Summary
Algorithms designed to identify canonical yeast prions predict that ~250 human proteins, including several RNA-binding proteins associated with neurodegenerative disease, harbor a distinctive prion-like domain (PrLD) enriched in uncharged polar amino acids and glycine. PrLDs in RNA-binding proteins are essential for the assembly of ribonucleoprotein granules. However, the interplay between human PrLD function and disease is not understood. Here, we define pathogenic mutations in PrLDs of hnRNPA2/B1 and hnRNPA1 in families with inherited degeneration affecting muscle, brain, motor neuron and bone, and a case of familial ALS. Wild-type hnRNPA2 and hnRNPA1 display an intrinsic tendency to assemble into self-seeding fibrils, which is exacerbated by the disease mutations. Indeed, the pathogenic mutations strengthen a ‘steric zipper’ motif in the PrLD, which accelerates formation of self-seeding fibrils that cross-seed polymerization of wild-type hnRNP. Importantly, the disease mutations promote excess incorporation of hnRNPA2 and hnRNPA1 into stress granules and drive the formation of cytoplasmic inclusions in animal models that recapitulate the human pathology. Thus, dysregulated polymerization caused by a potent mutant ‘steric zipper’ motif in a PrLD can initiate degenerative disease. Related proteins with PrLDs must be considered candidates for initiating and perhaps propagating proteinopathies of muscle, brain, motor neuron and bone.
Elucidating the genetic basis of rare, inherited diseases can provide valuable insights to the molecular pathogenesis of common diseases. Inclusion body myopathy with frontotemporal dementia (FTD), Paget’s disease of bone and amyotrophic lateral sclerosis (ALS) (sometimes called “IBMPFD/ALS”) is a rare disorder characterized by progressive degeneration of muscle, brain, motor neurons and bone accompanied by prominent TDP-43 pathology1. Patients with this rare, inherited syndrome experience features of IBM, FTD, ALS or PDB indistinguishable from familial and sporadic cases of these disorders, and the disease may manifest in multiple tissues in the same patient1,2. Recently the name “multisystem proteinopathy” (MSP) has been adopted to reflect the expanding phenotype and prominent proteinaceous pathology that characterizes this syndrome. Some, but not all, cases of MSP are caused by mutations in the VCP gene3, which encodes the AAA+-ATPase VCP, a ubiquitin-dependent segregase. The discovery that VCP mutations cause MSP led to the subsequent discovery of pathogenic VCP mutations in more common diseases such as sporadic or familial forms of ALS2, FTD4, IBM5, and PDB6. These rare MSP families represent a unique opportunity to identify fundamental molecular defects shared among age-related diseases; thus it is highly desirable to identify additional genetic mutations responsible for this syndrome.
Identification of a pathogenic mutation in hnRNPA2B1 in VCP-negative MSP
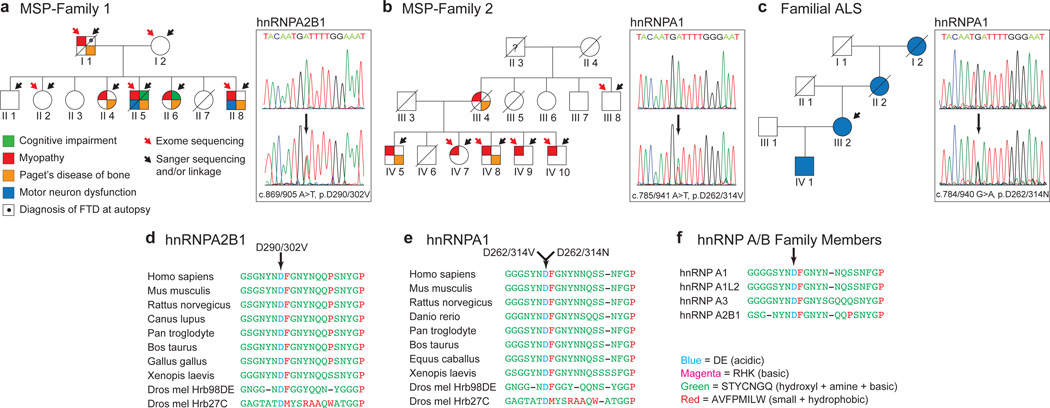
We identified a family (family 1) with dominantly inherited degeneration affecting muscle, bone, brain, and motor neurons that was clinically indistinguishable from prior families we have seen with VCP-related MSP (Fig. 1a and Supplementary Fig. 1 and Table 1). Sequencing the entire VCP gene including introns and exons in affected patients revealed no synonymous or non-synonymous variants. Genetic analysis of this family by exome sequencing and linkage analysis in parallel (Supplementary Fig. 1) identified a single novel variant (c.869/905A>T, p.D290V/302V) that co-segregated with disease and impacted the gene encoding hnRNPA2B1, a ubiquitously expressed RNA-binding protein (Fig. 1a). hnRNPA2B1 is expressed as two alternatively spliced isoforms: A2 and B1. The shorter hnRNPA2, which lacks 12 amino acids in the N-terminal region, is the major isoform accounting for ~90% of the protein in most tissues. This mutation substitutes valine for an aspartate residue that is evolutionarily conserved (Fig. 1d) and also centered in a motif that is conserved in multiple human paralogs in the hnRNP A/B family (Fig 1f and Supplementary Fig. 3).
Figure 1. Identification of novel disease mutations in MSP and ALS.
a. Family 1 pedigree indicating individuals affected by dementia, myopathy, PDB, and ALS. The causative mutation was p.D290V/302V in hnRNPA2B1. b. Family 2 pedigree indicating individuals affected by myopathy and PDB. The causative mutation was p.D262/314V in hnRNPA1. c. The pedigree of a family with ALS. The causative mutation was p.D262/314N in hnRNPA1. d–e. Sequence alignment of hnRNPA2/B1 (d) and hnRNPA1 (e) orthologs showing evolutionary conservation of the mutated aspartate and surrounding residues. f. Sequence alignment of 4 human paralogs of the hnRNP A/B family in which the disease-affected residue and surrounding residues are highly conserved.
Identification of pathogenic mutations in hnRNPA1
Additional validation of the pathogenicity of the hnRNPA2B1 mutation came from the analysis of family 2. The clinical features of this family with VCP-negative MSP have been previously described7. To identify the culprit mutation we took the same strategy as for family 1 (Supplementary Fig. 2 and Table 2). This analysis identified 5 novel SNVs and 1 indel that co-segregated with disease. Of these, a mutation in hnRNPA1 (c.785/941A>T, p.D262V/314V) stood out because it was the only variant that impacted a conserved residue, it was predicted to be deleterious, and hnRNPA1 is highly expressed in affected tissues (Fig. 1b and Supplementary Fig. 2). Moreover, this mutation in hnRNPA1 was identical to that found in hnRNPA2B1 in family 1, creating a substitution of valine for a highly conserved aspartate residue that is centered in a motif conserved in multiple human paralogs the hnRNP A/B family (Fig. 1e–f and Supplementary Fig. 3). Subsequent to the identification of these MSP-causing mutations we screened the exomes of 212 familial ALS cases for sequence variants affecting hnRNPA2B1 or hnRNPA1. In one dominantly inherited case in which known ALS genes were formally excluded we identified a mutation (c.784/940G>A; p.D262/314N) impacting the identical, conserved aspartate residue in hnRNPA1 (Fig. 1c, e–f).
Identification of disease mutations in hnRNPA2B1 and hnRNPA1 was intriguing for 3 reasons. First, hnRNPA2/B1 and hnRNPA1 directly interact with TDP-43 and function cooperatively to regulate RNA metabolism8. Second, an unbiased genetic screen previously identified fly homologs of TDP-43, hnRNPA2B1 and hnRNPA1 as suppressors of VCP-related degeneration in a Drosophila model of MSP9 (Supplementary Fig. 3). Finally, hnRNPA2/B1 was previously implicated in neurodegenerative disease. Specifically, hnRNPA2/B1 is sequestered in RNA foci in the fragile X-associated tremor ataxia syndrome (FXTAS)10, binds the expanded rCGG repeats that underlie this disease11,12, and is a genetic modifier in a Drosophila model of FXTAS11,12.
hnRNPA2B1 and hnRNPA1 pathology in MSP
Muscle biopsies from patients II5 (family 1) and IV9 (family 2) showed atrophic fibers, central nuclei and rimmed vacuoles characteristic of IBM (Supplementary Fig. 4a–c). Whereas in normal muscle hnRNPA2B1 and hnRNPA1 are exclusively nuclear (Fig. 2a, e and Supplementary Fig. 4d), analysis of muscle tissue from patient II5 (family 1) showed that hnRNPA2B1 cleared from many nuclei and accumulated in cytoplasmic inclusions in ~10% of fibers (Fig. 2b and Supplementary Fig. 4e). Muscle from this patient also exhibited TDP-43 pathology consisting of nuclear clearance and cytoplasmic inclusions, consistent with prior observations in VCP-related and sporadic IBM (Fig. 2j and Supplementary Fig. 4p)13. Interestingly, hnRNPA2B1 pathology was also observed in VCP-related and sporadic IBM (Fig. 2c, d and Supplementary Fig. 4h, i).
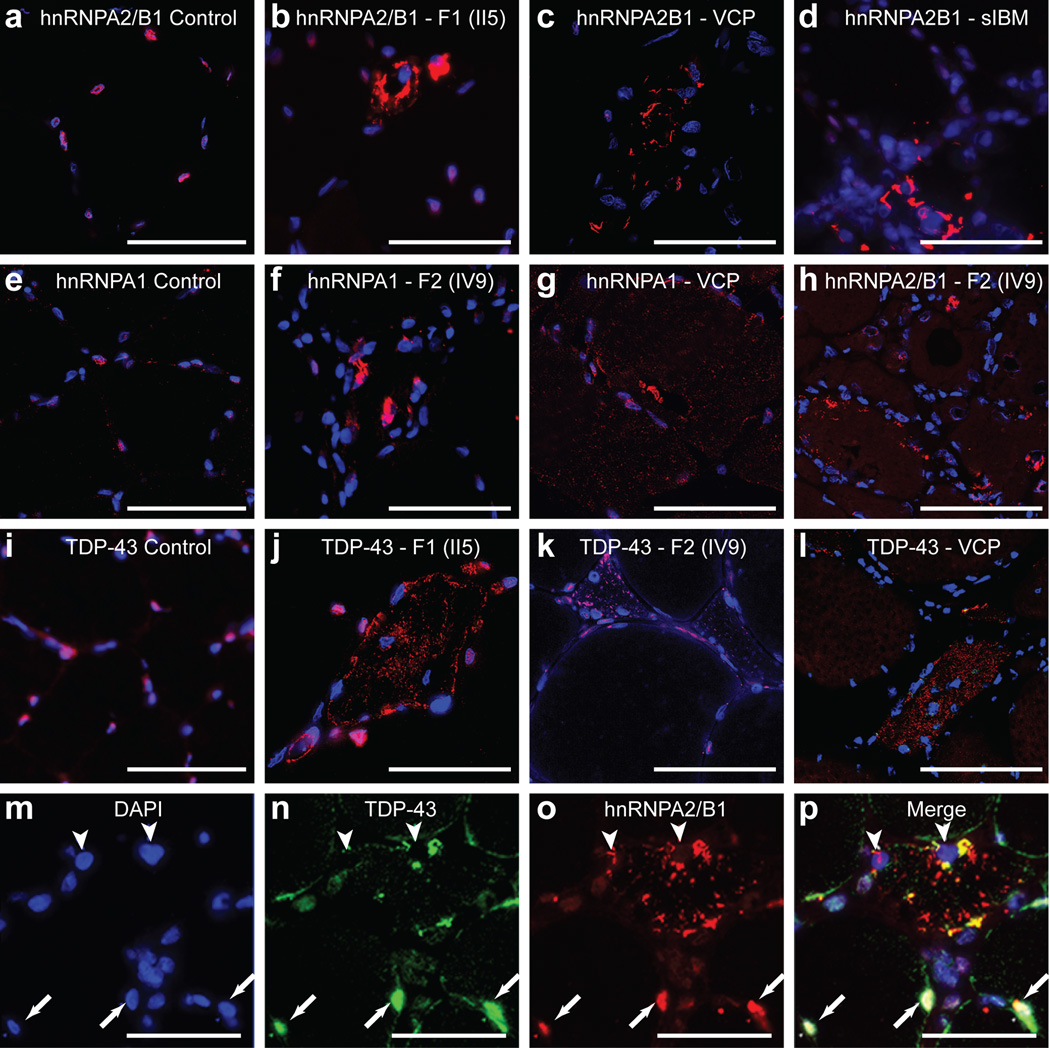
Figure 2. Cytoplasmic pathology of hnRNPA2/B1 and hnRNPA1.
a–d. Immunohistochemical analysis of hnRNPA2B1 (red) in normal muscle (a), muscle biopsy from patient II5 from family 1 (b), a patient with MSP caused by VCP mutation (R155H) (c), a patient with sporadic IBM (d). hnRNPA2/B1 (red) was cleared from DAPI-stained nuclei (blue) and accumulated in cytoplasmic inclusions (b–d). e–g. Immunohistochemical analysis of hnRNPA1 (red) in a normal muscle (e), a muscle biopsy from patient IV9 from family 2 (f), a patient with MSP caused by VCP mutation (R155H) (g). hnRNPA1 was cleared from nuclei and accumulated in cytoplasmic inclusions (f, g). h. In muscle tissue from patient IV9 (family 2), hnRNPA2/B1 was cleared from nuclei and accumulated in cytoplasmic inclusions. (i–l) Immunohistochemical analysis of TDP-43 (red) in a normal muscle (i), muscle biopsy from patient II5 from family 1 (j), muscle biopsy from patient IV9 from family 2 (k), and a patient with MSP caused by VCP mutation (R155H) (l). TDP-43 (red) was cleared from DAPI-stained nuclei (blue) and accumulated in cytoplasmic inclusions (j–l). m–p. Immunohistochemical analysis of hnRNPA2/B1 and TDP-43 colocalization in a patient with sporadic IBM. DAPI (m), TDP-43 (n), hnRNPA2/B1 (o) and merged images (p) are shown. hnRNPA2/B1 (red) and TDP-43 (green) were cleared from nuclei (arrowheads) in an atrophic muscle fiber, but found in nuclei of neighboring unaffected fibers (arrows). Scale bars represent 50 µM.
Analysis of muscle from patient IV9 (family 2) revealed nuclear clearance and cytoplasmic inclusions of hnRNPA1 also in ~10% of fibers (Fig. 2f and Supplementary Fig. 4j). hnRNPA2B1 and TDP-43 pathology were also observed in the muscle biopsy from patient IV9 (family 2) (Fig. 2h, k and Supplementary Fig. 4f, g). Moreover, hnRNPA1 pathology was also observed in VCP-related and sporadic IBM (Fig. 2g and data not shown). Therefore, regardless of etiology we have found hnRNPA2B1, hnRNPA1 and TDP-43 pathology in sporadic and familial IBM. Pathological redistribution of hnRNPA2B1, hnRNPA1, and TDP-43 was not observed in other types of muscle disease (Supplementary Fig. 4q). FUS/TLS pathology was not observed in these cases, thus pathological redistribution does not affect all hnRNPs. Double staining revealed that fibers with TDP-43 pathology typically also exhibited hnRNPA2B1 and hnRNPA1 pathology (Fig. 2m–p and Supplementary Fig. 4g, k). In these instances there was often partial co-localization of TDP-43, hnRNPA2B1 and hnRNPA1. We also observed limited immunopositivity for ubiquitin and p62 (Supplementary Fig. 4l–o).
Disease mutations impact a prion-like domain in hnRNPA2/B1 and hnRNPA1
hnRNPA2/B1 and hnRNPA1 each have C-terminal, glycine-rich domains that are essential for activity and mediate interaction with TDP-438. FoldIndex predicts these domains to be intrinsically unfolded (Fig. 3a–b). Interestingly, these low complexity domains have amino acid composition similar to that of yeast prion domains. Indeed, an algorithm devised by Alberti et al.14 predicted that the C-terminal regions of hnRNPA2 (residues 185–341) and hnRNPA1 (residues 186–320) are prion-like (Fig. 3a–d), whereas these domains narrowly missed the cutoff for prion propensity according to the Toombs et al. algorithm15. Interestingly, similar “prion-like domains” (PrLDs) are present in many hnRNPs, including TDP-43 and FUS (Supplementary Fig. 5 and Table 3). The disease-causing mutations fall at the center of the predicted PrLD, and, according to both algorithms, are expected to enhance prion-like behavior (Fig. 3a–b).
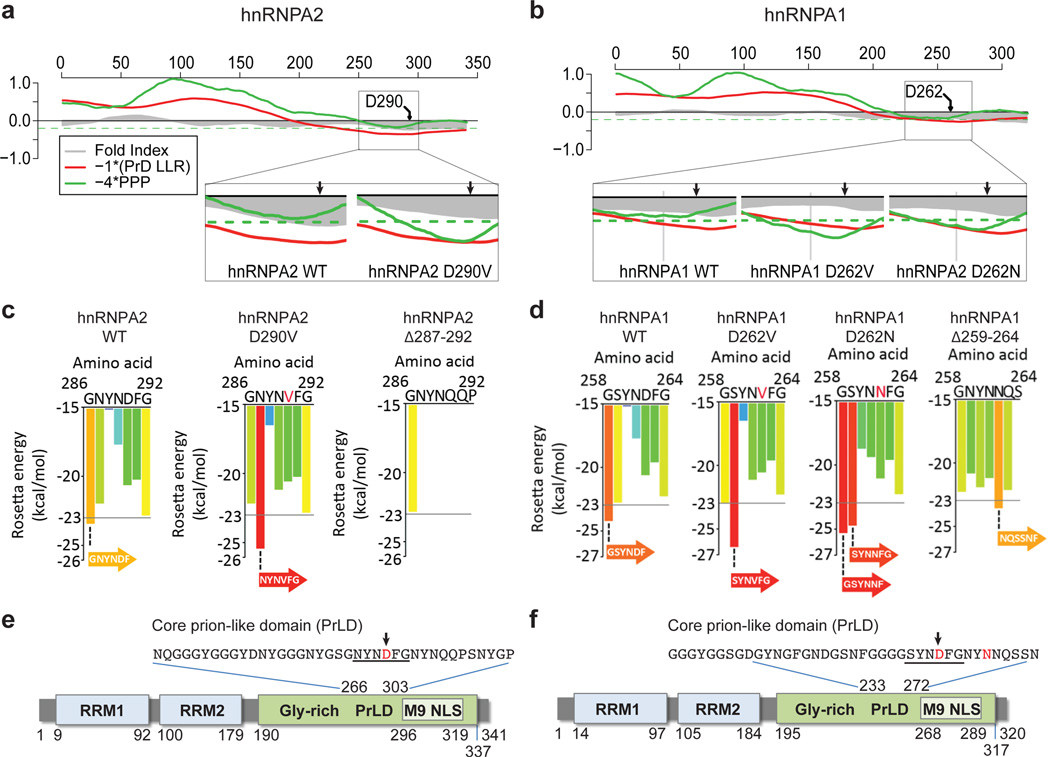
Figure 3. The disease mutations impact a PrLD in hnRNPA2/B1 and hnRNPA1.
a–b. FoldIndex predicts an extended intrinsically unfolded regions (grey curve less than zero) in the C-termini of hnRNPA2 and hnRNPA1. These regions were also predicted to be prion-like according to the algorithm of Alberti et al14 (red curve less than zero), and narrowly missed the cutoff for the prion propensity by algorithm of Toombs et al15 (green curve below the dashed green line). All curves represent averages of 41 consecutive windows of 41 amino acids, corresponding to the criteria of Toombs et al15 The disease mutations were predicted to make these domains more prionogenic (insets). c–d. ZipperDB detected 6-amino-acid stretches (underlined in e–f) within the core PrLDs for which the disease mutations increased the predicted amyloid fibril–forming potential beyond the Rosetta threshold. e–f. Domain architecture of hnRNPA2 and hnRNPA1 shows the RNA-recognition motifs (RRM1 and RRM2), the C-terminal glycine-rich domain, and an M9 nuclear localization signal. The PrLDs are centered in the C-terminal glycine-rich domain. Highly similar predictions were made for the minor isoforms of hnRNPA2B1 (hnRNPB1) and hnRNPA1 (hnRNPA1 isoform b).
We also examined hnRNPA2/B1 and hnRNPA1 with ZipperDB, a structure-based threading algorithm, which scores 6-amino acid segments for their propensity to form 2 self-complementary beta strands termed “steric zippers” that form the spine of amyloid fibrils16. Hexapeptides with Rosetta energy lower than −23kcal/mol are predicted to form steric zippers, with lower energy predicting higher amyloidogenicity. ZipperDB predicted that disease mutations increase the potency of steric zipper motifs present in the PrLDs of hnRNPA2/B1 and hnRNPA1 (Fig. 3c–f). This analysis is discussed in greater detail in Supplementary Fig. 6. Taken together, multiple algorithms predict that the PrLDs of hnRNPA2/B1 and hnRNPA1 are intrinsically disordered, but poised to access higher order self-templating structures. Notably, the disease mutations are centered within these PrLDs and predicted to promote transition to an ordered structure.
hnRNPA2 and hnRNPA1 are intrinsically fibrillization-prone and disease mutations accelerate fibrillization
We directly tested the predictions that (i) hnRNPA2 and hnRNPA1 are prone to fibrillization and (ii) this property is enhanced by disease-causing mutations. First, we experimentally assessed the ZipperDB prediction for the impact of disease mutations on steric zipper motifs found in hnRNPA2/B1 and hnRNPA1. Remarkably, the synthetic mutant hexapeptides of hnRNPA2/B1-D290V (NYNVFG) and hnRNPA1-D262V (SYNVFG) rapidly assembled into amyloid fibrils, as shown by thioflavin-T fluorescence and electron microscopy (EM), whereas the corresponding wild-type peptides did not even after several weeks (Fig. 4a–d). Thus, the disease mutations in hnRNPA2/B1 and hnRNPA1 generate highly amyloidogenic hexapeptides precisely as predicted. The more potent steric zippers that result from disease mutations are likely to be significant for two reasons. First, introduction of similarly potent steric zippers is sufficient to force fibril formation even in model proteins that do not ordinarily fibrillize17. Second, in hnRNPA1 and hnRNPA2 these potent steric zippers are centered in the intrinsically disordered PrLD and are thus available to make intermolecular contacts and drive fibril formation.
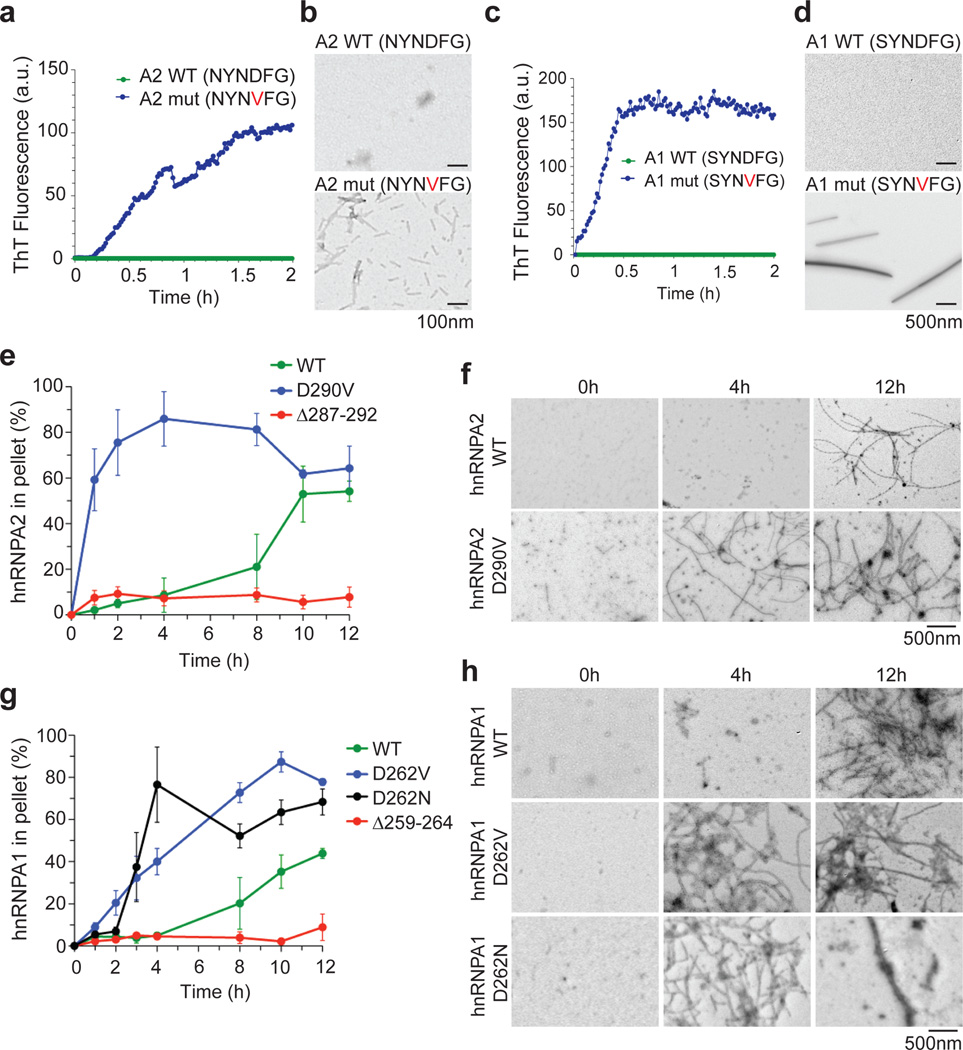
Figure 4. Disease mutations accelerate hnRNPA2 and hnRNPA1 fibrillization.
a. Synthetic hexapeptides A2 wild-type (NYNDFG) or mutant (NYNVFG) were incubated at 25°C for 2h. Fibrillization was monitored by ThT fluorescence. b. EM of A2 wild-type or mutant hexapeptides after 10min at 25°C. Bar, 0.1µm. c–d. Fibrillization analysis of A1 wild-type (SYNDFG) or mutant (SYNVFG) as in (a–b). e. Full-length hnRNPA2 WT, hnRNPA2 D290V, or hnRNP2Δ287–292 was incubated at 25 °C with agitation for 0–12h. At various times, the amount of aggregated hnRNPA2 was determined. Values represent means ± SEM (n=3). f. EM of hnRNPA2 fibrillization reactions after 0, 4, and 12h at 25°C. Note the absence of fibers after 4h for hnRNPA2 wild-type. Bar, 0.5µm. g–h. Fibrillization of full-length hnRNPA1 wild-type, hnRNPA1-D262V, hnRNPA1-D262N, or hnRNPA1Δ259–264 monitored as in (e, f).
We next assessed the fibrillization propensity of full-length proteins. We purified bacterially expressed, GST-tagged wild-type and mutant hnRNPA2 and hnRNPA1 as soluble proteins under native conditions and assessed fibrillization by sedimentation analysis and EM. Wild-type hnRNPA2 and hnRNPA1 were both intrinsically aggregation prone with a lag phase of ~4h (Fig. 4e, g). EM revealed that wild-type hnRNPA2 and hnRNPA1 form fibrils (Fig. 4f, h). The pronounced lag suggests that a critical rate-limiting step in hnRNPA2 and hnRNPA1 fibrillization is nucleation. Indeed, a small quantity (5% wt/wt) of preformed hnRNPA2 or hnRNPA1 fibrils greatly accelerated the assembly of hnRNPA2 or hnRNPA1, respectively (Supplementary Fig. 7a, e). By contrast, preformed hnRNPA2 fibrils did not seed assembly of hnRNPA1 (Supplementary Fig. 7e). Likewise, preformed hnRNPA1 fibrils did not seed assembly of hnRNPA2 (Supplementary Fig. 7a). Thus, wild-type hnRNPA1 and hnRNPA2 spontaneously form self-seeding fibrils.
Importantly, the disease mutations greatly accelerated hnRNPA2 and hnRNPA1 fibrillization (Fig. 4e–h). In all cases, the lag phase was curtailed and fibrillization was well advanced while the wild-type protein remained in lag phase. Thus, the disease mutations directly promote nucleation of hnRNPA2 and hnRNPA1 into fibrils. Moreover, fibrils formed by hnRNPA2-D290V, hnRNPA1-D262V, and hnRNPA1-D262N not only seeded their own assembly (Supplementary Fig. 7b, f, g), but also promoted fibrillization of their respective wild-type counterparts (Supplementary Fig. 7d, i). Neither hnRNPA2-D290V fibrils nor hnRNPA1-D262V fibrils seeded the assembly of TDP-43, another RNA-binding protein with a PrLD.
Finally, we assessed the importance of the steric zipper motifs in fibrillization. We deleted the steric zipper residues 287–292 from hnRNPA2 and 259–264 from hnRNAPA1 and assessed fibrillization in vitro. Importantly, neither hnRNPA2Δ287–292 nor hnRNPA1Δ259–264 formed fibrils (Fig. 4e, g), even after 24h with agitation. Moreover, hnRNPA2Δ287–292 did not fibrillize when seeded by fibrils of wild-type or mutant hnRNPA2 (Supplementary Fig. 7c), nor did hnRNPA1Δ259–264 fibrillize when seeded by fibrils of wild-type or mutant hnRNPA1 (Supplementary Fig. 7h). Thus, residues 287–292 of hnRNPA2 and 259–264 of hnRNPA1 are critical for spontaneous and seeded fibrillization of the full-length protein. Collectively, these findings indicate that steric zipper motifs centered in the PrLDs of wild-type hnRNPA2 and hnRNPA1 are critical to their intrinsic tendency to fibrillize and that disease mutations introduce more potent steric zippers, which accelerate nucleation and polymerization (Supplementary Fig. 6)
The PrLD of mutant hnRNPA2/B1 supports prion activity in yeast
Yeast prion proteins are generally modular, meaning that prion domains from one protein can be transferred to another, while retaining prion activity18. To determine whether the PrLD of hnRNPA2/B1 could support prion activity in yeast, we replaced the Sup35 nucleation domain (residues 3–40) with the core PrLD (residues 261–303) from either wild-type or mutant hnRNPA2/B1 and expressed these fusion proteins as the sole copies of Sup35 in the cell. Prion formation was detected by monitoring nonsense suppression of the ade2-1 allele and verified by monitoring a series of prion-defining criteria, including curability with guanidine, transferability with cytoduction and sensitivity to expression levels (detailed in Supplementary Fig. 8). This analysis showed that the hnRNPA2/B1 core PrLD can substitute for the Sup35 nucleation domain in supporting prion formation, and that the D290V mutation specifically promotes the nucleation activity. Moreover, full-length hnRNPA1 and A2 form cytoplasmic aggregates and are toxic in yeast (Supplementary Fig. 9).
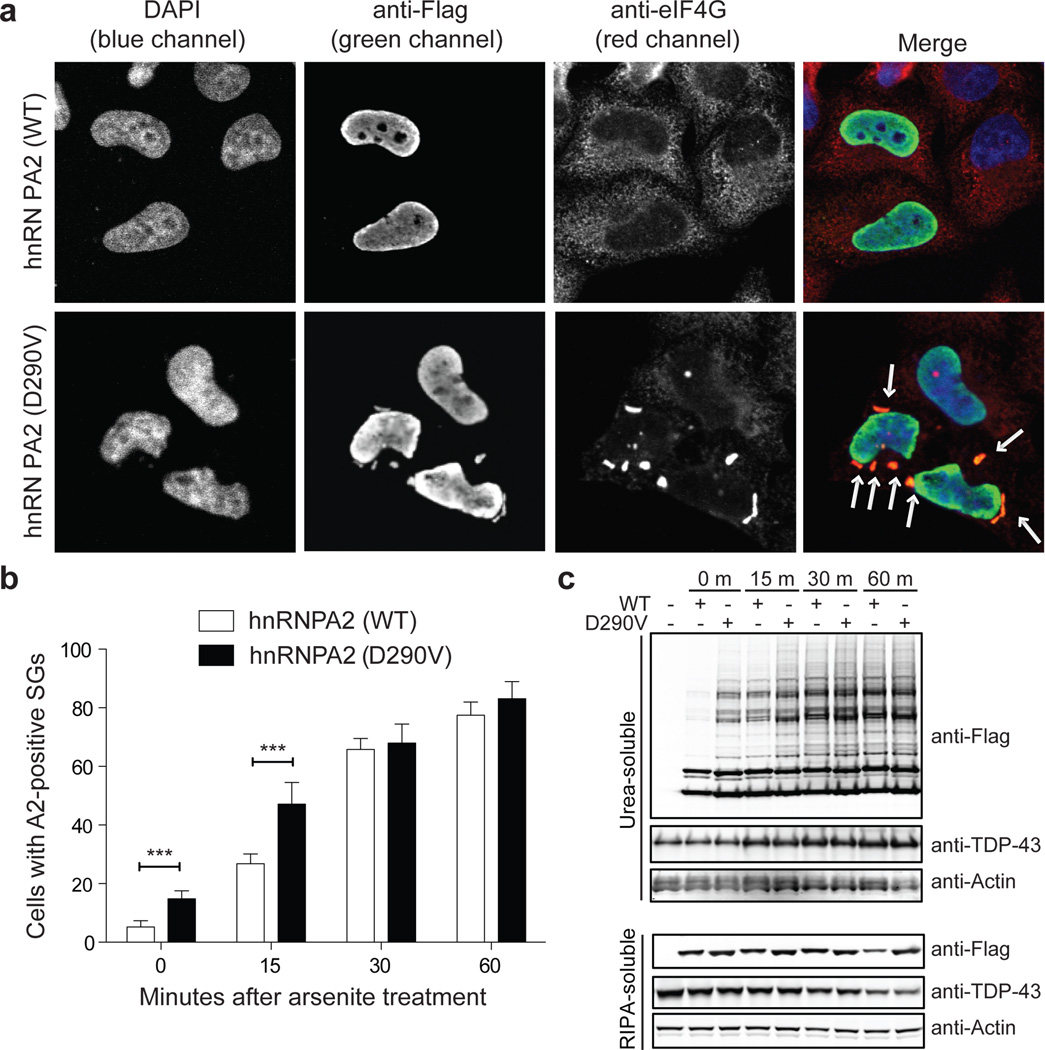
Disease mutations enhance recruitment of hnRNPA2 and hnRNPA1 to stress granules
Remarkably, the PrLDs in hnRNPA2B1 and hnRNPA1 correspond to the “low complexity sequences” found in various hnRNPs (including TDP-43 and FUS) that are essential determinants of RNA granule assembly19. Stress granules (SGs) are cytoplasmic ribonucleoprotein (RNP) granules composed of repressed translation complexes20,21. TDP-43 and FUS are recruited to SGs and this is enhanced by disease mutations20. Thus, we hypothesized that hnRNPA2B1 and hnRNPA1 would be recruited to SGs and that this would be enhanced by disease mutations. To test this, we first examined the subcellular localization of endogenous hnRNPA2/B1 in HeLa cells before and after SG induction with arsenite. At baseline, endogenous hnRNPA2/B1 was localized exclusively to nuclei (Supplementary Fig. 10). Following arsenite treatment hnRNPA2/B1 relocalized to eIF4G–positive cytoplasmic puncta, showing the recruitment of endogenous hnRNPA2/B1 to SGs (Supplementary Fig. 10).
To examine the impact of disease mutations, we expressed Flag-tagged versions of wild-type and mutant hnRNPA2. There was significantly greater incorporation of mutant hnRNPA2 into constitutive SGs than wild-type hnRNPA2 (Fig. 5a). Moreover, following arsenite treatment mutant hnRNPA2 was incorporated into SGs more rapidly than wild-type hnRNPA2, although by 30 min their degree of incorporation was similar (Fig. 5b). Both wild-type and mutant hnRNPA2 formed high molecular weight species that accumulated in the detergent-insoluble fraction (Fig. 5c). Similar to the kinetics of SG incorporation, mutant hnRNPA2 accumulated as high molecular weight species more rapidly than wild-type hnRNPA2, although by 30 min their accumulation was comparable (Fig. 5c). Similar results were obtained upon examination of endogenous hnRNPA1 as well as exogenous wild-type and mutant hnRNPA1 (Supplementary Fig. 11). Interestingly, hnRNPA2-positive and hnRNPA1-positive SGs were also immunopositive for TDP-43 (Supplementary Fig. 12). We also generated fibroblast cell lines from patients II4 and II6 (family 1). In these cells endogenous mutant hnRNPA2 accumulated in constitutive SGs that were immunopositive for eIF4G in addition to TDP-43 and VCP (Supplementary Fig. 13).
Figure 5. hnRNPA2 recruitment to SGs is accelerated by disease mutation.
a–b. HeLa cells were transfected with Flag-tagged wild-type or mutant hnRNPA2 and stained with anti-Flag (green), anti-eIF4G (red), and DAPI (blue). Arrows indicate hnRNPA2- and eIF4G–positive SGs. b. HeLa cells were transfected as in (a), treated with 0.5 mM sodium arsenite for the indicated time, and immunostained as in (a). The percentage of cells displaying hnRNPA2-positive SGs at indicated time points following treatment with arsenite cells are plotted. Data represent mean ± SEM (n=3) (*** P<0.001). c. HeLa cells were transfected and stimulated as in (b), and sequentially extracted with RIPA and urea buffer. Immunoblotting was conducted with anti-Flag and anti-TDP-43 antibodies.
Mutant hnRNPs accumulate in pathological inclusions in vivo
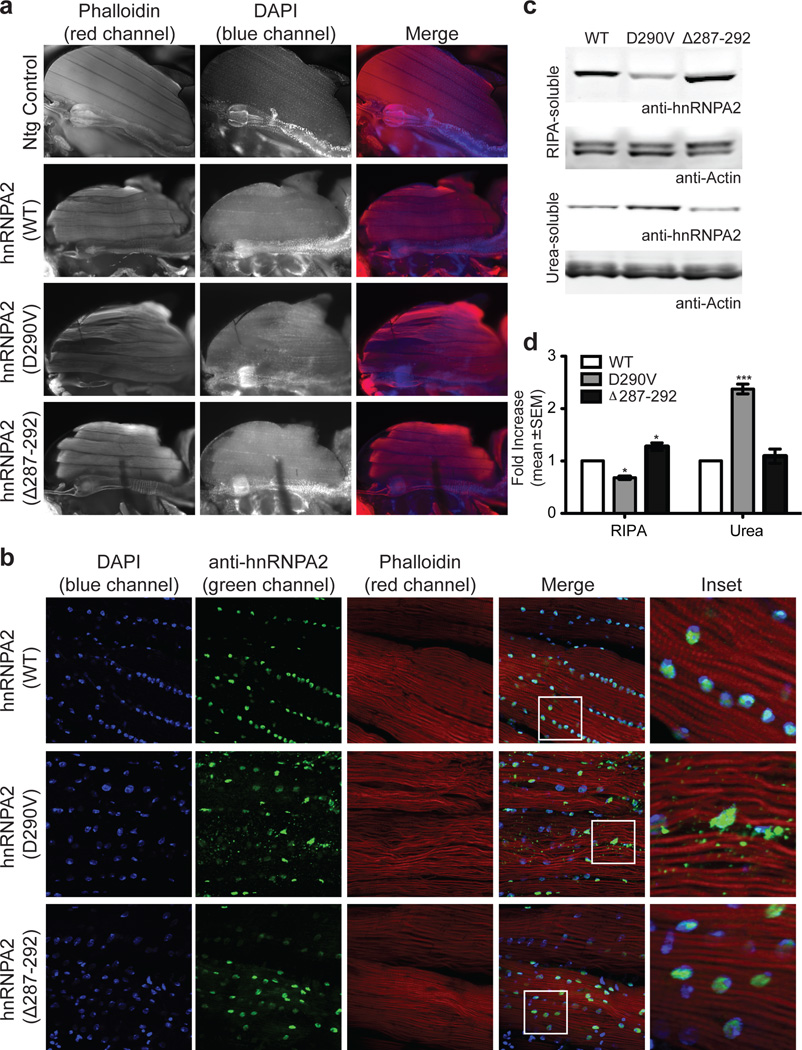
We generated transgenic Drosophila expressing wild-type or mutant forms of human hnRNPA2 and hnRNPA1, as well as wild-type or mutant forms of the fly homolog Hrb98DE (Supplementary Figs. 14 and 15). Expression of wild-type hnRNPA2 in Drosophila indirect flight muscle led to mild degeneration affecting the rostral portion of several muscles, whereas expression of hnRNPA2-D290V caused severe degeneration that affected all muscles (Fig. 6a). Importantly, flies over-expressing hnRNPA2Δ287–292, in which the hexapeptide (NYNDFG) in the PrLD was deleted, had virtually normal muscles (Fig. 6a). This result indicates that the severe toxicity found in flies over-expressing mutant hnRNPA2 requires the presence of this potent steric zipper motif.
Figure 6. Mutant hnRNPA2 forms cytoplasmic inclusions in Drosophila.
a. Adult flies were dissected to expose the dorsal longitudinal indirect flight muscle and stained with Texas Red-phalloidin (red), and DAPI (blue). Flies expressing human wild-type hnRNPA2 under control of the MHC-GAL4 driver showed mild degeneration, whereas flies expressing mutant human hnRNPA2 show severe degeneration affecting all muscles. Flies expressing hnRNPA2Δ287–292 show muscle histology similar to flies expressing wild-type hnRNPA2. b. wild-type hnRNPA2 localizes exclusively to nuclei, whereas hnRNPA2-D290V also accumulates extensively in cytoplasmic inclusions. hnRNPA2Δ287–292 localizes exclusively to nuclei. c. Thoraces of adult flies were dissected and sequential extractions were performed to examine the solubility profile of hnRNPA2. d. Quantification of the blot shown in c. Data represent mean ± SEM (n=3) (*P < 0.05, ***P < 0.001 by two-way ANOVA with Bonferroni’s post hoc test).
Immunohistochemical analysis showed that wild-type hnRNPA2 localized appropriately to nuclei, as did hnRNPA2Δ287–292, whereas hnRNPA2-D290V largely accumulated in cytoplasmic inclusions (Fig. 6b). We observed a correlation between cytoplasmic inclusion formation and solubility of hnRNPA2. Specifically, mutant hnRNPA2-D290V protein was largely recovered from the detergent-insoluble fraction, whereas wild-type hnRNPA2 and hnRNPA2Δ287–292 proteins were mostly found in the detergent-soluble fraction (Fig. 6c–d). Thus, the degree of muscle degeneration in flies expressing hnRNPA2 correlates with the extent of cytoplasmic inclusions and with hnRNPA2 solubility. Entirely consistent results were obtained upon expression of wild-type and mutant versions of hnRNPA1 and Hrb98DE (Supplementary Fig. 15).
Lastly, we evaluated the impact of the disease mutation on hnRNPA2 cellular localization in mammalian muscle. We electroporated mouse tibialis anterior with plasmid expressing Flag-tagged versions of wild-type or mutant hnRNPA2. Wild-type hnRNPA2 was localized appropriately within nuclei, whereas mutant hnRNPA2-D290V was excluded from nuclei and accumulated in cytoplasmic inclusions (Supplementary Fig. 16) highly reminiscent of the pathology seen in MSP patients.
Discussion
Polymerization of PrLDs underlies the orderly, regulated phase transition that drives assembly of nonmembrane-bound organelles including RNA granules, which serve as "reaction crucibles" in which many aspects of RNA metabolism occur22. Disease mutations introduce more potent steric zippers into the PrLDs of hnRNPA2 and hnRNPA1, dysregulating and accelerating nucleation and polymerization, altering the dynamics of RNA granule assembly, which likely has adverse consequences for RNA metabolism. In addition to hnRNPA2 and hnRNPA1, at least four other RNA-binding proteins that harbor PrLDs (TDP-43, FUS, EWSR1, and TAF15) accumulate in disease pathology23. By analogy to hnRNPA2/B1 and hnRNPA1, we speculate that these and perhaps other PrLD-containing proteins contribute to the initiation or propagation of disease by a similar mechanism. Indeed, ~250 human proteins are predicted to harbor PrLDs, many of which are RNA-binding proteins (Supplementary Fig. 5 and Table 3), and must be considered candidates for contributing to degenerative disease.
Thus, disease could ensue from unregulated polymerization initiated spontaneously by PrLDs upon environmental stress, by a mutation impacting the PrLD, or by a mutation impacting another factor (e.g. VCP) that regulates the ribonucleoprotein granule assembly with altered metabolism of RNA as a consequence.
Notably, diseases associated with pathological inclusions of PrLD-containing proteins frequently show “spreading” pathology, in which cellular degeneration with intracellular inclusions starts in one epicenter and subsequently spreads to neighboring tissue24. While not directly addressed here, this study suggests that cell-to-cell transmission of a self-templating conformer could contribute to the spreading pathology that is characteristic of these diseases.
The discovery of VCP mutations in MSP1 led to the subsequent discovery of pathogenic VCP mutations in sporadic or familial forms of ALS2, FTD4, IBM5, and PDB6. Therefore, subsequent to the discovery of mutations in hnRNPA2B1 and hnRNPA1 underlying MSP, we screened 212 familial ALS cases and identified a pathogenic hnRNPA1 mutation in one family, as described above. As this manuscript was being assembled for publication we completed screening of 305 sporadic ALS cases and identified a nonsynonymous variant in hnRNPA1 (c.800/956A>G, p.N267/319S) in one classic, late-onset case in which mutations in known ALS genes had been excluded (Supplementary Figure 17). Although not definitive, this variant is likely pathogenic since it is centered in the core PrLD and introduces a potent steric zipper similar to that introduced by D262V/N mutations. The frequency of hnRNPA2B1 and hnRNPA1 mutations in ALS, and the possibility that mutations in these genes underlie some sporadic and familial forms of FTD, IBM, and PDB, is will be important to address in the future.
Methods
Patients
Patients were examined by M.B. and J.P.T. except where indicated. Review Board (IRB) of the University of Miami approved the study protocol and all participants provided written informed consent.
Exome capture and next-generation sequencing
Exome enrichment of 3 µg of genomic DNA was performed by using the Agilent SureSelect Human All Exon kit capture library (G3362 for 5 DNA samples from family 1 and G3370 for 5 DNA samples from family 2) according to the manufacturer’s protocol (SureSelect Human All Exon Illumina Paired-End protocol version 1.0.1). The capture library, containing regions totaling approximately 38 Mb (G3362) or 50 Mb (G3370), is designed to target all human exons in the NCBI consensus coding sequence (CCDS) database. Captured DNAs were sequenced on an Illumina GAIIx sequencer (Illumina, San Diego, CA) with 76 bp (family 1) or 100 bp (family 2) paired-end reads. At least 2 lanes of sequencing data were collected for each sample to generate sufficient coverage. Image analyses and base calling were performed by using the Illumina Genome Analyzer Pipeline software (GAPipeline version 1.5 or higher) with default parameters. Reads were aligned to a human reference sequence (UCSC assembly hg19, NCBI build 37) and genotypes were called at all positions at which there were high-quality sequence bases (Phred-like score Q25 or greater) at minimum coverage of 8, using CLC Genomics Workbench v4.5.1 (CLC Bio, Cambridge, MA). For each sample, more than 50% of at least 100 million reads were uniquely mapped to the targeted human exon regions to give mean depth of coverage of 123. Under such coverage, approximately 94% of targeted regions were covered by 5 reads or more, and more than 85% were covered by more than 20 reads (Supplementary Fig. 1). To identify the pathogenic mutations, we performed a series of filtering steps (Supplementary Fig. 1). We first discarded the variants that did not change the amino acid sequence. We then generated a list of variants that are present in affected individuals but absent in nonaffected individuals. Variants previously reported in dbSNP132 were excluded, as the causative variants were uncommon and unlikely to be registered in the SNP database. The variants were further filtered by using the in-house database (a collection of variants from whole-exome sequencing of more than 625 individuals) by a similar methodology. Finally Sanger sequencing was performed on DNAs of 3 additional members (both affected and nonaffected) from each family to exclude the nonpathogenic mutations. Presence of mutations was confirmed in all candidate disease-causing variants by Sanger sequencing.
Linkage analysis
Eight DNA samples (parents and 6 siblings) from family 1 were genotyped using the Affymetrix Genome-Wide Human SNP Array 6.0 (Santa Clara, CA, USA). SNP genotype calls were performed according to the standard Affymetrix protocols. A subset of SNPs chosen on the basis of heterozygosity was used for linkage analysis, using dChip Software (www.biosun1.harvard.edu/complab/dchip/) under Parametric Linkage Analysis and a dominant model with 99% penetrance in heterozygotes and a disease allele frequency of 0.001. Five genomic segments with a size greater than 100 kb, at least 20 SNP markers, and an LOD score greater than 0 are shown in Supplementary Fig. 1.
Generation of phylogram for hnRNP A/B gene family
TBPH of D. melanogaster was used to BLAST against the RefSeq protein database including species of M. musculus, D. melanogaster, A. gambiae, H. sapiens, D. Rerio (zebrafish), C. elegans and G. gallus (jungle fowl). The top 100 sequences (according to BLAST E-Values) were obtained. Sequences sharing more than 98% similarity were filtered by UCLUST25 because they likely represent alternatively spliced variants of the same genes. Eighty-one sequences remaining after this process were aligned using the MAFFT multiple sequence alignment program26. The DNA coding sequences of all proteins were extracted and threaded onto the protein sequences to form an in-frame codon alignment. Squint27 was used to extract the DNA coding alignment of the two RRM domains present in each protein using the protein alignment as a guide. Phylogenetic trees of the codon alignment including twenty-one genes from only human and drosophila were generated using the genetic algorithm-based GARLI28 method.
Constructs
cDNAs encoding human hnRNPA2 (accession number NM_002137.2) or hnRNPA1 (accession number NM_002136.1) in the plasmid pCMV6-XL5 were obtained from OriGene. A Flag epitope tag was added to the 5’-end of hnRNPA2 or hnRNPA1 by PCR. The cDNAs were then subcloned into a pcDNA 3.1 plasmid, using restriction sites EcoRI and XhoI, creating pcDNA 3.1/Flag-hnRNPA2 or hnRNPA1. cDNAs encoding drosophila Hrb98DE (accession number NM_170373.1) was obtained from Drosophila Genomics Research Center. Missense mutations of hnRNPA2 (hnRNPA2 D290V), hnRNPA1 (hnRNPA1 D252V) and drosophila Hrb98DE (Hrb98DE D302V) were generated by site-directed mutagenesis. Sequence analysis was conducted on all plasmid constructs.
Antibodies
The following commercial antibodies were used in this study: mouse monoclonal anti-hnRNPA2/B1 antibodies (EF-67 and DP3B3) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); rabbit polyclonal anti-TDP-43 antibody (Protein Tech Group, Chicago, IL); anti-eIF4G antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); anti-hnRNPA1 antibody (Cell Signaling Technology, Inc., Danvers, MA); anti-VCP antibody (Epitomics, Inc., Burlingame, CA); anti-Flag M2 antibody (Sigma). Anti-hnRNPA1 (4B10 and 9H10) antibodies were generous gifts from Dr. G. Dreyfuss. Validation of these antibodies is shown in Supplementary Fig. 18.
Cell culture and transfection
HEK293 cells and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% L-glutamate. Human fibroblasts were grown in DMEM: nutrient mixture F-12 (DMEM/F12) supplemented with 20% FBS, 1% penicillin/streptomycin, and 1% L-glutamate. Cells were transfected using Lipofectamine LTX with Plus Reagent (Invitrogen), according to the manufacturer’s instructions.
RNAi
siRNAs against hnRNPA2/B1 were ordered from Santa Cruz Biotechnology (sc-43841, target sequences: CAGUUCCGUAAGCUCUUUAtt, GGAUGGCUAUAAUGGGUAUtt and GGAUCAUGGUGUAAUAAGAtt) and Dharmacon (ON-TARGETplus SMARTpool, Human HNRPA2B1 (3181), target sequences: CGGUGGAAAUUUCGGACCA, GCUGUUUGUUGGCGGAAUU, GGAGAGUAGUUGAGCCAAA and GAGGAGGAUCUGAUGGAUA). Control siRNAs were ordered from Dharmacon (ON-TARGETplus Non-targeting siRNA #1 (# D-001810-01–05). HeLa cells were transfected in 6-well plates with Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s protocol and collected after 48 and 72 h.
Immunofluorescence studies
Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.5% Triton X-100 in PBS for 10 min, blocked with 5% goat serum in PBS for 30 min, and incubated with primary antibody for 2 h at room temperature or overnight at 4 °C. Primary antibodies were visualized with secondary antibodies conjugated with Alexa Fluor 488 and Alexa Fluor 594 (Molecular Probes, Invitrogen), and nuclei were detected using DAPI. Stained cells were examined using a confocal microscope (Zeiss LSM 510 NLO Meta) with Zeiss ZEN software.
Solubility and biochemical analyses
Sequential extractions were performed to examine the solubility profile of hnRNPA2/B1. Cells were washed twice with PBS, lysed in cold RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS, and 1 mM EDTA), and sonicated. Cell lysates were cleared by centrifugation at 100,000 g for 30 min at 4 °C to generate RIPA-soluble samples. To prevent carry-overs, the resulting pellets were washed twice with PBS (i.e., resonicated and recentrifuged). Only supernatants from the first centrifugation were analyzed. RIPA-insoluble pellets were then extracted with urea buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, pH 8.5), sonicated, and centrifuged at 100,000 g for 30 min at 22 °C. Protease inhibitors were added to all buffers before use. Protein concentration was determined by the bicinchoninic acid method (Pierce), and proteins were resolved by NuPAGE Novex 4–12% Bis-Tris Gel (Invitrogen).
In vivo electroporation of mice
All animal work was performed with approval of the St. Jude Children’s Research Hospital Committee on Animal Care. Mice were anesthetized and the skin overlying the tibialis anterior (TA) muscle was shaved, and the animals were injected with endotoxin-free Flag-hnRNPA2 wild-type or D290V mutant diluted in sterile PBS to a volume of 50 µl by using a 0.5 ml syringe fitted with a 29-gauge needle. Two-needle array electrodes (BTX, San Diego, California) were inserted into the muscle immediately after DNA delivery for electroporation. The distance between the electrodes was 5 mm, and the array was inserted longitudinally relative to the muscle fibers. In vivo electroporation parameters were as follows: voltage, 75 V; pulse length, 50-ms; number of pulses, 6; pulse interval, 200 ms; desired field strength, 200 V/cm, given by a BTX ECM830 Electro Square Porator. After 7 days of recovery, mice were processed for immunofluorescence analysis.
Muscle histochemistry and immunohistochemistry
Human samples were processed for routine histochemical analysis as previously described13. For skeletal muscle immunohistochemistry, isolated skeletal muscle was mounted by using tragacanth gum and quick frozen in liquid nitrogen–cooled 2-methylbutane. Sections (8 µM) were placed on slides and fixed in 4% paraformaldehyde for 5 min and then ice-cold acetone for 5 min. Sections were blocked in PNB (Perkin Elmer) for 1 h and then incubated overnight in primary antibody diluted in PNB. After serial washes in 1X PBS, slides were incubated in secondary antibody diluted in PNB for 2 h. Sections were mounted with MOWIOL containing DAPI. Specimens were examined using a fluorescent microscope (80i upright; Nikon) and charge-coupled device camera (EZ monochrome; Roper Industries) with deconvolution software analysis (NIS Elements; Nikon). Nonfluorescent images were taken with a 5-megapixel color charge-coupled device (Nikon). Image processing and analysis were performed with NIS Elements 4.0 software and Photoshop CS3 (Adobe).
Yeast strains, media, and plasmids
Yeast cells were grown in rich media (YPD) or in synthetic media lacking uracil and containing 2% glucose (SD/-Ura), raffinose (SRaf/-Ura), or galactose (SGal/-Ura). The TDP-43 and FUS yeast expression constructs have been previously described29–31. WT and IBMPFD-associated mutant HNRNPA2B1 and HNRNPA1 Gateway® entry clones were generated by PCR, incorporating the flanking Gateway attB1 and attB2 sites along with a Kozak consensus sequence. Resulting PCR products were shuttled into pDONR221, using a Gateway BR reaction. Two versions of each entry clone (with or without stop codon) were then used in LR reactions with pAG416Gal-ccdB, pAG426Gal-ccdB, pAG416Gal-ccdB-GFP, or pAG426Gal-ccdB-GFP32 to generate the CEN and 2 µm untagged or YFP-fusion constructs. Primer sequences are available upon request. To prevent unwanted recombination events owing to unstable repetitive sequences in the HNRNPA2B1 and HNRNPA1 DNA sequence, we propagated plasmids in SURE-2 or STBL-3 E. coli at 30 °C. For testing the phenotype of WT and IBMPFD-associated mutant hnRNPA2 as Sup35-fusion proteins, PCR was used to amplify amino acids 261–303 of hnRNPA2 and insert it into Sup35 in the place of amino acids 3–40. The resulting PCR products were cotransformed with BamHI/HindIII-cut pJ526 (from Dan Masison, National Institutes of Health; Ref. 33) into strain 780-1D (MATα kar1-1 SUQ5 ade2-1 his3Δ202 leu2Δ1 trp1Δ63 ura3-52 sup35::KanMX) containing the SUP35 maintainer plasmid pJ533 (from Dan Masison; Ref. 34). Transformants were selected on medium lacking leucine, and then transferred to 5-fluoroorotic acid-containing medium to select for loss of pJ533. Sequences were confirmed by DNA sequencing. Overexpression plasmids were generated by PCR amplification, restriction digest, and cloning into compatible sites in pKT24 (from Kim Taylor, NABI, Rockville, MD), which contains a GAL1 promoter, as previously described 35.
Yeast transformation and spotting assays
Yeast procedures were performed according to standard protocols36. The PEG/lithium acetate method was used to transform yeast with plasmid DNA37. For spotting assays, yeast cells were grown overnight at 30 °C in liquid media containing raffinose (SRaf/-Ura) until they reached log or mid-log phase. Cultures were then normalized for OD 600, serially diluted, spotted onto synthetic solid media containing glucose or galactose lacking uracil, and grown at 30 °C for 2–3 days. HNRNPA2B1-Sup35 fusions were tested for prion formation and for stability and curability of the Ade+ phenotype as previously described35. Cytoductions were performed as previously described38. The recipient strain was YER746 (MATa kar1-1 SUQ5 ade2-1 his3 leu2 trp1 ura3 arg1::HIS3 sup35::KanMx), carrying plasmid pER697 (URA3), which expresses the D290V A2-Sup35 fusion from the SUP35 promoter.
Fluorescence microscopy for yeast
For fluorescence microscopy experiments, single colony isolates of yeast strains were grown to mid-log phase in SRaf/-Ura media at 30 °C. Cultures were spun down and resuspended in the same volume of SGal/-Ura to induce expression of the HNRNPA2B1 or HNRNPA1 constructs. Cultures were induced with galactose for 4–6 h and processed for microscopy. Images were obtained by an Olympus IX70 inverted microscope and a Photometrics CoolSnap HQ 12-bit CCD camera. Z-stacks of several fields were collected for each strain. The images were deblurred using a nearest neighbor algorithm in the Deltavision Softworx software and representative cells were chosen.
Hexapeptide assembly
The A2 WT (NYNDFG), A2 D290V (NYNVFG), A1 WT (SYNDFG) and A1 D262V (SYNVFG) hexapeptides were synthesized at the Keck Biotechnology Resource Laboratory at Yale University School of Medicine. A1 peptides were dissolved at 1 mM and A2 peptides at 5 mM in 150 mM KCl, 40 mM HEPES-KOH, pH 7.4, and 1 mM DTT and used immediately for assembly reactions. Fiber assembly reactions were monitored at λex/em 440/482 nm, in the presence of 25 µM Thioflavin-T (ThT), at room temperature on a Tecan Safire2 or Tecan Infinite M1000 plate reader. Alternatively, reactions were processed for electron microscopy as for the full-length hnRNPs (see below).
hnRNP purification
WT and mutant hnRNPA2 or hnRNPA1 were expressed and purified from Escherichia coli as GST-tagged proteins. Expression constructs were generated in pDuet to contain a TEV-cleavable site, resulting in a GST-TEV-hnRNP construct30,31. GST-TEV-hnRNP was overexpressed in E. coli BL21 (DE3) RIL cells (Agilent) and purified under native conditions using a glutathione-sepharose column (GE) according to the manufacturer’s instructions. Proteins were eluted from the glutathione sepharose with assembly buffer (AB: 40 mM HEPES-KOH, 150 mM KCl, 5% Glycerol, 20 mM glutathione, pH 7.4). Protein was centrifuged for 10 min at 16,100 g, and supernatant was separated from pellet to remove any protein aggregates. Protein concentration was determined by Bradford assay (Bio-Rad) in comparison to BSA standards.
In vitro fibril formation assay
For GST-tagged protein, aggregation was initiated by adding TEV protease (Invitrogen) to GST-TEV-hnRNPA2 (3µM) or GST-TEV-hnRNPA1 (5µM) in AB. Aggregation reactions were incubated at 25°C for 0–12 h with or without agitation at 1200 rpm in an Eppendorf Thermomixer. For self-seeded and cross-seeded reactions, hnRNPA2, hnRNPA2 D290V, hnRNPA1, or hnRNPA1 D262V fibrils were assembled at 5µM for 24h with agitation at 1400rpm in AB. These preformed fibrils then used to seed (5% wt/wt) the assembly of hnRNPA2 (2.5µM), hnRNPA2 D290V (2.5µM), hnRNPA1 (5µM), hnRNPA1 D262V (5µM), or hnRNPA1 D262N (5µM) in AB as indicated. Here, assembly reactions were agitated at 1,200rpm at 25°C. For hnRNPA2 cross-seeding reactions the amount of preformed fibrils added was increased to 10% wt/wt and reactions were not agitated. In hnRNPA1 cross-seeding reactions, preformed fibrils were briefly sonicated to fragment fibrils prior to addition to the assembly reaction. For hnRNPA1 cross-seeding reactions the amount of preformed fibrils added was 5% wt/wt and reactions were not agitated. For sedimentation analysis, samples were centrifuged at 16,100 g for 10 min at 25 °C. Pellet fractions were resolved by SDS-PAGE and stained with Coomassie Blue. The amount of protein in the pellet fraction was determined by densitometry in comparison to known amounts of hnRNP. For electron microscopy (EM) of in vitro aggregation reactions, samples (10 µl) were adsorbed onto glow-discharged 300-mesh Formvar/carboncoated copper grid (Electron Microscopy Sciences) and stained with 2% (w/v) aqueous uranyl acetate. Excess liquid was removed, and grids were allowed to air dry. Samples were viewed by a JEOL 1010 transmission electron microscope.
Fly stocks and culture
The hnRNPA2 wild-type and D290V cDNAs were subcloned into pUASTattB plasmid, using restriction sites EcoRI and XhoI, creating pUASTattB/hnRNPA2 WT or D290V. Hrb98DE wild-type and D302V were subcloned into pUASTattB plasmid using NotI and XhoI sites. Flies carrying pUASTattB transgenes were generated by a standard injection and φC31 integrase-mediated transgenesis technique. To express a transgene in muscles, MHC-GAL4 was used (from Dr. Guillermo Marqués). All Drosophila stocks were maintained in a 25 °C incubator with a 12 h day/night cycle.
Adult fly muscle preparation and immunohistochemistry
Adult flies were embedded in a drop of OCT compound (Sakura Finetek) on a slide glass, frozen with liquid nitrogen, and bisected sagitally by a razor blade. After fixing with 4% paraformaldehyde in PBS, hemithoraces were stained by Texas Red-X-Phalloidin (Invitrogen) and DAPI according to manufacturer’s instructions. Stained hemithoraces were mounted in 80% glycerol, and the musculature was examined by DMIRE2 (Leica, 10X). For hnRNPA2 staining, hemithoraces were permeabilized with PBS containing 0.2% Triton X-100 and stained with anti-hnRNPA2/B1 (EF-67) antibody (Santa Cruz Biotechnology) and Alexa 488 conjugated secondary antibody (Invitrogen). Stained muscle fibers were dissected and mounted in Fluormount-G (Southern Biotech) and imaged with a Marianas confocal microscope (Zeiss, 63X)
Fly western blotting
Thoraces of adult flies were prepared and ground in PBS containing 0.2% Triton X-100. After adding SDS sample buffer, samples were boiled for 5 min and analyzed by the standard Western blotting method provided by Odyssey system (LI-COR) with 4–12% NuPAGE Bis-Tris gel (Invitrogen) and anti-hnRNPA2/B1 antibody (Santa Cruz, 1:1000).
Supplementary Material
Acknowledgements
We thank the patients whose participation made this work possible. We thank the St. Jude Pediatric Cancer Genome Project and Jinghui Zhang in particular for providing access to control sequencing data. We thank Cinzia Gellera, Bob Baloh, Matt Harms, Sabine Krause, Gideon Dreyfuss and Tim Cundy for sharing reagents. We thank Sandra Donkervoort and Steven Mumm for coordinating samples, and Ana Taylor for editorial assistance. JPT was supported by ALSAC, the Packard Foundation and NIH NS053825; JPT and MB were supported by the ALS Association; JQT was supported by NIH AG032953; JS was supported by NIH DP2OD002177 and NS067354 and the Ellison Medical Foundation; EDR was supported by NSF MCB-1023771.
Footnotes
Author Contributions
H.J.K., N.C.K., E.D.R., C.C.W., J.S. and J.P.T. designed experiments. H.J.K., N.C.K., E.A.S., J.M., Z.D., K.S.M., B.F., S.L., A.M., A.P.K., Y.R.L. and A.F.F. performed the experiments. K.B.B., A.M.W., R.R., J.L.P., S.A.G., J.Q.T., B.N.S., S.T., A.S.G., J.M., C.E.S., M.K., J.K., A.P., M.B. and V.E.K. provided patient clinical material, clinical evaluation, or evaluation of patient clinical material. H.J.K., N.C.K., Y.D.W., R.C., B.J.T., A.D.G., O.D.K., E.D.R., J.S. and J.P.T. contributed to data analysis. E.D.R., O.D.K. and C.C.W. contributed to manuscript preparation. H.J.K., J.S. and J.P.T. wrote the manuscript.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Nalbandian A, et al. The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J Mol Neurosci. 2011;45:522–531. doi: 10.1007/s12031-011-9627-y. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 4.Neumann M, Tolnay M, Mackenzie IR. The molecular basis of frontotemporal dementia. Expert Rev Mol Med. 2009;11:e23. doi: 10.1017/S1462399409001136. [DOI] [PubMed] [Google Scholar]
- 5.Shi Z, et al. Characterization of the Asian myopathy patients with VCP mutations. Eur J Neurol. 2011 doi: 10.1111/j.1468-1331.2011.03575.x. [DOI] [PubMed] [Google Scholar]
- 6.Chung PY, et al. Indications for a genetic association of a VCP polymorphism with the pathogenesis of sporadic Paget’s disease of bone, but not for TNFSF11 (RANKL) and IL-6 polymorphisms. Mol Genet Metab. 2011;103:287–292. doi: 10.1016/j.ymgme.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Kottlors M, et al. Late-onset autosomal dominant limb girdle muscular dystrophy and Paget’s disease of bone unlinked to the VCP gene locus. J Neurol Sci. 2010;291:79–85. doi: 10.1016/j.jns.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Buratti E, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 9.Ritson GP, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwahashi CK, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 11.Sofola OA, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin P, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salajegheh M, et al. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve. 2009;40:19–31. doi: 10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng PK, Eisenberg D. Short protein segments can drive a non-fibrillizing protein into the amyloid state. Protein Eng Des Sel. 2009;22:531–536. doi: 10.1093/protein/gzp037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 19.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Neumann M, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 25.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 26.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 27.Goode MG, Rodrigo AG. SQUINT: a multiple alignment program and editor. Bioinformatics. 2007;23:1553–1555. doi: 10.1093/bioinformatics/btm128. [DOI] [PubMed] [Google Scholar]
- 28.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph. D. dissertation, The University of Texas at Austin. 2006 [Google Scholar]
- 29.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberti S, Gitler AD, Lindquist S. A suite of Gateway((R)) cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast (Chichester, England) 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Y, et al. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell. 2005;4:289–297. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross CD, McCarty BM, Hamilton M, Ben-Hur A, Ross ED. A promiscuous prion: Efficient induction of [URE3] prion formation by heterologous prion domains. Genetics. 2009;183:929–940. doi: 10.1534/genetics.109.109322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guthrie C, Fink GR. Methods in Ezymology: Guide to Yeast Genetics and Molecular and Cell Biology. Academic Press. 2002;169 [Google Scholar]
- 37.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.