SUMMARY
Objective
Pro-inflammatory cytokines play an important role in inducing cartilage degradation during osteoarthritis pathogenesis. Muscle is a tissue that lies near cartilage in situ. However, muscle’s non-loading biochemical effect on cartilage has been largely unexplored. Here, we tested the hypothesis that muscle cells can regulate the response to pro-inflammatory cytokine-mediated damage in chondrocytes derived from human bone marrow-derived mesenchymal stem cells (hMSCs).
Method
hMSCs were allowed to undergo chondrogenic differentiation in porous silk scaffolds in the typical chondrogenic medium for 12 days. For the next 9 days, the cells were cultured in chondrogenic medium containing 50% conditioned medium derived from C2C12 muscle cells or fibroblast control cells, and were subject to treatments of pro-inflammatory cytokines IL-1β or TNFα.
Results
Both IL-1β and TNFα-induced strong expression of multiple MMPs and hypertrophic markers Runx2 and type X collagen. Strikingly, culturing hMSC-derived chondrocytes in C2C12 muscle cell conditioned medium strongly inhibited the expression of all these genes, a result further confirmed by GAG content and histological evaluation of matrix protein. To determine whether these effects were due to altered chondrocyte growth and survival, we assayed the expression of cell proliferation marker Ki67, cell cycle arrest markers p21 and p53, and apoptosis marker caspase 3. Muscle cell-conditioned medium promoted proliferation and inhibited apoptosis, thereby suggesting a possible decrease in the cellular aging and death that typically accompanies cartilage inflammation.
Conclusion
Our findings suggest the role of muscle in cartilage homeostasis and provide insight into designing strategies for promoting resistance to pro-inflammatory cytokines in hMSC-derived chondrocytes.
Keywords: Cartilage tissue engineering, Osteoarthritis, Pro-inflammatory, Cytokines, Myokines, Stem cells
Introduction
Articular cartilage serves a load-bearing role in diarthrodial joints1, 2. However, degenerative erosion of this tissue can lead to osteoarthritis (OA) which is estimated to affect 71 million North Americans by the year 20303. Due to the narrow reparative potential of cartilage, along with its avascular and aneural nature4, current treatments are mainly restricted to alleviating symptoms and pain5. Cartilage tissue engineering has emerged as an attractive therapeutic option for restoring and regenerating cartilage in situ. Such strategies rely on the combination of repair cells with the appropriate three-dimensional (3D) biomaterial scaffolding and biochemical cues necessary to create a regenerative microenvironment. Engineered constructs can then be implanted at the site of injury to facilitate healing and restoration of tissue function.
For adequate healing and regeneration, engineered cartilage requires appropriate coupling of progenitor repair cells with biocompatible scaffolding that facilitates the generation of a tissue construct similar to native cartilage. With respect to progenitor repair cells, matrix-secreting primary chondrocytes are limited in quantity and capacity for expansion, and are susceptible to de-differentiation in vitro4, 6. In comparison, human bone marrow-derived mesenchymal stem cells (hMSCs) have emerged as an attractive cell source for cartilage tissue engineering4, 6. These multipotent cells can be obtained in relatively large amounts, rapidly expanded ex vivo, and differentiated to adopt the chondrogenic cell fate7. In terms of 3D scaffolding, multiple materials have been explored for engineering cartilage tissue. Among them, silk, derived from the Bombyx mori (silkworm), stands out as a biomaterial whose biocompatibility, slow degradation, and robust mechanical properties make it suitable for cartilage tissue engineering8–11.
The hostile pathological environment in the arthritic joint imposes a major challenge on cartilage tissue engineering. For successful cartilage repair, the engineered tissue construct must also be able to maintain its stability in the inflammatory microenvironment in vivo during OA pathogenesis12, 13. In the joint, pro-inflammatory cytokines, most notably IL-1β and TNFα, cause cartilage damage in OA by stimulating the expression of a number of proteolytic enzymes, including matrix metalloproteinases (MMPs) and aggrecanases5, 14–16. Additionally, to further drive the degradation cascade, IL-1β and TNFα also promote chondrocyte hypertrophy, cellular senescence, and cell death, thereby reducing the proliferation and viability potential for chondrocytes5, 17, 18.
Muscle lies in close proximity to cartilage during development19 and throughout life. This tissue provides cartilage with a biomechanical stimulation that promotes nutrient distribution and maintains homeostasis20. For example, age-associated muscle loss is linked to the progression of OA in the elderly21 and reduced muscle strength has been shown to be a risk factor for knee OA22. As a result, muscle strengthening has been explored as an intervention to prevent or delay the onset of OA23. Furthermore, muscle paralysis, which leads to muscle atrophy, induces joint abnormality and cartilage degradation24–26. On the other hand, bone morphogenetic protein 2 (BMP-2)-expressing muscle tissue was found to serve as a bridge in enhancing and accelerating femur fracture healing in a rat model27. While these studies have demonstrated the significance of muscle tissue on skeletal development, it is unclear whether this regulation involves merely biomechanical stimuli, biochemical mediators, or both28, 29.
Recently, our group investigated the non-loading biochemical effect of muscle cells on cartilage gene expression. Our results showed that a rat chondrocyte cell line co-cultured with muscle cells or cultured in muscle cell-conditioned medium as monolayers had significantly higher expression levels of cartilage matrix proteins in the presence of IL-1β and TNFα30, 31. However, for the purpose of tissue engineering, it is critical to evaluate the effect of muscle cell-derived factors on primary cells in a 3D microenvironment under such inflammatory stimuli.
In this study, we subjected hMSCs-derived differentiating hondrocytes grown in 3D silk scaffolds to C2C12 muscle cell-conditioned media as well as pro-inflammatory cytokines IL-1β and TNFα. Administration of IL-1β and TNFα strongly induced the gene expression of matrix degrading enzymes (MMPs) and hyper-trophic markers such as Runx2 and type X collagen (Col X) in hMSC-derived chondrocytes. In contrast, culturing in conditioned medium derived from C2C12 muscle cells resulted in strong inhibition of the expression of these genes, a result further confirmed by quantification of proteoglycan content and histological assessment. To determine whether muscle cell-derived factors altered chondrocyte growth and survival, we assayed the expression of cell proliferation and cell cycle arrest markers as well as apoptosis markers. We found that culturing in muscle cell-conditioned medium promoted chondrocyte proliferation and inhibited cell death under inflammatory stimuli. Together, our findings demonstrate that muscle cell-derived factors, or myokines, provide enhanced resistance to pro-inflammatory cytokine-mediated cartilage degradation, hypertrophy, and cell growth arrest in hMSC-derived chondrocytes and suggest that such factors may be utilized to improve the stability of bioengineered cartilage in the hostile microenvironment of arthritic joints.
Methods
Biomaterial scaffolds
Silk fibroin scaffolds were fabricated as previously described8. Briefly, B. mori cocoons were boiled in 0.02 M Na2CO3 to extract sericin proteins. Silk fibroin solution was then lyophilized and resuspended in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP). NaCl particles (size: 106–212 µm) were added to 12% silk-HFIP solution (w/v) to generate porous scaffolds with a pore size of 106-212 µm. Scaffolds were then punched into 5 mm × 3 mm (diameter × height) disks for our experiments. Scaffolds of the same dimensions, made of d,d-l,l polylactic acid (PLA) and bovine type I and type III collagen, were obtained from BD Biosciences (Franklin Lakes, NJ).
Cell culture
hMSCs were purchased from Lonza Walkersville, Inc (Walkersville, MD). The lot numbers purchased were 1-OF3825 (donor #1), 8F3520 (donor #2), and 3F4287 (donor #3). Cells were expanded in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Grand Island, NY) containing 10% fetal bovine serum (FBS) (Hyclone, Rockford, IL), 1% antibiotic-antimycotic (Gibco), 0.1 mM non-essential amino acids (Gibco), and 1 ng/mL basic fibroblast growth factor (Peprotech, Rocky Hill, NJ). For chondrogenic differentiation, cells were cultured DMEM containing 1% antibiotic-antimycotic, ITS + supplement (BD Biosciences), 0.1 mM ascorbic acid 2-phosphate (Sigma Aldrich, St. Louis MO), 1.25 mg/mL human serum albumin (Sigma), 10−7 M dexamethasone (Sigma), and 10 ng/mL transforming growth factor beta 3 (TGF-β3; R&D Systems).
Murine myoblast cell line (C2C12) and murine fibroblast cell line (NIH3T3) were purchased from ATCC (Rockville, MD). Cell lines were cultured in DMEM with 10% FBS and 1% antibiotic-antimycotic. Cells were maintained at a 75–80% confluence for 24 h. Conditioned medium was sterile-filtered using a 0.22-µm syringe filter for subsequent use.
hMSC seeding and differentiation
Prior to cell seeding, scaffolds were immersed in sterile DMEM. 0.5 × 106 hMSCs in 15 µL of chondrogenic media were seeded into scaffolds and allowed to attach for 2 h. hMSC-seeded scaffolds were then transferred into 1 mL of fresh media in 24-well tissue culture plates at 37°C under static culturing conditions for 2 days before being placed on an incubated dynamic shaker (5–6 RPM) for remainder of study.
For the study of hMSC differentiation on different scaffolds, hMSCs (passage 4) were seeded into PLA, collagen, or silk scaffolds, and maintained in chondrogenic media for 21 days. For hMSC differentiation in the presence of pro-inflammatory cytokines, hMSCs (passage 5) were differentiated in silk scaffolds for 12 days using chondrogenic media. At this time, scaffolds were divided into six groups [Fig. 1(A)] and cultured for an additional 9 days in: 1:1 chondrogenic media and C2C12 conditioned media, 5 ng/mL IL-1β in a 1:1 mixture of chondrogenic media and C2C12 conditioned media, and 5 ng/mL TNFα in a 1:1 mixture of chondrogenic media and C2C12 conditioned media. A control group of scaffolds were cultured with NIH3T3, rather than C2C12, conditioned media. In all 21-day studies, media was changed every 3 days. Unless stated otherwise, data shown represents a sample size of n ≥ 3 independent samples (cell-seeded scaffolds) from one experiment using one donor.
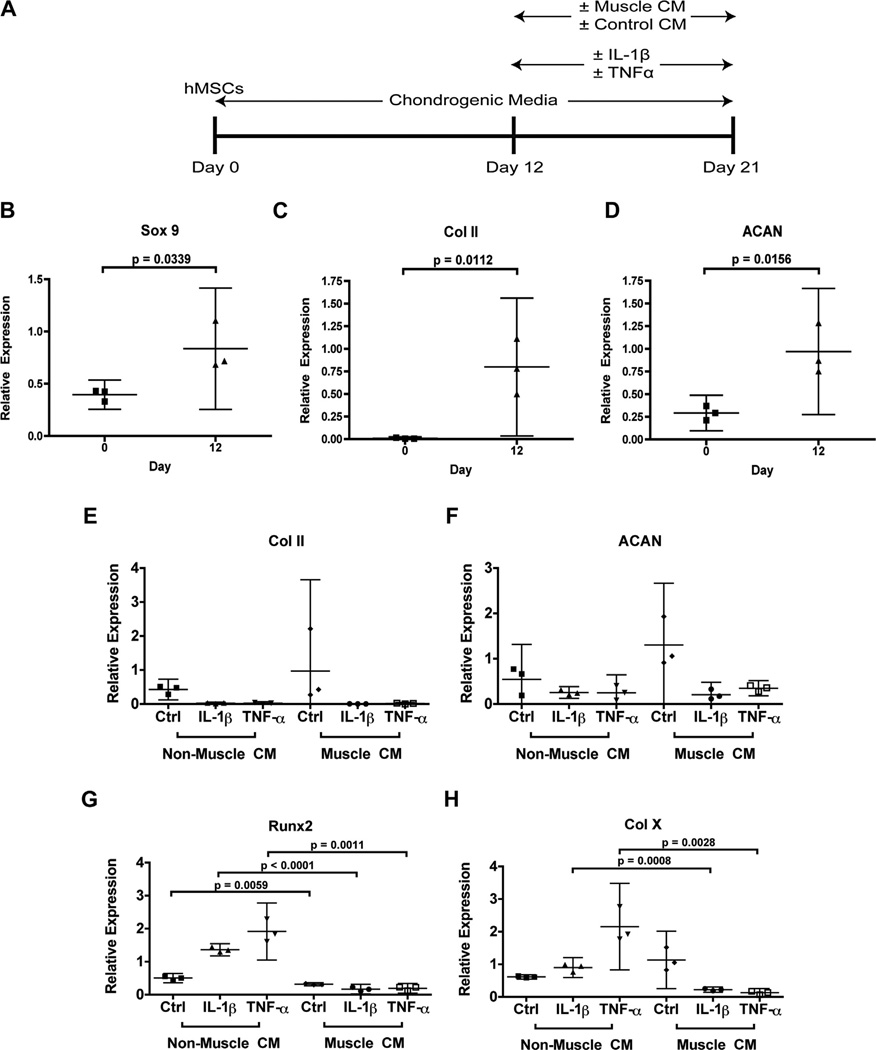
Fig. 1.
Study design and mRNA expression of chondrogenic genes. hMSCs were differentiated for 21 days into chondrocytes using chondrogenic media, with treatment of pro-inflammatory cytokines and muscle cell- or non-muscle control cell-conditioned media after Day 12 (A). Relative expression of markers for cartilage matrix production at Day 12 included transcription factor SOX9 (B), collagen II (Col II) (C), and ACAN (D), as compared to undifferentiated hMSCs at Day 0. Matrix markers at Day 21 included collagen II (E) and ACAN (F), while hypertrophic markers included Runx2 (G) and collagen X (Col X) (H). Expression was normalized to GAPDH. Data are presented as mean with 95% confidence intervals (n = 3 independent samples (cell-seeded scaffolds) from one experiment completed using one donor’s cells).
RT-PCR analysis
Total RNA was isolated from scaffolds using the RNeasy Mini kit (Qiagen; Valencia, CA) as per the manufacturer’s protocol. RNA was then transcribed to cDNA using random primers (Invitrogen), MMLV reverse transcriptase kit (Invitrogen), and dNTPs (New England BioLabs; Ipswich, MA) and stored at −20°C until RT-PCR analyses. For RT-PCR, cDNA was mixed with gene-specific primers (sequences available upon request) and SYBR Green Supermix (Quanta Biosciences, Inc., Gaithersburg, MD), quantified using BioRad’s iQ5 Real Time PCR Detection System and optical software (Hercules, CA), and normalized against gylceraldehyde-3-phosphate-dehydrogenase (GAPDH). For all analyses, at least three independent samples (cell-seeded scaffolds) from one experiment were analyzed (n ≥ 3).
Histology
Constructs were fixed in formalin, dehydrated in graded ethanol solutions, cleared with xylene, embedded in paraffin, and sectioned into 5- or 10-µm thick sections. After deparaffinization, sections were stained with hemotoxilin and 1% eosin (H&E) and toluidine blue. Diaminobenzidine (DAB) immunocytochemistry was carried out using the DAB staining kit (Vector Laboratories; Burlingame CA). Primary antibodies for type II collagen (Abcam; Cambridge, MA), caspase 3 (Abcam), and Ki67 (Vector Laboratories) were used. When appropriate, sections were counterstained with methyl green, prior to imaging and counting.
Sulfated glycosaminoglycans (GAG) and DNA content quantification
Constructs were cut into pieces and digested in papain solution (10 U/mg in PBS containing 5 mM cysteine-HCl and 5 mM EDTA) at 60°C overnight, sonicated, and then stored at −20°C. Sulfated glycosaminoglycan (GAG) content was quantified based on the absorbance at 525 nm using dimethylmethylene blue (DMMB) dye and a shark chondroitin sulfate standard32. DNA content was quantified using Picogreen (Invitrogen). At least three independent samples were analyzed for each condition.
Statistical analysis
Each experiment was prepared from a single donor’s cells. Data was calculated from at least three independent samples (cell-seeded scaffolds) from one experiment using one donor’s cells and is reported as mean with 95% confidence intervals. Data was further analyzed using Prism 4 (Graphpad; La Jolla, CA) and a student’s t-test or one-way analysis of variance (ANOVA) with Tukey post hoc assessment. Statistical significance was assigned where appropriate with P < 0.05.
Results
Muscle cell-derived factors inhibit hypertrophy in hMSC-derived chondrocytes under inflammatory stimuli
To assess the effects of muscle cell-derived factors on the response of hMSC-derived chondrocytes to inflammatory cytokines, hMSCs were seeded onto silk scaffolds and allowed to undergo chondrogenic differentiation for 12 days using established chondrogenic medium11. We chose silk scaffolds for hMSC seeding based on our comparison study on the chondrogenic potentials of hMSCs seeded into silk, collagen, or PLA scaffolds, where we found that hMSCs in silk scaffolds produced the highest content of GAG and DNA, as well as higher levels of aggrecan (ACAN) mRNA and lower levels of mRNA for caspase 3 (CASP3) (Fig. S1). For studying hMSC differentiation in the presence of pro-inflammatory cytokines and muscle-derived factors, we first assessed the expression of cartilage-specific genes at Day 12 using RT-PCR and compared to undifferentiated cells at Day 0. While chondrogenesis is still ongoing at Day 12, a level of chondrogenic differentiation has been achieved prior to the addition of cytokines and conditioned media. This finding is signified by the induction of SOX 9, a transcription factor that acts as a master regulator in chondrogenic differentiation, as well as cartilage matrix genes type II collagen (Col II) and ACAN [Fig. 1(B–D)].
Over the next 9 days, hMSC-derived chondrocytes were subject to IL-1β or TNFα treatment. In the presence of pro-inflammatory cytokines, the expression of cartilage matrix genes Col II and ACAN was significantly reduced while expression of hypertrophic cartilage markers Col X and Runx2 was induced, as compared with controls [Fig. 1(E–H)]. It was also noted that TNFα had a stronger effect in inducing chondrocyte hypertrophy than IL-1β [Fig. 1 (G–H)]. Culturing with C2C12 muscle cell-conditioned medium did not rescue IL-1β- and TNFα-induced Col II and ACAN reduction [Fig. 1 (E–F)]. Strikingly however, culturing with muscle cell-conditioned medium led to a significant decrease in the expression of hypertrophic markers Col X and Runx2, suggesting that muscle cell-derived factors inhibited pro-inflammatory cytokine-induced chondrocyte hypertrophy [Fig. 1 (G–H)].
Muscle cell-derived factors strongly inhibit the induction of multiple MMPs by inflammatory stimuli in hMSC-derived chondrocytes
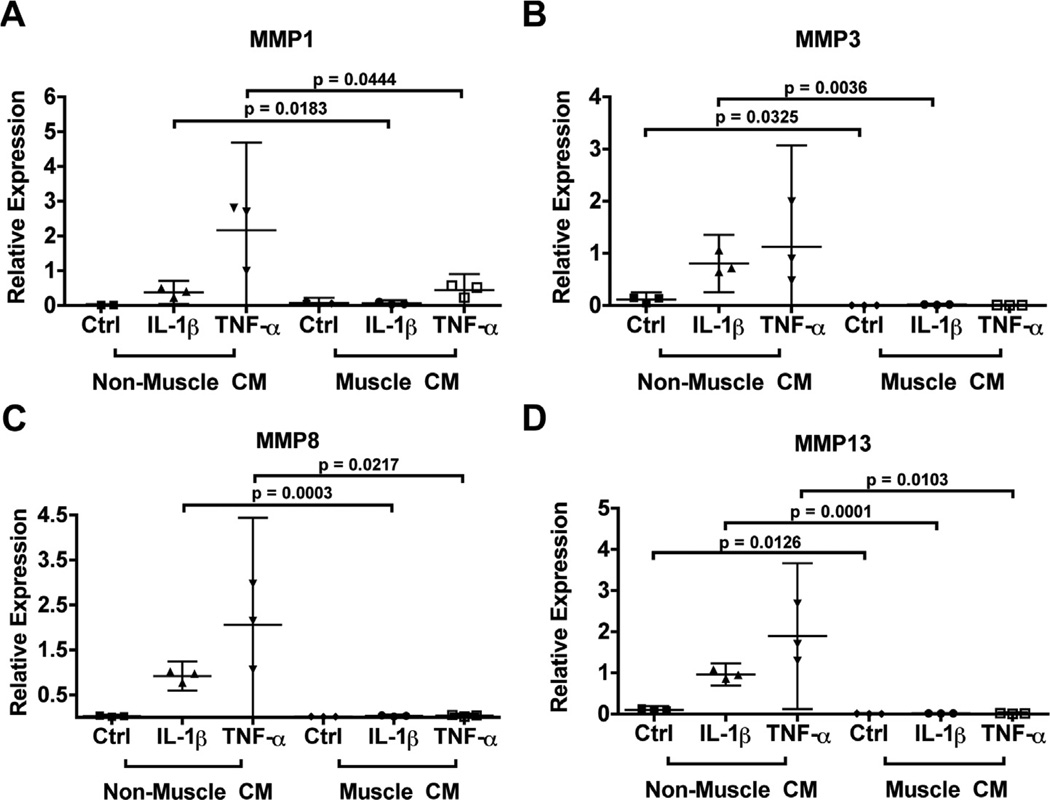
Matrix metalloproteinases MMP1, MMP8, and MMP13 are involved in the proteolytic degradation of cartilage matrix protein Col II, while MMP3 activates MMP1 as well as degrades matrix proteoglycans33. Both IL-1β and TNFα induced a significant and consistent upregulation of MMP1 [Fig. 2(A)], MMP3 [Fig. 2(B)], MMP8 [Fig. 2(C)], and MMP13 [Fig. 2(D)] in hMSC-derived chondrocytes cultured in the control condition. Strikingly, culturing in C2C12 muscle cell-conditioned medium essentially abolished the induction of all these MMPs. Even in the absence of pro-inflammatory cytokines, muscle cell-conditioned medium significantly reduced expression of MMP3 and MMP13.
Fig. 2.
Relative expression of matrix degradation markers in differentiated hMSCs at Day 21 after treatment of pro-inflammatory cytokines and muscle cell-derived factors as compared with controls. Matrix degradation markers included MMP1 (A), MMP3 (B), MMP8 (C), and MMP13 (D). Expression was normalized to GAPDH. Data are presented as mean with 95% confidence intervals (n = 3 independent samples (cell-seeded scaffolds) from one experiment completed using one donor’s cells).
Muscle cell-derived factors promote GAG production and Col II protein expression in the presence of inflammatory stimuli
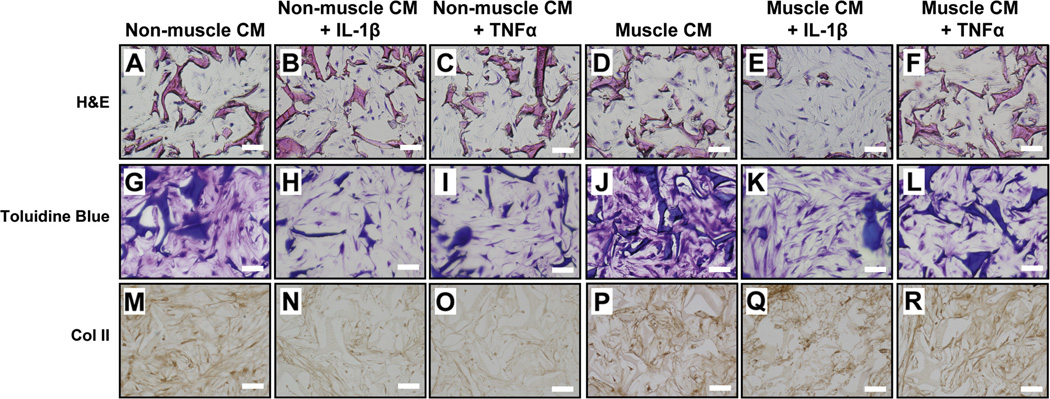
H&E histological analysis showed that the cells were distributed uniformly throughout scaffolds [Fig. 3 (A–F)]. To determine the levels of cartilage matrix deposition in the cartilage constructs, we performed toluidine blue staining [Fig. 3 (G–L)], as well as immunocytochemical staining for Col II [Fig. 3 (M–R)]. While both toluidine blue staining and Col II staining were diminished with the addition of pro-inflammatory cytokines in cells grown in control conditioned media, staining remained consistent for all cells cultured with muscle cell-conditioned media, regardless of inflammatory cytokine treatment.
Fig. 3.
Representative histological evaluation of hMSC-derived chondrocytes within silk scaffolds after Day 21 of culture in the presence of pro-inflammatory cytokines and muscle cell-derived factors as compared to controls. H&E staining (A–F). Toluidine blue staining for proteoglycans (G–L) and immunocytochemical staining for type II collagen (M–R). (Scale bars: 250 µm).
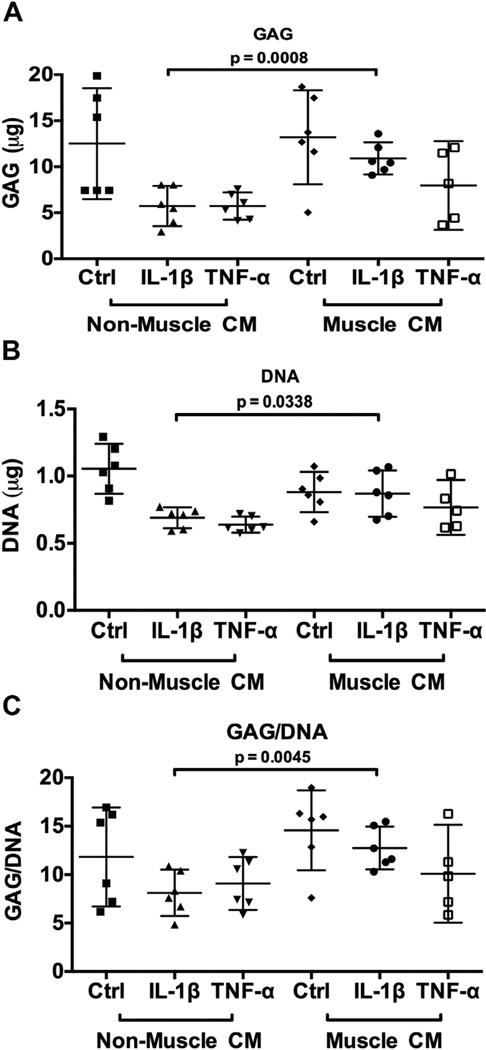
To further assess whether muscle cell-derived factors promoted cartilage matrix production, we quantified the amount of sulfated GAG within these constructs using the DMMB assay32. While there were no observable differences in GAG [Fig. 4(A)] and DNA [Fig. 4(B)] content among the samples in the absence of cytokine treatment, constructs cultured in the presence of muscle cell-derived factors yielded significantly higher levels of GAG and DNA content, as well as GAG/DNA ratio [Fig. 4(C)] under IL-1β, but not TNFα treatment as compared to the control. This result suggests that under the inflammatory condition of IL-1β, muscle cell-derived factors promoted the maintenance of cartilage matrix.
Fig. 4.
Biochemical quantification of glycosaminoglycan (A) and DNA (B) content, as well as ratio of GAG/DNA (C), in differentiated hMSCs at Day 21 after treatment of pro-inflammatory cytokines and muscle cell-derived factors as compared with controls. Data are presented as mean with 95% confidence intervals (n = 6 independent samples (cell-seeded scaffolds) using one donor’s cells).
Muscle cell-derived factors promote cell proliferation and inhibit apoptosis under IL-1β treatment
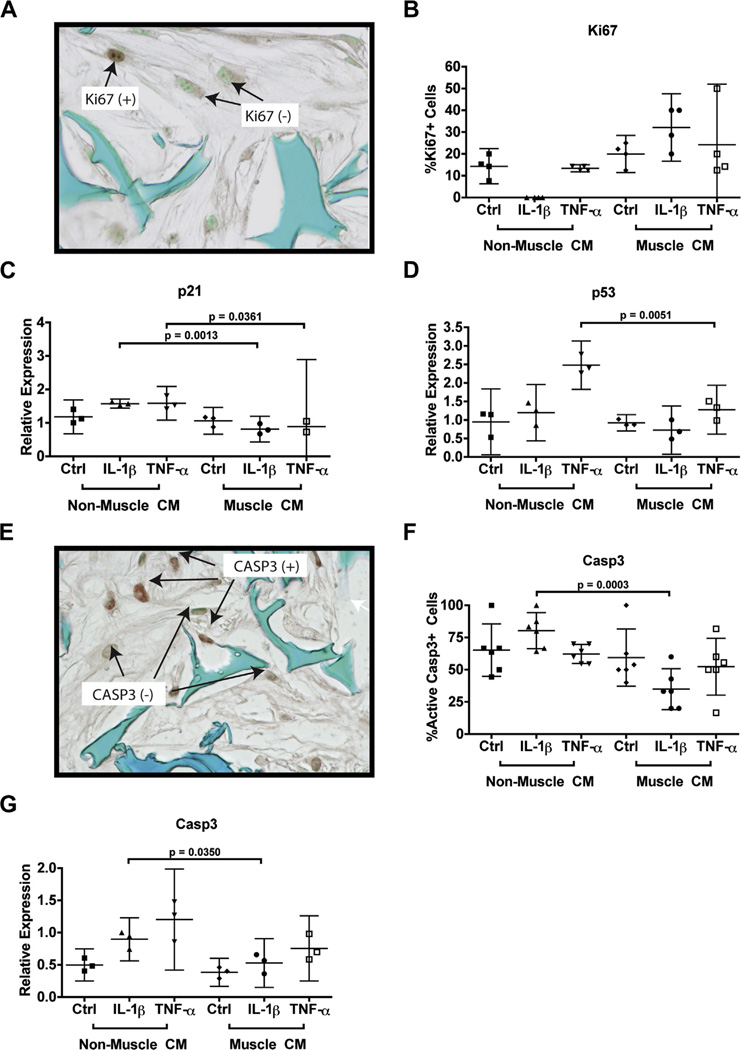
Because we observed that cells cultured in muscle cell-conditioned medium showed increased DNA content under IL-1β treatments, we next investigated whether this increase was due to increased cell proliferation or decreased cell death. To examine cell proliferation status, we used immunocytochemistry to quantify the number of cells that were stained positive for Ki67, a marker for cell proliferation [Fig. 5(A–B)]. Muscle cell-conditioned medium did not enhance cell proliferation under non-inflammatory conditions, which is consistent with the quantification of total DNA level. IL-1β treatment completely abolished Ki67 staining, suggesting a strong inhibition of cell proliferation. Remarkably, under IL-1β treatment, muscle cell-derived factors induced a stark increase in the percentage of Ki67-positive cells. Unlike IL-1β, TNFα did not cause a significant reduction in cell proliferation as compared to the control condition. Furthermore, no changes in cell proliferation were observed for TNFα treatment in the presence of muscle cell-derived factors.
Fig. 5.
Biochemical quantification and relative gene expression analysis of cell proliferation, cell cycle regulation, and apoptosis. Cells were stained via immunocytochemistry for proliferative marker Ki67 (A) and the percentage of Ki67 + stained cells were quantified (B). mRNA expression for markers for cell cycle regulation and inhibition included p21 (C) and p53 (D). Cells were stained via immunocytochemistry for active caspase 3 (CASP3) (E) And the percentage of positively stained cells were quantified (F). mRNA expression for CASP3 was also assessed (G). All mRNA expression was normalized to GAPDH. For immunocytochemistry, sections were counterstained with methyl green. Data are presented as mean with 95% confidence intervals (n = 4 sections for quantification of Ki67 + cells, n = 3 independent samples (cell-seeded scaffolds) for RT-PCR, and n = 6 sections for quantification of caspase 3 + cells).
To assay if the change of cell proliferation was caused by altered expression of cell cycle regulators, we then assessed the mRNA expression of cell cycle inhibitors p21 [Fig. 5(C)] and p53 [Fig. 5(D)]. Culturing with muscle cell-conditioned medium did not significantly alter the expression of these two markers under the non-inflammatory conditions. On the other hand, both IL-1β and TNFα induced the expression of p21, while TNFα also significantly induced p53 expression. Significantly, culturing with muscle cell-conditioned media inhibited IL-1β- and TNFα-induced p21, as well as TNFα-induced p53 expression.
To determine the effect of muscle cell-derived factors on pro-inflammatory cytokine-induced apoptosis, we then examined the expression of caspase 3 (CASP3), an enzyme activated in apoptotic cells [Fig. 5(E–F)]. The number of cells positively-stained for active CASP3 were significantly reduced with the addition of muscle cell-conditioned medium under IL-1β treatment, a finding that was further correlated with the reduction of CASP3 mRNA expression [Fig. 5(G)]. Muscle cell-conditioned medium did not appear to alter CASP3 mRNA or protein expression under TNFα treatment, which is consistent with the DNA quantifications. Nonetheless, our data clearly suggests that muscle cell-derived factors promote cell proliferation and survival under the treatment of IL-1β.
Discussion
Catabolic mediators, in particular pro-inflammatory cytokines IL-1β and TNFα, have been demonstrated to inhibit chondrogenesis and promote chondrocyte hypertrophy and degradation in OA joints, thus leading to cartilage instability14. This is especially important for immature engineered tissues composed of progenitor stem cells actively undergoing chondrogenesis as such constructs may not have generated enough protective extracellular matrix (ECM) at the time of construct implantation and consequently, may be especially more susceptible to degradation12, 34. Indeed, the therapeutic viability of engineered cartilage relies on methods that promote resistance to these catabolic inducers. Here, we addressed this challenge by investigating the effects of muscle cell-derived factors on the stability of cartilage constructs under inflammatory conditions.
Our findings demonstrated that muscle cell-derived factors inhibited the mRNA expression of MMPs as well as hypertrophic markers in bioengineered cartilage. Pro-inflammatory cytokines IL-1β and TNFα cause cartilage destruction by inducing the expression of MMPs, promoting chondrocyte hypertrophy, and reducing the synthesis of cartilage matrix genes. Although the induction of hypertrophic markers by IL-1β and TNFα were about 2–4 fold, we believe this is biologically meaningful as OA chondrocytes are known to exhibit higher levels of hypertrophy35. For all the MMPs we examined (MMP1, MMP3, MMP8, and MMP13), muscle cell-derived factors abolished IL-1β- and TNFα-induced expression at the mRNA level, indicating a strong impairment of the cartilage-destructive ability of these pro-inflammatory cytokines. Furthermore, we found a significant inhibition of hypertrophic markers Col X and Runx2 in constructs cultured with muscle cell-conditioned medium, suggesting that muscle cell-derived factors inhibited the terminal differentiation of hMSCs, which is one of the major obstacles for the translational application of hMSC chondrogenesis36. These results were consistent among all three donors tested (Figs. 1 and 2 for donor #1; Fig. S2 for donors #2 and #3). Although there were no differences in the mRNA expression of Col II and ACAN in hMSC-derived chondrocytes cultured with muscle cell-conditioned medium, the dramatic reduction of MMPs and the inhibition of chondrocyte hypertrophy could result in the maintenance of protein expression of cartilage matrix components under inflammatory conditions.
Thus, to substantiate our RT-PCR gene expression analyses, we performed biochemical and histological studies to quantitatively and qualitatively evaluate the expression of cartilage matrix proteins. Our immunocytochemistry and toluidine blue staining results showed that cartilage matrix proteins, mainly Col II and total proteoglycan, produced by constructs exposed to muscle cell-conditioned medium were maintained as compared to non-muscle cell-conditioned medium under IL-1β or TNFα stimulation. Quantification of sulfated GAG content revealed only statistically significant differences in GAG levels between muscle-treated samples and controls in the case of IL-1β stimulation. For TNFα treatments, it is not clear why there are no quantifiable significant differences in GAG content as compared to the proteoglycan histological staining. Such difference in results may be likely due to the fact that while the DMMB dye, which was used for GAG quantification, only binds to sulfated proteoglycans, toluidine blue stains both sulfated and non-sulfated proteoglycans, such as hyaluronan37. Further investigation of individual proteoglycan components may help to understand whether muscle cell-derived factors promote additional cartilage matrix components in maintaining the stability of hMSC-derived chondrocytes under inflammatory conditions. It should be noted that TNFα led to a higher level of cartilage destruction as evidenced by its stronger induction of chondrocyte hypertrophic markers Col X and Runx2, as well as all MMPs, as compared to IL-1β. Consequently, to overcome the inflammatory damage induced by TNFα, a larger amount of muscle cell-derived factors may be required. Finally, these results are consistent with our prior study using rat chondrosarcoma (RCS) chondrocytes in 2D cultures, which showed that muscle cell-secreted factors promoted cartilage matrix protein expression under inflammatory cytokine treatments30, 31.
To further explore the mechanisms by which muscle cell-derived factors regulate chondrocyte stability under inflammatory conditions, we analyzed cell proliferation and cell death. Muscle cell-derived factors strongly rescued IL-1β-induced growth arrest, which may be mediated in part by downregulating the expression of cell cycle inhibitor p21. Although p53 mRNA expression was not induced by IL-1β or inhibited by muscle cell-conditioned medium, it is not clear whether the level of p53 activation was altered in these conditions. Interestingly, TNFα did not change the proliferation level in these cells as assayed by Ki67 staining; however, it did significantly induce p21 and p53 expression. It has been established that these two transcription factors, in addition to their function as cell cycle inhibitors, also play a significant role in cellular aging and senescence38, two events that are involved in the pathogenesis of OA39. Thus, it possible that IL-1β and TNFα induce cellular aging and senescence as well, and it will be interesting to determine the effect of muscle cell-derived factors on these pro-inflammatory cytokine-induced events.
We also observed that muscle cell-derived factors inhibited IL-1β-induced CASP3 mRNA expression but did not inhibit TNFα-induced expression. As noted previously, higher levels of muscle cell-conditioned medium might be required to overcome TNFα-induced CASP3 expression, as TNFα typically is a stronger inducer of apoptosis40, 41. Furthermore, it is worth noting that while cytokine-induced increases in CASP3 mRNA expression were observed, there was no similar trend observed in cytokine-induced protein expression of active CASP3, a cleaved product of pro-CASP3. Here, it is not clear whether the enzymatic activity of CASP3 was altered by IL-1β and TNFα. What our data clearly shows is that muscle cell-conditioned medium significantly inhibited the protein expression of activated CASP3 with respect to IL-1β treatment. These findings parallel the observed increase in DNA content by muscle cell-derived factors during IL-1β treatment and further propose the role of muscle in promoting cell survival under the condition of IL-1β treatment.
Several future studies can stem from the results presented here. It is important to determine the effective component(s) in the muscle cell-conditioned medium that are contributing to the observed prevention of matrix degradation. Recently, muscle has been recognized as an endocrine organ and is found to secrete multiple myokines42. Although muscle lies outside the fibrous tissue of the joint capsule, it is in the position to influence cartilage gene expression locally and systemically, as these myokines can potentially pass through this capsule. In fact, it was reported that synovial fibroblasts of RA joints could even spread from one joint to another43. However, we are not clear which myokines regulate the response of chondrocytes toward inflammatory stimuli. Identification of relevant myokines will provide further insight into cartilage development and may help develop a new platform for tissue engineering technologies, as key factors could be coupled with the biomaterial scaffolding or can be expressed locally in situ. Additionally, in our study, we did not selectively isolate muscle cells at a specific stage of myogenesis (myoblast, myocyte, or myotube) but used a mixed population of muscle cells. As each stage has a variable gene expression profile44, further studies are warranted to identify what secreted factors in which specific stage contribute to the stability of hMSC-derived chondrocytes. Furthermore, this work involves the use of factors derived from the rodent C2C12 muscle cell line. For therapeutic relevance, additional study is recommended using primary human muscle cells to ensure that the effect observed is indeed due to myokines and not to non-muscle specific murine factors. Finally, it is still unclear how muscle cell-derived factors may alter the activity of IL-1β and TNFα. The complete inhibition of chondrocyte hypertrophy as well as the expression of all of the MMPs by muscle cell-derived factors is particularly striking. The fact that the mRNA levels of collagen II and ACAN were unaffected suggests that this effect is very specific to the degradation cascade elicited by these pro-inflammatory cytokines. As NF-κB activation is the key event that mediates the subsequent upregulation of MMPs by both IL-1β and TNFα during chondrogenic inhibition34, 45, it would be interesting to further explore the effect of muscle cell-derived factors on NF-κB activation.
In summary, our findings reveal an important role of muscle in regulating cartilage homeostasis and offer a mechanism to promote stability and resistance in engineered cartilage to pro-inflammatory cytokines. We hope this study will lead to a deeper understanding of the control of cartilage development, which may eventually be harnessed to improve tissue engineering strategies.
Supplementary Material
Acknowledgments
LZ and DK were supported by grants from NSF (CBET-0966920) and NIH (R01AR059106 and P41 EB002520). RR was supported by a postdoctoral fellowship from NIH/NIGMS (K12GM074869).
Role of the funding source
The NSF, NIH, and NIGMS had no role in the design, analysis or interpretation of the data or in the writing of the manuscript.
Footnotes
Author contributions
Conception and design: Rainbow, Zeng.
Acquisition of data: Rainbow, Kwon, Foote.
Provision of study materials: Preda, Kaplan.
Analysis and interpretation of data: Rainbow, Zeng.
Drafting of article: all authors.
Final approval of article: all authors.
Conflict of interest
None of the authors have any conflict of interest related to this work.
Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.joca.2013.04.011.
References
- 1.Little CJ, Bawolin NK, Chen X. Mechanical properties of natural cartilage and tissue-engineered constructs. Tissue Eng Part B Rev. 2011 Aug;17(4):213–227. doi: 10.1089/ten.TEB.2010.0572. [DOI] [PubMed] [Google Scholar]
- 2.Huang AH, Yeger-McKeever M, Stein A, Mauck RL. Tensile properties of engineered cartilage formed from chondrocyte- and MSC-laden hydrogels. Osteoarthritis Cartilage. 2008 Sep;16(9):1074–1082. doi: 10.1016/j.joca.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hootman JM, Helmick CG. Projections of US prevalence of arthritis and associated activity limitations. Arthritis Rheum. 2006 Jan;54(1):226–229. doi: 10.1002/art.21562. [DOI] [PubMed] [Google Scholar]
- 4.Mahmoudifar N, Doran PM. Chondrogenesis and cartilage tissue engineering: the longer road to technology development. Trends Biotechnol. 2012 Mar;30(3):166–176. doi: 10.1016/j.tibtech.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Wieland HA, Michaelis M, Kirschbaum BJ, Rudolphi KA. Osteoarthritis e an untreatable disease? Nat Rev Drug Discov. 2005 Apr;4(4):331–344. doi: 10.1038/nrd1693. [DOI] [PubMed] [Google Scholar]
- 6.Gadjanski I, Spiller K, Vunjak-Novakovic G. Time-Dependent processes in stem cell-based tissue engineering of articular cartilage. Stem Cell Rev. 2011 Oct 21; doi: 10.1007/s12015-011-9328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wescoe KE, Schugar RC, Chu CR, Deasy BM. The role of the biochemical and biophysical environment in chondrogenic stem cell differentiation assays and cartilage tissue engineering. Cell Biochem Biophys. 2008;52(2):85–102. doi: 10.1007/s12013-008-9029-0. [DOI] [PubMed] [Google Scholar]
- 8.Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, Kaplan DL. Materials fabrication from Bombyx mori silk fibroin. Nat Protoc. 2011 Oct;6(10):1612–1631. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meinel L, Hofmann S, Karageorgiou V, Zichner L, Langer R, Kaplan D, et al. Engineering cartilage-like tissue using human mesenchymal stem cells and silk protein scaffolds. Biotechnol Bioeng. 2004 Nov 5;88(3):379–391. doi: 10.1002/bit.20252. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann S, Knecht S, Langer R, Kaplan DL, Vunjak-Novakovic G, Merkle HP, et al. Cartilage-like tissue engineering using silk scaffolds and mesenchymal stem cells. Tissue Eng. 2006 Oct;12(10):2729–2738. doi: 10.1089/ten.2006.12.2729. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Kim UJ, Blasioli DJ, Kim HJ, Kaplan DL. In vitro cartilage tissue engineering with 3D porous aqueous-derived silk scaffolds and mesenchymal stem cells. Biomaterials. 2005 Dec;26(34):7082–7094. doi: 10.1016/j.biomaterials.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Lima EG, Tan AR, Tai T, Bian L, Stoker AM, Ateshian GA, et al. Differences in interleukin-1 response between engineered and native cartilage. Tissue Eng Part A. 2008 Oct;14(10):1721–1730. doi: 10.1089/ten.tea.2007.0347. [DOI] [PubMed] [Google Scholar]
- 13.Francioli S, Cavallo C, Grigolo B, Martin I, Barbero A. Engineered cartilage maturation regulates cytokine production and interleukin-1beta response. Clin Orthop Relat Res. 2011 Oct;469(10):2773–2784. doi: 10.1007/s11999-011-1826-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldring MB, Otero M, Tsuchimochi K, Ijiri K, Li Y. Defining the roles of inflammatory and anabolic cytokines in cartilage metabolism. Ann Rheum Dis. 2008 Dec;67(Suppl 3):iii75–iii82. doi: 10.1136/ard.2008.098764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelletier JP, Martel-Pelletier J, Abramson SB. Osteoarthritis, an inflammatory disease: potential implication for the selection of new therapeutic targets. Arthritis Rheum [Review] 44(6):1237–1247. doi: 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011 Sep;23(5):471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of pro-inflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011 Jan;7(1):33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Wang X, Kaplan DL. A 3D cartilage e inflammatory cell culture system for the modeling of human osteoarthritis. Biomaterials. 2011 Aug;32(24):5581–5589. doi: 10.1016/j.biomaterials.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, et al. The formation of skeletal muscle: from somite to limb. J Anat. 2003 Jan;202(1):59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knobloch TJ, Madhavan S, Nam J, Agarwal S, Jr, Agarwal S. Regulation of chondrocytic gene expression by biomechanical signals. Crit Rev Eukaryot Gene Expr. 2008;18(2):139–150. doi: 10.1615/critreveukargeneexpr.v18.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott D, Blizzard L, Fell J, Jones G. Prospective study of self-reported pain, radiographic osteoarthritis, sarcopenia progression, and falls risk in community-dwelling older adults. Arthritis Care Res (Hoboken) 2012 Jan;64(1):30–37. doi: 10.1002/acr.20545. [DOI] [PubMed] [Google Scholar]
- 22.Slemenda C, Heilman DK, Brandt KD, Katz BP, Mazzuca SA, Braunstein EM, et al. Reduced quadriceps strength relative to body weight: a risk factor for knee osteoarthritis in women? Arthritis Rheum. 1998 Nov;41(11):1951–1959. doi: 10.1002/1529-0131(199811)41:11<1951::AID-ART9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Roos EM, Herzog W, Block JA, Bennell KL. Muscle weakness, afferent sensory dysfunction and exercise in knee osteoarthritis. Nat Rev Rheumatol. 2011 Jan;7(1):57–63. doi: 10.1038/nrrheum.2010.195. [DOI] [PubMed] [Google Scholar]
- 24.Drachman D, Sokoloff L. The role of movement in embryonic joint development. Dev Biol. 1966;14:401. [Google Scholar]
- 25.Hao Y, Ma Y, Wang X, Jin F, Ge S. Short-term muscle atrophy caused by botulinum toxin-A local injection impairs fracture healing in the rat femur. J Orthop Res. 2012 Apr;30(4):574–580. doi: 10.1002/jor.21553. [DOI] [PubMed] [Google Scholar]
- 26.Rehan Youssef A, Longino D, Seerattan R, Leonard T, Herzog W. Muscle weakness causes joint degeneration in rabbits. Osteoarthritis Cartilage. 2009 Sep;17(9):1228–1235. doi: 10.1016/j.joca.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Evans CH, Liu FJ, Glatt V, Hoyland JA, Kirker-Head C, Walsh A, et al. Use of genetically modified muscle and fat grafts to repair defects in bone and cartilage. Eur Cell Mater. 2009;18:96–111. doi: 10.22203/ecm.v018a09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knapp JR, Davie JK, Myer A, Meadows E, Olson EN, Klein WH. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006 Feb;133(4):601–610. doi: 10.1242/dev.02249. [DOI] [PubMed] [Google Scholar]
- 29.Elkasrawy M, Immel D, Wen X, Liu X, Liang LF, Hamrick MW. Immunolocalization of myostatin (GDF-8) following musculoskeletal injury and the effects of exogenous myostatin on muscle and bone healing. J Histochem Cytochem. 2012 Jan;60(1):22–30. doi: 10.1369/0022155411425389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns DM, Lee PG, Uchimura T, Seufert CR, Kwon H, Zeng L. The role of muscle cells in regulating cartilage matrix production. J Orthop Res. 2010 Apr;28(4):529–536. doi: 10.1002/jor.21014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cairns DM, Uchimura T, Kwon H, Lee PG, Seufert CR, Matzkin E, et al. Muscle cells enhance resistance to pro-inflammatory cytokine-induced cartilage destruction. Biochem Biophys Res Commun. 2010 Jan 29;392(1):22–28. doi: 10.1016/j.bbrc.2009.12.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldberg RL, Kolibas LM. An improved method for determining proteoglycans synthesized by chondrocytes in culture. Connect Tissue Res. 1990;24(3–4):265–275. doi: 10.3109/03008209009152154. [DOI] [PubMed] [Google Scholar]
- 33.Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 2002;4(3):157–164. doi: 10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehling N, Palmer GD, Pilapil C, Liu F, Wells JW, Muller PE, et al. Interleukin-1beta and tumor necrosis factor alpha inhibit chondrogenesis by human mesenchymal stem cells through NF-kappaB-dependent pathways. Arthritis Rheum. 2009 Mar;60(3):801–812. doi: 10.1002/art.24352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage. 2012 Mar;20(3):223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Steinert AF, Ghivizzani SC, Rethwilm A, Tuan RS, Evans CH, Noth U. Major biological obstacles for persistent cell-based regeneration of articular cartilage. Arthritis Res Ther. 2007;9(3):213. doi: 10.1186/ar2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston JB. A simple, nondestructive assay for bound hyaluronan. J Biomed Mater Res. 2000;53(2):188–191. doi: 10.1002/(sici)1097-4636(2000)53:2<188::aid-jbm9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H. Molecular signaling and genetic pathways of senescence: Its role in tumorigenesis and aging. J Cell Physiol. 2007 Mar;210(3):567–574. doi: 10.1002/jcp.20919. [DOI] [PubMed] [Google Scholar]
- 39.Loeser RF. Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthritis Cartilage. 2009 Aug;17(8):971–979. doi: 10.1016/j.joca.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carames B, Lopez-Armada MJ, Cillero-Pastor B, Lires-Dean M, Vaamonde C, Galdo F, et al. Differential effects of tumor necrosis factor-alpha and interleukin-1beta on cell death in human articular chondrocytes. Osteoarthritis Cartilage. 2008 Jun;16(6):715–722. doi: 10.1016/j.joca.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-Armada MJ, Carames B, Lires-Dean M, Cillero-Pastor B, Ruiz-Romero C, Galdo F, et al. Cytokines, tumor necrosis factor-alpha and interleukin-1beta, differentially regulate apoptosis in osteoarthritis cultured human chondrocytes. Osteoarthritis Cartilage. 2006 Jul;14(7):660–669. doi: 10.1016/j.joca.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen BK. Muscles and their myokines. J Exp Biol. 2011 Jan 15;214(Pt 2):337–346. doi: 10.1242/jeb.048074. [DOI] [PubMed] [Google Scholar]
- 43.Lefevre S, Knedla A, Tennie C, Kampmann A, Wunrau C, Dinser R, et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat Med. 2009 Dec;15(12):1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kislinger T, Gramolini AO, Pan Y, Rahman K, MacLennan DH, Emili A. Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics. 2005 Jul;4(7):887–901. doi: 10.1074/mcp.M400182-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Goldring MB, Otero M, Plumb DA, Dragomir C, Favero M, El Hachem K, et al. Roles of inflammatory and anabolic cytokines in cartilage metabolism: signals and multiple effectors converge upon MMP-13 regulation in osteoarthritis. Eur Cell Mater. 2011;21:202–220. doi: 10.22203/ecm.v021a16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.