Abstract
Peptidylarginine deiminases are a family of enzymes that mediate post-translational modifications of protein arginine residues by deimination or demethylimination to produce citrulline. In vitro, the activity of PADs is dependent on calcium and reductive reagents carrying a free sulfhydryl group. The discovery that PAD4 can target both arginine and methyl-arginine for citrullination about 10 years ago renewed our interest in studying this family of enzymes in gene regulation and their physiological functions. The deregulation of PADs is involved in the etiology of multiple human diseases, including cancers and autoimmune disorders. There is a growing effort to develop isoform specific PAD inhibitors for disease treatment. However, the regulation of the activity of PADs in vivo remains largely elusive, and we expect that much will be learned about the role of these enzymes in normal life cycle and under pathology conditions.
Keywords: peptidylarginine deiminase, histone, gene regulation, cancer, autoimmunity
1. INTRODUCTION
1.1 The early discovery of PADs as protein citrullination enzymes
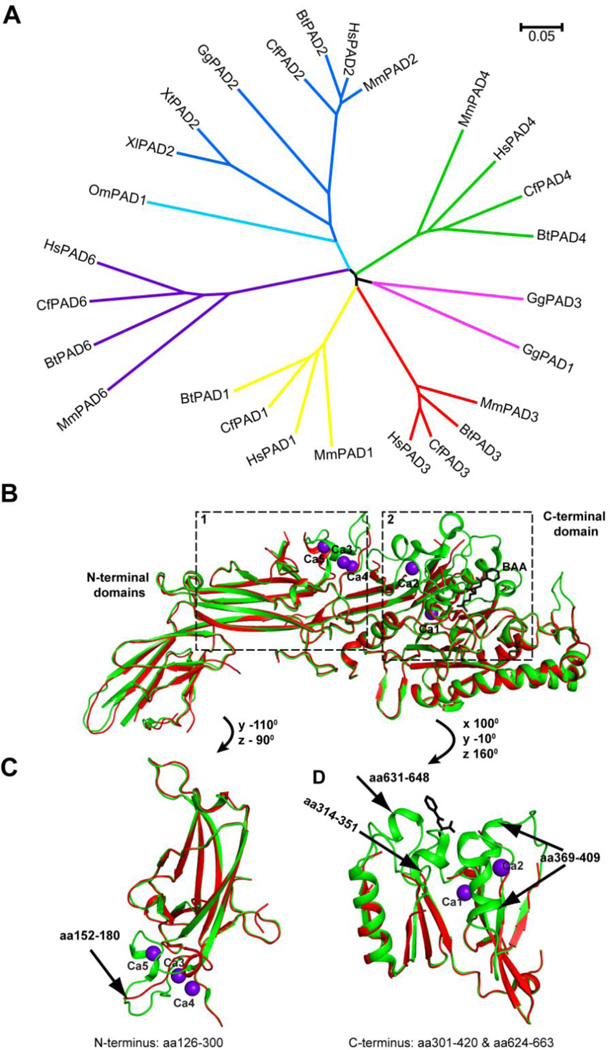
Citrulline residues were detected in early 1960s from polypeptide hydrolysates of hair inner root sheath cells and medullary cells [1]. Since there is no citrulline tRNA in vivo, citrulline can only be produced by enzymatic modifications after protein synthesis. Subsequently, the peptidylarginine deiminases (PADs, also called PADIs) were identified, which convert protein arginine residues to citrulline in a calcium dependent manner ([2], reviewed in [3]). PADs are involved nomal functions in the immune and reproduction systems as well as in the skin (see recent review in these particular areas in [4–6]). In each of the mammalian vertebrate genomes, five highly conserved PADs exist, including PAD1, 2, 3, 4 and 6. (Fig. 1A). In the last decade or so, more research on the gene features, tissue-specific distribution and preferred substrates of PADs has been performed. Human PAD4 can bind five calcium ions (Fig. 1B). Several flexible parts of PAD4 form stable secondary structures after the binding of calcium and substate, indicating that calcium stablizes the conformation of PAD4 and may facilitate the formation of the active site cleft (Fig. 1C and 1D). The calcium binding sites in PAD4 are conserved in PAD1, −2, and −3 except for PAD6. Thus, calcium is an important regulator of the active PAD enzymes [7].
Fig 1. The PAD family of proteins and the effect of calcium on the PAD4 structure.
(A) Phylogenetic analyses of 26 PAD proteins from several vertebrate species. Sequences of mammalian PADs were derived from databases for Human (Hs: Homo sapiens), mouse (Mm: Mus musculus), cow (Bt: Bos taurus), dog (Cf: Canis lupis familiaris). Also included are PADs from other species, such as chicken (Gg: Gallus gallus), trout (Om: Oncorhynchus mykiss) and frog (Xl: Xenopus laevis; Xt: Xenopus Silurana tropicalis). The unrooted phylogenetic tree was generated using ClustalW2 on EMBL-EBI website and visualized by MEGA 5.2. Scale bar represents 0.05 amino acid substitutions per site. Accession number for PADs in alignment are as follows: HsPAD1 (NP_037490.2), HsPAD2 (NP_031391.2), HsPAD3 (NP_057317.2), HsPAD4 (NP_036519.2), HsPAD6 (NP_997304.3), MmPAD1 (NP_035189.1), MmPAD2 (NP_032838.2), MmPAD3 (NP_035190.3), MmPAD4 (NP_035191.2), MmPAD6 (NP_694746.2), BtPAD1 (NP_001094742.1), BtPAD2 (NP_001098922.1), BtPAD3 (XP_003585850.1), BtPAD4 (NP_001179102.1), BtPAD6 (XP_002685843.1), CfPAD1 (XP_851932.1), CfPAD2 (XP_544539.2), CfPAD3 (XP_535391.2), CfPAD4 (XP_848494.1), CfPAD6 (NP_001091016), GgPAD1 (XP_425729.3), GgPAD2 (XP_425730.3), GgPAD3 (NP_990374), OmPAD1 (NP_001153973.1), XlPAD2 (NP_001080369), XtPAD2 (NP_001096490.1).
(B) Ribbon presentation of superimposed structures of Ca2+ and substrate bound PAD4 (green, 1WDA) and free PAD4 (Red, 1WD8). Five Ca2+ ions bound to PAD4 are shown as purple spheres. Substrate BAA (benzoyl-L-arginine amide) is in a stick model.
(C) Superimposed structure of Ca2+ and substrate bound PAD4 (green) and free PAD4 (red) in boxed region 1 (N-terminal amino acids 126–300). The arrow denotes the region with major conformational changes after calcium binding.
(D) Superimposed structure of Ca2+ and substrate bound PAD4 (green) and free PAD4 (red) in boxed region 2 (C-terminal amino acids 301–420 and 624–663). Arrows denote the regions with major conformational changes.
1.2 The five PADs
1.2.1
PAD1 is mainly expressed in epidermis and uterus [8, 9]. PAD1 deiminates keratin K1 and is involved in the cornification of epidermal tissues [10].
1.2.2
PAD2 is widely expressed in multiple tissues, including secretory glands, brain, uterus, spleen, pancreas, skeletal muscle [11–16]. The expression of PAD2 can be regulated at both mRNA splicing and protein translation levels [15, 16]. Myelin basic protein of the central nervous system and vimentin in skeletal muscle and macrophages are long known substrates of PAD2 [16, 17]. Recently, β and γ-actins were identified as PAD2 substrates in human neutrophils [18]. PAD2 is mainly a cytoplasmic protein, but a fraction of PAD2 may become nuclear in canine and human mammary epithelial cells [19, 20]. Nuclear PAD2 may citrullinate histones H3 and H4 [19, 21–23], suggesting a role of this protein in gene regulation.
1.2.3. PAD3 is localized to epidermis and hair follicles [24–26]
PAD3 is colocalized with trichohyalin, a structural protein in the inner root sheath and medulla of hair follicles [25, 26]. In addition, PAD3 colocalizes with profilaggrin and filaggrin in the epidermis [26]. PAD3 targets filaggrin, which interacts with keratin intermediate filament to regulate epidermal homeostasis in the granular layer and lower stratum corneum of human epidermis [2, 25, 26]. Deimination of filaggrin and trichohyalin in vitro by recombinant PAD3 further supports that PAD3 is involved in regulating epidermis functions [25, 26].
1.2.4
PAD4 (also called PADV and PADI4) is detected mainly in white blood cells including granulocytes and monocytes under normal physiological conditions [16, 27, 28]. However, in a wide range of tumors of various tissue origins, the overexpression of PAD4 was detected, suggesting that PAD4 plays a role in tumorigenesis [29, 30]. PAD4 is localized primarily in the nucleus and contains a nuclear localization signal sequence at its N-terminus. PAD4 citrullinates a range of nuclear proteins, such as histones H2A, H3 and H4, ING4, p300/CBP, nucleophosmin and nuclear lamin C, thereby playing an important role in nuclear functions [31–36].
1.2.5
PAD6 was originally identified from mouse eggs and embryos and was named ePAD (egg PAD) [37]. PAD6 regulates oocyte cytoskeletal sheet formation and female fertility [38]. Recently, it is found that PAD6 localizes to the cytoplasmic lattices and regulates the function of microtubules during early embryo development [39]. In human tissues, PAD6 is mainly restricted to ovary, testis and peripheral blood leucocytes [40]. Interestingly, unlike the other PADs, PAD6 have lost some of the conserved Ca2+ binding residues and the active center cysteine residue is also different from other PADs [41], suggesting that PAD6 is likely not an active deiminase.
Recent studies have revealed the role of PADs in physiological and pathological conditions. In the following sessions, we will focus on the role of PADs in gene regulation, innate immunity, cancers and autoimmune diseases. We will further discuss the potential of PADs as druggable targets for disease treatment.
2. PAD4 and PAD2 IN GENE REGULATION
2.1. PAD4 in gene regulation
In eukaryotic cells, nuclear DNA is organized with two of each histones H3, H4, H2B, and H2A to form a nucleosome core particle, which is further organized with the linker DNA and histone H1 to form the 10 nm chromatin fiber and folded to form the higher order chromatin structures. The nuclear structure and 3D organization is under continuous remodeling to adapt to the physiological and environmental changes that the cells are exposed to. Because of this structural organization, histone modifications, including methylation, acetylation, phosphorylation and citrullination, work as a signaling network to provide the on the off signal for gene expression and/or a landing platform for effector protein binding [42–44].
Histone modifying enzymes with opposite activities counteract each other’s effect, such as histone acetyltransferases (HATs) and histone deacetylases (HDACs), kinases and phosphatases [45, 46]. Until the Lys demethylases, such as LSD1 and JmjC domain-containing dioxygenase, as well as the Arg deiminase PAD4 were identified [47, 48], histone methylation on Arg and Lys residues was considered as static rather than dynamic because of the low turnover rate of the methyl groups [49]. PAD4 antagonizes CARM1 (also called PRMT4) and PRMT1 mediated histone H3 and H4 Arg methylation through a reaction dubbed as demethylimination in reflecting the removal of the methyl-imine group from monomethyl-arginine residues [33]. The activity of PAD4 on asymmetrical dimethyl-arginine residues is very low. We have observed that PAD4 prefers methyl-Arg in histone proteins over methyl-Arg in short peptides as substrates, suggesting the substrate has an allosteric effect on PAD4 [33]. CARM1 and PRMT1 function as transcription coactivators by catalyzing histone Arg monomethylation and asymmetrical dimethylation [42, 50–53]. By antagonizing Arg methylation, PAD4 functions as a transcription corepressor. In the case of ER target genes in the breast cancer MCF-7 cells, PAD4 regulates histone Arg methylation via its citrullination activity on the gene promoters [33, 54]. Interestingly, the modification of histone Arg residues on the ER (estrogen receptor) target promoters fluctuates over time after estradiol treatment, whereby the increase in histone citrullination correlates with the decrease in histone Arg methylation, indicating that opposite enzymes are alternatively working on the ER target gene promoters [55].
In addition to the ER target genes, our group has found that PAD4 interacts with the tumor suppressor and transcription factor p53 and functions as a corepressor to regulate the expression of multiple p53 target genes [56, 57]. Before DNA damage, a high level of histone citrullination and PAD4 was detected on the promoter of p53 target genes, such as p21/CIP1/WAF1, GADD45 and PUMA [56, 57]. After DNA damage, PAD4 association and histone citrullination decreases on these gene promoters with a concomitant increase in histone Arg methylation, suggesting that citrullination and arginine methylation counteract each other’s function to regulate gene expression.
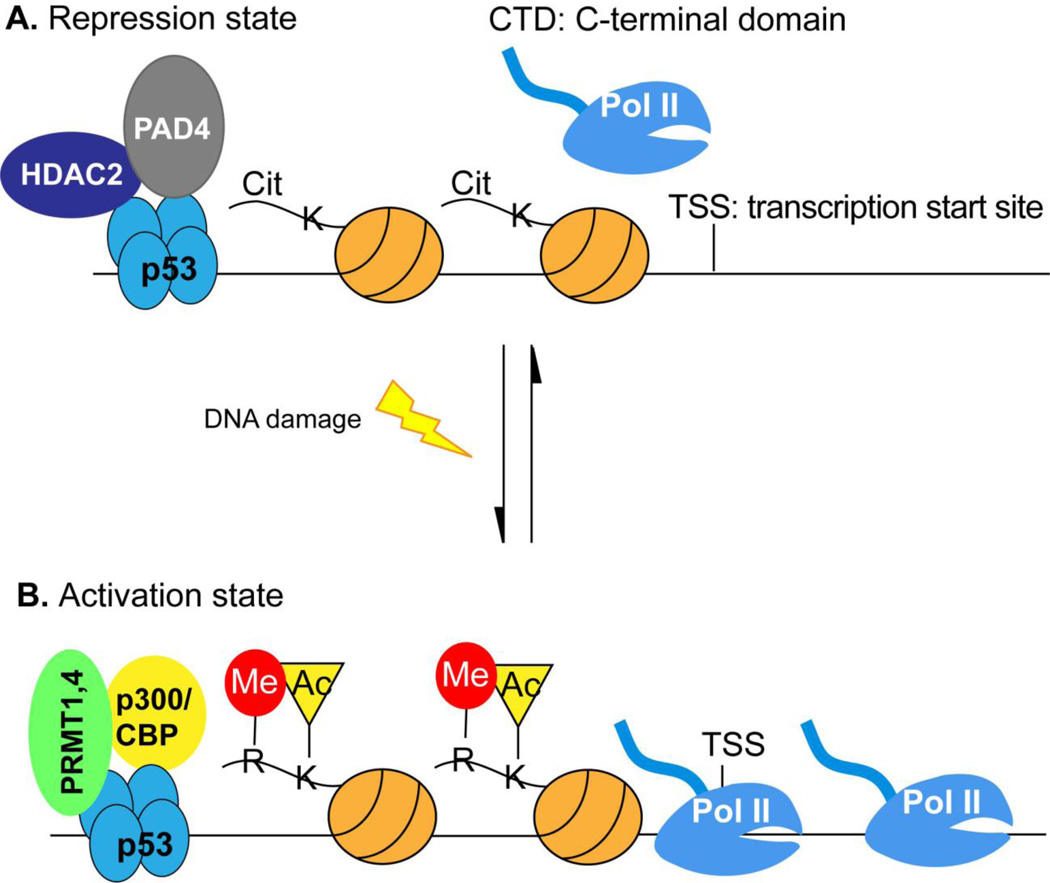
The role of histone acetylation in gene activation has been long established. A landmark 2004 paper with rich in vitro biochemical analyses showed that protein Arg methyltransferases and histone acetyltransferase p300/CBP cooperatively activate the p53-mediated transcription [42]. Reversely, we found that PAD4 and HDAC2 associate with p53 in a dynamic fashion [56]. Before DNA damage, both PAD4 and HDAC2 are associated with p53 target gene promoters, while they dissociate from the gene promoters after DNA damage allowing the activation of the p53 target genes, such as p21, GADD45 and PUMA [56]. Taken together, we propose that histone Arg modification in concert with histone Lys acetylation forms a molecular switch on the p53 target gene promoters for gene regulation (Fig. 2).
Fig 2. A molecular switch operated on histone Arg and Lys residues of p53 target promoters for gene regulation.
This molecular switch model predicts that p53 target genes can be activated through PAD4 and HDAC2 inhibition.
(A) Before p53 target gene activation, PAD4 and HDAC2 function at the promoters to citrullinate Arg residues and deacetylate Lys residues on histone proteins to suppress gene expression. RNA Pol II is not recruited to the promoter or paused at the promoter under these conditions.
(B) Upon activation signal such as DNA damage is sensed by p53, it switches its interaction partners from PAD4 and HDAC2 to PRMT1/4 and p300/CBP. These later two histonemodifying enzymes generate histone Arg methylation and Lys acetylation at target gene promoters, such as p21, GADD45 and PUMA to facilitate gene activation.
Although the corepressor function of PAD4 has been well established, it may also play a coactivator role in a promoter context dependent manner. A genome wide ChIP-chip study of PAD4 promoter association in MCF-7 cells found that PAD4 is enriched on the promoter regions of actively transcribed genes [58]. Motif analyses found that many of the PAD4 bound genes contain potential binding sites for Elk-1, a member of the ETS oncogene family [58]. It was proposed that PAD4 interacts with and citrullinates Elk-1 thereby facilitating Elk-1 phosphorylation to activate transcription [58]. Additionally, PAD4 can target histone H3 Arg8 for citrullination and subsequently affects the binding of HP1 (heterochromatin protein 1) to the nearby H3 Lys9 methylation site [59, 60]. The dissociation of HP1 from its binding cognate sites after citrullination activates transcription in multiple sclerosis patients [60]. In addition, the dissociation of HP1 likely regulates chromatin decondensation during the formation of neutrophil extracellular traps (NETs) [59].
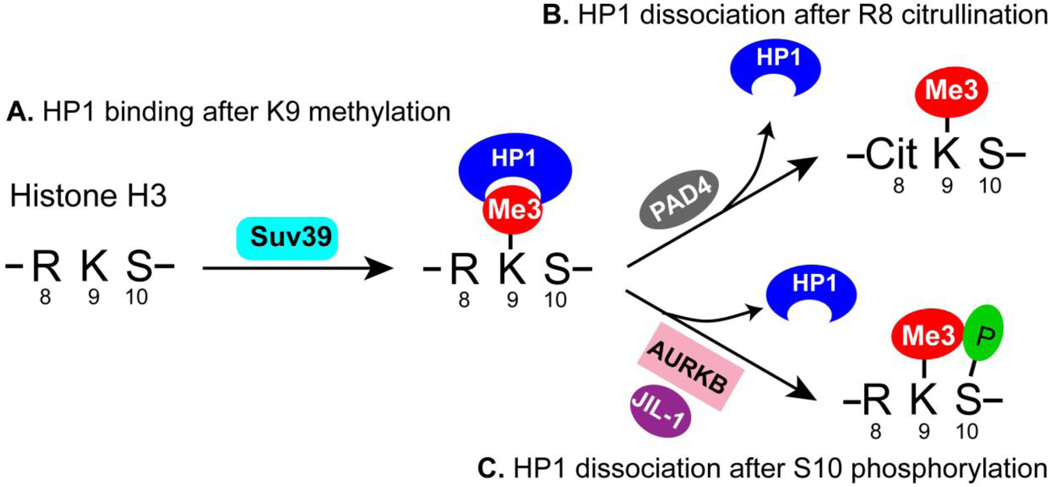
The hypothesis of the “histone code” predicts that histone modifications may function in a combinatorial manner to regulate chromatin biology [61]. Two adjacent modification sites were proposed to form a binary code to antagonize or synergize the function of each other [62]. The effect of histone H3 Ser10 phosphorylation on the function of H3 Lys9 methylation and HP1 in cell cycle and gene regulation was well studied [63, 64]. The new finding of the effect of H3 Arg8 citrullination on HP1 binding to H3 Lys9 methylation greatly enriched the binary code concept and highlights the dense cell signaling information that a cluster of histone residues carries (Fig. 3).
Fig 3. Cit-methyl and methyl-phos binary codes regulate the function of HP1.
(A) The trimethylation of histone H3 Lys9 (K9me3) by the Suv39 histone methyltransferase leads to the binding of heterochromatin protein 1 (HP1) to regulate heterochromatin formation and/or to repress gene expression.
(B) The citrullination of H3 Arg8 by PAD4 produces a Cit-methyl binary code to decrease the binding of HP1 to K9me3.
(C) The phosphorylation of H3 Ser10 by aurora B or JIL-1 produces a methyl-phos binary code to decreases the binding of HP1 to K9me3.
2.2. PAD2 in gene regulation
That PAD2 can citrullinate histone H4 Arg3 in vitro raises a role of PAD2-mediated histone citrullination in transcriptional regulation [21]. The expression of PAD2 is regulated by estrogen in vertebrate uterus and pituitary gland [65–67]. Recent data from the Coonrod’s group support that PAD2 responses to cellular signals to regulate transcription via histone citrullination. Only during the diestrus phase of the reproduction cycle, PAD2 in mammary gland epithelial cells was found to citrullinate the histone H3 N-terminus [19]. Moreover, PAD2 is detected in human breast luminal epithelial cells, and associates with target gene promoters in the ERα positive breast cancer MCF-7 cells to regulate histone H3 Arg26 citrullination and transcription [20]. Estradiol stimulates the recruitment of PAD2 to the estrogen-response element of estrogen receptor alpha (ERα) target gene promoters. Since PAD2 does not have a nuclear localization signal, its association with ERα may facilitate its translocation from the cytosolic to the nuclear compartment [20]. As such, PAD2 likely mediates chromatin decondensation and activation of target gene transcription during cellular response to estrogen stimulation [23]. These studies indicate that PAD2 functions as an epigenetic regulator of gene activity and plays a potential role in breast cancer progression.
3. PAD4 and NETs
3.1. NETs, a highly decondensed form of chromatin
Peripheral blood neutrophils serve as the first line of defense against microbial infection [68–70]. Although neutrophils are well known for their antimicrobial function via phagocytosis, it was reported in 2004 that a distinct mechanism of killing bacteria in the extracellular space using a meshwork of highly decondensed chromatin in association with antimicrobial proteins (e.g., myeloperoxidase and neutrophil elastase) [68]. Upon encountering bacteria, a certain percentage of neutrophils release their chromatin into the extracellular space, forming chromatin-based structures called neutrophil extracellular traps (NETs) (reviewed by [71]). NET formation can be induced by IL-8, PMA, LPS and diverse microbes, such as bacteria and fungi, as well as protozoan parasites [68, 72]. As a mechanism of immune defense, NETs could mediate the microbial death and/or limits pathogen spread in host [68, 73, 74]. Although the role of NETs in innate immunity and human disease has gained much attention, the mechanism regulating the NET formation is less clear. The activation of the intracellular signaling pathways, such as the MAPK pathway and the ROS (reactive oxygen species) signaling pathways, has been implicated in the formation of NETs [75–77]. Since NET formation directly leads to cell death (dubbed netosis) and the release of nuclear and cytoplasmic components into the extracellular space has harmful consequences to the surrounding normal tissues, this process needs deliberate regulation.
3.2. Histone hypercitrullination in NET formation
Chromatin in NETs is extremely decondensed, with a diameter ranging between 10–30 nm [68]. This degree of chromatin decondensation indicates that the neutrophil chromatin can be unfolded extensively to form the polymers of nucleosomes, i.e., the 10 nm chromatin fibers. A proteomic study of the NET protein components has found that even linker histone H1 is diminished from NET chromatin [78]. However, the chromatin decondensation mechanisms underlying NET formation remain further explored.
Retrospectively, our finding that PAD4 and histone hypercitrullination are involved in the formation of neutrophil extracellular traps is a serendipitous story. In the early months of 2004, we found that prolonged treatment of HL-60 granulocytes with calcium ionophore produced sticky chromatin that glues many of cells in a cluster. The gluy chromatin was stained with the DNA dye and very strongly stained with the histone H4Cit3 antibody, suggesting that there is a link between the high-degree of histone citrullination and the formation of NETs. The link of PAD4 and NET formation was eventually published in 2009 [79]. Around the time we were preparing to publish our report, the Radic group published immunostaining images to show that NET chromatin can be stained by a histone citrullination antibody [80]. It is now clear that one the normal physiological function of PAD4 in peripheral blood neutrophils is to regulate the formation of NETs.
Histone hypercitrullination is an important intermediate step for the formation of NETs. In HL-60 granulocytic cells, the inhibition of PAD4 and histone hypercitrullination decreases chromatin decondensation and the formation of NET like structures [79]. Importantly, citrullination of a 12-mer nucleosome array decreased the chromatin folding by the linker histone H1 [79]. The role of PAD4 in NET formation is further supported by genetic studies in mice. The PAD4 knockout mice can survive to adulthood and are fertile with a normal number of the neutrophil counts [81]. In PAD4 null neutrophils, the basal level of histone citrullination is undetectable. After stimulation with LPS, H2O2, PMA and bacteria, histone citrullination is greatly increased in wild type neutrophils but remains undetectable in the null cells [81]. The lack of histone citrullination is correlated with a lack of NET formation in the null cells, indicating the PAD4 is required for NET formation under these experimental conditions [81]. Furthermore, elevated PAD4 activity is sufficient for the chromatin decondensation to form NET-like structures. First, treatment of detergent permeated cells with GST-PAD4 but not its catalytic inactive mutant GST-PAD4 C645S induced the release of chromatin from HL-60 cells to form decondensed chromatin structures [79]. Surprisingly, forced overexpression of PAD4 in osteosarcoma U2OS cells was also sufficient for the formation of NET-like chromatin structures [59]. These results indicate that PAD4 activity is required and sufficient for the formation of decondensed chromatin, a process that is essential for the formation of NETs.
How does citrullination mechanistically unfold chromatin? First, the degree of histone citrullination associated with NET formation is very high. Based on Edman degradation experiments to analyze histone citrullination, we estimate that over 50% of histone H3 Arg8 can be converted to citrulline [33]. Since citrullination eliminates the positive charge of histones, this modification may weaken the interaction of histones and DNA thereby decreasing chromatin fiber compaction to facilitate NET formation. Second, PAD4 mediated histone hypercitrullination could exclude HP1β from binding to chromatin, offering a new mechanism for PAD4 mediated NET formation [59]. Third, citrullinated nucleosome arrays are not folded properly by the linker histone H1. Moreover, many other proteins can be targeted by PAD4 for citrullination. These other PAD4 substrates could paly a modulatory role for the process of NET formation.
3.3.3 PAD4 in NET-mediated killing of bacteria and many inflammatory conditions
Evolution is a driving force behind the adaption of pathogenic bacteria to the host’s immune defense system, while the host constantly creates mechanisms to eliminate invading microbes. Indeed, many pathogenic bacteria that are infectious to humans produce extracellular DNase to facilitate them to evade killing by NETs. For example, the EndA gene in Streptococcus pneumonia encodes a membrane bound and extracellular faced endonuclease and the Nuc gene of Streptococcus aureus encodes the secreted micrococcal nuclease (MNase) [82]. Moreover, certain serotypes of the group A Streptococcus pyogenes secret a DNase called Sda1, which facilitates the bacteria to generate a local skin infection called fasciitis [73]. Consistent with the role of NETs in bacteria killing, the PAD4 knockout mice are more susceptible to bacterial infections in the experimental fasciitis model, and neutrophils from the PAD4 deficient mice showed significantly decreased bacteria killing activity in vitro [81].
Like many other immune defense mechanisms in the human body, NETs serve as an antimicrobial defense weapon as well as fan the fire for inflammation, leading to tissue damages. NETs are implicated in many pathological conditions, such as appendicitis, cystic fibrosis, tuberculosis, sepsis, lupus and nephritis [4, 68, 83–86]. A common theme is that neutrophils recruited to the inflammation sites will die by netosis to form NETs and release their nuclear and cytoplasmic components into the extracellular space. The PAD4 protein released from neutrophils may citrullinate proteins and produce autoimmune antigens to cause chronic autoimmune diseases, such as rheumatoid arthritis (see below). Furthermore, NETs may have a quick impact on thrombosis by contributing the chromatin threads as a building material for the formation of thrombus [87]. Conversely, the lack of NET formation in the PAD4 null mice decreases the formation and maintenance of thrombi in the blood veins [6].
4. PAD IN HUMAN DISEASES
In addition to the above-mentioned pathological conditions associated with NETs, PADs and its citrullinated products have been associated with many human diseases. In this section, we will discuss the potential involvement of PAD4 in rheumatoid arthritis (RA) and PAD2 in multiple sclerosis (MS). We will then discuss the involvement of PAD4 in cancers.
4.1 Rheumatoid Arthritis
RA is a chronic autoimmune disease featured with inflammatory synovium and infiltration of activated macrophages. The anti-citrullinated protein antibodies are the most specific autoantibodies present in the RA sera [88–90]. Most of these autoantibodies can be detected in the early stage of the disease [91, 92], which makes them useful diagnostic markers of RA [93–95]. Those citrullinated proteins, including fibrin, fibrinogen and vimentin, were produced in synovial fluid of the inflammatory joints [96–101]. The presence of citrulline residues in these proteins sends the immune cells a false alarm and initiates immune responses to generate anticitrulline antibodies against these proteins [102, 103]. Many PAD2 and PAD4 expressing leucocytes infiltrate into the inflammatory synovial tissues in RA patients and release large amount of PAD2 and PAD4 in the synovial fluid, which in turn produce high levels of citrullinated proteins [104, 105]. Since the calcium concentration in the extracellular space is at the millimolar levels, PAD4 released from the dying neutrophils during the NET formation process can be super-activated to citrullinate joint proteins [16]. In addition to these pathology links, the involvement of PAD4 in rheumatoid arthritis is also supported by human genetic studies, in particular in the oriental ethnic groups [106–108].
4.2 Multiple Sclerosis and Alzheimer’s
MS is mainly a PAD2 related inflammatory disorder in the central nervous system associated with excess citrullination of myelin basic protein (MBP), resulting in the demyelination of the myelin sheath and affecting the nerve signal transduction [109, 110]. About 18% of MBP protein is citrullinated at 6 out of its 19 Arg residues in healthy individuals, while 45% or more of MBP is citrullinated in MS patients [111]. Hypercitrullination of MBP weakens its interactions with phospholipid and disrupts the formation of normal multilayer myelin sheath structures. In addition, hypercitrullinated MBP is more susceptible to protein degradation by the protease cathepsin D [112–114]. Elevated PAD2 in the myelin sheath is related to an increase in MBP citrullination prior to the onset of MS symptoms [109, 115]. PAD2 overexpression in transgenic mice increases the amount of citrullinated MBP and accelerates the development of demyelination [116–118]. Moreover, upregulation of PAD2 and the presence of the inflammatory signals at the affected axon regions might further increase PAD4 locally to exacerbate the inflammatory disease [115, 119]. It is surprising that knockout of PAD2 did not produce a significant phenotype in the nervous system given the importance of MBP citrullination in the axon electrical signal transmission [120]. Whether other PAD enzymes, such as PAD4 play a redundant role with PAD2 in the central nervous system remain to be solved.
In addition to multiple sclerosis, PAD2 and PAD4 are also expressed in neurons [121]. Protein citrullination in the central nervous system may cause protein denaturation and precipitation, which in turn lead to the Alzheimer’s disease [121–123]. These studies highlight the possibility of PAD2 as a drug target for the treatment of nervous system diseases.
4.3 Cancer
Epigenetic modification plays an important role in tumorigenesis [124, 125]. Two major epigenetic alterations in cancer cells are histone modifications and DNA methylation. In this session we will focus on the effect of histone modifications on aberrant tumor suppressor gene silencing during tumorigenesis [126, 127]. Tumor suppressor p53 is one key transcription factor in maintaining cellular homeostasis. In response to diverse upstream signals, such as starvation, DNA damage and various stress signals, p53 regulates its downstream genes to cope with stress and control the cell fate [128–131]. Many of the p53 target genes are tumor suppressor genes that regulate the cell cycle arrest, programed cell death, and autophagy.
Under normal conditions, PAD4 is mainly expressed in the peripheral blood leukocytes. In pathology studies using a large cohort of human patient samples, PAD4 is overexpressed in a majority of tumor tissues, including osteosarcoma, colon adenocarcinoma, esophagus adenocarcinoma, ovary adenocarcinoma, pancreas adenocarcinoma, and stomach adenocarcinoma [29, 30]. Given that PAD4 functions as a corepressor of p53 to regulate its downstream tumor suppressor genes, the overexpression of PAD4 in tumor tissues suggests that it might be involved in tumorigenesis. Interestingly, the expression of PAD4 is directly regulated by p53, suggesting that PAD4 may form a negative feedback loop to regulate p53 [132]. As we have discussed in the previous section, PAD4 catalyzed histone citrullination coupled with HDAC2 catalyzed deacetylation represses p53 target gene expression [56]. Since HDACs are targets for cancer drug development, we recently devoted a solid effort to test if PAD4 is a druggable target for cancer treatment (see below).
5. PAD proteins as druggable targets for disease treatment
A benzoyl-arginine-derived compound Cl-amidine has been reported as a pan PAD inhibitor in vitro by forming covalent bond with the active center Cys of the enzyme thereby blocking the binding of natural substrates to the active site [133–135]. However, Cl-amidine inhibits cancer cell growth with an IC50 of ~150–200 µ.∈ [56, 57], which limits its clinical value in cancer treatment. Recently, we performed medicinal modification of Cl-amidine and developed novel PAD inhibitors with low micromolar IC50 for PAD4 enzymatic activity inhibition and cancer cell growth inhibition [22]. Compared with Cl-amidine, the novel inhibitors show increased efficacy in PAD4 inhibition and more importantly increased membrane penetration [22]. The lead compound YW3–56 alters the expression of a cohort of genes, including many p53 target genes, to control the cell cycle and cell death [22]. Treatment of human cancer cells with YW3–56 induces the expression of SESN2, a recently identified p53 target gene that serves as an upstream inhibitor of the mTORC1-signaling pathway [22, 136, 137]. YW3–56 disturbs the autophagy flux and leads to cell death [22]. YW3–56 has demonstrated anti-tumor activity in mouse xenograft studies with no overt adverse effect to vital organs, whereas a combination of YW35-6 with HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) further improved the tumor growth inhibition [22]. Future studies to explore the spectrum of cancers that can be targeted and the molecular mechanisms underlying the anticancer effect of YW3–56 will be key to evaluate its potential as an anticancer drug.
In addition to the chloroacetamidine-based inhibitors, other types of PAD inhibitors have been developed [113, 138–142]. The efficacy and specificity of these inhibitors will need to be further improved. Interestingly, the mitotic inhibitor paclitaxel used in cancer chemotherapy can inhibit PAD2 mediated MBP citrullination in vitro [143]. Paclitaxel attenuated demyelination symptoms and induced the remyelination of the damaged myelin sheaths [144], suggesting that paclitaxel is a possible anti-MS compound. Recently, fluorescence-based PAD4 activity assay approaches were reported thus opening the door for future large-scale screening of PAD4 specific inhibitors [145, 146]. Furthermore, given that a combination of DNA methyltransferase inhibitor and HDAC inhibitor like SAHA can synergistically activate tumor suppressors in cancer cells and animal tumor models [147–150], the combined usage of PAD4 and HDAC inhibitors might be a promising strategy in future cancer treatment.
6. Conclusions and future perspectives
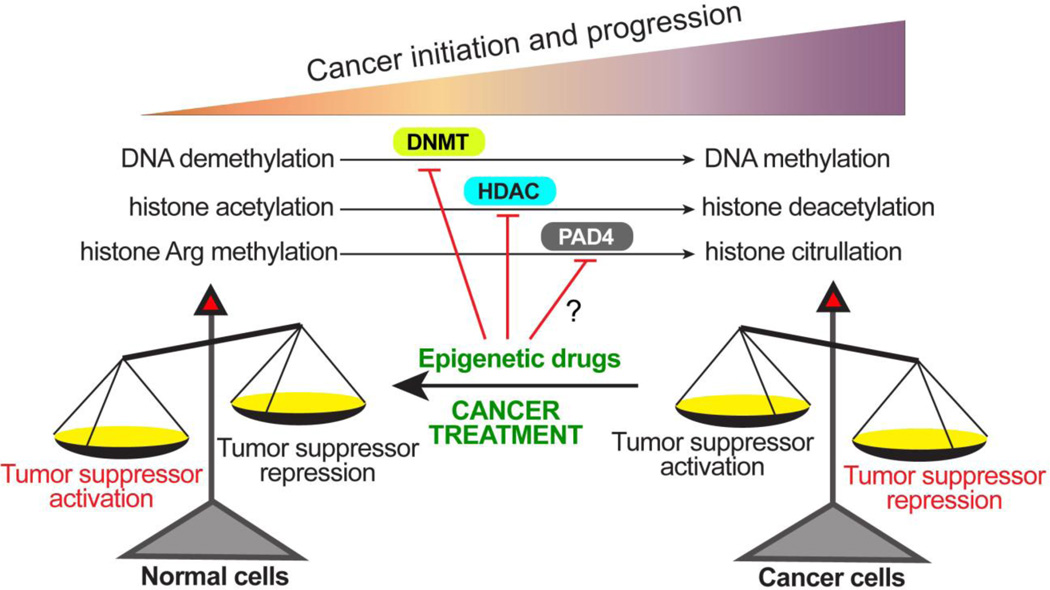
The citrullination of structural protein in the epidermis and neuronal system by peptidylarginine deiminase likely plays important roles in regulating the structure and function of these tissues. In addition, histone citrullination by PAD4 and PAD2 may influence the expression of many genes that are important for physiological functions. Deregulated citrullination has profound impact on human disease etiology. The role of histone and protein citrullination in cancers is emerging in recent years. Tumorigenesis is featured with many spontaneous driver and standby mutations. Targeting individual mutations one at a time may not be efficient. Therefore, it is still an attractive idea to reverse the tumorigenesis by altering the gene expression profile of cancer cells to that of the normal cells. One view of the tumorigenesis is the gradual silencing of the tumor suppressor genes. Many chromatin DNA and histone modifying enzymes are involved in this process, such the DNA methyltransferases, histone deacetylases, and peptidylarginine deiminases (Fig. 4). Others and us have shown that inhibition of PAD4 by small molecules can turn on the expression of the tumor suppressor genes in cancer cells, raising a possibility that PAD4 is druggable target for cancer treatment. In the field of cancer epigenetics, inhibitors for both DNA methyltransferases and histone deacetylases have been successfully used for cancer treatment. Only time will tell if inhibitors of PAD4 will eventually be effective in clinical tests.
Fig 4. An epigenetic view of tumorigenesis and cancer treatment.
During the process of cancer initiation and progression, DNA methylation catalyzed by the DNA methyltransferases (DNMTs), histone deacetylation catalyzed by histone deacetylases (HDACs), and histone citrullination catalyzed by PAD4 work singularly or synergistically to epigenetically silence tumor suppressor genes thereby leading to untamed growth of cancer cells and tissues. Based on this theory, cancer treatment can be achieved with inhibitors targeting these epigenetic modifiers to restore histone modification patterns that are favorable for tumor suppressor gene expression thereby promoting cell cycle arrest and cell death.
Table 1.
Tissue distribution, substrates, biological functions and related diseases of PADs.
| Protein distribution | Substrates | Biological functions | Diseases | |
|---|---|---|---|---|
| PAD1 | Epidermis; Uterus |
Keratin K1; Filaggrin [26] |
Cornification of epidermal tissues |
Psoriasis [151] |
| PAD2 | Broadly expressed: Pituitary gland; Brain; Uterus; Spleen; Spinal cord; Skeletal muscle |
MBP Vimentin β and γ-actins Histones H3 and H4 |
Plasticity of the CNS; Transcription regulation; Innate immune defense [152]; Female reproduction [153] |
Multiple sclerosis; Rheumatoid arthritis; Alzheimer’s disease [118, 119, 154, 155]; Prion disease [122] |
| PAD3 | Epidermis; Hair follicles |
Filaggrin; Trichohyalin |
Regulation of epidermal functions |
Unknown |
| PAD4 | Neutrophils; Monocytes; Macrophages; Mammary gland Epithelial cells; Tumors; |
Histones H2A, H3 and H4; ING4; p300/CBP; Nucleophosmin; Nuclear lamin C |
Chromatin decondensation; transcription regulation; Tumorigenesis; Innate immunity; NET formation |
Rheumatoid arthritis; Multiple sclerosis [156]; Cancers |
| PAD6 | Eggs; Ovary; Early embryo; |
Unknown | Oocyte cytoskeletal sheet formation and female fertility; early embryo development (mouse) |
Unknown |
Highlights.
-
-
PADs are a family of enzymes that citrullinate protein Arg residues.
-
-
PADs are involved in regulating p53 and nuclear receptor target genes.
-
-
PAD4 plays a role in the formation of NETs and antibacterial innate immunity.
-
-
Human cancers and autoimmune disorders involve deregulation in protein citrullination.
-
-
PADs are promising drug targets for disease treatment.
Acknowledgements
We thank members of the Wang lab as well as the Center for Eukaryotic Gene Regulation for helpful discussions. Shu Wang was partially supported by the BMMB graduate program. Research is supported by NIH R01 CA136856.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rogers GE. Occurrence of Citrulline in Proteins. Nature. 1962;194:1149–1151. doi: 10.1038/1941149a0. [DOI] [PubMed] [Google Scholar]
- 2.Rogers G, Taylor L. The enzymic derivation of citrulline residues from arginine residues in situ during the biosynthesis of hair proteins that are cross-linked by isopeptide bonds. 1977 doi: 10.1007/978-1-4684-3282-4_17. [DOI] [PubMed] [Google Scholar]
- 3.Vossenaar ER, Zendman AJW, van Venrooij WJ, Pruijn GJM. PADa, growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 4.Baka Z, Gyorgy B, Geher P, Buzas EI, Falus A, Nagy G. Citrullination under physiological and pathological conditions. Joint, bone, spine : revue du rhumatisme. 2012;79:431–436. doi: 10.1016/j.jbspin.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Horibata S, Coonrod SA, Cherrington BD. Role for peptidylarginine deiminase enzymes in disease and female reproduction. The Journal of reproduction and development. 2012;58:274–282. doi: 10.1262/jrd.2011-040. [DOI] [PubMed] [Google Scholar]
- 6.Rohrbach AS, Slade DJ, Thompson PR, Mowen KA. Activation of PAD4 in NET formation. Frontiers in immunology. 2012;3:360. doi: 10.3389/fimmu.2012.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y-L, Tsai IC, Chang C-W, Liao Y-F, Liu G-Y, Hung H-C. Functional Roles of the Non-Catalytic Calcium-Binding Sites in the N-Terminal Domain of Human Peptidylarginine Deiminase 4. PloS one. 2013;8:e51660. doi: 10.1371/journal.pone.0051660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rus’d AA, Ikejiri Y, Ono H, Yonekawa T, Shiraiwa M, Kawada A, Takahara H. Molecular cloning of cDNAs of mouse peptidylarginine deiminase type I, type III and type IV, and the expression pattern of type I in mouse. European Journal of Biochemistry. 1999;259:660–669. doi: 10.1046/j.1432-1327.1999.00083.x. [DOI] [PubMed] [Google Scholar]
- 9.Guerrin M, Ishigami A, Méchin M-C, Nachat R, Valmary S, Sebbag M, Simon M, Senshu T, Serre G. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem. J. 2003;370:167–174. doi: 10.1042/BJ20020870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishida-Yamamoto A, Senshu T, Eady RAJ, Takahashi H, Shimizu H, Akiyama M, Iizuka H. Sequential Reorganization of Cornified Cell Keratin Filaments Involving Filaggrin-Mediated Compaction and Keratin 1 Deimination. 2002;118:282–287. doi: 10.1046/j.0022-202x.2001.01671.x. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe A, Hikichi AKK, Ohtsuka R, Okuyama A, Senshu T. Combined biochemical and immunochemical comparison of peptidylarginine deiminases present in various tissues. Biochim Biophys Acta. 1988;966:375–383. doi: 10.1016/0304-4165(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe K, Senshu T. Isolation and characterization of cDNA clones encoding rat skeletal muscle peptidylarginine deiminase. Journal of Biological Chemistry. 1989;264:15255–15260. [PubMed] [Google Scholar]
- 13.Nagata S, Senshu T. Peptidylarginine deiminase in rat and mouse hemopoietic cells. Experientia. 1990;46:72–74. doi: 10.1007/BF01955420. [DOI] [PubMed] [Google Scholar]
- 14.Urano Y, Watanabe K, Sakaki A, Arase S, Watanabe Y, Shigemi F, Takeda K, TAkiyama K, Senshu T. Immunohistochemical demonstration of peptidylarginine deiminase in human sweat glands. 1990 doi: 10.1097/00000372-199006000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Vossenaar ER, Nijenhuis S, Helsen MMA, van der Heijden A, Senshu T, van den Berg WB, van Venrooij WJ, Joosten LAB. Citrullination of synovial proteins in murine models of rheumatoid arthritis. Arthritis & Rheumatism. 2003;48:2489–2500. doi: 10.1002/art.11229. [DOI] [PubMed] [Google Scholar]
- 16.Vossenaar ER, Radstake TRD, van der Heijden A, van Mansum MAM, Dieteren C, de Rooij D-J, Barrera P, Zendman AJW, van Venrooij WJ. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Annals of the Rheumatic Diseases. 2004;63:373–381. doi: 10.1136/ard.2003.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamensa JWE, Moscarello MA. Deimination of Human Myelin Basic Protein by a Peptidylarginine Deiminase from Bovine Brain. Journal of Neurochemistry. 1993;61:987–996. doi: 10.1111/j.1471-4159.1993.tb03612.x. [DOI] [PubMed] [Google Scholar]
- 18.Darrah E, Rosen A, Giles JT, Andrade F. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Annals of the Rheumatic Diseases. 2012;71:92–98. doi: 10.1136/ard.2011.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherrington BD, Morency E, Struble AM, Coonrod SA, Wakshlag JJ. Potential role for peptidylarginine deiminase 2 (PAD2) in citrullination of canine mammary epithelial cell histones. PloS one. 2010;5:e11768. doi: 10.1371/journal.pone.0011768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherrington BD, Zhang X, McElwee JL, Morency E, Anguish LJ, Coonrod SA. Potential role for PAD2 in gene regulation in breast cancer cells. PloS one. 2012;7:e41242. doi: 10.1371/journal.pone.0041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarmento OF, Digilio LC, Wang Y, Perlin J, Herr JC, Allis CD, Coonrod SA. Dynamic alterations of specific histone modifications during early murine development. Journal of cell science. 2004;117:4449–4459. doi: 10.1242/jcs.01328. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, Fisher M, Oshaben K, Zhao N, Gu Y, Wang D, Chen G, Wang Y. Anticancer Peptidylarginine Deiminase (PAD) Inhibitors Regulate the Autophagy Flux and the Mammalian Target of Rapamycin Complex 1 Activity. Journal of Biological Chemistry. 2012;287:25941–25953. doi: 10.1074/jbc.M112.375725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL, Thompson PR, Mancini MA, Lis JT, Coonrod SA. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13331–13336. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavanas S, Mechin MC, Nachat R, Adoue V, Coudane F, Serre G, Simon M. Peptidylarginine deiminases and deimination in biology and pathology: relevance to skin homeostasis. J Dermatol Sci. 2006;44:63–72. doi: 10.1016/j.jdermsci.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Kanno T, Kawada A, Yamanouchi J, Yosida-Noro C, Yoshiki A, Shiraiwa M, Kusakabe M, Manabe M, Tezuka T, Takahara H. Human Peptidylarginine Deiminase Type III: Molecular Cloning and Nucleotide Sequence of the cDNA, Properties of the Recombinant Enzyme, and Immunohistochemical Localization in Human Skin. 2000;115:813–823. doi: 10.1046/j.1523-1747.2000.00131.x. [DOI] [PubMed] [Google Scholar]
- 26.Nachat R, Mechin M-C, Charveron M, Serre G, Constans J, Simon M. Peptidylarginine Deiminase Isoforms Are Differentially Expressed in the Anagen Hair Follicles and Other Human Skin Appendages. J Investig Dermatol. 2005;125:34–41. doi: 10.1111/j.0022-202X.2005.23763.x. [DOI] [PubMed] [Google Scholar]
- 27.Asaga H, Nakashima K, Senshu T, Ishigami A, Yamada M. Immunocytochemical localization of peptidylarginine deiminase in human eosinophils and neutrophils. Journal of Leukocyte Biology. 2001;70:46–51. [PubMed] [Google Scholar]
- 28.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. The Journal of biological chemistry. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 29.Chang X, Han J. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Molecular Carcinogenesis. 2006;45:183–196. doi: 10.1002/mc.20169. [DOI] [PubMed] [Google Scholar]
- 30.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakashima K, Hagiwara T, Ishigami A, Nagata S, Asaga H, Kuramoto M, Senshu T, Yamada M. Molecular Characterization of Peptidylarginine Deiminase in HL-60 Cells Induced by Retinoic Acid and 1α,25-Dihydroxyvitamin D3. Journal of Biological Chemistry. 1999;274:27786–27792. doi: 10.1074/jbc.274.39.27786. [DOI] [PubMed] [Google Scholar]
- 32.Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem Biophys Res Commun. 2002;290:979–983. doi: 10.1006/bbrc.2001.6303. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Wysocka J, Sayegh J, Lee Y-H, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 Regulates Histone Arginine Methylation Levels via Demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. The Journal of biological chemistry. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 35.Guo Q, Fast W. Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. The Journal of biological chemistry. 2011;286:17069–17078. doi: 10.1074/jbc.M111.230961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y, Matsuda K. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun. 2012;3:676. doi: 10.1038/ncomms1676. [DOI] [PubMed] [Google Scholar]
- 37.Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, Hao Z, Jayes FC, Bush LA, Shetty J, Shore AN, Reddi PP, Tung KS, Samy E, Allietta MM, Sherman NE, Herr JC, Coonrod SA. ePAD, an oocyte and early embryoabundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003;256:73–88. doi: 10.1016/s0012-1606(02)00126-4. [DOI] [PubMed] [Google Scholar]
- 38.Esposito G, Vitale AM, Leijten FPJ, Strik AM, Koonen-Reemst AMCB, Yurttas P, Robben TJAA, Coonrod S, Gossen JA. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Molecular and Cellular Endocrinology. 2007;273:25–31. doi: 10.1016/j.mce.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, Gosden R, Coonrod SA. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol. 2011;350:311–322. doi: 10.1016/j.ydbio.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavanas S, Méchin M-C, Takahara H, Kawada A, Nachat R, Serre G, Simon M. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene. PADI6, Gene. 2004;330:19–27. doi: 10.1016/j.gene.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 41.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nature structural & molecular biology. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 42.An W, Kim J, Roeder RG. Ordered Cooperative Functions of PRMT1, p300, and CARM1 in Transcriptional Activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Fischer JJ, Toedling J, Krueger T, Schueler M, Huber W, Sperling S. Combinatorial effects of four histone modifications in transcription and differentiation. Genomics. 2008;91:41–51. doi: 10.1016/j.ygeno.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annual review of biochemistry. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 45.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 46.Nowak SJ, Pai CY, Corces VG. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Molecular and cellular biology. 2003;23:6129–6138. doi: 10.1128/MCB.23.17.6129-6138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 49.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 50.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO reports. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 52.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Current biology : CB. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 54.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 55.Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F. Functional connection between deimination and deacetylation of histones. Molecular and cellular biology. 2009;29:4982–4993. doi: 10.1128/MCB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li P, Wang D, Yao H, Doret P, Hao G, Shen Q, Qiu H, Zhang X, Wang Y, Chen G, Wang Y. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene. 2010;29:3153–3162. doi: 10.1038/onc.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Molecular and cellular biology. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR, Roberson MS, Kraus WL, Coonrod SA. Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLoS genetics. 2011;7:e1002112. doi: 10.1371/journal.pgen.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leshner M, Wang S, Lewis C, Zheng H, Chen XA, Santy L, Wang Y. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Frontiers in immunology. 2012;3:307. doi: 10.3389/fimmu.2012.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma P, Azebi S, England P, Christensen T, Moller-Larsen A, Petersen T, Batsche E, Muchardt C. Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLoS genetics. 2012;8:e1002934. doi: 10.1371/journal.pgen.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 62.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 63.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 64.Zhang W, Deng H, Bao X, Lerach S, Girton J, Johansen J, Johansen KM. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development. 2006;133:229–235. doi: 10.1242/dev.02199. [DOI] [PubMed] [Google Scholar]
- 65.Arai T, Kusubata M, Kohsaka T, Shiraiwa M, Sugawara K, Takahara H. Mouse uterus peptidylarginine deiminase is expressed in decidual cells during pregnancy. Journal of cellular biochemistry. 1995;58:269–278. doi: 10.1002/jcb.240580302. [DOI] [PubMed] [Google Scholar]
- 66.Senshu T, Akiyama K, Nagata S, Watanabe K, Hikichi K. Peptidylarginine deiminase in rat pituitary: sex difference, estrous cycle-related changes, and estrogen dependence. Endocrinology. 1989;124:2666–2670. doi: 10.1210/endo-124-6-2666. [DOI] [PubMed] [Google Scholar]
- 67.Takahara H, Kusubata M, Tsuchida M, Kohsaka T, Tagami S, Sugawara K. Expression of peptidylarginine deiminase in the uterine epithelial cells of mouse is dependent on estrogen. The Journal of biological chemistry. 1992;267:520–525. [PubMed] [Google Scholar]
- 68.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 69.Segal AW. How neutrophils kill microbes. Annual review of immunology. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nathan C. Neutrophils and immunity: challenges and opportunities. Nature reviews. Immunology. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 71.Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? The Journal of cell biology. 2012;198:773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaplan MJ, Radic M. Neutrophil extracellular traps: double-edged swords of innate immunity. J Immunol. 2012;189:2689–2695. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Current biology : CB. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 74.Menegazzi R, Decleva E, Dri P. Killing by neutrophil extracellular traps: fact or folklore? Blood. 2012;119:1214–1216. doi: 10.1182/blood-2011-07-364604. [DOI] [PubMed] [Google Scholar]
- 75.Hakkim A, Fuchs TA, Martinez NE, Hess S, Prinz H, Zychlinsky A, Waldmann H. Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nature chemical biology. 2011;7:75–77. doi: 10.1038/nchembio.496. [DOI] [PubMed] [Google Scholar]
- 76.Bianchi M, Hakkim A, Brinkmann V, Siler U, Seger RA, Zychlinsky A, Reichenbach J. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 2009;114:2619–2622. doi: 10.1182/blood-2009-05-221606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Kockritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med (Berl) 2009;87:775–783. doi: 10.1007/s00109-009-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS pathogens. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. The Journal of cell biology. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol. 2008;180:1895–1902. doi: 10.4049/jimmunol.180.3.1895. [DOI] [PubMed] [Google Scholar]
- 81.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. The Journal of experimental medicine. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Kockritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. Journal of innate immunity. 2010;2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braian C, Hogea V, Stendahl O. Mycobacterium tuberculosis- Induced Neutrophil Extracellular Traps Activate Human Macrophages. Journal of innate immunity. 2013 doi: 10.1159/000348676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hakkim A, Furnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V, Herrmann M, Voll RE, Zychlinsky A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol. 2013;190:1217–1226. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marcos V, Zhou Z, Yildirim AO, Bohla A, Hector A, Vitkov L, Wiedenbauer EM, Krautgartner WD, Stoiber W, Belohradsky BH, Rieber N, Kormann M, Koller B, Roscher A, Roos D, Griese M, Eickelberg O, Doring G, Mall MA, Hartl D. CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nature medicine. 2010;16:1018–1023. doi: 10.1038/nm.2209. [DOI] [PubMed] [Google Scholar]
- 87.Fuchs TA, Brill A, Duerschmied D, Schatzberg D, Monestier M, Myers DD, Jr., Wrobleski SK, Wakefield TW, Hartwig JH, Wagner DD. Extracellular DNA traps promote thrombosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boggs JM, Rangaraj G, Koshy KM. Analysis of the membrane-interacting domains of myelin basic protein by hydrophobic photolabeling. Biochim Biophys Acta. 1999;1417:254–266. doi: 10.1016/s0005-2736(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 89.Raptopoulou A, Sidiropoulos P, Katsouraki M, Boumpas DT. Anti-citrulline antibodies in the diagnosis and prognosis of rheumatoid arthritis: evolving concepts. Critical reviews in clinical laboratory sciences. 2007;44:339–363. doi: 10.1080/10408360701295623. [DOI] [PubMed] [Google Scholar]
- 90.Ying S, Dong S, Kawada A, Kojima T, Chavanas S, Mechin MC, Adoue V, Serre G, Simon M, Takahara H. Transcriptional regulation of peptidylarginine deiminase expression in human keratinocytes. J Dermatol Sci. 2009;53:2–9. doi: 10.1016/j.jdermsci.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 91.Vasishta A. Diagnosing early-onset rheumatoid arthritis: the role of anti-CCP antibodies. American clinical laboratory. 2002;21:34–36. [PubMed] [Google Scholar]
- 92.van Boekel MA, Vossenaar ER, van den Hoogen FH, van Venrooij WJ. Autoantibody systems in rheumatoid arthritis: specificity, sensitivity and diagnostic value. Arthritis research. 2002;4:87–93. doi: 10.1186/ar395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Visser H, le Cessie S, Vos K, Breedveld FC, Hazes JM. How to diagnose rheumatoid arthritis early: a prediction model for persistent (erosive) arthritis. Arthritis and rheumatism. 2002;46:357–365. doi: 10.1002/art.10117. [DOI] [PubMed] [Google Scholar]
- 94.Meyer O, Labarre C, Dougados M, Goupille P, Cantagrel A, Dubois A, Nicaise-Roland P, Sibilia J, Combe B. Anticitrullinated protein/peptide antibody assays in early rheumatoid arthritis for predicting five year radiographic damage. Ann Rheum Dis. 2003;62:120–126. doi: 10.1136/ard.62.2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trouw LA, Mahler M. Closing the serological gap: promising novel biomarkers for the early diagnosis of rheumatoid arthritis. Autoimmun Rev. 2012;12:318–322. doi: 10.1016/j.autrev.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 96.Masson-Bessiere C, Sebbag M, Durieux JJ, Nogueira L, Vincent C, Girbal-Neuhauser E, Durroux R, Cantagrel A, Serre G. In the rheumatoid pannus, anti-filaggrin autoantibodies are produced by local plasma cells and constitute a higher proportion of IgG than in synovial fluid and serum. Clinical and experimental immunology. 2000;119:544–552. doi: 10.1046/j.1365-2249.2000.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, Serre G. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 98.Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S, Yamamoto K. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology. 2005;44:40–50. doi: 10.1093/rheumatology/keh414. [DOI] [PubMed] [Google Scholar]
- 99.van Beers JJ, Raijmakers R, Alexander LE, Stammen-Vogelzangs J, Lokate AM, Heck AJ, Schasfoort RB, Pruijn GJ. Mapping of citrullinated fibrinogen B-cell epitopes in rheumatoid arthritis by imaging surface plasmon resonance. Arthritis Res Ther. 2010;12:R219. doi: 10.1186/ar3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vossenaar ER, Despres N, Lapointe E, van der Heijden A, Lora M, Senshu T, van Venrooij WJ, Menard HA. Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res Ther. 2004;6:R142–R150. doi: 10.1186/ar1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smeets TJ, Vossenaar ER, van Venrooij WJ, Tak PP. Is expression of intracellular citrullinated proteins in synovial tissue specific for rheumatoid arthritis? Comment on the article by Baeten et al. Arthritis and rheumatism. 2002;46:2824–2826. doi: 10.1002/art.10473. author reply 2826–2827. [DOI] [PubMed] [Google Scholar]
- 102.Mechin MC, Sebbag M, Arnaud J, Nachat R, Foulquier C, Adoue V, Coudane F, Duplan H, Schmitt AM, Chavanas S, Guerrin M, Serre G, Simon M. Update on peptidylarginine deiminases and deimination in skin physiology and severe human diseases. International journal of cosmetic science. 2007;29:147–168. doi: 10.1111/j.1467-2494.2007.00377.x. [DOI] [PubMed] [Google Scholar]
- 103.Wegner N, Lundberg K, Kinloch A, Fisher B, Malmstrom V, Feldmann M, Venables PJ. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunol Rev. 2010;233:34–54. doi: 10.1111/j.0105-2896.2009.00850.x. [DOI] [PubMed] [Google Scholar]
- 104.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, Saxne T, Malmstrom V, Venables PJ. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis and rheumatism. 2008;58:2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 105.Vossenaar ER, Zendman AJ, Van Venrooij WJ. Citrullination, a possible functional link between susceptibility genes and rheumatoid arthritis. Arthritis Res Ther. 2004;6:1–5. doi: 10.1186/ar1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nature genetics. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 107.Perricone C, Ceccarelli F, Valesini G. An overview on the genetic of rheumatoid arthritis: a never-ending story. Autoimmun Rev. 2011;10:599–608. doi: 10.1016/j.autrev.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 108.Cheng J, Zhang H, Zhuang C, Liu R. Peptidylarginine deiminase type 4 and methyl-CpG binding domain 4 polymorphisms in Chinese patients with rheumatoid arthritis. The Journal of rheumatology. 2012;39:1159–1165. doi: 10.3899/jrheum.120007. [DOI] [PubMed] [Google Scholar]
- 109.Moscarello MA, Pritzker L, Mastronardi FG, Wood DD. Peptidylarginine deiminase: a candidate factor in demyelinating disease. Journal of Neurochemistry. 2002;81:335–343. doi: 10.1046/j.1471-4159.2002.00834.x. [DOI] [PubMed] [Google Scholar]
- 110.Moscarello MA, Wood DD, Ackerley C, Boulias C. Myelin in multiple sclerosis is developmentally immature. The Journal of clinical investigation. 1994;94:146–154. doi: 10.1172/JCI117300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wood DD, Bilbao JM, O'Connors P, Moscarello MA. Acute multiple sclerosis (Marburg type) is associated with developmentally immature myelin basic protein. Annals of neurology. 1996;40:18–24. doi: 10.1002/ana.410400106. [DOI] [PubMed] [Google Scholar]
- 112.Boggs JM, Rangaraj G, Koshy KM, Ackerley C, Wood DD, Moscarello MA. Highly deiminated isoform of myelin basic protein from multiple sclerosis brain causes fragmentation of lipid vesicles. Journal of neuroscience research. 1999;57:529–535. [PubMed] [Google Scholar]
- 113.Pritzker LB, Joshi S, Gowan JJ, Harauz G, Moscarello MA. Deimination of myelin basic protein. 1. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry. 2000;39:5374–5381. doi: 10.1021/bi9925569. [DOI] [PubMed] [Google Scholar]
- 114.Cao L, Goodin R, Wood D, Moscarello MA, Whitaker JN. Rapid release and unusual stability of immunodominant peptide 45–89 from citrullinated myelin basic protein. Biochemistry. 1999;38:6157–6163. doi: 10.1021/bi982960s. [DOI] [PubMed] [Google Scholar]
- 115.Wood DD, Ackerley CA, Brand Bvd, Zhang L, Raijmakers R, Mastronardi FG, Moscarello MA. Myelin localization of peptidylarginine deiminases 2 and 4: comparison of PAD2 and PAD4 activities. Lab Invest. 2008;88:354–364. doi: 10.1038/labinvest.3700748. [DOI] [PubMed] [Google Scholar]
- 116.Johnson RS, Roder JC, Riordan JR. Over-expression of the DM-20 myelin proteolipid causes central nervous system demyelination in transgenic mice. J Neurochem. 1995;64:967–976. doi: 10.1046/j.1471-4159.1995.64030967.x. [DOI] [PubMed] [Google Scholar]
- 117.Mastronardi FG, Mak B, Ackerley CA, Roots BI, Moscarello MA. Modifications of myelin basic protein in DM20 transgenic mice are similar to those in myelin basic protein from multiple sclerosis. The Journal of clinical investigation. 1996;97:349–358. doi: 10.1172/JCI118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Musse AA, Li Z, Ackerley CA, Bienzle D, Lei H, Poma R, Harauz G, Moscarello MA, Mastronardi FG. Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis Model Mech. 2008;1:229–240. doi: 10.1242/dmm.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carrillo-Vico A, Leech MD, Anderton SM. Contribution of Myelin Autoantigen Citrullination to T Cell Autoaggression in the Central Nervous System. The Journal of Immunology. 2010;184:2839–2846. doi: 10.4049/jimmunol.0903639. [DOI] [PubMed] [Google Scholar]
- 120.Raijmakers R, Vogelzangs J, Raats J, Panzenbeck M, Corby M, Jiang H, Thibodeau M, Haynes N, van Venrooij WJ, Pruijn GJ, Werneburg B. Experimental autoimmune encephalomyelitis induction in peptidylarginine deiminase 2 knockout mice. The Journal of comparative neurology. 2006;498:217–226. doi: 10.1002/cne.21055. [DOI] [PubMed] [Google Scholar]
- 121.Acharya NK, Nagele EP, Han M, Coretti NJ, DeMarshall C, Kosciuk MC, Boulos PA, Nagele RG. Neuronal PAD4 expression and protein citrullination: possible role in production of autoantibodies associated with neurodegenerative disease. J Autoimmun. 2012;38:369–380. doi: 10.1016/j.jaut.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 122.Jang B, Ishigami A, Maruyama N, Carp RI, Kim YS, Choi EK. Peptidylarginine deiminase and protein citrullination in prion diseases: strong evidence of neurodegeneration. Prion. 2013;7:42–46. doi: 10.4161/pri.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fau JB, Fau IA, Fau MN, Fau CR, Fau KY, Choi EK. Peptidylarginine deiminase and protein citrullination in prion diseases: strong evidence of neurodegeneration. doi: 10.4161/pri.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 125.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annual review of medicine. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 126.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Current opinion in genetics & development. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 128.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 129.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 131.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 132.Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Regulation of Protein Citrullination through p53/PADI4 Network in DNA Damage Response. Cancer Research. 2009;69:8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 133.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jones JE, Slack JL, Fang P, Zhang X, Subramanian V, Causey CP, Coonrod SA, Guo M, Thompson PR. Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem Biol. 2012;7:160–165. doi: 10.1021/cb200258q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 138.Bozdag M, Dreker T, Henry C, Tosco P, Vallaro M, Fruttero R, Scozzafava A, Carta F, Supuran CT. Novel small molecule protein arginine deiminase 4 (PAD4) inhibitors. Bioorganic & medicinal chemistry letters. 2013;23:715–719. doi: 10.1016/j.bmcl.2012.11.102. [DOI] [PubMed] [Google Scholar]
- 139.Moscarello MA, Lei H, Mastronardi FG, Winer S, Tsui H, Li Z, Ackerley C, Zhang L, Raijmakers R, Wood DD. Inhibition of peptidyl-arginine deiminases reverses protein-hypercitrullination and disease in mouse models of multiple sclerosis. Dis Model Mech. 2013;6:467–478. doi: 10.1242/dmm.010520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moscarello MA, Pritzker L, Mastronardi FG, Wood DD. Peptidylarginine deiminase: a candidate factor in demyelinating disease. J Neurochem. 2002;81:335–343. doi: 10.1046/j.1471-4159.2002.00834.x. [DOI] [PubMed] [Google Scholar]
- 141.Teo CY, Shave S, Chor AL, Salleh AB, Rahman MB, Walkinshaw MD, Tejo BA. Discovery of a new class of inhibitors for the protein arginine deiminase type 4 (PAD4) by structure-based virtual screening. BMC bioinformatics. 2012;13(Suppl 17):S4. doi: 10.1186/1471-2105-13-S17-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wei L, Wasilewski E, Chakka SK, Bello AM, Moscarello MA, Kotra LP. Novel Inhibitors of Protein Arginine Deiminase with Potential Activity in Multiple Sclerosis Animal Model. J Med Chem. 2013 doi: 10.1021/jm301755q. [DOI] [PubMed] [Google Scholar]
- 143.Pritzker LB, Moscarello MA. A novel microtubule independent effect of paclitaxel: the inhibition of peptidylarginine deiminase from bovine brain. Biochim Biophys Acta. 1998;1388:154–160. doi: 10.1016/s0167-4838(98)00175-7. [DOI] [PubMed] [Google Scholar]
- 144.Moscarello MA, Mak B, Nguyen TA, Wood DD, Mastronardi F, Ludwin SK. Paclitaxel (Taxol) attenuates clinical disease in a spontaneously demyelinating transgenic mouse and induces remyelination. Mult Scler. 2002;8:130–138. doi: 10.1191/1352458502ms776oa. [DOI] [PubMed] [Google Scholar]
- 145.Wang Q, Priestman MA, Lawrence DS. Monitoring of Protein Arginine Deiminase Activity by Using Fluorescence Quenching: Multicolor Visualization of Citrullination. Angewandte Chemie International Edition. 2013;52:2323–2325. doi: 10.1002/anie.201208464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wildeman E, Pires MM. Facile Fluorescence-Based Detection of PAD4-Mediated Citrullination. Chembiochem : a European journal of chemical biology. 2013 doi: 10.1002/cbic.201300173. [DOI] [PubMed] [Google Scholar]
- 147.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, Baylin SB. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 148.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature genetics. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 149.Steele N, Finn P, Brown R, Plumb JA. Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. British journal of cancer. 2009;100:758–763. doi: 10.1038/sj.bjc.6604932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 2001;61:7025–7029. [PubMed] [Google Scholar]
- 151.Ishida-Yamamoto A, Senshu T, Takahashi H, Akiyama K, Nomura K, Iizuka H. Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. The Journal of investigative dermatology. 2000;114:701–705. doi: 10.1046/j.1523-1747.2000.00936.x. [DOI] [PubMed] [Google Scholar]
- 152.Lee HJ, Joo M, Abdolrasulnia R, Young DG, Choi I, Ware LB, Blackwell TS, Christman BW. Peptidylarginine deiminase 2 suppresses inhibitory {kappa}B kinase activity in lipopolysaccharide-stimulated RAW 264.7 macrophages. The Journal of biological chemistry. 2010;285:39655–39662. doi: 10.1074/jbc.M110.170290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Horibata S, Coonrod SA, Cherrington BD. Role for Peptidylarginine Deiminase Enzymes in Disease and Female Reproduction. Journal of Reproduction and Development. 2012;58:274–282. doi: 10.1262/jrd.2011-040. [DOI] [PubMed] [Google Scholar]
- 154.Asaga H, Akiyama K, Ohsawa T, Ishigami A. Increased and type II-specific expression of peptidylarginine deiminase in activated microglia but not hyperplastic astrocytes following kainic acid-evoked neurodegeneration in the rat brain. Neuroscience letters. 2002;326:129–132. doi: 10.1016/s0304-3940(02)00334-8. [DOI] [PubMed] [Google Scholar]
- 155.Asaga H, Ishigami A. Protein deimination in the rat brain after kainate administration: citrulline-containing proteins as a novel marker of neurodegeneration. Neuroscience letters. 2001;299:5–8. doi: 10.1016/s0304-3940(00)01735-3. [DOI] [PubMed] [Google Scholar]
- 156.Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P, Moscarello MA. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]