Novel septin 9 repeat motifs interact with the acidic C-terminal tails of β-tubulin to promote microtubule bundling and asymmetric neurite growth.
Abstract
Septin 9 (SEPT9) interacts with microtubules (MTs) and is mutated in hereditary neuralgic amyotrophy (HNA), an autosomal-dominant neuropathy. The mechanism of SEPT9 interaction with MTs and the molecular basis of HNA are unknown. Here, we show that the N-terminal domain of SEPT9 contains the novel repeat motifs K/R-x-x-E/D and R/K-R-x-E, which bind and bundle MTs by interacting with the acidic C-terminal tails of β-tubulin. Alanine scanning mutagenesis revealed that the K/R-R/x-x-E/D motifs pair electrostatically with one another and the tails of β-tubulin, enabling septin–septin interactions that link MTs together. SEPT9 isoforms lacking repeat motifs or containing the HNA-linked mutation R88W, which maps to the R/K-R-x-E motif, diminished intracellular MT bundling and impaired asymmetric neurite growth in PC-12 cells. Thus, the SEPT9 repeat motifs bind and bundle MTs, and thereby promote asymmetric neurite growth. These results provide the first insight into the mechanism of septin interaction with MTs and the molecular and cellular basis of HNA.
Introduction
Septins are hetero-oligomeric GTP-binding proteins that assemble into nonpolar filamentous structures, which are essential for microtubule (MT)-dependent cell processes such as mitosis and vesicle transport (Spiliotis, 2010; Saarikangas and Barral, 2011; Mostowy and Cossart, 2012). In mammalian cells, septin depletion affects MT organization, dynamics, and post-translational modifications (Spiliotis et al., 2008; Bowen et al., 2011), but how septins interact with MTs is unknown. Septin 9 (SEPT9) is a ubiquitously expressed septin that caps the ends of septin heteromers (Füchtbauer et al., 2011; Kim et al., 2011; Sellin et al., 2011). Alternative splicing and translation start sites give rise to SEPT9 isoforms, which differ in the length and sequence of their N terminus, which is critical for the association of septin heteromers with MTs (Sellin et al., 2012). Missense mutations in the N terminus of the SEPT9 are genetically linked to hereditary neuralgic amyotrophy (HNA), a hereditary neuralgic disorder with symptoms (shoulder/arm pain and atrophy) similar to those of the idiopathic Parsonage-Turner syndrome (Kuhlenbäumer et al., 2005; Hannibal et al., 2009; van Alfen, 2011). Abnormal expression of SEPT9 isoforms has also been reported in many cancers (Connolly et al., 2011a). Overexpression of SEPT9 isoforms induces oncogenic phenotypes including resistance to the anti-cancer drug paclitaxel, which stabilizes and bundles MTs (Orr et al., 2003; Gonzalez et al., 2007; Chacko et al., 2012). Unraveling the mechanism of SEPT9 interaction with MTs could point to the molecular roles of SEPT9 in these disorders and lead to therapeutic strategies.
Results and discussion
The basic N-terminal domain of SEPT9 binds and bundles MTs by interacting with the acidic C-terminal tails of β-tubulin
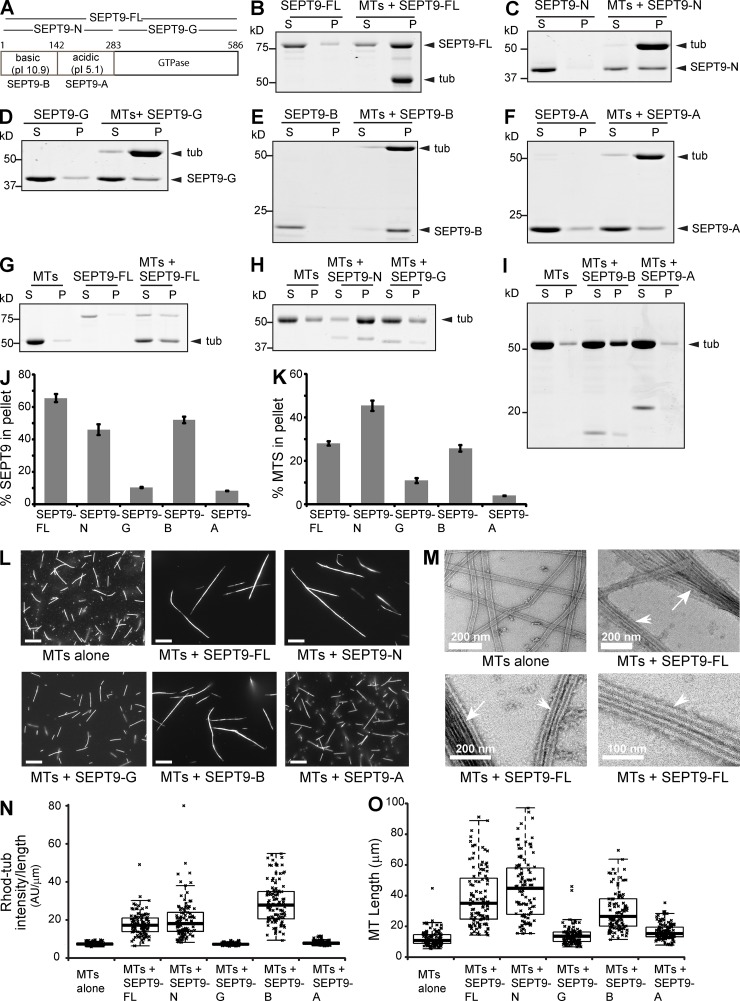
To determine how SEPT9 interacts with MTs, we first sought to identify the SEPT9 domain that binds MTs using in vitro co-sedimentation assays. Recombinant His-tagged fragments were made from the longest SEPT9_i1 isoform (SEPT9-FL; Fig. 1 A), which consists of the GTP-binding domain (aa 283–586; SEPT9-G) and a structurally disordered N-terminal tail (aa 1–283; SEPT9-N; Fig. S1 A) with a basic region (aa 1–142; SEPT9-B) and a proline-rich acidic domain (aa 143–283; SEPT9-A). SEPT9-FL co-sedimented with MTs (Fig. 1, B and J) and the apparent Kd value was 3.2 µM (Fig. S1 B). In contrast to SEPT9-G and SEPT9-A, which pelleted with MTs weakly (Fig. 1, D–F and J), SEPT9-N and SEPT9-B bound MTs similarly to SEPT9-FL (Fig. 1, C, E, and J).
Figure 1.
SEPT9 binds and bundles MTs via a basic N-terminal domain. (A) Sequence and domains of SEPT9_i1. (B–F) Coomassie-stained SDS-PAGE gels of the supernatant (S) and pellet (P) fractions after high speed (39,000 g) sedimentation of pre-polymerized paclitaxel-stabilized MTs with domains of SEPT9_i1. (G–I) Low speed (8,000 g) sedimentation of MTs in the presence of SEPT9_i1 domains. (J and K) Graphs show percentages of total protein pelleted with MTs at 39,000 g (J) and percentage of total MTs pelleted at 8,000 g (K) in three independent experiments. (L) Images show X-rhodamine–labeled MTs after mixing with recombinant SEPT9 fragments. Bars, 10 µm. (M) Negative stain EM images of MTs before and after mixing with SEPT9_i1. Arrows and arrowheads point to MT bundles and doublets, respectively. (N and O) Plots show the intensity of X-rhodamine fluorescence per micron of MT (O; n = 100) and the length of MTs (P; n = 100) per condition.
Next, low speed MT pelleting and visual MT assays were used to examine whether SEPT9 can bundle MTs. At low speed (8,000 g), MT sedimentation was increased by SEPT9-FL, SEPT9-N, and SEPT9-B (Fig. 1, G–I and K), but SEPT9-G and SEPT9-A had no effect (Fig. 1, H, I, and K). Incubation of fluorescent MTs with SEPT9-FL showed a marked increase in the length, thickness, and brightness of MT bundles (Fig. 1, L, N, and O). This bundling effect was independently confirmed by negative stain EM. MTs were arranged in bundles and doublets of parallel tubules that made tight contacts (Fig. 1 M). Elongated MT bundles were also observed with fluorescent MTs that were stabilized with the nonhydrolyzable GTP analogue GMP-CPP (Fig. S1 C). Decoration of paclitaxel- and GMP-CPP–stabilized MT bundles with recombinant GFP-tagged SEPT9-FL, but not SEPT2, demonstrated that SEPT9 binds and bundles MTs strongly and specifically (Fig. S1, D–I). In agreement with the MT sedimentation assays, significant bundling of fluorescent MTs was observed for only SEPT9-N and SEPT9-B (Fig. 1, L, N, and O). These data indicate that SEPT9 binds and bundles MTs via its basic N-terminal domain. Because SEPT9 forms hetero-oligomeric complexes with other septins (e.g., SEPT7, SEPT6), we purified SEPT9 in the presence of SEPT6/7 and tested how SEPT6/7/9 complexes affect MT bundling compared with SEPT9 alone or SEPT2/6/7. SEPT6/7/9 induced low speed sedimentation of MTs at a concentration (0.2 µM) that neither SEPT9 nor SEPT2/6/7 had an effect (Fig. S1, J and K). Thus, SEPT9 appears to be a key subunit for the bundling of MTs by septin complexes.
Electrostatic interactions between positively charged domains and the negatively charged C-terminal tails (CTTs) of tubulin underlie the mechanism by which some microtubule-associated proteins (MAPs) and kinesin motors associate with MTs (Amos and Schlieper, 2005; Marx et al., 2006). We hypothesized that electrostatics could similarly be involved in SEPT9-MT binding and bundling. Consistent with this possibility, MT bundling by SEPT9 was attenuated with increasing ionic strength (Fig. S1 L). We therefore tested whether the acidic CTTs of tubulin are required for MT-SEPT9 binding and aimed to identify the tubulin subunit(s) that associate with SEPT9.
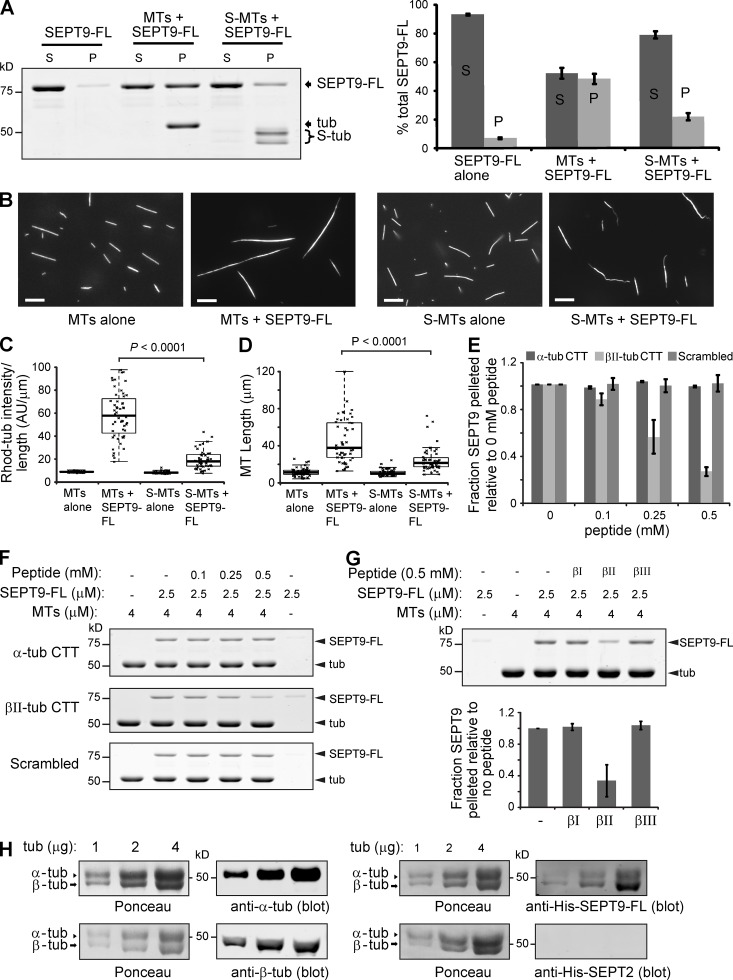
We first repeated the MT-SEPT9 co-sedimentation and visual assays using pre-polymerized MTs treated with subtilisin, which removes the CTTs of tubulin; ∼50% of α-tubulin and ∼90% β-tubulin tails were cleaved. Pelleting of SEPT9_i1 with MTs decreased by >50% after subtilisin-treatment (Fig. 2 A). Fluorescent MT elongation and bundling by SEPT9_i1 were also attenuated after subtilisin treatment (Fig. 2, B–D). To further test whether SEPT9-MT binding involves the CTTs of tubulin, we performed MT-SEPT9 pelleting assays in the presence of synthetic peptides, whose sequences corresponded to the CTTs of α- and βII-tubulin and a scrambled control. MT-SEPT9 binding decreased in a dose-dependent manner by βII, but not scrambled or α-tubulin CTT (Fig. 2, E and F). Interestingly, the CTTs of βI- and βIII-tubulin isotypes did not have an effect, indicating that SEPT9 interacts preferentially with βII-tubulin (Fig. 2 G). Preferential interaction of SEPT9 with β-tubulin was also confirmed with a blot-overlay assay, in which α- and β-tubulin subunits were separated by SDS-PAGE (Fig. 2 H). We concluded that electrostatic interactions between the acidic CTT of β-tubulin and the basic N-terminal domain of SEPT9 play a central role in SEPT9-MT binding and bundling.
Figure 2.
SEPT9 interacts with the acidic C-terminal tails of β-tubulin. (A) Gel shows supernatant (S) and pellet (P) fractions after sedimentation (39,000 g) of SEPT9-FL with untreated and subtilisin-treated MTs (S-MTs). Graph shows percentage of total SEPT9-FL in the S and P fractions. (B) Images show untreated and subtilisin-treated X-rhodamine–labeled MTs after mixing with SEPT9-FL. Bars, 10 µm. (C and D) Plots show the fluorescence intensity per micron of MT (C; n = 50) and the length of MTs (D; n = 50). (E and F) Graph shows fraction of SEPT9-FL pelleted with MTs in the presence of increasing concentrations of α-tubulin, βII-tubulin, and scrambled CTT peptide relative to no peptide in three independent experiments. Gels show the pellet fractions after MT pelleting with SEPT9-FL in the presence of increasing concentrations of peptides. (G) Gels show the pellet fractions after sedimentation of MTs with SEPT9-FL in the presence of βI-, βII-, and βIII-tubulin CTT peptides. Graph shows the fraction of SEPT9-FL pelleted in the presence of peptides relative to no peptide in three independent experiments. (H) Increasing amounts of bovine brain tubulin was separated by 7.5% SDS-PAGE and transferred to nitrocellulose membranes, which were stained with Ponceau S red. Membranes were blotted with DM1A and TUB2.1 antibodies against α- and β-tubulin, respectively, and overlaid with His-tagged SEPT9-FL and SEPT2, which were detected with anti-His antibodies.
Novel SEPT9 repeat motifs K/R-x-x-E/D and R/K-R-x-E provide a mechanism for MT bundling
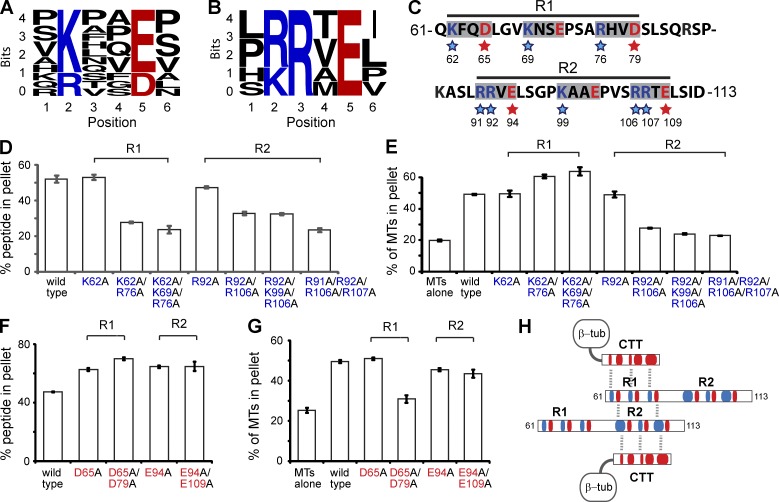
To hone in on the mechanism of SEPT9-MT interaction, we sought to identify domains or repeat motifs similar to the positively charged MT-binding domains of MAPs, MT plus end–tracking proteins and motors (Amos and Schlieper, 2005; Marx et al., 2006; Akhmanova and Steinmetz, 2008). Although we were not able to identify any known motifs, the N terminus of SEPT9_i1 contains multiple repeats of the tetrapeptide sequences K/R-x-x-E/D and R/K-R-x-E (Fig. 3, A and B), which are often flanked by proline or serine residues. By truncating the basic N-terminal domain of SEPT9_i1, we determined that aa 61–113 comprise the most minimal region of SEPT9_i1 that binds and bundles MTs (Fig. S2, A–C) and contains multiple repeats of the identified motifs.
Figure 3.
MT bundling by novel SEPT9 repeat motifs K/R-x-x-E/D and R/K-R-x-E. (A and B) WebLogo alignments of eleven hexapeptide sequences containing the K/R-x-x-E/D motif (A) and six sequences containing the R/K-R-x-E motif (B) within the N terminus (aa 1–286) of SEPT9_i1. The height of each residue indicates the frequency of its presence at the indicated positions. (C) Sequence of aa 61–113 of SEPT9_i1. The blue and red stars highlight the basic and acidic residues mutated to alanine. (D and E) Graphs show percentage of total SEPT9_i1(61–113) (wild-type and basic residue mutants) pelleted with MTs at 39,000 g (D), and percentage of total tubulin pelleted at 8,000 g (E) in three independent experiments. Representative gels are shown in Fig. S2. (F and G) Graphs show percentage of total 61–113 peptide (wild-type and acidic residue mutants) pelleted with MTs at 39,000 g (F), and percentage of total tubulin pelleted at 8,000 g (G) in three independent experiments. Representative gels are shown in Fig. S2. (H) Schematic shows a model of electrostatic interactions between the acidic (red) CTTs of tubulin and the basic (blue) residues of the SEPT9 repeat motifs. MT cross-linking is achieved by interactions between the acidic and basic residues of the K/R-x-x-E/D and R/K-R-x-E motifs, respectively.
We set out to elucidate the role of these novel motifs in MT binding and bundling by alanine scanning mutagenesis. The first half of the SEPT9_i1(61–113) peptide contains a region (R1) of three K/R-x-x-E/D motifs and the second half (R2) consists of a K-x-x-E motif flanked by two R-R-x-E motifs (Fig. 3 C); neither R1 nor R2 alone bound or bundled MTs. Alanine substitution of the basic residues in two or all three of the R1 motifs resulted in an ∼50% decrease in MT binding (Fig. 3 D and Fig. S2 D). A similar decrease was observed by mutating the basic residues of the R2 motifs (Fig. 3 D and Fig. S2 E). We assessed the effects of the same mutations in MT bundling using a low speed sedimentation assay. Strikingly, alanine substitutions of the basic residues of the R-R-x-E motifs of R2 decreased MT bundling, but mutating the basic residues of the K/R-x-x-E/D motifs of R1 did not decrease MT bundling (Fig. 3 E; Fig. S2, F and G). These data indicate that although the basic residues of both K/R-x-x-E/D and R-R-x-E contribute to MT binding, the arginine residues of the R-R-x-E motifs are uniquely critical for MT bundling.
Next, we assessed the role of the acidic residues. Alanine substitutions of the acidic residues in one or two of the motifs of either R1 or R2 resulted in a moderate (∼30%) increase of MT-SEPT9_i1(61–113) binding (Fig. 3 F and Fig. S2 H). MT bundling, however, decreased by ∼50% when the acidic residues in two of the K/R-x-x-E/D motifs of the R1 region were mutated (Fig. 3 G and Fig. S2 I). In contrast, alanine substitutions of glutamate in the R-R-x-E motifs of the R2 region resulted in only a marginal decrease in MT bundling (Fig. 3 G and Fig. S2 I). These results show that although the acidic residues dampen somewhat the electrostatic affinity of all motifs for the CTTs of β-tubulin, the acidic residues of the K/R-x-x-E/D motifs (R1) are uniquely critical for MT bundling. Because the basic residues of the R-R-x-E motif are also uniquely involved in MT bundling, we posit that the extra arginine residues of the R-R-x-E motifs allow for additional interactions with the acidic residues of the K/R-x-x-E/D motifs, enabling homophilic trans interactions between SEPT9_i1(61–113) peptides that cross-link MTs into bundles (Fig. 3 H). Consistent with this model, a GST-tagged version of the N terminus of SEPT9 bound His-tagged SEPT9-N and SEPT9-FL (Fig. S2, J and K). Thus, homophilic trans interactions between the N termini of SEPT9 are likely to facilitate septin and MT cross-linking.
SEPT9 repeat motifs are required for intracellular MT bundling and the generation of neurite asymmetry
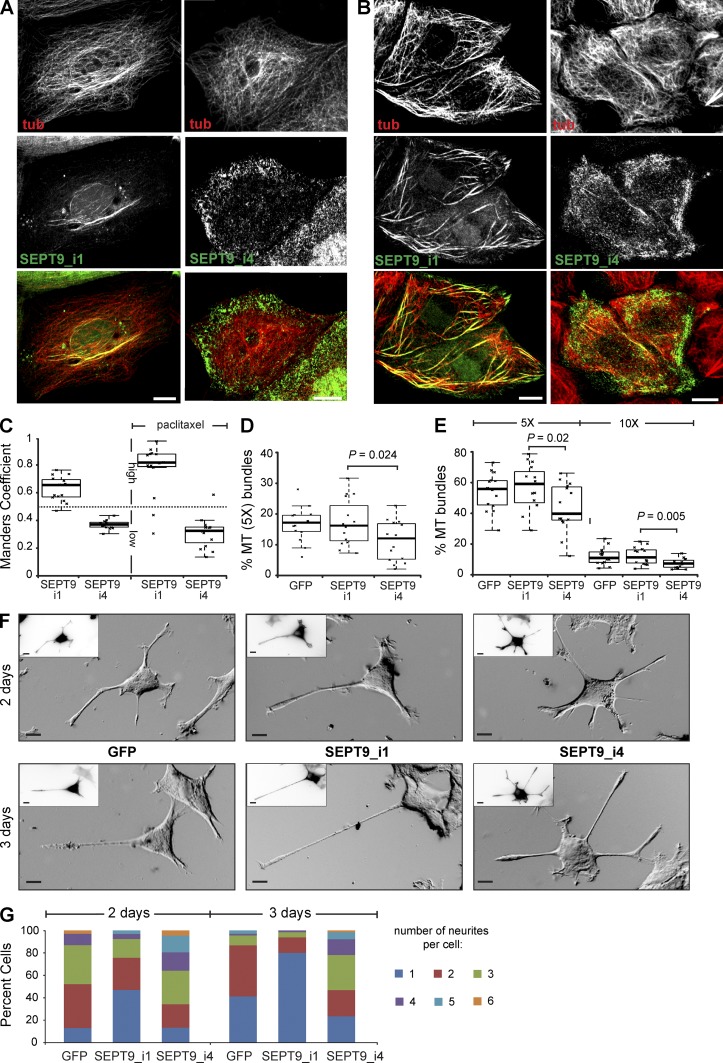
Given the mechanistic role of the K/R-R/x-x-E/D motifs in MT binding and bundling in vitro, we examined the functional significance of these motifs for intracellular MT bundling and neuronal morphogenesis, which requires MT bundling by MAPs (e.g., tau, MAP1B) that support the asymmetric growth of neuronal protrusions termed neurites (Caceres and Kosik, 1990; Teng et al., 2001; Feltrin et al., 2012). We hypothesized that SEPT9 isoforms that lack K/R-R/x-x-E/D motifs from their N terminus could affect MT bundling and asymmetric neurite growth. Among the known SEPT9 protein isoforms, SEPT9_i4 lacks the N-terminal 164 aa and therefore most of the MT-binding and -bundling repeat motifs of SEPT9_i1. To test if the absence of these motifs affects intracellular MT bundling, we expressed GFP-tagged chimeras of these isoforms in MDCK cells, in which septins were previously shown to colocalize with MTs (Spiliotis et al., 2008; Bowen et al., 2011). We found that SEPT9_i1 colocalized strongly with perinuclear MT bundles, whereas SEPT9_i4 was distributed at peripheral lamellae showing little to no colocalization with MTs (Fig. 4, A and C). Upon treatment with paclitaxel, an MT-stabilizing drug that induces MT bundling, SEPT9_i1 colocalized with MT bundles extensively, whereas SEPT9_i4 showed little colocalization with MTs (Fig. 4, B and C). Interestingly, paclitaxel-treated cells that expressed SEPT9_i4 cells appeared to have less number of long and straight MT bundles (Fig. 4 B). This effect was quantified by measuring the relative percentage of MTs with fluorescence intensities 5× or 10× greater than the mean intensity of the dimmest single peripheral MTs, using a segmentation analysis of 3D confocal image stacks as previously published (Sammak and Borisy, 1988; Bowen et al., 2011). MDCK cells that expressed SEPT9_i4-GFP had consistently lower percentage of 5× or 10× MTs compared with control and SEPT9_i1-GFP in the presence or absence of paclitaxel (Fig. 4, D and E). A lower percentage of MT bundles was also observed in fibroblasts from Sept9 knockout mice at steady-state and after treatment with paclitaxel (Fig. S3). Thus, SEPT9 is critical for the intracellular organization of MTs into bundles.
Figure 4.
SEPT9 repeat motifs are required for MT bundling and asymmetric neurite growth. (A and B) Maximal projections of 3D confocal microscopy images of MDCK cells expressing GFP-tagged SEPT9_i1 and SEPT9_i4 before (A) and after (B) treatment with 5 µM paclitaxel for 1.5 h. (C) Manders coefficients for the colocalization of GFP-tagged SEPT9_i1 and SEPT9_i4 with MTs in MDCK cells (n = 15). High and low colocalization are indicated by coefficients >0.5 and <0.5, respectively. (D and E) Plots show the fluorescence intensity of putative MT bundles with 5× and 10× the mean intensity of single MTs as percentage of total MT intensity in MDCK cells (n = 15) before (D) or after (E) treatment with paclitaxel. (F) Phase-contrast images show PC12 cells transfected with GFP and GFP-tagged SEPT9_i1 and SEPT9_i4 after a 2- and 3-d NGF treatment. Insets show GFP fluorescence in inverted monochrome. Bars, 10 µm. (G) Graph shows percentage of PC12 cells (n = 90) with one or more neurites. Pooled data from three independent experiments are shown.
During neuronal morphogenesis, MT bundling supports the asymmetric growth of a neurite, which extends faster and longer than other neurites to become the presumptive axon. To test whether SEPT9_i1 and the K/R-R/x-x-E/D motifs affect neurite growth, we assayed for NGF-induced neurite formation in PC12 cells. After 48 and 72 h of NGF treatment, the percentage of GFP-SEPT9_i1–expressing cells with single neurites was respectively fivefold and twofold higher than control cells (Fig. 4, F and G). In contrast to this amplification of asymmetric neurite growth by SEPT9_i1, expression of SEPT9_i4 had the opposite effect, increasing the percentage of cells with more than three neurites by twofold after 48 h of NGF treatment (Fig. 4, F and G). Notably, after 72 h of NGF exposure, the percentage of SEPT9_i4–expressing cells with single neurites was twofold lower than control cells (Fig. 4 G) and the average neurite length was shorter than the length of single neurites in SEPT9_i1–expressing cells (2.3 vs. 3.3 times the cell width). These results indicate that the MT-bundling properties of SEPT9 promote asymmetric neurite growth, which depends on the presence of K/R-R/x-x-E/D motifs within the N-terminal sequence of SEPT9.
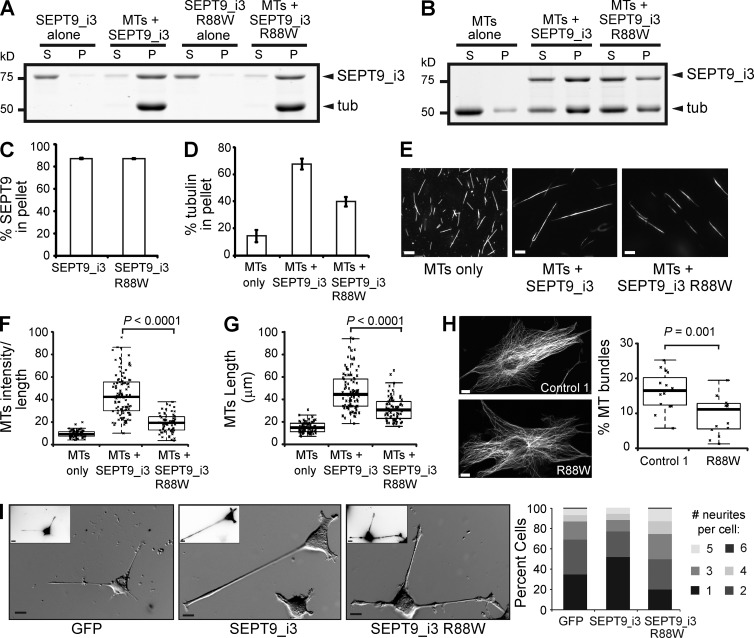
HNA-linked mutation R88W impairs MT bundling and asymmetric neurite growth
Alterations in the N-terminal sequence of SEPT9 have been genetically linked to HNA, a neuralgic disorder whose molecular etiology remains unknown (Kuhlenbäumer et al., 2005; Hannibal et al., 2009; van Alfen, 2011). Several HNA patients carry the mutation R88W in isoform 3 of SEPT9 (Hannibal et al., 2009), which has one K/R-x-x-E/D motif less than SEPT9_i1 and contains seven unique residues in place of aa 1–25 of SEPT9_i1. Because the R88W mutation of SEPT9_i3 maps to the first arginine of an R-R-x-E motif, we asked whether this mutation affects the MT-binding and -bundling properties of SEPT9_i3. The missense mutation did not affect high speed pelleting of SEPT9_i3 with MTs (Fig. 5, A and C), but low speed sedimentation of MTs decreased by ∼30% (Fig. 5, B and D). In fluorescent MT-bundling assays, the R88W mutation weakened the MT-bundling effects of SEPT9_i3 (Fig. 5, E–G). To determine if the R88W mutation alters MT bundling in vivo, we obtained dermal fibroblasts from healthy control individuals and HNA patients with the R88W genotype. We found that the fraction of MTs with 5× the mean intensity of single MTs was lower in the HNA patient fibroblasts compared with cells from a healthy individual (Fig. 5 H). Given the role of SEPT9_i1 and its repeat motifs in asymmetric neurite growth, we tested how the wild-type and R88W mutant versions of SEPT9_i3 affect neurite outgrowth in PC12 cells. Expression of GFP-SEPT9_i3 increased the percentage of cells with a single neurite by 50% relative to GFP-expressing cells (Fig. 5 I). The R88W mutant, however, increased the overall percentage of cells with more than three neurites (Fig. 5 I). Taken together, these data show that the HNA-linked R88W mutation has adverse effects on MT bundling and asymmetric neurite growth.
Figure 5.
HNA-linked mutation R88W impairs MT bundling and neurite asymmetry. (A and B) Gels show supernatant (S) and pellet (P) fractions from high (A; 39,000 g) and low (B; 8,000 g) speed MT-pelleting assays. (C and D) Graphs show percentages of total protein co-pelleted with MTs (C) and percentage of total tubulin pelleted (D) in three independent experiments. (E) Images show X-rhodamine–labeled MTs after mixing with recombinant SEPT9_i3 and SEPT9_i3(R88W). Bars, 10 µm. (F and G) Plots show the intensity of X-rhodamine fluorescence per micron of MT (F; n = 100) and the length of MTs (G; n = 100) per condition. (H) Images of dermal cells from healthy individual (control 1) and HNA patient with the R88W genotype stained for α-tubulin. Bars, 10 µm. Plot shows intensity of putative MT bundles with 5× the mean intensity of single MTs as percentage of total MT intensity (n = 15). (I) Phase-contrast images show PC12 cells transfected with GFP and GFP-tagged wild-type and R88W SEPT9_i3. Insets show GFP fluorescence in inverted monochrome. Graph shows percentage of cells (n = 90) with one or more neurites. Pooled data from three independent experiments are shown. Bars, 10 µm.
Our results provide the first insight into the mechanism of septin interaction with MTs. We have shown that the N terminus of SEPT9 binds and bundles MTs via repeat motifs similar to those of conventional MAPs (Noble et al., 1989; Butner and Kirschner, 1991; Cravchik et al., 1994; Ferralli et al., 1994). Formation of uniform linear arrays of bundled MTs is likely to involve highly ordered cis and trans interactions between SEPT9 containing hetero-oligomers. Cis interactions between the GTP-binding domains of septins mediate their assembly into heteromeric filaments (Sirajuddin et al., 2007). Conversely, trans interactions between the C-terminal coiled-coil domains of septins mediate the pairing of septin filaments (John et al., 2007; Bertin et al., 2008; DeMay et al., 2011; de Almeida Marques et al., 2012). SEPT9 molecules lack C-terminal tails, but our data indicate that their N-terminal extensions could mediate trans interactions by electrostatic pairing between the acidic and basic residues of the K/R-x-x-E/D and R/K-R-x-E motifs, respectively. SEPT9-mediated MT cross-linking could be further reinforced by the trans interactions between the C-terminal coiled-coil domains of SEPT2, SEPT6, and SEPT7, which hetero-oligomerize with SEPT9 (Kim et al., 2011; Sellin et al., 2011). Although SEPT2 does not bind MTs (Fig. S1), previous work indicates that SEPT7 and possibly SEPT6 interact with MTs (Hu et al., 2012; Moon et al., 2013). These subunits, however, lack the repeat motifs of SEPT9.
Septins colocalize with MTs in various cell types, but the physiological significance of this interaction is poorly understood. Here, we have found that SEPT9 and its N-terminal repeat motifs affect intracellular MT bundling and asymmetric neurite growth. Importantly, expression of SEPT9 isoforms with deletions (SEPT9_i4) and mutations (HNA-linked R88W) in the repeated K/R-R/x-x-E/D sequence motifs impair MT bundling and asymmetric neurite growth. Interestingly, cancer cells that overexpress SEPT9_i4 have increased resistance to the drug paclitaxel (Chacko et al., 2012). It is unknown if paclitaxel-mediated MT bundling contributes to the cytotoxicity of the drug, but our data indicate that SEPT9 and its repeat motifs are partly required for paclitaxel-induced MT bundling. This role of SEPT9, however, could vary between different cell types. Given the preferential interaction of SEPT9 with βII-tubulin, variations in the expression of β-tubulin isotypes could affect septin association with MTs. In summary, our results suggest that alterations in SEPT9 isoform expression trigger changes in the intracellular organization and function of MTs, and thereby could contribute to the pathology of HNA and cancer.
Materials and methods
Cells, peptides, and plasmids
MDCKII/G cells were maintained in low glucose DME media (Sigma-Aldrich) supplemented with 10% FBS (Cell Generation) and 1 g/liter NaHCO3. MDCK cells were transfected with the plasmids pEGFP-SEPT9_v1 and pEGFP-SEPT9_v4, which encode respectively for GFP-tagged SEPT9_i1 and SEPT9_i4 and were constructed by PCR amplification of SEPT9_v1 (AF189713) and SEPT9_v4 (AJ312322) from normal breast tissue cDNA and insertion into pEGFP-C2 (Connolly et al., 2011b). PC-12 cells were maintained in high glucose DME supplemented with 6% donor-defined equine serum (Hyclone), 6% defined bovine calf serum (Hyclone), and 1 g/liter NaHCO3. PC-12 differentiation and neurite growth were induced after 24 h of transfection by incubation in low serum (1% horse serum, 1% bovine calf serum) DME containing 100 ng/ml 2.5S NGF (Harlan Biosciences). Embryonic fibroblasts derived from Sept9cond/cond and Sept9del/del mice were provided by E.-M. Füchtbauer (University of Aarhus, Aarhus, Denmark) and maintained in DME with 10% FBS as described previously (Füchtbauer et al., 2011). Dermal cells were derived from skin biopsies taken from a healthy individual (control 1) and an HNA patient with the SEPT9_i3_R88W mutation (c.262C>T). Both subjects provided written informed consents and the biopsy protocol was approved by the University of Kiel ethics committee. Under local anesthesia, a standard skin-biopsy punch (diameter 3 mm; Kai Industries) was used to take a skin sample from the extensor surface of the upper arm. After removal of the subcutaneous fatty tissue, skin explants were cultured in high glucose DME media with 20% FBS for 5–7 d. Skin fibroblasts that grew out of the explants were isolated and passaged. Experiments with R88W and control 1 fibroblasts were performed at passages 7–11 and 9–13, respectively. The peptides NH2-CEVGVDSVEGEGEEEGEEY-COOH (α-tubulin CTT), NH2-CQETAEEYQDEEQGEADAEDFG-COOH (control, scrambled βII-tubulin CTT), and NH2-CQYQDATAEEEEDFGEEAEEEA-COOH (βI tubulin CTT) were purchased at >95% purity from GenScript. The peptides NH2-CQYQDATADEQGEFEEEEGEDEA-COOH (βII-tubulin CTT) and NH2-CQYQDATAEEEGEMYEDDEEESEAQGPK-COOH (βIII-tubulin CTT) were purchased at >95% purity from LifeTein, LLC.
His-SEPT9_i1-expressing plasmid was constructed by PCR amplifying human SEPT9_i1 (NP_001106963) using the primers 5′-CGTAAGCTTGCATGAAGAAGTCTTACTC-3′ and 5′-GTACTCGAGCTACATCTCTGGGGC-3′ and cloning the amplified fragment into the HindIII and XhoI sites of pET-28a(+). His-GFP-SEPT9_i1-expressing plasmid was constructed by PCR amplifying GFP-SEPT9_i1 from pEGFP-C2-SEPT9_v1 (see above) using the primers 5′-TCGAAGCTTCCATGGTGAGCAAGGGC-3′ and 5′-GTACTCGAGCTACATCTCTGGGGC-3′ and inserting the fragment into the HindIII and XhoI sites of pET-28a(+). His-SEPT9_i3-expressing plasmid was made by PCR amplifying human SEPT9_v3 (NP_006631) using the primers 5′-CATGCTAGCATGGAGAGGGACCGG-3′ and 5′-ACTAAGCTTTTACATCTCTGGGGC-3′ and cloning the amplified fragment into the NheI and HindIII sites of pET-28a(+). His-GFP-SEPT2-expressing plasmid was constructed by PCR amplifying human GFP-SEPT2 (NP_006146) from pEGFP-C1-SEPT2 using primers 5′-TCGGGATCCATGGTGAGCAAGGGC-3′ and 5′-TCGAAGCTTTCACACATGCTGCCCGAG-3′ and cloning the amplified fragment into the BamHI and HindIII sites of pET-28a(+). Plasmids expressing His-tagged SEPT9-N (aa 1–283 of SEPT9_i1), SEPT9-B (aa 1–142 of SEPT9_i1), SEPT9-A (aa 143–283 of SEPT9_i1), SEPT9-G (aa 284–586 of SEPT9_i1), SEPT9_i3 (R88W), SEPT9_1(61–113), and its mutants were made with the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies) using the pET-SEPT9_v1 and pET-SEPT9_v3 plasmids. pGEX-SEPT9-N– and pGEX-SEPT9-G–expressing GST-tagged SEPT9-N and SEPT9-G were constructed by PCR amplifying SEPT9-N and SEPT9-G and inserting into HindIII and XhoI sites of pGEX-KT-ext.
For the bacterial expression of septin heterotrimers, the bicistronic plasmid SEPT6-H-SEPT7 was constructed for the simultaneous expression of nontagged (NT) SEPT6 and His-tagged SEPT7. First, His-SEPT7 expressing plasmid was constructed by PCR amplifying the rat SEPT7 transcript variant 2 (NM 0011137.4) using the primer 5′-TCGGGATCCATGTCGGTCAGTGCG-3′ and 5′-GTAAAGCTTTTAAAAGATCTTGCC-3′ and cloning the amplified fragment into the BamHI and HindIII sites of pET-28a(+). Second, a His-tagged SEPT6 was constructed by PCR amplifying the human SEPT6_v3 (NP_665798) using the primer 5′-TCGGGATCCATGGCAGCGACCGATATAG-3′ and 5′-TCGAAGCTTTTAATTTTTCTTCTC-3′ and the amplified fragment was inserted into the BamHI and HindIII sites of pET-28a(+). Using the QuikChange II Site-Directed Mutagenesis kit, this plasmid was modified by removing the His tag in front of SEPT6 and inserting a SpeI site in front of the stop codon of SEPT6. Subsequently, the SEPT7 gene and the upstream ribosome-binding site were excised from pET-His-SEPT7 by digesting with XbaI and HindIII, and the fragment was subcloned into the SpeI and HindIII sites of the modified pET-SEPT6 to create a SEPT6-H-SEPT7–expressing plasmid. The plasmids encoding for NT-SEPT2 and NT-SEPT9_i1 were made from pET plasmids expressing His-SEPT9 (see above) and His-SEPT2, which was constructed by PCR amplifying mouse SEPT2 (NP_006146) using the primer 5′-TCGGGATCCATGTCTAAGCAACAACC-3′ and 5′-ATCCTCGAGTCACACATGCTGCCCG-3′ and cloning into the BamHI and XhoI sites of pET-28a(+). The His-SEPT9 and His-SEPT2 sequences and their upstream ribosome binding sites were then excised using XbaI and XhoI, and subcloned into pET-15b, which contains an ampicillin resistance marker. His tag sequences were then truncated with the QuikChange II Site-Directed Mutagenesis kit to create NT-SEPT2– and NT-SEPT9_i1–expressing plasmid.
Expression and purification of recombinant proteins
Plasmids encoding for recombinant proteins were transformed into Escherichia coli BL21(DE3) (Invitrogen). After bacterial cultures reached an OD600 of 0.8, protein expression was induced with 0.5 mM IPTG for 16 h at 18°C. Bacteria were centrifuged at 5,000 rpm for 5 min at 4°C. Pellets were resuspended in buffer containing 1% Triton X-100, 50 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, and 10 mM imidazole, and lysed using a French pressure cell (1,280 psi). Cell lysates were clarified by centrifuging at 14,000 g for 30 min at 4°C. Supernatants were loaded on columns containing 500 µl Ni-NTA beads (QIAGEN). Columns were washed with 10 ml washing buffer (50 mM Tris, pH 8, 300 mM NaCl, 10% glycerol, and 10 mM imidazole). Proteins were eluted with elution buffer (50 mM Tris, pH 8, 300 mM NaCl, 10% glycerol, and 250 mM imidazole) and dialyzed overnight in buffer containing 50 mM Tris, pH 8.0, 150 mM NaCl, and 10% glycerol. The heterotrimeric complexes SEPT2/6/7 and SEPT6/7/9_i1 were purified from E. coli BL21(DE3) by co-transforming the pET-28a(+) plasmid encoding for SEPT6-H-SEPT7 and the pET-15b plasmid encoding for NT-SEPT2 or NT-SEPT9_i1. The bacteria were selected on LB plates containing both kanamycin and ampicillin. The heterotrimeric complexes were purified using a Ni-NTA column. GST-tagged proteins were purified by lysing bacteria in GST binding buffer (25 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 0.5% Trition X-100) using a French press. Supernatants were clarified by centrifuging at 14,000 g for 30 min at 4°C and loaded on columns containing 500 µl Protino glutathione agarose 4B beads (Macherey Nagel). Columns were extensively washed with GST binding buffer and eluted with elution buffer (25 mM Tris HCl, pH 7.5, 150 mM NaCl, 5% glycerol, and 50 mM glutathione) and dialyzed overnight.
High and low speed MT cosedimentation and protein binding assays
Bovine brain tubulin (10 µM; >99% pure; Cytoskeleton, Inc.) was polymerized in G-PEM (80 mM Pipes, pH 6.9, 1 mM EGTA, 1 mM MgCl2, 1 mM GTP, and 10% glycerol) plus 80 µM paclitaxel for 30 min at 37°C. Subtilisin-cleaved MTs (S-MTs) were prepared by incubating pre-polymerized MTs with 30 µg/ml subtilisin (Sigma-Aldrich) at 37°C for 3 h. The reaction was terminated by addition of 2 mM PMSF. S-MTs were pelleted at 16,000 g on a cushion buffer (80 mM K-Pipes, pH 6.9, 1 mM MgCl2, 1 mM EGTA, 60% glycerol, and 20 µM paclitaxel) for 10 min at room temperature and then resuspended in 20 µl G-PEM with 20 µM paclitaxel. S-MTs were obtained from the supernatant fraction after re-centrifugation on 40 µl of cushion buffer at 12,000 g for 5 min. In high speed sedimentation assays, untreated, mock-, or subtilisin-treated MTs were incubated with recombinant SEPT9 proteins (10 µM) or SEPT9(61–113) peptides (30 µM) for 20 min at room temperature. Each reaction was placed on cushion buffer and centrifuged at 39,000 g (high speed) for 20 min at 25°C in an ultracentrifuge (Optima TL100; Beckman Coulter). In low speed sedimentation assays, MTs were incubated with recombinant SEPT9 proteins (2 µM), SEPT9(61–113) peptides (30 µM), or SEPT2/6/7, SEPT6/7/9_i1 complexes and SEPT9_i1 (0.2 µM; Fig. S1, J and K) for 10 min at room temperature. Each reaction was placed on cushion buffer and centrifuged at 8,000 g for 5 min. Pellets were resuspended in PBS of the same volume with the supernatants. Equal volumes of pellet and supernatant were loaded onto 10% SDS-PAGE and gels were stained with Coomassie Brilliant Blue. Gels were scanned and protein band densities were quantified with the Odyssey infrared scanning system (LI-COR Biosciences).
Binding assays between GST- and His-tagged proteins were performed (Fig. S2, J and K) by incubating 25 µg of purified GST-SEPT9-N or GST-SEPT9-G with 20 µl glutathione agarose 4B beads for 15 min. After washing three times with GST binding buffer, beads were incubated with 20 µg His-SEPT2, His-SEPT9, His-SEPT9-N, or His-SEPT9-G for 1.5 h. Beads were washed with GST binding buffer five times before resuspended with loading buffer and boiled. Samples were loaded into 10% SDS-PAGE gels and stained with Coomassie Brilliant Blue.
Visualization of MT bundling by fluorescence and electron microscopy
X-rhodamine–labeled bovine brain tubulin (10 µM; Cytoskeleton, Inc.) was polymerized in G-PEM buffer containing 80 µM paclitaxel. MTs were incubated with recombinant proteins (20 nM) in G-PEM buffer containing 20 µM paclitaxel for 10 min at room temperature. An aliquot (5 µl) of each reaction mix was mounted on a slide and sealed with a glass coverslip and nail polish. Slides were imaged on a fluorescent microscope (Axio Observer; Carl Zeiss) equipped with a Plan-Apo 63×/1.40 NA objective, a deep-cooled CCD camera (ORCA-AG; Hamamatsu Photonics), and Slidebook 5.0 software (Intelligent Imaging Innovations). All images were taken in the TRITC channel at 50-ms exposures. MT lengths and intensities were quantified by masking individual MTs (>5 µm). MT length and average fluorescence intensity per pixel after background subtraction were measured using Slidebook 5.0 software.
For visualization of MT bundling by negative stain electron microscopy, pre-polymerized MTs (Cytoskeleton, Inc.) were mixed with recombinant SEPT9-FL and incubated with glow-discharged 300 mesh Holey carbon copper grids covered with a thin layer of carbon. Grids were washed with 2% uranyl formate and subsequently blotted and air dried. Images were collected with a transmission electron microscope (Tecnai T12; FEI) equipped with a CCD camera (UltraScan 4000; Gatan, Inc.) operating at accelerating voltage of 120 kV, defocus value of −950 nm, and magnification of 52,000.
Western blots and overlay assays
Bovine brain tubulin (>99% pure; Cytoskeleton, Inc.) was separated by 7.5% SDS-PAGE and transferred to PROTRAN B85 nitrocellulose membranes (Whatman), which were stained with Ponceau S (Sigma-Aldrich) and scanned with a CanoScan LiDE 210 (Canon). After extensive wash with ddH2O, membranes were blotted with mouse antibody DM1A against α-tubulin (1:100,000; Sigma-Aldrich) or TUB2.1 antibody against β-tubulin (1:100,000; Sigma-Aldrich), and secondary Alexa Fluor 680 goat anti–mouse IgG (1:15,000; Invitrogen). For blot overlay assays, membranes were incubated for 1 h at 4°C in blocking buffer (10 mM Tris HCl, pH 6.8, 150 mM NaCl, 1 mM DTT, 0.1% Tween 20, 5% nonfat dry milk, and 0.5% BSA). Subsequently, membranes were overlayed with His-SEPT9_i1 (200 nM) in blocking buffer for 2 h. Membranes were washed with TBS containing 0.1% Tween 20 and incubated in the same buffer containing 2% BSA and mouse antibody against 6×His tag (1:5,000; R&D Systems) followed by secondary Alexa Fluor 680 goat anti–mouse IgG (1:15,000; Invitrogen). Blots were imaged with the Odyssey infrared imaging system (LI-COR Biosciences).
Immunofluorescence microscopy and image analysis
Cells (MDCK and skin fibroblasts) treated with paclitaxel (Sigma-Aldrich) or carrier (DMSO; Sigma-Aldrich) were fixed with PHEM buffer (60 mM Pipes-KOH, pH 6.9, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2) containing 3% paraformaldehyde (Electron Microscopy Sciences), 0.05% glutaraldehyde (Electron Microscopy Sciences), and 0.5% Triton X-100. Microtubules were stained with mouse antibody DM1A against α-tubulin (Sigma-Aldrich) and donkey DyLight 594–conjugated F(ab’)2 to mouse IgG (Jackson ImmunoResearch Laboratories, Inc.). Samples were mounted with FluorSave (EMD Millipore) and imaged on a laser-scanning confocal microscope (FluoView 1000; Olympus) using a Plan Apochromat 60×/1.42 NA objective. Serial optical sections were acquired from the bottom to the top of each cell at 0.2-µm steps. Each z-stack was imported into Slidebook 5.0 software for colocalization and MT fluorescence analyses. Background fluorescence was removed and fluorescence intensity segmentation was used to mask MTs and septin filaments, and colocalization values were automatically calculated using the Manders algorithm of the Slidebook 5.0 software. To determine the fraction of MT bundles relative to total MTs, we first measured the mean fluorescence intensity per pixel for the dimmest single MTs found in the periphery of each cell. Subsequently, MTs and MT regions with pixel values over 5 or 10 times the single MT pixel value were masked. The sum fluorescent intensity of these putative MT bundles was calculated as a fraction of the total MT fluorescence intensity in a 3D stack of images using Slidebook 5.0 software.
PC-12 cells were fixed with PHEM buffer (60 mM Pipes-KOH, pH 6.9, 25 mM Hepes, 10 mM EDTA, and 2 mM MgCl2) containing 3% paraformaldehyde (Electron Microscopy Sciences) and 5% sucrose. Cells were permeabilized with 0.5% Triton X-100 and stained for α-tubulin and imaged on a fluorescent microscope (Axio Observer; Carl Zeiss) equipped with a Plan-Apo 63×/1.40 NA objective, a deep-cooled CCD camera (ORCA-AG; Hamamatsu Photonics), and Slidebook 5.0 software. Microtubule-positive protrusions with lengths longer than the diameter of the cell soma were scored as neurites.
Statistical analysis and prediction of intrinsic disorder
Datasets were plotted in box-and-whisker diagrams. The bold horizontal line marks the median value and the bottom and top of each box corresponds respectively to the 25th (Q1) and 75th (Q3) percentiles of the range of values shown. Whisker ends correspond to the minimum and maximum values of each dataset. Values 1.5 times more than the Q3 value or 1.5 times less than the Q1 value were considered statistical outliers and were plotted outside the whisker portions of the diagram. Kolmogorov-Smirnov tests were performed to assess the normal distribution of each dataset and unpaired Student’s t tests were used to derive P-values for normally distributed datasets with equal standard deviations (SDs). The Welch t test was used to compare datasets with unequal SDs and the Mann-Whitney test was used to compare datasets, which were not normally distributed.
The prediction of intrinsic disorder of SEPT9_i1 was performed by combining results from GlobProt (http://globplot.embl.de), DisEMBL (http://dis.embl.de), DISOPRED2 (http://bioinf.cs.ucl.ac.uk/psipred/?disopred=1), IUPred (http://iupred.enzim.hu), DISpro (http://www.ics.uci.edu/∼baldig/dispro.html), OnD-CRF (http://babel.ucmp.umu.se/ond-crf/), and DRIP-PRD (http://www.sbc.su.se/∼maccallr/disorder). The predicted intrinsic disorder of every amino acid was set to 1 or 0 for values that were above or below the intrinsic disorder cut-off points of each algorithm (Seeger et al., 2012).
Online supplemental material
Fig. S1 shows the predicted intrinsic disorder of SEPT9_i1 and its dissociation constant, and the effects of GMP-CPP and ionic strength on the interaction of SEPT9_i1 with MTs. Fig. S2 shows the MT-binding and -bundling properties of SEPT9(61–113) and all of its mutants. Fig. S3 shows that SEPT9 is partly required for taxol-mediated MT bundling. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308068/DC1.
Supplementary Material
Acknowledgments
We thank Cristina Montagna (Albert Einstein Medical College, Bronx, NY); Ernst-Martin Füchtbauer (Aarhus University, Aarhus, Denmark) for cells and reagents; and Almudena Pacheco, Napoleão Valadares, and Jacob Lazarus for their advice. All microscopy was performed at Drexel University’s Cell Imaging Center.
This work was supported by National Institutes of Health grant GM097664 to E.T. Spiliotis and Deutsche Forschungsgemeinschaft grant KU1194/4-1 to G. Kuhlenbäumer and M. Pendziwiat.
Footnotes
Abbreviations used in this paper:
- CTT
- C-terminal tail
- HNA
- hereditary neuralgic amyotrophy
- MAP
- microtubule-associated protein
- MT
- microtubule
- SEPT9
- septin 9
References
- Akhmanova A., Steinmetz M.O.. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Amos L.A., Schlieper D.. 2005. Microtubules and maps. Adv. Protein Chem. 71:257–298. 10.1016/S0065-3233(04)71007-4 [DOI] [PubMed] [Google Scholar]
- Bertin A., McMurray M.A., Grob P., Park S.S., Garcia G., III, Patanwala I., Ng H.L., Alber T., Thorner J., Nogales E.. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 105:8274–8279. 10.1073/pnas.0803330105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.R., Hwang D., Bai X., Roy D., Spiliotis E.T.. 2011. Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J. Cell Biol. 194:187–197. 10.1083/jcb.201102076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butner K.A., Kirschner M.W.. 1991. Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115:717–730. 10.1083/jcb.115.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres A., Kosik K.S.. 1990. Inhibition of neurite polarity by tau antisense oligonucleotides in primary cerebellar neurons. Nature. 343:461–463. 10.1038/343461a0 [DOI] [PubMed] [Google Scholar]
- Chacko A.D., McDade S.S., Chanduloy S., Church S.W., Kennedy R., Price J., Hall P.A., Russell S.E.. 2012. Expression of the SEPT9_i4 isoform confers resistance to microtubule-interacting drugs. Cell Oncol (Dordr). 35:85–93. 10.1007/s13402-011-0066-0 [DOI] [PubMed] [Google Scholar]
- Connolly D., Abdesselam I., Verdier-Pinard P., Montagna C.. 2011a. Septin roles in tumorigenesis. Biol. Chem. 392:725–738. 10.1515/BC.2011.073 [DOI] [PubMed] [Google Scholar]
- Connolly D., Yang Z., Castaldi M., Simmons N., Oktay M.H., Coniglio S., Fazzari M.J., Verdier-Pinard P., Montagna C.. 2011b. Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression. Breast Cancer Res. 13:R76. 10.1186/bcr2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravchik A., Reddy D., Matus A.. 1994. Identification of a novel microtubule-binding domain in microtubule-associated protein 1A (MAP1A). J. Cell Sci. 107:661–672. [DOI] [PubMed] [Google Scholar]
- de Almeida Marques I., Valadares N.F., Garcia W., Damalio J.C., Macedo J.N., de Araújo A.P., Botello C.A., Andreu J.M., Garratt R.C.. 2012. Septin C-terminal domain interactions: implications for filament stability and assembly. Cell Biochem. Biophys. 62:317–328. 10.1007/s12013-011-9307-0 [DOI] [PubMed] [Google Scholar]
- DeMay B.S., Bai X., Howard L., Occhipinti P., Meseroll R.A., Spiliotis E.T., Oldenbourg R., Gladfelter A.S.. 2011. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J. Cell Biol. 193:1065–1081. 10.1083/jcb.201012143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltrin D., Fusco L., Witte H., Moretti F., Martin K., Letzelter M., Fluri E., Scheiffele P., Pertz O.. 2012. Growth cone MKK7 mRNA targeting regulates MAP1b-dependent microtubule bundling to control neurite elongation. PLoS Biol. 10:e1001439. 10.1371/journal.pbio.1001439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferralli J., Doll T., Matus A.. 1994. Sequence analysis of MAP2 function in living cells. J. Cell Sci. 107:3115–3125. [DOI] [PubMed] [Google Scholar]
- Füchtbauer A., Lassen L.B., Jensen A.B., Howard J., Quiroga Ade.S., Warming S., Sørensen A.B., Pedersen F.S., Füchtbauer E.M.. 2011. Septin9 is involved in septin filament formation and cellular stability. Biol. Chem. 392:769–777. 10.1515/BC.2011.088 [DOI] [PubMed] [Google Scholar]
- Gonzalez M.E., Peterson E.A., Privette L.M., Loffreda-Wren J.L., Kalikin L.M., Petty E.M.. 2007. High SEPT9_v1 expression in human breast cancer cells is associated with oncogenic phenotypes. Cancer Res. 67:8554–8564. 10.1158/0008-5472.CAN-07-1474 [DOI] [PubMed] [Google Scholar]
- Hannibal M.C., Ruzzo E.K., Miller L.R., Betz B., Buchan J.G., Knutzen D.M., Barnett K., Landsverk M.L., Brice A., LeGuern E., et al. 2009. SEPT9 gene sequencing analysis reveals recurrent mutations in hereditary neuralgic amyotrophy. Neurology. 72:1755–1759. 10.1212/WNL.0b013e3181a609e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Bai X., Bowen J.R., Dolat L., Korobova F., Yu W., Baas P.W., Svitkina T., Gallo G., Spiliotis E.T.. 2012. Septin-driven coordination of actin and microtubule remodeling regulates the collateral branching of axons. Curr. Biol. 22:1109–1115. 10.1016/j.cub.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John C.M., Hite R.K., Weirich C.S., Fitzgerald D.J., Jawhari H., Faty M., Schläpfer D., Kroschewski R., Winkler F.K., Walz T., et al. 2007. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 26:3296–3307. 10.1038/sj.emboj.7601775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Froese C.D., Estey M.P., Trimble W.S.. 2011. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 195:815–826. 10.1083/jcb.201106131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlenbäumer G., Hannibal M.C., Nelis E., Schirmacher A., Verpoorten N., Meuleman J., Watts G.D., De Vriendt E., Young P., Stögbauer F., et al. 2005. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat. Genet. 37:1044–1046. 10.1038/ng1649 [DOI] [PubMed] [Google Scholar]
- Marx A., Müller J., Mandelkow E.M., Hoenger A., Mandelkow E.. 2006. Interaction of kinesin motors, microtubules, and MAPs. J. Muscle Res. Cell Motil. 27:125–137. 10.1007/s10974-005-9051-4 [DOI] [PubMed] [Google Scholar]
- Moon I.S., Lee H., Walikonis R.S.. 2013. Septin 6 localizes to microtubules in neuronal dendrites. Cytotechnology. 65:179–186. 10.1007/s10616-012-9477-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Cossart P.. 2012. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13:183–194. [DOI] [PubMed] [Google Scholar]
- Noble M., Lewis S.A., Cowan N.J.. 1989. The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J. Cell Biol. 109:3367–3376. 10.1083/jcb.109.6.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr G.A., Verdier-Pinard P., McDaid H., Horwitz S.B.. 2003. Mechanisms of Taxol resistance related to microtubules. Oncogene. 22:7280–7295. 10.1038/sj.onc.1206934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., Barral Y.. 2011. The emerging functions of septins in metazoans. EMBO Rep. 12:1118–1126. 10.1038/embor.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammak P.J., Borisy G.G.. 1988. Direct observation of microtubule dynamics in living cells. Nature. 332:724–726. 10.1038/332724a0 [DOI] [PubMed] [Google Scholar]
- Seeger M.A., Zhang Y., Rice S.E.. 2012. Kinesin tail domains are intrinsically disordered. Proteins. 80:2437–2446. 10.1002/prot.24128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Sandblad L., Stenmark S., Gullberg M.. 2011. Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell. 22:3152–3164. 10.1091/mbc.E11-03-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Stenmark S., Gullberg M.. 2012. Mammalian SEPT9 isoforms direct microtubule-dependent arrangements of septin core heteromers. Mol. Biol. Cell. 23:4242–4255. 10.1091/mbc.E12-06-0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kühlmann D., Macara I.G., Weyand M., Stark H., Wittinghofer A.. 2007. Structural insight into filament formation by mammalian septins. Nature. 449:311–315. 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Spiliotis E.T. 2010. Regulation of microtubule organization and functions by septin GTPases. Cytoskeleton (Hoboken). 67:339–345. [DOI] [PubMed] [Google Scholar]
- Spiliotis E.T., Hunt S.J., Hu Q., Kinoshita M., Nelson W.J.. 2008. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J. Cell Biol. 180:295–303. 10.1083/jcb.200710039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J., Takei Y., Harada A., Nakata T., Chen J., Hirokawa N.. 2001. Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J. Cell Biol. 155:65–76. 10.1083/jcb.200106025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alfen N. 2011. Clinical and pathophysiological concepts of neuralgic amyotrophy. Nat Rev Neurol. 7:315–322. 10.1038/nrneurol.2011.62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.