmt1 constructs, Dnmt1(291–1620) (291) and Dnmt1(602–1620) (602) used in this study are indicated. B, DNA length-dependent DNA methylation activity of Dnmt1(291–1620) (left panel, 291) and Dnmt1(602–1620) (right panel, 602) toward hemi-methylated DNA of 12, 16, 20, 30, and 42 bp with one hemi-methylated CpG is shown. C, Dnmt1(602–1620) was incubated with the RFTS, and then the DNA methylation activity toward 12-bp (blue) and 42-bp hemi-methylated DNA (red) was determined. Specific activities of DNA methylation (mol of CH3 transferred/h/mol of Dnmt1) were determined. The average activities ± S.D. (error bars; n = 3) are shown. The inhibition of Dnmt1(602–1620) was statistically significant at 50, 100, and 200 nm RFTS (p < 0.001).
For the propagation of DNA methylation patterns in vivo during replication, Uhrf1 (ubiquitin-like with PHD and ring finger domains; also known as Np95 and ICBP90) was reported to be a prerequisite factor (16). The SRA (SET and RING finger-associated) domain of Uhrf1 specifically recognizes hemi-methylated CpG and flips the methylated cytosines out of double-stranded DNA (17–19). Recently, it was reported that Dnmt1 recognizes ubiquitylated lysine 23 of histone H3, which is catalyzed by the RING finger E3 ligase activity of Uhrf1 and is a necessary step for maintenance methylation (20).
In the present study, we have demonstrated that the direct interaction between the SRA of Uhrf1 and Dnmt1 contributes to the removal of the RFTS from the catalytic pocket to allow access of the substrate hemi-methylated DNA to the catalytic center.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant Dnmt1
Truncated and mutated mouse Dnmt1 cDNAs were subcloned into pFastBac-Htb (Invitrogen) with the GST cDNA inserted in-frame at the 5′ end. The baculovirus harboring Dnmt1 cDNAs coding 291–1620 and 602–1620 was expressed in Sf9 cells and purified as described elsewhere (11, 14). The mutant Dnmt1(291–1620) with E531A/D532A, E531K/D532K, and D554K were purified as for the wild-type Dnmt1. All of the constructs were confirmed by dideoxy sequencing (21).
As for preparation of the recombinant RFTS, 291–601 of Dnmt1 was subcloned into the EcoRI and XhoI sites of pGEX6P-1. The wild-type and the Dnmt1 harboring mutations E531A/D532A and D55K were expressed in Escherichia coli BL21(DE3) CodonPlus-RIL with 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 16 °C for 24 h. The expressed proteins were purified with a GSH-Sepharose column. To remove the GST tag, the eluate was treated with PreScission protease (GE Healthcare) and reloaded onto the GSH-Sepharose column, and the flow-through fractions were collected.
Preparation of Recombinant SRA of Uhrf1
Mouse Uhrf1 coding SRA(409–617) or with mutation D474A/R496A was subcloned into the BamHI and XhoI sites of pGEX6P-1 or pET30a with a GST sequence added at the 5′ end in-frame. DNA sequences were confirmed by the dideoxy method (21). The SRA protein was induced for expression in E. coli BL21(DE3) CodonPlus-RIL with 0.5 mm isopropyl β-d-1-thiogalactopyranoside and then cultured at 18 °C for 16 h. The expressed proteins were purified with a GSH-Sepharose column. To remove the GST tag, PreScission protease was used as described for the RFTS.
Protein Concentration
The concentrations of purified recombinant Dnmt1, RFTS, and SRA were determined from the absorption at 280 nm with a Nanodrop 2000 spectrophotometer (Thermo Scientific). The protein concentrations used for the pulldown assaying were determined with a BCA Protein Assay kit (Pierce).
Oligonucleotide DNA
The following DNA sequences are the methylated strands of the hemi-methylated DNA used for methylation activity measurements (Gene Design, Mino, Osaka); 12-bp, 5′-GCAATCMGGTAG-3′; 16-bp, 5′-CGGCAATCMGGTAGAC-3′; 20-bp, 5′-GACGGCAATCMGGTAGACGA-3′; 30-bp, 5′-ACGACGACGGCAATCMGGTAGACGACGACG-3′; 42-bp, 5′-GATCCGACGACGACGGCAATCMGGTAGACGACGACGACGATC-3′; 42 bp-12CpG, 5′-GATCMGAMGAMGAMGAMGAMGAMGAMGAMGAMGAMGAMGATC-3′. M in the sequences denotes 5-methylcytosine. As for the 12-bp F-DNA, 5-methylcytosine was replaced by 5-fluorocytosine. Equal amounts of the methylated and complementary strand oligomers (100 μm each) in 50 mm NaCl, 1 mm EDTA, and 10 mm Tris-HCl, pH 8.0, were incubated at 97 °C for 5 min and then gradually targeted to 70 °C after 2 h, 50 °C after 4 h, and then to 15 °C after 2 h.
DNA Methylation Reaction
DNA methylation activities were determined as described elsewhere (10), in brief, in a total volume of 25 μl of reaction buffer comprising 5 mm EDTA, 50 mm NaCl, 2.7 m glycerol, 0.2 mm PMSF, and 20 mm Tris-HCl, pH 7.4. As for the standard assay, 6.4 nm Dnmt1, 66 nm DNA, and 2.2 μm 3H-labeled-S-adenosyl-l-methionine (10 or 15 Ci/mmol) (PerkinElmer Life Sciences) were used unless otherwise stated. Reaction mixtures were incubated at the indicated temperatures for several different times, and then the specific activity (mol of CH3 transferred to DNA/h/mol of Dnmt1 enzyme) was determined within a linear time course range. The activation energies of DNA methylation activities of Dnmt1 were determined from the slopes of Arrhenius plot; the logarithms of DNA methylation activities obtained (ln(DNA methylation activity), ordinate axis) against inverse temperatures (1/T, abscissa). The activation energy, which is −(ln(DNA methylation activity)·T)·R (gas constant; 8.314 J/T·mol), was calculated from the slope of a linear regression. As for determination of the effect of the SRA on the DNA methylation activity, the SRA was premixed with DNA and then incubated for 30 min on ice, unless otherwise indicated in the figure legends.
The 5′ end of 12-bp F-DNA was labeled with T4 polynucleotide kinase (Toyobo) and [γ-32P]ATP (6000 Ci/mmol; MP Biomedicals). Dnmt1(291–1620) or Dnmt1(602–1620) (6.4 pmol) was incubated with 10 pmol of labeled F-DNA and 60 pmol of S-adenosyl-l-methionine, in the absence or presence of 5 or 20 pmol of the SRA in a 25-μl reaction mixture comprising 50 mm NaCl, 20% glycerol (w/v), 0.2 mm EDTA, 0.2 mm PMSF, 1 mm DTT, and 20 mm Tris-HCl, pH 7.4, followed by incubation at 37 °C for 1 h. After the incubation, the mixtures were subjected to SDS-polyacrylamide gel electrophoresis in a 7.5% gel. The protein bands were visualized with Coomassie Brilliant Blue R-250 (CBB) (Nacalai Tesque) staining and a BAS2000 Bioimage analyzer (Fuji Film), and then exposed to x-ray film (Fuji Film). The amount of F-DNA bound to Dnmt1 was quantitated with Image-Gauge software (Fuji Film), and the protein band stained with CBB was quantitated with Quantity One (Bio-Rad).
Pulldown Assaying of Dnmt1 with Uhrf1
SRA or SRA(D474A/R496A) (1.5 μg) was mixed with GST-Dnmt1(291–1620) (1.2 μg), GST-Dnmt1(602–1620) (1 μg), or GST-RFTS (2.3 μg) coupled to GSH-Sepharose, respectively, and then incubated at 25 °C for 30 min in a 40 μl of binding buffer comprising 50 mm NaCl, 10% glycerol, 0.1% (w/v) Nonidet P-40, and 50 mm Tris-HCl, pH 7.4. Dnmt1(291–1620) (1 μg) or Dnmt1(602–1620) (0.8 μg) was mixed with GST-SRA (3 μg) coupled to GSH-Sepharose, respectively, and then incubated under the same conditions as above. Then, the beads were washed three times with binding buffer. The bound, unbound, and washed fractions were precipitated with 10% (w/v) trichloroacetic acid. The protein bands were separated by SDS-polyacrylamide gel electrophoresis in a 12% gel. The protein bands were visualized with CBB.
Gel Shift Assaying
The indicated amounts of the RFTS, SRA, or SRA and RFTS were incubated with 12-bp or 42-bp DNA with one hemi-methylated CpG (0.2 μm) in a solution comprising 50 mm NaCl, 20% glycerol (w/v), 0.2 mm EDTA, 0.2 mm PMSF, 1 mm DTT, and 20 mm Tris-HCl, pH 7.4, at 4 °C for 30 min. After the incubation, the mixtures were subjected to 0.8% agarose gel electrophoresis in 0.2× TBE and 2% glycerol at 4 °C with 150 V for 10 min. DNA was stained with GelGreen (Biotium) and visualized with a fluoro-imager, Typhoon FLA 9500 (GE Healthcare).
RESULTS
Dnmt1 Containing the RFTS Cannot Methylate Short Hemi-methylated DNA
In the previous study on the x-ray crystal structure of mouse Dnmt1, we found that the RFTS plugs the catalytic pocket (14). This indicates that the RFTS has to be removed from this position for the DNA methylation. Interestingly, however, even in the presence of the RFTS, Dnmt1 can methylate hemi-methylated DNA (11, 14). On the contrary, Syeda et al. reported that recombinant Dnmt1 containing the RFTS exhibited no DNA methylation activity when a short hairpin hemi-methylated DNA was used as the methyl acceptor (22). We assumed that the difference between our and Syeda's results may be the length of the methyl group acceptor DNA. Short DNA may not be able to gain access to the catalytic center when the RFTS is plugging the catalytic pocket.
To determine whether or not the DNA length is the determinant for the accession of DNA to the catalytic pocket for DNA methylation activity, we prepared 12-, 16-, 20-, 30-, and 42-bp double-stranded hemi-methylated DNA, each of which contained one hemi-methylated CpG site. Recombinant Dnmt1(291–1620) and Dnmt1(602–1620) (Fig. 1A), containing and lacking the RFTS, respectively, were purified, and we determined the DNA methylation activity toward these substrates. As shown in Fig. 1B, Dnmt1(291–1620) could not methylate 12-bp hemi-methylated DNA, but showed significant activity toward the DNA longer than 16-bp hemi-methylated DNA, and then the activity increased as the DNA length increased. Different from Dnmt1(291–1620), Dnmt1(602–1620) lacking the RFTS showed significant DNA methylation activity even when 12-bp hemi-methylated DNA was used as the methyl group acceptor. Increasing DNA length only mildly increased the DNA methylation activity of Dnmt1(602–1620). These results indicate that the 12-bp short hemi-methylated DNA used in the present study cannot gain access to the catalytic center by displacing the RFTS from the catalytic pocket to be methylated. For access to the catalytic center, the DNA length must be >12 bp.
The RFTS coding 291–601 of Dnmt1 was separately purified and added to Dnmt1(602–1620), and then the DNA methylation activity toward 12-bp and 42-bp hemi-methylated DNA was determined. The RFTS added to the reaction mixture did not affect the DNA methylation activity when 42-bp hemi-methylated DNA was used as the methyl group acceptor. However, the RFTS domain inhibited the DNA methylation activity toward 12-bp hemi-methylated DNA in a dose-dependent manner (Fig. 1C). This is consistent with the report that the DNA methylation activity of Dnmt1 lacking the RFTS domain is inhibited on the addition of the RFTS when a short hemi-methylated hairpin DNA is used as the substrate (22). This suggests that short DNA such as 12-bp hemi-methylated DNA cannot efficiently gain access to the catalytic center when the RFTS domain occupies the catalytic pocket, whereas long DNA can.
Mutant Dnmt1(291–1620) That Impairs the Hydrogen Bonds between the RFTS and Catalytic Domain Significantly Methylates 12-bp Hemi-methylated DNA
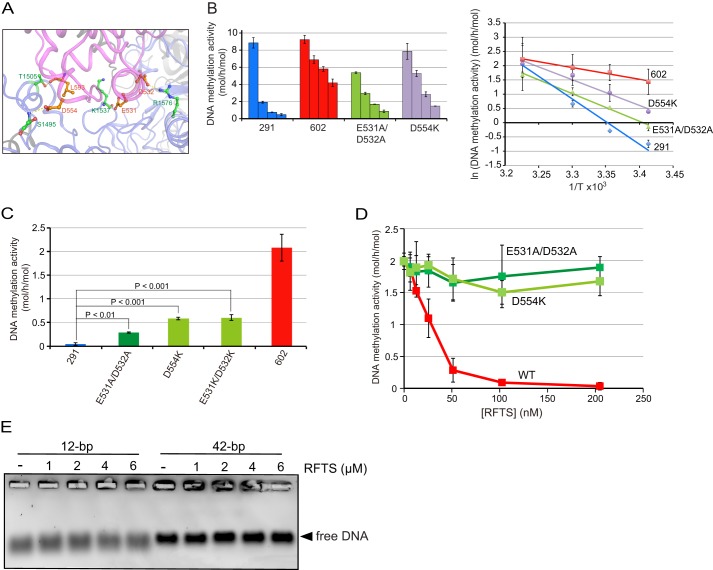
The RFTS is anchored to the catalytic pocked through four hydrogen bonds between Glu-531, Asp-532, Asp-554, and Leu-593 in the RFTS and Lys-1537, Arg-1576, Ser-1495, and Thr-1505 in the catalytic domain, respectively (14) (Fig. 2A). For this, there is an energy barrier for removal of the RFTS domain from the catalytic pocket to make DNA accessible to the catalytic center. The difference in activation energy between Dnmt1(291–1620) and Dnmt1(602–1620) as to DNA methylation is about 80 kJ/mol (14). It is, therefore, expected that impairment of the hydrogen bonds formed between the RFTS and catalytic domains makes it rather easy to remove the RFTS from the catalytic pocket through lowering of the activation energy, which means that even the 12-bp hemi-methylated DNA can gain access to the catalytic center. We replaced amino acid residues Glu-531 and Asp-532, or Asp-554 in the RFTS of Dnmt1(291–1620), which form hydrogen bonds through their side chains, purified the recombinants, and then determined the DNA methylation activity. As shown in Fig. 2B, the Dnmt1(291–1620) mutations harboring E531A and D532A (E531A/D532A), and D554K in the RFTS significantly lowered the activation energy for the DNA methylation activity, the activation energies being calculated to be 97 and 90 kJ/mol, respectively. Under identical conditions, the activation energies of Dnmt1(291–1620) and Dnmt1(602–1620) were 133 and 34 kJ/mol, respectively. Expectedly, the replacement of Glu-531 and Asp-532 with alanine or lysine, or Asp-554 with lysine in the RFTS in Dnmt1(291–1620) partially but significantly exhibited the DNA methylation activity toward 12-bp hemi-methylated DNA (Fig. 2C). This further supports that the 12-bp short hemi-methylated DNA cannot remove the RFTS from the catalytic pocket of wild-type Dnmt1(291–1620) and thus cannot gain access to the catalytic center of Dnmt1.
FIGURE 2.
Dnmt1(291–1620), which disrupted the hydrogen bonds between the RFTS and catalytic domains, exhibits significant DNA methylation activity toward 12-bp hemi-methylated DNA. A, image of the four hydrogen bonds between the RFTS (light violet) and catalytic domains (light blue) of Dnmt1 (taken from Protein Data Bank ID code 3AV6). The hydrogen bonds between the side chains of Glu-531, Asp-532, and Asp-554, and the main chain of Leu-593 in the RFTS and Lys-1537, Arg-1576, Ser-1495, and Thr-1505, respectively, are shown. B, DNA methylation activities of Dnmt1(291–1620) (291), Dnmt1(602–1620) (602), and Dnmt1(291–1620) with E531A/D532A, and D554K determined at 37, 30, 25, and 20 °C (bars from left to right), respectively, toward 42-bp DNA with 12 hemi-methylated CpG. Average activities ± S.D. (error bars; n = 3) are shown (left panel). The logarithms of DNA methylation activities obtained (ln(DNA methylation activity), ordinate axis) against inverse temperatures (1/T, abscissa) are plotted (Arrhenius plot) (right panel). C, DNA methylation activities of Dnmt1(291–1620) (291), Dnmt1(602–1620) (602), and Dnmt1(291–1620) with the mutations E531A and D532A (E531A/D532A), D554A (D554K), and E531K and D532A (E531A/D532K) determined toward 12-bp hemi-methylated DNA as in Fig. 1. D, Dnmt1(602–1620) incubated with the RFTS domain without any mutation (WT, red) or with mutation E531A/D532A (dark green) or D544K (light green). Then the DNA methylation activity toward 12-bp hemi-methylated DNA was determined. The average values ± S.D. (n = 3) are plotted. E, the indicated amounts of the RFTS incubated with 12-bp or 42-bp DNA with one hemi-methylated CpG (0.2 μm). After the incubation, the mixtures were subjected to gel electrophoresis, and then DNA was visualized.
As shown in Fig. 1C, the addition of separately prepared RFTS to the reaction mixture inhibited the DNA methylation activity of Dnmt1(602–1620) toward 12-bp hemi-methylated DNA. The RFTS carrying the E531A/D532A or D554K mutations showed almost no effect on the DNA methylation activity of Dnmt1(602–1620) compared with that of the wild-type RFTS fragment (Fig. 2D). Considering that the RFTS could not bind to the substrate DNA by itself (Fig. 2E), this result could be a reflection of the fact that the RFTS mutants were less stably anchored to the catalytic pocket due to their impairment in the hydrogen bond formation with the catalytic domain.
The SRA Domain of Uhrf1 Stimulates DNA Methylation
Uhrf1 is a prerequisite factor for maintenance DNA methylation in vivo (16). The SRA of Uhrf1 specifically binds hemi-methylated CpG-containing DNA and flips the methylated cytosine out of the double-stranded DNA (17–19). Because the RFTS is the sequence that brings Dnmt1 to the replicating region (13), and Uhrf1 is co-localized with Dnmt1 in the replication region (16), it is reasonable to speculate that direct or indirect interaction of the RFTS and SRA contributes to the removal of the RFTS from the catalytic pocket, thus allowing DNA access to the catalytic center. According to an in vitro binding study, the SRA bound to Dnmt1(291–1620) containing the RFTS and the RFTS itself (Dnmt1(291–601)) (Fig. 3A). However, the SRA did not bind to the Dnmt1(602–1620) lacking the RFTS (Fig. 3B). These results indicate that the RFTS interacts directly with the SRA.
FIGURE 3.
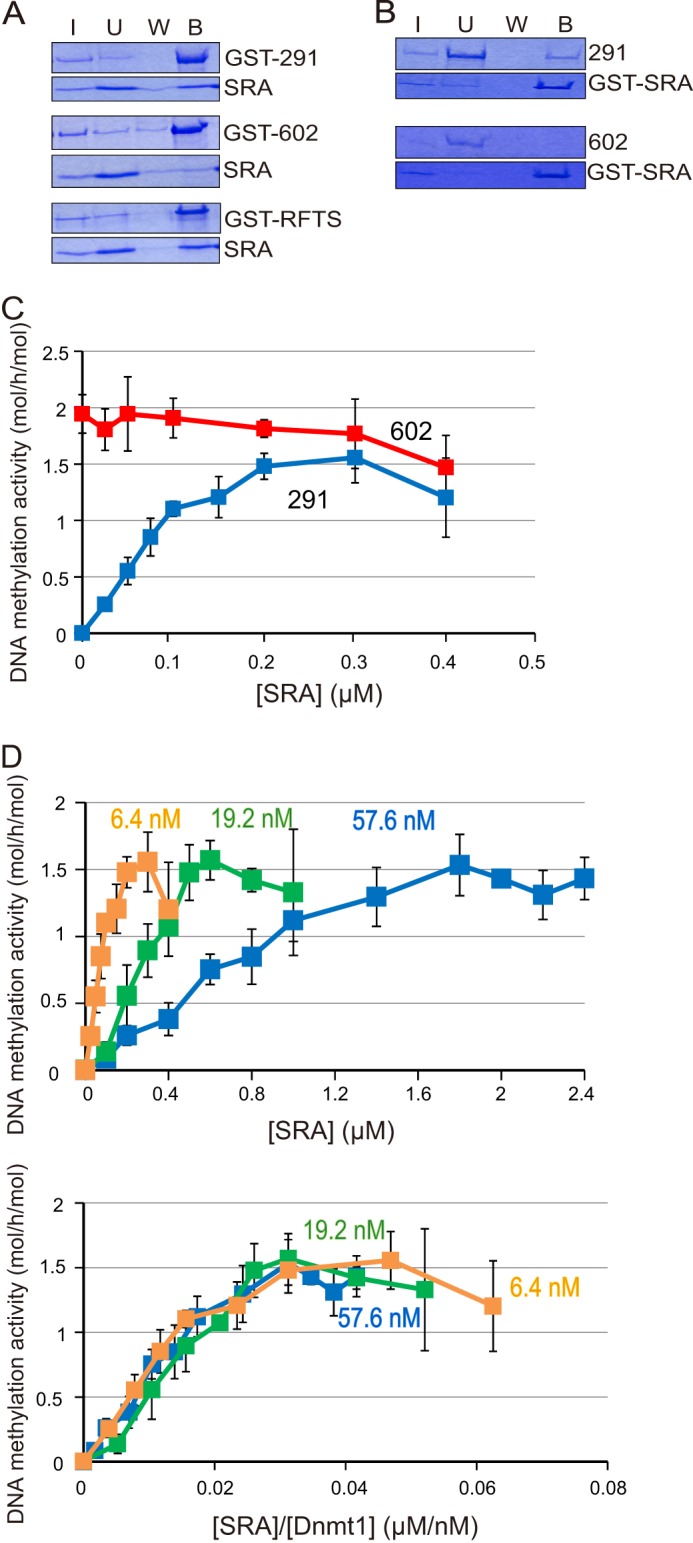
The SRA binds to the RFTS of Dnmt1 and induces DNA methylation activity toward 12-bp hemi-methylated DNA. A, the SRA was incubated with GST-tagged Dnmt1(291–1620) (GST-291), Dnmt1(602–1620) (GST-602), or Dnmt1(291–602) (GST-RFTS) bound to GSH-Sepharose. After the incubation, input (I), unbound (U), wash (W), and bound (B) fractions were analyzed by SDS-polyacrylamide gel electrophoresis. The protein bands were visualized by CBB staining. Equivalent amounts of samples were loaded, other than for the input (I) fractions, the amounts loaded being one fourth of those of the other fractions. B, Dnmt1(291–1620) (291) or Dnmt1(602–1620) (602) was incubated with GST-tagged SRA (GST-SRA) bound to GSH-Sepharose. Input (I), unbound (U), wash (W), and bound (B) fractions were analyzed as in A. C, DNA methylation activities of Dnmt1(291–1620) (blue) and Dnmt1(602–1620) (red) toward 12-bp hemi-methylated DNA were titrated with the SRA. The specific activities of DNA methylation (mol of CH3 transferred/h/mol of Dnmt1) were determined, and the average activities ± S.D. (error bars; n = 3) are shown. The stimulation of the DNA methylation activities of Dnmt1(291–1620) was statistically significant at all of the SRA concentrations examined (p < 0.001), except for 0.05 μm SRA where p < 0.01. D, DNA methylation activities toward 12-bp hemi-methylated DNA were determined by titrating the SRA with a fixed amount of DNA and three different concentrations of Dnmt1(291–1620): 6.4 (light brown), 19.2 (light green), and 57.6 nm (blue). Average activities ± S.D. (n = 3) are shown (upper panel). The concentrations of SRA in the upper panel were normalized with Dnmt1 and the DNA methylation activities are replotted (lower panel).
As described above, Dnmt1(291–1620) could not methylate 12-bp hemi-methylated DNA, but Dnmt1(602–1620) could. If the SRA contributes to the removal of the RFTS from the catalytic pocket, Dnmt1(291–1620) containing the RFTS may exhibit DNA methylation activity toward the 12-bp DNA on the addition of the SRA. As shown in Fig. 3C, Dnmt1(291–1620) exhibited a significant level of DNA methylation activity depending on the addition of the SRA in a dose-dependent manner. Under these conditions, the DNA methylation activity of Dnmt1(602–1620) was not stimulated by the addition of the SRA. This indicates that the SRA contributes to allowing 12-bp hemi-methylated DNA to gain access to the catalytic center, possibly by removing the RFTS from the catalytic pocket.
SRA-dependent Short DNA Methylation Activity of Dnmt1(291–1620) Is Due to Direct Interaction between the SRA and Dnmt1
Dnmt1(291–1620) activity was titrated with the SRA with three different concentrations of Dnmt1 and a fixed amount of 12-bp hemi-methylated DNA (Fig. 3D, upper panel). With higher Dnmt1(291–1620) concentrations, the SRA concentrations necessary for the maximum activation were higher. Interestingly, when the SRA concentration was normalized to that of Dnmt1(291–1620) and replotted, the titration curves with different concentrations of Dnmt1(291–1620) almost fit an identical curve (Fig. 3D, lower panel). These results strongly suggest that direct interaction between the SRA and Dnmt1 is crucial for the 12-bp hemi-methylated DNA methylation activity.
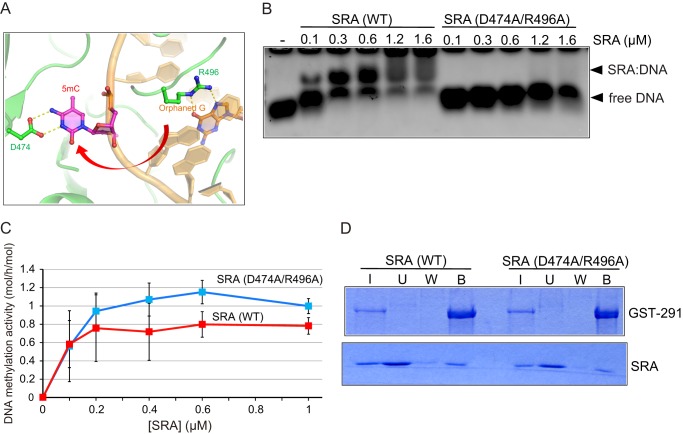
Because it is known that the SRA domain binds specifically to hemi-methylated DNA to flip the methylated cytosine out of the double-stranded DNA (17–19), we next examined whether or not the SRA-12-bp hemi-methylated DNA complex is a functional substrate for Dnmt1(291–1620). It was reported that Asp-474 and Arg-496 of Uhrf1 in the SRA recognize the methyl group of the flipped out methylated cytosine and orphaned guanine, respectively (17) (Fig. 4A). The mutant SRA (D474A/R496A) did not show significant 12-bp hemi-methylated DNA binding activity (Fig. 4B). The DNA methylation activity of Dnmt1(291–1620) was titrated against the mutant SRA (Fig. 4C). Despite the lack of DNA binding activity, the mutant SRA stimulated the DNA methylation activity of Dnmt1(291–1620) toward 12-bp hemi-methylated DNA. As shown in Fig. 4D, the mutant SRA significantly bound to Dnmt1(291–1620) although its amount was apparently less compared with that of the wild-type SRA. The results clearly indicate that the interaction of Dnmt1 and the DNA-free SRA solely affects the stimulation of the DNA methylation activity.
FIGURE 4.
The mutant SRA as the amino acid residues responsible for flipping of the methylated cytosine out of double-stranded DNA cannot bind to 12-bp hemi-methylated DNA but can stimulate DNA methylation activity. A, image of the structure of the SRA bound to hemi-methylated DNA and flipping out of the methylated cytosine (5mC) of the double-stranded DNA is shown. Asp-474 is holding 5mC, and Arg-496 is stabilizing the orphaned Gly. The red arrow indicates the flipping of 5mC. The structure data were taken from PDB accession number 2ZKD. B, gel-shift assaying of the SRA and that with D474A/R496A was performed as in Fig. 2E. 12-bp hemi-methylated DNA (0.2 μm) was incubated with the indicated amounts of the SRA or the mutant. Free DNA (arrows) and DNA-bound SRA (arrowheads) are shown. C, DNA methylation activities of Dnmt1(291–1620) (12.8 μm) toward 12-bp hemi-methylated DNA were determined by titrated with SRA (WT, red) or SRA(D474A/R496A, blue). The average activities ± S.D. (n = 4) are shown. The stimulation of the DNA methylation activities of Dnmt1(291–1620) was statistically significant at all of the SRA (D474A/R496A) concentrations examined (p < 0.001), except for 0.1 μm SRA (D474A/R496A) where p < 0.05. Error bars, S.D. D, the SRA or mutant was incubated with GST-tagged Dnmt1(291–1620) (GST-291) bound to GSH-Sepharose. After the incubation, input (I), unbound (U), wash (W), and bound (B) fractions were analyzed by SDS-polyacrylamide gel electrophoresis. The protein bands were visualized by CBB staining. Equivalent amounts of samples were loaded, other than for the input (I) fractions, the amounts loaded being one fourth of those of the other fractions.
The Interaction between the SRA and Dnmt1 Promotes Accession of DNA to the Catalytic Center
Because the effect of the SRA was examined as DNA methylation activity, the assay system comprised many steps; removal of the RFTS domain from the catalytic pocket, accession of hemi-methylated DNA to the catalytic center, and transfer of a methyl group to cytosine. Reportedly, the SRA binds to hemi-methylated DNA, and the SRA and Dnmt1 cannot bind to the same hemi-methylated DNA at the same time due to steric hindrance (17). To methylate hemi-methylated DNA bound to the SRA, the SRA must release the DNA. To this end, the SRA binding to DNA was competed for by the RFTS. As shown in Fig. 5A, the RFTS significantly inhibited the DNA binding of the SRA in a dose-dependent manner. This suggests that the direct interaction between the RFTS and SRA forces the release of the hemi-methylated DNA from the SRA. The interaction between the RFTS and SRA domains may facilitate not only the removal of the RFTS domain from the catalytic pocket but also access of the substrate hemi-methylated DNA to the catalytic center.
FIGURE 5.
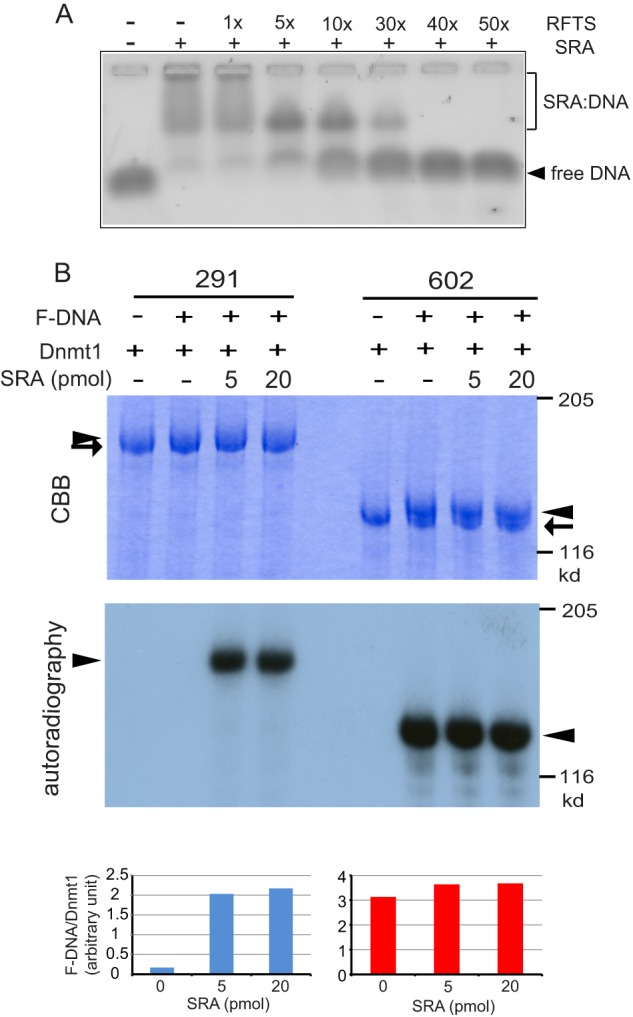
Interaction between the SRA and RFTS domains facilitates accession of 12-bp hemi-methylated DNA to the catalytic center of Dnmt1. A, the SRA binding to 12-bp hemi-methylated DNA was competed for by the RFTS in a dose-dependent manner. SRA (0.6 μm) and 12-bp hemi-methylated DNA (0.2 μm) binding was competed for with 1–50-fold higher concentrations of RFTS and then analyzed by gel shift assaying. B, Dnmt1(291–1620) or Dnmt1(602–1620) was incubated with 12-bp 32P-labeled F-DNA in the absence or presence of the SRA. After the reaction, samples were subjected to SDS-polyacrylamide gel electrophoresis, and the protein bands and radiolabeled F-DNA were visualized with CBB (top panel) and autoradiography (middle panel), respectively. Arrows and arrowheads indicate free Dnmt1 and Dnmt1 bound to F-DNA, respectively. The radioactive bands visualized with a BAS2000 were quantitated and normalized to those of the CBB-stained Dnmt1 bands (bottom panel).
It was reported that the cysteine residue at the catalytic center of Dnmt1 covalently traps F-DNA, in which the methylcytosine of hemi-methylated DNA was replaced with fluorocytosine (23). To evaluate directly the accession of DNA to the catalytic center, 12-bp F-DNA was synthesized. DNA methylation reactions were performed for Dnmt1(291–1620) and Dnmt1(602–1620) using 12-bp F-DNA, of which the 5′ end was labeled with 32P, in the absence and presence of the SRA. The reaction mixtures were subjected to SDS-polyacrylamide gel electrophoresis, and the radioactive bands associated with Dnmt1 were quantitated. As shown in Fig. 5B, F-DNA scarcely bound to Dnmt1(291–1620), but the binding significantly increased in the presence of the SRA. On the contrary, Dnmt1(602–1620), which lacks the RFTS, gave F-DNA-bound bands regardless of the presence or absence of the SRA. The results clearly indicate that the interaction between the RFTS and SRA allows the short 12-bp DNA to gain access to the catalytic center.
DISCUSSION
As found in the previous study, there is an energy barrier as to removal of the RFTS from the catalytic pocket to provide room for DNA (14). This energy barrier is due to the hydrogen bonds formed between the RFTS and catalytic domains. In the present study, DNA longer than 16-bp and the interaction of the RFTS of Dnmt1 with the SRA domain in Uhrf1 were shown to contribute to the displacement of the RFTS from the catalytic pocket and to the allowance of access of hemi-methylated DNA to the catalytic center.
Dnmt1(291–1620) but not Dnmt1(602–1620) scarcely methylated 12-bp short hemi-methylated DNA (Fig. 1B). Because Dnmt1(602–1620), which lacks the RFTS, methylated 12-bp hemi-methylated DNA, the catalytic domain of Dnmt1(602–1620) by itself was able to recognize 12-bp hemi-methylated DNA as a methyl group acceptor. On the contrary, hemi-methylated DNA longer than 16-bp was able to be methylated even by Dnmt1(291–1620). Dnmt1(291–1620) harboring mutations of the RFTS involved in the hydrogen bonds with the catalytic domain showed significant DNA methylation activity toward 12-bp hemi-methylated DNA (Fig. 2C), indicating that impairment of the hydrogen bonds between the RFTS and catalytic domains cannot inhibit completely the accession of 12-bp hemi-methylated DNA to the catalytic center. Regarding longer hemi-methylated DNA, the DNA was apparently able to remove the RFTS from the catalytic pocket, yielding the DNA methylation activity. This is supported by the observation that exogenously added RFTS effectively inhibited the DNA methylation activity of Dnmt1(602–1620) only when 12-bp hemi-methylated DNA was used as the methyl group acceptor, i.e. not that DNA toward 42-bp (Fig. 1C). In addition, the mutated RFTS as to the residues involved in the hydrogen bond formation was less effective in inhibiting the DNA methylation activity (Fig. 2D). Apparently, the DNA methylation activity of Dnmt1(291–1620) was saturated when the DNA was longer than 30 bp (Fig. 1B). It was found that 12-bp DNA tightly interacts with the Dnmt1 catalytic domain on co-crystallization of the F-DNA and the catalytic domain of Dnmt1 (15). The present study suggests that hemi-methylated DNA of longer than 16 bp can interact directly with Dnmt1(291–1620) to remove the RFTS from the catalytic pocket. Considering that Dnmt1(291–1620) can processively methylate hemi-methylated DNA (11), the extra DNA sequence of longer than 16-bp, which was determined to interact with the Dnmt1 catalytic pocket, may contribute to the processive methylation property of Dnmt1 as to DNA.
The RFTS is responsible for recruitment of Dnmt1 to the replication region (13) and interacts with Uhrf1 during replication (16). Because Uhrf1 is a prerequisite factor for maintenance methylation in vivo (16), the removal of the RFTS from the catalytic pocket should be dependent on the interaction of Dnmt1 with Uhrf1 in the replicating region. The interaction of the SRA of Uhrf1 with the RFTS of Dnmt1 to remove the RFTS from the catalytic pocket and to provide hemi-methylated DNA to the catalytic center could be a fail-safe mechanism that contributes to the faithful inheritance of the methylation patterns by the next generation. Because the SRA binds selectively to hemi-methylated DNA (17–19), the function of the SRA may not be limited to the displacement of the RFTS from the catalytic pocket of the Dnmt1 but also to the processivity of the methylation of hemi-methylated DNA. The RFTS and SRA interaction-dependent release of hemi-methylated DNA (Fig. 5A) following the removal of the RFTS from the catalytic pocket is an important step for the correct feeding of the hemi-methylated DNA from the SRA to the catalytic center of Dnmt1.
Acknowledgments
We thank Yumiko Yamagami for constructing the baculovirus coding Dnmt1 and expression in sf9 cells and Keiko Shinohara for purification of the Dnmt1.
This work was supported in part by grants-in-aid for scientific research B (to S. T.) and scientific research C (to I. S.) from the Japan Society for the Promotion of Science, and CREST by Japan Science and Technology Agency (to M. S. and I. S.).
- RFTS
- replication foci targeting sequence
- CBB
- Coomassie Brilliant Blue R-250
- F
- 5-fluorocytosine
- SRA
- SET and RING finger-associated
- Uhrf1
- ubiquitin-like with PHD and ring finger 1.
REFERENCES
- 1. Li E. (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 [DOI] [PubMed] [Google Scholar]
- 2. Reik W., Lewis A. (2005) Co-evolution of X chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 6, 403–410 [DOI] [PubMed] [Google Scholar]
- 3. Goll M. G., Bestor T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 4. Suetake I., Hayata D., Tajima S. (2006) The amino terminus of mouse DNA methyltransferase 1 forms an independent domain and binds to DNA with the sequence involving PCNA binding motif. J. Biochem. 140, 763–776 [DOI] [PubMed] [Google Scholar]
- 5. Rountree M. R., Bachman K. E., Baylin S. B. (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25, 269–277 [DOI] [PubMed] [Google Scholar]
- 6. Kim G. D., Ni J., Kelesoglu N., Roberts R. J., Pradhan S. (2002) Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21, 4183–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pradhan S., Kim G. D. (2002) The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 21, 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F. (1997) Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000 [DOI] [PubMed] [Google Scholar]
- 9. Kameshita I., Sekiguchi M., Hamasaki D., Sugiyama Y., Hatano N., Suetake I., Tajima S., Sueyoshi N. (2008) Cyclin-dependent kinase-like 5 binds and phosphorylates DNA methyltransferase 1. Biochem. Biophys. Res. Commun. 377, 1162–1167 [DOI] [PubMed] [Google Scholar]
- 10. Sugiyama Y., Hatano N., Sueyoshi N., Suetake I., Tajima S., Kinoshita E., Kinoshita-Kikuta E., Koike T., Kameshita I. (2010) The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1δ/ϵ. Biochem. J. 427, 489–497 [DOI] [PubMed] [Google Scholar]
- 11. Vilkaitis G., Suetake I., Klimasauskas S., Tajima S. (2005) Processive methylation of hemi-methylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 280, 64–72 [DOI] [PubMed] [Google Scholar]
- 12. Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 22, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leonhardt H., Page A. W., Weier H. U., Bestor T. H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873 [DOI] [PubMed] [Google Scholar]
- 14. Takeshita K., Suetake I., Yamashita E., Suga M., Narita H., Nakagawa A., Tajima S. (2011) Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. U.S.A. 108, 9055–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song J., Teplova M., Ishibe-Murakami S., Patel D. J. (2012) Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335, 709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T. A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 [DOI] [PubMed] [Google Scholar]
- 17. Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 [DOI] [PubMed] [Google Scholar]
- 18. Avvakumov G. V., Walker J. R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C. H., Dhe-Paganon S. (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825 [DOI] [PubMed] [Google Scholar]
- 19. Hashimoto H., Horton J. R., Zhang X., Bostick M., Jacobsen S. E., Cheng X. (2008) The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F., Koseki H., Nakanishi M. (2013) Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253 [DOI] [PubMed] [Google Scholar]
- 21. Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. (1977) Nucleotide sequence of bacteriophage ΦX174 DNA. Nature 265, 687–695 [DOI] [PubMed] [Google Scholar]
- 22. Syeda F., Fagan R. L., Wean M., Avvakumov G. V., Walker J. R., Xue S., Dhe-Paganon S., Brenner C. (2011) The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. J. Biol. Chem. 286, 15344–15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brank A. S., Van Bemmel D. M., Christman J. K. (2002) Optimization of baculovirus-mediated expression and purification of hexahistidine-tagged murine DNA(cytosine-C5)-methyltransferase-1 in Spodoptera frugiperda 9 cells. Protein Expr. Purif. 25, 31–40 [DOI] [PubMed] [Google Scholar]