Summary
The development of an effective vaccine has been hindered by the enormous diversity of human immunodeficiency virus-1 (HIV-1) and its ability to escape a myriad of host immune responses. In addition, conserved vulnerable regions on the HIV-1 envelope glycoprotein are often poorly immunogenic and elicit broadly neutralizing antibody responses (BNAbs) in a minority of HIV-1-infected individuals and only after several years of infection. All of the known BNAbs demonstrate high levels of somatic mutations and often display other unusual traits, such as a long heavy chain complementarity determining region 3 (CDRH3) and autoreactivity, traits that can be limited by host tolerance controls. Nonetheless, the demonstration that HIV-1-infected individuals can make potent BNAbs is encouraging, and recent progress in isolating such antibodies and mapping their immune pathways of development is providing new strategies for vaccination.
Keywords: monoclonal antibodies (mAbs), broadly neutralizing antibodies (BNAb), unmutated ancestor (UA), B-cell lineages, immune tolerance
Introduction
Protection against viral pathogens is mediated by an intricate interaction of innate and adaptive immune responses that work together to prevent infection or control viral replication. Despite this complexity, a necessary component of most effective viral vaccines is the induction of virus-specific antibodies that inactivate or neutralize the invading pathogen (1). Since human immunodeficiency virus-1 (HIV-1) was first identified in 1983 (2, 3) and shown to be the cause of acquired immunodeficiency syndrome (AIDS) in 1984 (4), there has been a major focus on the development of a vaccine to prevent infection and on the specific viral proteins that would be the target of potentially protective antibodies. Intensive early investigation led to the elucidation of the genomic organization of HIV-1 (5) and the recognition that it was an enveloped lentivirus consisting of a gp120 surface unit and gp41 transmembrane domain (5-7). Further biochemical characterization demonstrated that the HIV-1 envelope glycoprotein (Env) was a heavily glycosylated trimer of gp120 and gp41 heterodimers and that gp120 interacted with the CD4+ T-cell receptor to initiate viral entry (8, 9). Upon binding CD4, gp120 undergoes conformational changes that result in the formation of a stable intermediate form which binds to a secondary cellular co-receptor, usually the chemokine receptor CCR5 or CXCR4 (10-13). Cellular entry is mediated by rearrangements of gp41, leading to viral-cell fusion (9, 14). Based on this understanding of viral entry and the requirement for functionally conserved regions of Env, it has long been presumed that antibodies targeting the appropriate regions of gp120 or gp41 could block viral entry and potentially prevent infection (15, 16).
But investigators have also recognized that HIV-1 is different from other viruses for which successful vaccines have been made. HIV-1 infects CD4+ T-helper lymphocytes that are critical for an effective immune response to the pathogen and upon responding can also become infected (8, 17). HIV-1 also integrates into the genome and rapidly forms a latent pool of resting cells that are largely invisible to the immune system (18, 19). These attributes are obstacles to immune responses that would completely prevent HIV-1 infection. During the phase of chronic infection, the infidelity of the HIV-1 encoded reverse transcriptase (20, 21) and the propensity for in vivo recombination result in a continually evolving viral quasispecies that evades the autologous immune response (22, 23). The ability of the HIV-1 Env to shield vulnerable regions from neutralizing antibodies (NAbs) provides yet another obstacle for vaccine design (24-28). Thus, HIV-1 poses a unique set of impediments to the induction of fully protective immune responses.
Serum neutralizing antibodies: the early days
HIV-1, initially termed lymphadenopathy-associated virus (LAV) (29) and HTLV-III (4), was first isolated by co-cultures of infected patient samples with neoplastic T-cell lines such as H9 or CEM cells (2, 4, 30). By 1985, investigators had demonstrated that sera from infected subjects contained antibodies capable of blocking infection of cells or inhibiting cell to cell spread in vitro (31, 32), and later that these antibodies targeted the gp120/gp160 Env proteins (33). These findings, together with the success of the recombinant subunit hepatitis B virus vaccine, provided encouragement for the first wave of HIV-1 candidate vaccines consisting of vaccinia and related pox-virus vectors encoding gp160 (34, 35) and purified recombinant gp120 and gp160 proteins (36-38). Remarkably, these first phase I trials were initiated in the late 1980s within 5-6 years of the discovery of HIV-1 (39). Early phase I trials included pox-virus vectors to prime the immune response and recombinant proteins as a boost or recombinant proteins alone formulated with various adjuvants. Initial results were encouraging, demonstrating the elicitation of high titer anti-gp160 antibodies and robust NAb responses (35-37). However, the enthusiasm for these candidate vaccines was quickly tempered when it was appreciated that viral isolates grown on neoplastic cell lines underwent a process of adaptation that resulted in the selection of a highly neutralization-sensitive subset of viruses (40, 41). The neutralization sensitivity of these T-cell line-adapted (TCLA) viruses was sharply contrasted by relative neutralization resistance of virus isolates isolated by cultures with primary human CD4+ T cells and thus termed primary HIV-1 isolates (42, 43). It was subsequently shown that vaccine recipients developed antibody responses that were effective against TCLA viral isolates but exhibited little or no neutralization activity against primary HIV-1 isolates (44, 45). Serological mapping indicated that the vaccine-induced antibody response was often preferentially directed against linear epitopes on the viral Env, including the variable region 3 (V3) loop (46-50). Thus, the V3 loop was initially termed the principal neutralizing determinant (PND) (46-48), until it became clear that the V3 regions of Env was usually shielded within the trimeric structure of the viral spike and was relatively inaccessible to NAbs (51-54).
The lack of primary isolate neutralization induced by recombinant gp120 vaccines led to controversy regarding advanced testing of these vaccines, but eventually two phase III efficacy trials of gp120 vaccines were conducted. The VAX004 and VAX003 trials were initiated in 1998 and 1999, respectively. The VAX004 vaccine consisted of two clade B gp120s and was conducted in North America and the Netherlands. The study population was men who have sex with men and women at high risk for heterosexual transmission. The VAX003 vaccine also contained two gp120s, one from a clade B strain and the other from a clade E (CRF01-AE) viral strain (AIDSVAX® B/E). This trial was conducted in Thailand among intravenous drug users. The results of both studies were reported in 2005 (55-57). Neither vaccine resulted in a reduced risk of HIV-1 infection, nor did either have any impact on the level of plasma viremia upon infection. These results were widely believed to indicate that a different quality of antibody, perhaps neutralizing activity against primary strains of HIV-1, would be required for vaccine protection (39, 58-60).
The first human broadly neutralizing monoclonal antibodies
In contrast to vaccine sera, several groups of investigators reported that serum from some HIV-1-infected donors was able to neutralize diverse primary strains of HIV-1 (40, 45, 53, 61, 62). Initially, the mechanism of this serum neutralization was poorly understood; in some cases adsorption with gp120 could remove most of the neutralizing activity, while for other sera neutralizing activity was evident in the gp120 flow through fractions (63). Our early knowledge of important viral neutralization epitopes came from the isolation of a handful of neutralizing human monoclonal antibodies (mAbs) that were first reported between 1993 and 1994. These mAbs targeted three distinct regions of the viral Env: the CD4-binding site of gp120 (mAb b12), surface glycans on the outer domain of gp120 (mAb 2G12) and the membrane proximal external region (MPER) of gp41, just prior to the transmembrane spanning sequence (mAbs 2F5, 4E10, and Z13e) (64-66). Numerous other mAbs were also reported, including those to the V1V2 and V3 regions (67), the coreceptor binding region of gp120 and the heptad repeat-1 region of gp41, but these antibodies were generally weakly neutralizing or highly strain specific (24). Importantly, in vivo passive infusion studies using chimeric SIV/HIV viral infection of macaque monkeys demonstrated that NAbs could completely block viral infection (16, 68-72), thus highlighting the importance of understanding the molecular structure of each of these neutralization epitopes as a means to pursue the design of improved vaccine immunogens. In 1998, the co-crystal structure of the gp120 core protein liganded with soluble CD4 and the coreceptor binding mAb 17b was solved (73, 74). While the gp120 core included N and C-terminal truncations and removal of V1V2 and V3 loops, this structure provided new insights into the mechanism of the gp120-CD4 interaction and set the stage for additional co-crystal structures of NAbs. Between 2003 and 2009, investigators solved liganded structures of mAbs 2G12, b12, 2F5, and 4E10 (75-78). These antibody structures not only provided a new level of knowledge about potential vulnerable viral epitopes but also unveiled some rather daunting characteristics of the known BNAbs. The 2G12 mAb that binds a cluster of surface glycans was shown to adopt an unusual domain swap configuration in which the two heavy chains interact to form a large monovalent binding surface (75). No prior antibody with a similar configuration had been described, and it was unclear how such an antibody was generated or might be induced via immunization. The CD4bs mAb b12 was derived from a phage display library such that its natural light chain pair was not known. In addition, it demonstrated a rather unusual heavy chain only binding interface (no light chain binding) that was mediated by a moderately long CDRH3 loop (78). Finally, the gp41 membrane-proximal external region (MPER)-directed mAbs 2F5 and 4E10 were both shown to display features of polyreactivity and autoreactivity (79, 80). In addition, both 2F5 and 4E10 have long hydrophobic HCDR3 regions as well as excess somatic mutations, additional traits that can predispose antibodies to tolerance control mechanisms, such as clonal deletion (81).
As additional HIV-1 isolates from geographically diverse regions of the world were studied, it became clear that none of the above BNAbs was able to neutralize the majority of diverse HIV-1 strains. The glycan-targeted mAb 2G12 neutralized less than 50% of clade B isolates and was substantially weaker against non-clade B strains. The neutralization pattern of b12 similarly displayed a preference for clade B isolates with an overall breadth of reactivity of less than 50%. Among the two Env gp41 MPER antibodies, 4E10 displayed impressive breadth (~ 90% of viruses tested), but its potency was weak, especially when tested in assays with primary target cells. MAb 2F5 displayed moderate potency, but was generally weak against clade C isolates that were prevalent in many regions of Africa, India and China (53, 82-85).
Antibodies during natural HIV-1 infection and the next generation of BNAbs
The structural and functional analysis of HIV-1 NAbs b12, 2F5 and 4E10 led to some pessimism about the potential of the immune system to generate BNAbs against HIV-1 (86). This concern was highlighted by the observed lack of natural immunity to HIV-1 (87) and by studies demonstrating the ability of HIV-1 Env to continuously evolve to evade the autologous NAb response (22, 23). Additionally, as the full genetic diversity of HIV-1 was appreciated, we also began to understand the formidable immune evasion mechanisms that are intrinsic to the HIV-1 Env. These include carbohydrates that make up more than half the molecular weight of the HIV-1 Env and variable loops that act in concert with surface glycans to mask important neutralization epitopes (24, 25, 27). In addition, key epitopes such as the receptor binding sites or the MPER of gp41 are either recessed, transiently exposed or have limited spatial accessibility to antibody molecules (24, 27).
This rather bleak view of the HIV-1 humoral immune response began to change with the observation that some HIV-1 infected donors mounted NAb responses that were more potent and cross-reactive than those generated by current vaccine immunogens (40, 45, 53, 61, 62). Several groups of investigators reported that sera obtained after HIV-1-infection could potently neutralize primary isolates of HIV-1, and in some cases, were able to neutralize genetically diverse HIV-1 strains (88-92). Early work assessing serum mediated HIV-1 neutralization was often performed using primary HIV-1 isolates derived from cultures of human peripheral blood mononuclear cells (PBMCs) (40, 41, 93-97). These assays also used PBMCs or CD4+ T cells as targets cells for infection, and infection was often quantified after several rounds of viral replication. While these PBMC-based neutralization assays were critical for our appreciation of the difference between T-cell line adapted and primary HIV-1 isolates, the assays were generally not highly accurate or reproducible. PBMC culture-generated viral stocks contained a quasispecies of viruses, and variation in donor PBMC target cells further contributed to assay variability. Starting in 2005, there was a concerted effort to develop accurate high throughput neutralization assays based on the construction of recombinant Env-pseudoviruses that mediated a single round of viral infection (98-100). The advantage of such assays were that each pseudovirus contained a well-defined clonal sequence and that large panels of diverse HIV-1 Env’s could be rapidly constructed, thus better representing the global genetic diversity of HIV-1. These assays allowed the screening of large panels of HIV-1 sera and the identification of individual donors whose sera were able to neutralize the majority of HIV-1 strains. Overall, these data revealed that there was a heterogeneous spectrum of serum neutralization with some sera displaying weak neutralization and others able to neutralize most HIV-1 strains tested (53, 62, 88-92, 101-103). Depending on the cohort tested and the definitions used, these studies revealed that between 10% and 25% of HIV-1 infected subjects generate relatively potent cross-reactive NAbs (91, 92, 101, 102, 104-106).
Initially, the difficulty in defining the specific antibodies responsible for mediating broad serum neutralization hindered our ability to understand the nature of this immune response. It was clear that the handful of known neutralizing mAbs did not explain the more potent cross-reactive antibody response found in some donors, but it was unclear if serum neutralization resulted from the activity of polyclonal mixtures of antibodies or from a limited subset of antibodies targeting conserved epitopes. Although investigators expended substantial effort to isolate additional neutralizing mAbs, several factors limited the discovery of such mAbs. These included the small fraction of B cells that secrete HIV-1 specific neutralizing mAbs (107, 108) and the inefficiency of traditional methods of mAbs isolation. Several methodological advances however, created the framework for the subsequent isolation of numerous new HIV-1 neutralizing mAbs. These methods included the successful large scale culture of memory B-cells at close to limiting dilution concentrations (2-4 B cells per well) under conditions that resulted in sufficient IgG secretion to screen wells directly for HIV-1 neutralization (82, 109, 110). The specific heavy and light chain of individual B-cells were then recovered by PCR amplification and cloned into an IgG expression vector (111, 112). An additional approach was to use fluorescently labeled Env specific protein probes to identify and sort HIV-1 specific B cells and to recover the antibody genes by PCR as described above (84, 107, 108, 113-115). Beginning in 2009, these methodological advances led to the isolation and structural characterization of numerous neutralizing mAbs that have redefined our understanding of the architecture of the HIV-1 Env and vulnerable epitopes that could be targeted by NAbs. The major epitopes defined by these new mAbs included regions of the CD4 binding site of gp120 (84, 114, 116-119) and the MPER of gp41 (110, 115, 120), and two new peptide-glycan epitopes on gp120, one in the region of the V1V2 loop (82, 109) and the other in the region of V3 (83). While additional mAbs are likely to be discovered and possibly new viral epitopes defined, the current set of potent and broadly reactive mAbs comes close to recapitulating the potency and breadth of observed serum neutralizing activity (Table 1).
Table 1.
Genetic characteristics of HIV-1 broadly neutralizing monoclonal antibodies
| Viral Epitope | Antibody binding characteristicsa |
Antibody clonal family |
Year Published |
Isotype and subclassb |
Heavy chain V-gene |
Light chain V-gene (K or L) |
CDRH3 length (Kabat AA)c |
VH mutation (nt %)d |
VH mutation (AA %)d |
Neutralization Breadthe |
Neutralization Potencyf |
Polyrectiveg | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MPER of gp41 | contiguous sequence | 2F5 | 1993 | IgG3 | 2-5 | K1-13 | 22 | 14 | 15 | ++ | ++ | yes | 53, 65, 66, 77, 85 |
| contiguous sequence | 4E10 | 1994 | IgG3 | 1-69 | K3-20 | 18 | 14 | 20 | ++++ | ++ | yes | 53, 65, 66, 76, 85 | |
| contiguous sequence | M66.6 | 2011 | NR | 5-51 | K1-39 | 21 | 4.3 | 10 | + | ++ | yes | 120 | |
| contiguous sequence | CAP206-CH12 | 2011 | IgG1 | 1-69 | K3-20 | 15 | 12 | 19 | + | ++ | yes | 115 | |
| contiguous sequence | 10e8* | 2012 | IgG3 | 3-15 | L 3-19 | 20 | 21 | 26 | ++++ | +++ | no | 110 | |
| V1V2-glycan | peptido-glcyan | PG9, PG16 | 2009 | NR | 3-33 | L2-14 | 28 | 12-13 | 17 - 20 | +++ | +++ | no | 82 |
| peptido-glcyan | CH01 - 04 | 2011 | IgG1 | 3-20 | K3-20 | 24 | 14 - 17 | 23 - 29 | + | ++ | nog1 | 109, 170 | |
| peptido-glcyan | PGT 141-145 | 2011 | NR | 1-8 | K 2-28 | 31 - 32 | 12-18 | 21 - 29 | +++ | +++ | NR | 83 | |
| Outer domain glycan | glycan only | 2G12 | 1994 | IgG1 | 3-21 | K 1-5 | 14 | 21 | 32 | + | ++ | NR | 53, 65, 75 |
| V3-glycan | peptido-glycan | PGT121-123 | 2011 | NR | 4-59 | L 3-21 | 24 | 17 - 21 | 21 - 27 | ++ | ++++ | NR | 83 |
| peptido-glycan | PGT125-131 | 2011 | NR | 4-39 | L 2-8 | 19 | 15 - 23 | 23 - 33 | ++ | ++++ | NR | 83 | |
| peptido-glycan | PGT135-137 | 2011 | NR | 4-39 | K 3-15 | 18 | 17 - 20 | 25 - 29 | + | ++ | NR | 83 | |
| CD4 binding site | CDRH3 loop | b12 | 1994 | NR | 1-3 | K 3-20 | 18 | 13 | 20 | + | ++ | nog2 | 53, 64, 78 |
| no liganded structure | HJ16 | 2010 | NR | 3-30 | K4-1 | 19 | 15 | 37 | + | ++ | NR | 116 | |
| CDRH3 loop | CH103 - 106 | 2013 | IgG1 | 4-59 | L 3-1 | 13 | 14 - 16 | 22 | ++ | ++ | yesg3 | 118 | |
| mimics CD4 via CDRH2 | VRC01 - 03 | 2010 | IgG1 | 1-2 | K 3-20# | 12-14 | 30 - 32 | 40-48 | ++++ | +++ | no | 84, 119 | |
| mimics CD4 via CDRH2 | VRC-PG04, 04b | 2011 | IgG1 | 1-2 | K 3-20 | 14 | 30 | 38-39 | +++ | +++ | no | 114 | |
| mimics CD4 via CDRH2 | VRC-CH30 - 34 | 2011 | IgG1 | 1-2 | K1-33 | 13 | 23 - 24 | 36-40 | +++ | +++ | no | 114, 170 | |
| no liganded structure | 3BNC117, 3BNC60* | 2011 | NR | 1-2 | K1-33 | 10 | 27 | 37-40 | +++ | +++ | yesg4 | 117 | |
| mimics CD4 via CDRH2 | NIH45-46* | 2011 | IgG1 | 1-2 | K 3-20 | 16 | 30 | 41 | ++++ | +++ | yes | 117, 125 | |
| no liganded structure | 12A12, 12A21* | 2011 | NR | 1-2 | K1-33 | 13 | 23 - 25 | 38-40 | ++++ | +++ | yes | 117 | |
| no liganded structure | 8ANC131, 134* | 2011 | NR | 1-46 | K 3-20 | 16 | 27 | 37-38 | +++ | ++ | yes | 117 | |
| no liganded structure | 1NC9, 1B2530* | 2011 | NR | 1-46 | L 1-47 | 16 - 19 | 24 - 26 | 36-38 | + | ++ | yes | 117 |
Major binding characteristics indicated by co-crystal structural analysis. mAb 2G12 utilizes unusual domain swap structure - see indicated references. mAb b12 was isolated from phase display library and natural light chain pair was not retained; b12 displays heavy chain only binding in co-crystal structure with gp120.
Isotype indicates the natural isotype and subclass of the isolated antibody. In some cases, this was not reported. In many cases, the PCR amplified heavy and light chain genes are expressed in an IgG1 expression vector even though the natural antibody was a different subclass.
Reported CDRH3 lengths are often based on IMGT or Kabat nomenclature. The Kabat definition is generally used for structural studies and is often 2 AA residues shorter than the IMGT definition. For explanation of the differences in nomenclature, see www.imgt.org.
The percent mutations (nt) of an antibody heavy chain are based on comparison to the inferred germline gene. Percent mutations are sometime also reported based on translated AA sequences (AA).
Neutralization breadth is indicated for antibodies tested on large panels of > 100 tier 2 Env-pseudoviruses and the values shown are the percent of viruses neutralized at an IC50 of < 50 ug/ml. ++++ = > 90%; +++ = 75 - 90%; ++ = 50 - 74%; + = < 50%. If there are several members of a clonal family, the broadest clonal member was used.
Neutralization potency indicated by median or geometric mean IC50 (ug/ml) of neutralized viruses (i.e. excluding non-neutralized viruses); ++++, IC50 < 0.05; +++ , IC50 = 0.05 - 0.49; ++, IC50 = 0.5 - 4.9; +, IC50 = 5.0 - 50.
Poly or self reactivity as assessed by a combination of common autoimmune assays, including cardiolipin ELISA, Hep2 cell IFA and Athena assay for antinuclear antibodies.
One antibody in this clonal lineage, CH103 is polyreactive.
Some have found b12 polyreactive, but studies in b12 VHDJH + VLJL knock-in mice have demonstrated the degree of polyreactivity for this antibody is not sufficient to predispose these antibodies to tolerance deletion (D. Nemazee, personal communication).
CH103, CH104, CH106 are autoreactive, CH105 is not autoreactive.
3BNC117 is reported to 3y polyreactive, but not 3BNC60.
Additional members of the clonal family also reported in the primary publication. NIH45-45 is same donor and clonal family as VRC01. For mAbs 1NC9 and 1B2530, neutralization data only available for 1B2530
VRC01 and 02 were initially reported to derive from inferred VK3-11, but additional analysis of the antibody lineages strongly suggests VK3-20. Also, VRC03 was initially inferred to be from different clonal family than VRC01, but more detailed analysis suggests that VRC01, 02 and 03 are clonal relatives (J. Mascola, P. Kwong, unpublished data)
Notes: NR, not reported.
Eliciting broadly reactive neutralizing antibodies: gaps in our knowledge
Structural and genetic features of neutralizing mAbs
Remarkably, investigators have recently solved liganded crystal structures of mAbs bound to each of the four major neutralization epitopes, and the structural features of these mAbs has been recently reviewed in detail (28, 121). Here, we focus on some of the key features of these new mAbs and the implications of this knowledge for vaccine design, including key gaps in our knowledge that still limit our ability to elicit HIV-1 specific BNAbs. A review of the four major neutralization epitopes on the HIV-1 Env reveals some intriguing structural, genetic, and immunological characteristics of the antibodies to each epitope. The CD4-binding site would seem to be an obvious functionally conserved region of Env, yet most of the initial antibodies isolated to this site were poorly neutralizing. Structural studies of neutralizing and non-neutralizing CD4-binding site mAbs revealed that precise targeting of this region by antibody b12 was necessary to avoid steric and conformational clashes that would hinder binding to the native viral spike (122). Hence, many CD4-binding site mAbs display high affinity binding to recombinant gp120 but are unable to access the CD4-binding site on gp120 (123, 124). The isolation and structural characterization of the broadly reactive VRC01 mAb provided a fascinating solution to this problem: the antibody partially mimics binding by cellular receptor CD4 to gp120 (84, 119). Specifically, a region of the heavy chain of VRC01 is arranged in an antiparallel B sheet configuration that is similar to the interaction of domain 1 of CD4 with gp120. Structures of VRC01-like antibodies from a total of six different donors reveal a highly similar mode of recognition of the CD4-binding site (114, 119, 125, P. Kwong, personal communications). Light chain interactions with somewhat more variable regions of V5 and the D-loop regions of gp120 explain the few cases of viral resistance to this class of mAbs. A rather unique feature of VRC01 class antibodies is that heavy chain binding is mediated largely by CDRH2, a region encoded by the V-gene, rather than CDRH3 which is encoded by V-D-J recombination. Interestingly, all known VRC01 type antibodies derive from VH1-2 or the closely related VH1-46 gene, suggesting these two germline genes are required to generate these antibodies (84, 114, 117, 126). Also, these VRC01 class antibodies are highly somatically mutated (~ 20% – 30% nt changes in heavy chain compared to germline), and it is difficult to detect any gp120 binding when the amino acid sequence of the heavy chain V-region is reverted to germline (114, 117, 119, 127). This has raised questions about how naive B cells are triggered to generate VRC01 class antibodies (128-130).
Two additional neutralization epitopes on gp120 were identified when antibodies to conserved glycan-containing regions of V1V2 and V3 respectively were described. mAbs PG9 and PG16, and subsequent mAbs in this category, such as CH01-04 and PGT141-145 (82, 83, 109) bind to relatively conserved residues within V2 and require major interactions with surface glycans, most importantly a N-linked glycan at position 160 (131, 132). Similarly, the PGT mAbs described in 2011 bind to residues at the base of V3 and require specific glycan interactions, particularly at position N332 (83, 133). These glycan reactive antibodies are some of the most potent HIV-1 mAbs isolated and they share a common feature of a long CDRH3 loop that interacts with surface glycans and reaches past these glycans to make key contacts with either V2 or V3 residues on gp120. These antibodies are also highly somatically mutated, and hence, a characteristic of HIV-1 glycan reactive antibodies appears to be a long CDRH3 loop together with a high level of affinity maturation to acquire breadth of reactivity. In the case of V1V2 BNAbs, the germline reverted versions both CH01 and PG16 have been found to neutralize the same few HIV-1 strains, suggesting a common structural requirement for their induction (109, 134). Finally, a new MPER directed mAb, 10e8, was recently isolated and its co-crystal structure solved (110). This antibody binds to a region of the MPER that overlaps with the 4E10 epitope but includes C-terminal residues close to the transmembrane spanning domain. 10e8 is highly somatically mutated, is 5-10 fold more potent than 4E10 and can neutralize ~ 95% of viruses tested. Interestingly, mAb 10e8 does not possess the polyreactivity that is characteristic of mAbs 2F5 and 4E10, but it does have a long HCDR3 and a high number of somatic mutations (110).
Immunological roadblocks to BNAb elicitation
Despite the detailed knowledge of the major neutralizing mAbs described above and the associated atomic level descriptions of their viral epitopes, we are left with rather formidable barriers to their elicitation. Our best attempts to construct vaccine immunogens that present these key epitopes to the immune system have failed to generate antibodies that neutralize most strains of HIV-1 (28, 39, 44, 45, 58). An example of this phenomenon is observed with monomeric recombinant gp120 used as a vaccine. We have known since 1994 that the neutralizing mAb b12 binds with high affinity to the CD4-binding site on recombinant gp120 (64), yet numerous studies of gp120 immunization have shown that this does not result in the generation of NAbs like b12 or newer CD4-binding site antibodies like VRC01 (39, 44, 135, 136). This highlights a long standing dichotomy between antigenicity and immunogenicity that has been described by numerous investigators (24, 27, 63, 137, 138). While additional structural knowledge of new or existing antibodies will likely continue to refine our understanding of these epitopes, we have a wealth of information to provide momentum for our structure-based vaccine design efforts (28, 139, 140). However, a structure-based approach in and of itself will likely not solve the HIV-1 vaccine problem. Rather, the correct antigenic structure must be formulated and delivered in such a way as to overcome the immunological road blocks that appear to limit effective antibody response to the HIV-1 Env (129, 140-142). Thus, the design of improved HIV immunization strategies will likely require a better understanding of epitope dominance and subdominance, immune tolerance and anergy, and the immunogenetics of antibody induction and maturation, including the role of selected germline genes and the process of affinity maturation (Table 2).
Table 2.
Obstacles to the elicitation of HIV-1 broadly neutralizing antibodies
| Obstacles and Questions | Approach |
|---|---|
Epitope dominance/subdominance
|
|
Tolerance: deletion/anergy
|
|
Selected BCRs and Affinity Maturation
|
|
Regarding epitope subdominance, we need to understand why certain regions or subregions of the HIV-1 Env are poorly immunogenic and how to engage rare BNAb B-cell precursors and foster their expansion to yield a dominant plasma antibody response (28, 129). As noted previously, the HIV-1 Env is highly immunogenic and induces antibodies to gp41 and gp120 shortly after infection. The hierarchy of the humoral immune response during acute HIV-1 infection has recently been studied in detail (141, 143). The first HIV-1 Env specific antibodies are observed as early as 8 days after plasma viremia is first directed (143). These initial antibodies target immunodominant regions of gp41, followed by the development of non-neutralizing antibodies to gp120, including antibodies to variable loop regions, the CD4-binding site and the CD4-induced co-receptor sites on gp120. Some investigators have speculated that this early antibody response is driven by both surface-exposed immunodominant regions of gp120 and gp41 and by shed gp120 and gp41 stalks that may be a natural part of the viral life cycle (24, 144). Such gp120 and gp41 epitopes may not be accessible on the functional trimeric viral spike, explaining the lack of neutralization by these antibodies.
Over time, virus-neutralizing antibodies develop in most HIV-1 infected individuals, demonstrating that the humoral immune system can target epitopes on the native viral spike. The initial NAb response is directed to the autologous infecting virus and is generally not detected until more than 12 weeks of infection (22, 23, 91, 102, 141). Though these autologous NAbs exert immune pressure on the virus, they are highly strain specific; i.e., the can neutralize the autologous virus potently, but display little breadth of activity against heterologous viruses. Initial cross-reactive NAb may arise after several months of infection but the more broadly reactive NAb that target highly conserved epitopes are generally not found until more than two years of infection (91, 102). The kinetics of the HIV-1 Env antibody response raises numerous questions. Why is there a delay of several months or more before viral NAbs arise? When these antibodies do arise, what accounts for their initial strain-specificity and what characteristics confer neutralization breadth? Do strain-specific NAbs target more variable epitopes in contrast to BNAbs, or does a specific NAb lineage mature over time to gain the necessary affinity and precision to target conserved residues within a neutralizing epitopes? The answers to these questions have clear implications or vaccines design, as discussed below.
There is also evidence that the BNAb response is limited by self-tolerance and clonal deletion. Structural studies of mAbs 2F5 and 4E10 with respective gp41 peptides show that only a small part of their CDRH3s bind the gp41 epitope (76, 77). Additional studies suggested that remaining hydrophobic CDRH3 regions are important for antibody interactions with the viral membrane, to allow the antibody to lie in close proximity to the membrane. This appears to position the mAbs for high-affinity binding to the gp41 MPER (145-148). Interestingly, both 2F5 and 4E10 react with anionic lipids such as cardiolipin, consistent with the observations that the antibodies bind both to gp41 and to the nearby virion membrane (79, 80, 149). Alam and colleagues (80, 148) demonstrated that both 2F5 and 4E10 encounter the viral membrane followed by induction of a gp41 conformation change that leads to high affinity mAb docking to the MPER epitope on gp41. In this process the ‘hinge’ region of the MPER is extracted from contact with the viral membrane (145, 150). Importantly, in vitro polyreactivity of HIV-1 mAbs may be of clinical significance, as passive infusion of mAbs 4E10, 2F5, and 2G12 resulted in a prolonged partial thromboplastin time (PTT coagulation test) in subjects to whom they were administered (151, 152), although no clinical adverse events were noted. This prolonged PTT test, due to lipid-reactive antibodies that mediate lupus anticoagulant activity, can be detected in vitro for 4E10, but not for 2F5, 2G12, or other mAbs such as b12 or VRC01 (79, 153), suggesting that the prolonged PTT effect was the result of infusion of mAb 4E10.
The polyreactivity of some HIV-1 BNAbs raised the important concept of tolerance and immunoregulatory host controls as one set of factors limiting the frequent elicitation of these antibodies (142). To test this hypothesis, Verkoczy and colleagues (142, 154, 155) made homologous recombinant knock-in mice that expressed the mature and germline versions of the 2F5 antibody heavy and light chains. The polyreactivity of both the germline and mature 2F5 was sufficient to result in deletion of 95% of the antibody precursors in bone marrow at the first tolerance checkpoint. Importantly for vaccine development, not all 2F5 B cells were deleted but could be found in peripheral LN and spleen in an anergic state (155). These anergic BNAb B cells could be awakened by repetitive immunization with the gp41 MPER peptide epitope embedded in lipids (L. Verkoczy and B. Haynes, unpublished data). A similar deletion at the first tolerance check point was found in 4E10 knock-in mice (L. Verkoczy L and B Haynes, unpublished data). Thus, overcoming tolerance controls that limit BNAb induction has emerged as a key concept in HIV-1 vaccine development. Nussenzwieg and colleagues (108, 117, 156, 157) evaluated hundreds of HIV-1 Env specific mAbs from chronically HIV-1 infected donors and showed that these mAb were often highly affinity matured and polyreactive. Interestingly, the plasma cell repertoire of patients acutely infected with HIV-1 also contains autoreactive antibodies. Liao and colleagues found that initial gp41 directed antibodies were often highly mutated and polyreactive, and some were shown to react with gut flora antigens (158). These data raised the hypothesis that B cells were initially activated by gut flora prior and that cross-reactivity with gp41 may further shape the memory B-cell repertoire after HIV-1 infection.
A similar tolerance obstacle may affect BNAbs that interact with peptide-glycan epitopes in the V1V2 and V3 regions. These antibodies have extended CDRH3 structures that are required to reach peptide residues obstructed by glycans (Table 1). Both mouse and human studies have demonstrated a selection against long CDRH3 loops, presumably due to their increased auto or polyreactivity. Mouse studies suggest that many B cells encoding long CDRH3 regions are eliminated before reaching the periphery (159, 160), and human studies show a decreased prevalence of CDRH3 sequences in IgG memory compared to naive B-cell subsets (161) and a selection bias against long CDRH3 loops, particularly those with hydrophobic patches (162). Thus, a key question for eliciting these glycan-reactive BNAb is whether their long CDRH3 loops are encoded during germline recombination or whether an iterative affinity maturation process could result in short insertion events that gradually lengthen the CDRH3. This latter process would require immunization to drive an unprecedented level of affinity maturation. Since we know that a relatively small fraction of B-cell receptors (BCRs) encode for long CDRH3 lengths (161-163), perhaps the more likely scenario is that these long CDRH3 BCRs are selected by specific circulating Env immunogens with high enough affinity to drive their expansion and to overcome their potential autoreactive properties.
Another potential immunological roadblock arises when considering antibodies such as VRC01 that potently neutralize via binding to a conserved region of the CD4-binding site. As noted above, VRC01-type antibodies from several different donors display a high level of structural similarity and predominant gp120 contacts are made via the heavy chain CDR2 which is encoded by the VH1-2 gene (114, 119, 125). Hence, it appears that this potent class of antibodies derive from a highly restricted set of VH genes and subsequently undergo extensive affinity maturation to achieve effective broad and potent neutralization (121). Investigators have made the rather disconcerting observation that reversion of V-gene mutations to germline VH1-2, even with an intact mature CDRH3, abolished detectable binding to gp120 (117, 119). This appears true even when testing large panels of diverse gp120s (127) and has raised the question about the nature of the antigen that stimulated the naive VH1-2 BCR, e.g. was it an HIV-1 Env antigen or some unrelated antigen (128, 129)? Several groups of investigators have begun to use structural information to design variants of HIV Env immunogens that would bind to germline (or V-gene reverted) versions of VRC01. Finally and has already been noted, BNAbs appear to take months to years to evolve and have high levels of somatic mutation. This suggests a long and potentially tortuous pathway of evolution. Recent studies have shown that vaccination with HIV-1 gp120 induces anti-gp120 antibodies with ~3-5% somatic mutations and an upper range of 8% (164-166). In contrast, the mean level of somatic mutations among BNAbs is 16% with some BNAbs accumulating 32% mutations compared to germline nucleotide sequence (121, 129) (Table 1). Fortunately, newer BNAbs have recently been described that have normal CDRH3 lengths, fewer somatic mutations, and arise much earlier in HIV-1 infection (118). Thus, studies of BNAb lineages and their evolution from early infection through the process of maturation to potent neutralization may provide insights for vaccine immunogen design and immunization strategies.
Nature as a guide: the study of B-cell lineages to guide vaccine design
As noted above, immunization with antigenic HIV-1 Envs has not resulted in induction of subdominant BNAb B-cell responses similar to those that arise in some HIV-1-infected individuals. Rather, new approaches will likely be required to induce otherwise disfavored antibody lineages. We know that naive B cells compete for survival in germinal centers, and those with the highest affinity occupy the germinal center and expand to manifest plasma antibody levels (167-169). However, we know little about the early events that trigger the development of BNAbs responses in HIV-1 infection. Thus, several groups of investigators have begun to study these events in detail to gain insights for HIV vaccine design.
B-cell lineages from natural infection
The recent isolation and sequencing of numerous BNAbs provides opportunities to study how these antibodies develop and evolve. When more than one BNAb is isolated from an HIV-1 infected donor, either by antigen-specific B-cell sorting or by memory B-cell cultures, the isolated antibodies are often part of the same clonal lineage, suggesting that one lineage can account for a major fraction of the serum NAb response (82-84, 109, 114, 170). Once the genetic sequence of a BNAb is known, next generation sequencing provides a way to dramatically expand the view of an antibody clonal family (114, 162, 171-173). Bioinformatics based analyses allows the identification of intermediate antibody (IA) sequences and the potential to infer the unmutated ancestor (UA) of the BNAb lineage, i.e. the naive BCR sequence. Wu and colleagues (114) used 454 pyrosequencing of antibody gene transcripts to study the heavy and light chain antibody lineages of mAbs VRC01 and VRC-PG04 in their respective donors. Novel bioinformatics analysis allowed the identification of thousands of sequences that formed a specific BNAb lineage. While these studies included a single time point from a chronically infected donor, the investigators were able to identify heavy and light chain clonal relatives with far fewer somatic mutations than the mature isolated antibody. This allowed the expression and testing of antibody intermediates and the demonstration that high levels of affinity maturation were required for the full potency and breadth of the antibody. In spite of the success of elucidating BNAb clonal lineages from chronically infected subjects, these initial studies were limited by lack of samples from the time of transmission and therefore lack of the inciting transmitted-founder virus Env that was involved in stimulating the antibody lineage.
From 2005 to 2012, the Center for HIV/AIDS Vaccine Immunology (CHAVI) enrolled acute HIV-1-infected subjects and followed them for 3-5 years. A subset of these subjects developed cross-reactive Nabs, and their samples are now being studied for NAb and virus co-evolution. Numerous studies have shown that the initial NAb response is directed primarily to the autologous virus and that there is an ongoing pattern of viral escape and further maturation of the NAb response (22, 23, 93, 94, 102, 174, 175). The co-evolution of HIV-1 and the polyclonal antibody response can be thought of as an ‘arms race’, wherein the infecting virus induces an autologous NAb response that is initially limited in breadth. Viral escape mutants appear to drive the antibody response and ~20% of HIV-1 infected subjects develop cross-reactive NAbs (91, 102, 175). In a recent study, Moore and colleagues (175) reported two examples of HIV-1-infected donors whose transmitted-founder viruses did not have an N-linked glycosylation site at position 332 (a critical glycan for binding by the PGT group of mAbs). The transmitted viruses in each donor were initially neutralized by their autologous plasma, but later escaped by adding a glycan at position 332 (175). The development of N332-glycan containing viruses preceded the development of more broadly NAbs that targeted this N332 site. Thus, a BNAb response arose in response to viral escape from earlier autologous NAbs.
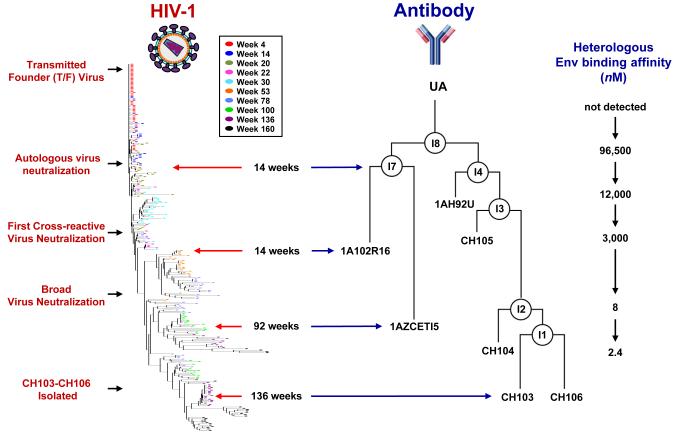
Liao and colleagues (118) recently isolated several closely related CD4-binding site NAbs from a CHAVI cohort seroconverter who had been HIV-1 infected for approximately 2.5 years, and solved the structure of one of these mAbs (CH103) bound to the outer domain of gp120. Using samples derived within several weeks of infection and sequentially thereafter, they were able to establish the initial transmitted-founder viral Env sequence and use antibody gene sequencing to infer the full antibody clonal lineage, including the UA and IA sequences of the CH103 lineage. Notably, the UA of CH103 avidly bound the transmitted-founder Env gp140, and early intermediate antibodies predominantly neutralized the early autologous virus strains. Importantly, the evolution of neutralization breadth was preceded by extensive viral diversification in and near the defined CH103 binding epitope. This was the first demonstration that a specific clonal HIV-1 antibody lineage (in this case to the CD4-binding site) evolves from restricted autologous virus neutralization to broad cross reactive neutralization (Fig. 1). Further studies of the co-evolution of the infecting Env sequences in relation to specific BNAb lineages are likely to provide additional key information on the maturation pathways of BNAbs.
Fig. 1. Coevolution of virus and a single antibody lineage in an HIV-1 seroconverter.
Mature CD4-binding site antibodies CH103-106 were isolated from circulating memory B-cells at week 136. Longitudinal sampling allowed inference and reconstruction of the evolution of the infecting viral sequence and of the specific neutralizing antibody lineage. The heavy chain antibody lineage was augmented with sequences derived from B-cell gene sequencing and bioinformatics analyses were used to infer early intermediates (IA) and the unmutated ancestor (UA). The left part of the figure displays a phylogenetic tree of Env sequences derived from week 4 through week 160. The UA and IA heavy chain sequences of the CH103 antibody lineages are shown alongside viral evolution. This antibody lineage evolved to gain high affinity Env binding, and virus neutralization evolved from strain-specific autologous virus activity to cross-reactive neutralization of heterologous viruses.
Strategies to elicit BNAbs: guiding the immune response
We have described several major obstacles to the elicitation of broadly reactive NAbs against HIV-1. These include epitope subdominance, tolerance and clonal deletion, gene-restricted antibody lineages, and high levels of affinity maturation. While each of these obstacles is formidable, our recent structural understanding of antibody mode of recognition together with genetic studies of antibody evolution are beginning to elucidate the pathways for BNAb development. This knowledge has raised additional questions to investigate but also provides new approaches to BNAb elicitation (Table 2).
There are at least two related approaches to address epitope subdominance: one is to remove or mask dominant epitopes (28, 176-178), and the other involves the structure-based design of immunogens that present specific conserved epitopes to the immune system (139, 179, 180). Numerous studies with variable region deleted Envs (181-183) or with glycan-masked immunogens (184) suggest that these approaches alone do not solve the problem, though they may have utility as part of a more complex approach. For example, native deglycosylation of gp140 Env exposes gp41 epitopes and enables the UA of some gp41 mAbs to bind HIV-1 Env and has the potential to improve Env immunogenicity (185). It may be possible to focus the immune response by initially immunizing with a restricted epitope and boosting with a native form of gp120 or gp140 (28, 179, 186). This has the potential to stimulate the correct naïve BCR and to affinity mature the response against the same epitope as it exists in its native form on the Env trimer. In this regard, there has been recent progress in the design of protein scaffolds on which specific HIV-1 epitopes can be transplanted and retain their native configuration (187-190), and several studies have shown proof-of-concept that such protein scaffolds can induce an epitope specific antibody response (187, 188). There has also progress in the design of cleaved gp140 molecules that retain antigenic reactivity to BNAbs (191-193). Investigators are just beginning to take advantage of these new immunogens and immunization strategies.
We have already discussed the obstacle of self-tolerance and clonal deletion in some detail. Antibodies with long hydrophobic CDRH3 regions may be disfavored during the process of affinity maturation and this may impact elicitation of glycan reactive BNAbs to the V1V2 and V3 regions (e.g. PG9, PGT128). Further studies should inform our understanding of how these V1V2 and V3 glycan BNAbs arise, if the long CHRH3 is selected at the germline level, and if glycan reactivity results from affinity maturation. This information would inform the design of vaccines to elicit these glycan-reactive antibodies. For example, the PG9 (V1V2-glycan) and PGT128 (V3-glcyan) groups of BNAbs make both protein and glycan contacts on gp120. If the early forms of these antibodies make mainly protein contacts (i.e. glycan reactivity evolves over time with affinity maturation), the approach to elicitation might favor presentation of the appropriate protein contacts surface first. However, if the early antibodies bind both protein and glycan, elicitation may require initial immunization with an appropriately glycosylated native trimer. Self-reactivity may be a particular problem for elicitation of antibodies to the MPER of gp41. BNAbs 2F5 and 4E10 are thought to have hydrophobic CDRH3 regions to close approximation to viral membrane, which favors antibody interaction with the gp41 MPER epitope. However, the recent discovery of broadly neutralizing 10e8 antibody demonstrates that the immune system can target the MPER without engendering substantial lipid reactivity. The isolation and characterization of additional antibodies such as the non-polyreactive 10e8 may offer insights into common features of this immune response. Strategies to overcome these disfavored antibody lineages include structure-based design of immunogens for high affinity binding to the BCR, high avidity multivalent antigen presentation, potent adjuvants and specific modulators (e.g. Toll-like receptor ligands) to enhance the innate and adaptive immune response (28, 129, 139, 140).
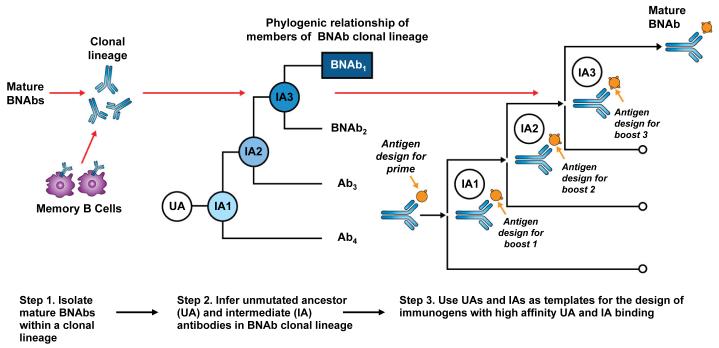
The high level of affinity maturation of most BNAbs presents a major potential obstacle to their elicitation by traditional immunization methods. It is clear from over 20 years of study of HIV-1 Env immunogenicity that immunization with a current Env vaccines will not induce broad NAbs (39, 194). In addition, only modest levels of affinity maturation have been reported with subunit protein immunization in humans (164-166). To overcome these limitations, we may have to employ new approaches to stimulate suitable B-cell lineages and guide affinity maturation. Studies such as those discussed above, have begun to trace the co-evolution of virus and BNAb lineages to carefully define the Envs immunogen associated with BNAb development (118) (Fig. 1). For vaccine design, the expression and study of these unmutated and early ancestor antibodies may be critical for defining Env immunogens with binding affinity for the appropriate naive B-cell receptor. This approach of B-cell lineage immunogen design has three steps (Fig. 2). The first step is the isolation of one or more clonally related broadly neutralizing mAbs from an HIV-1 infected donor. Second, using the sequences of the known mAb and next generation sequencing to expand the clonal family, one reconstructs the antibody lineage back to the UA. Third, using recombinant antibody technology, the members of the BNAb lineage are expressed and Env constructs are selected that optimally bind to each clonal lineage member at each stage of the lineage. These specific Env immunogens can be used in a prime and boost fashion to engage the naïve BCR and to further stimulate the evolution of the NAb response. (Fig. 2). Recently, Env antigens that bind to the unmutated and early intermediate antibodies of a number of B-cell lineages have been defined (109, 195), and in one case, those Envs with higher binding to the 2F5 UA have proved to induce the highest MPER antibodies (185). Importantly, detailed mapping of the co-evolution of a BNAb lineage and Env escape provides key information for informed design of either sequential or polyvalent mixtures of Envs or Env subunits. Thus, in addition to the recently defined CH103 BNAb linages, it will be important to define additional antibody lineages, including those directed to other conserved neutralization epitopes such as the MPER, V1V2-glycan, and V3-glycan sites. In addition to a highly lineage directed approach, it is also likely that the most effective vaccine-induced antibody response will be polyclonal and be directed to multiple neutralizing epitopes, thus future vaccine strategies may also include polyvalent immunogens as a way to stimulate multiple B-cell lineages or as a means to focus the response on common conserved regions (196-198).
Fig. 2. B-cell lineage-based approach to vaccine design.
Mature BNAbs can be isolated from HIV-1 infected donors using modern methods such as memory B-cell culture or sorting of antigen-specific B-cells. Based on the known BNAb sequence, next generation sequencing can be used to find numerous clonal relatives of the mature BNAb. If appropriate longitudinal samples are available, it is possible to infer the full antibody lineage, including the UA and IA. The expressed UA and IA sequences can then be used as templates for the design of HIV-1 immunogens with high affinity binding. Since the antibody lineage is known to evolve in response to viral evolution, it may be possible to design sequential immunogens with high affinity binding for the UA and IA, thus guiding the antibody response toward the mature antibody with broad neutralizing activity.
The observation that some HIV-1 BNAbs are restricted to BCRs that derive from specific VH genes has raised the additional question of how to stimulate these specific B cells. VRC01 type antibodies engage the CD4-binding site mainly via CDRH2 interactions, which is encoded by the VH gene. Thus, it is interesting that the UA (or even just V-gene reverted) version of VRC01 antibodies do not bind to heterologous Envs (117, 119, 127). This has led to the hypothesis that stimulation of such antibodies will require the rational design of modified Env immunogens to engage the appropriate BCR (121, 128-130, 139, 199). Several groups of investigators are pursuing variants of the core of gp120 (78, 179, 186, 200) or the more restricted outer domain of gp120 to stimulate CD4-binding site antibodies (201-203). Schief and colleagues (204) have recently reported on the design of an outer domain construct that can bind the V-gene reverted versions of VRC01 and related antibodies. These highly specific germline binding immunogens can be tested empirically for elicitation of VRC01 like antibodies. However, mice and presumably most other small animal species do not have a close homologue to the human VH1-2 gene. Even rhesus macaques that share a similar IgG locus and an average 93% genetic homology to humans (136) may not be the ideal model to test such immunogens, since small changes in the germline sequence may alter key contact sites (114, 119, 126). It may be possible to test such immunogens in mice engineered to have a human IgG locus, though we do not know if these mice can generate the level of affinity maturation required to achieve detectable HIV-1 neutralization (205). In contrast to the VRC01 class of antibodies, HIV-1 Env constructs have been found to bind the germline reverted versions of V1V2 (109, 134) and gp41 membrane proximal external region BNAbs (195). Likewise, the UA of the recently isolated CD4-binding site CH103 mAb can bind its donor autologous transmitted-founder Env with high affinity (118). Thus, lack of reported Env binding by germline reverted HIV-1 specific NAb appears to be a particular characteristic of the highly somatically mutated VRC01 class of CD4-binding site antibodies that bind predominantly via the V-gene encoded CDRH2. Even for these antibodies, investigators have not yet studied their development in enough detail to know if the germline antibodies can bind to the transmitted-founder Env.
Human vaccine studies: lessons from clinical efficacy studies
Early HIV-1 vaccine efficacy trials using gp120 envelope proteins demonstrated that these vaccines were not protective (56, 57). Similarly, a recombinant adenovirus type 5 vector based vaccine that expressed HIV-1 Gag, but not Env, induced antigen-specific CD8+ T cells but provided no protection (206). Taken together these data suggested a better quality of antibody responses would be needed to achieve some level of protection in a HIV-1 efficacy trial. In 2009, the results of the fourth HIV-1 vaccine efficacy trial, the RV144 trial in Thailand, was published (207). This trial used a canarypox vector (ALVAC) expressing HIV-1 clade B Gag, protease and CRF01_AE Env gp120 as a prime, and used both the ALVAC plus the same AIDSVAX® B/E gp120s used in the failed VAX003 trial as boosts. Unlike the prior two gp120 trials, this RV144 study demonstrated an estimated vaccine efficacy for prevention of transmission of 31%. While this level of efficacy was not sufficient to deploy this vaccine, it gave vaccine developers hope that a preventive vaccine is possible and that improved vaccines can be designed.
An extensive immune correlates analysis was performed on samples of RV144 vaccinees that received the vaccine. This involved a comparison of vaccine elicited immune responses in those who did not get infected versus those that did get infected (208). These analyses revealed two correlates of HIV-1 infection. First, it was shown that higher plasma levels of vaccine-elicited antibodies to the V1V2 loop was associated with a reduced risk of transmission, suggesting that these antibodies may have played a role in preventing transmission. Surprisingly, the second correlate was that high envelope plasma IgA levels correlated with infection risk, such that the higher the plasma IgA Env antibody level, the higher the transmission risk. Since there was no enhancement of overall infection risk due to the vaccine, these data suggested that high IgA levels might mitigate the effect of other potentially protective antibodies. In a separate study, investigators found a selection, or sieve effect, in the V2 region of viruses that infected RV144 vaccine recipients (209). Specifically, viruses in vaccine recipients were more likely to have an amino acid alteration at position 169 or 181 in the Env V2 region. These data complimented the finding of an association between high V1V2-binding antibodies and reduced risk of HIV-1 acquisition and further suggested that antibodies to the V2 region may have played a role in the vaccine mediated protection. To further understand the potential and mechanism of vaccine-induced V1V2 antibodies, Liao, Haynes, and colleagues (166) used an unbiased B-cell culture method to isolate mAbs from RV144 vaccine recipients. Among the 25 mAbs isolated, four were antibodies to the V2 region. Co-crystal structures and binding analysis demonstrated that these V2 mAbs recognized residue K169 within the V2 region, the same residue implicated in the sieve analysis mentioned above. Additional studies demonstrated that V1V2 antibodies that were induced by the RV144 vaccine could capture virions and neutralize infection of neutralization sensitive (tier 1 or TCLA) viruses but could not capture virions and neutralize the more-difficult-to neutralize (tier 2 or primary) viruses. However, these V1V2 antibodies could bind to tier 2 primary virus Envs on the surface of virus infected CD4 T cells and mediate their killing by natural killer cells through the process of antibody-dependent cellular cytotoxicity (ADCC). Interestingly, it has been demonstrated that RV144-induced IgA antibodies against the same epitopes as are bound by IgG, can indeed block the mediation of ADCC by IgG Env antibodies (210). A recent repeat of the immune correlates of protection in RV144 confirmed the correlation of antibody against the V1V2 region as a significant correlate of decreased transmission risk (211). Thus, these data suggest that protection in the RV144 study was mediated by a vaccine elicited antibody response, likely including antibodies to the V1V2 region of gp120. The specific in vivo mechanism of protection is not clear but could involve neutralization of a subset of sensitive viral strains, virion capture at the mucosal surface (212), and Fc receptor-mediated ADCC or related mechanisms by which antibody could kill cells expressing HIV-1 Env on the cell surface. Additional information regarding these possible mechanisms may come from passive antibody protection studies of the RV144 vaccine elicited V1V2 mAbs using in the rhesus chimeric simian-human immunodeficiency viruses (SHIVs) infection model. These immune correlates will also serve as markers for study in future vaccine efficacy trials. Efforts to improve on the RV144 immunogen by designing formulations of Env and adjuvants that can improve the quantity of potentially protective antibodies are underway. Obviously, these efforts will proceed in parallel with continued efforts to design vaccines that can elicit more robust and cross-reactive NAbs.
Closing remarks
The discovery HIV-1 broadly neutralizing mAbs and their specific modes of recognition on the viral Env provide an initial framework for improved vaccine design. Building on this knowledge, our recent understanding of immune pathways for BNAb development has provided insights for both vaccine design and immunization strategies. Together with the proof of concept that a vaccine can prevent HIV-1 infection in humans, this new knowledge has invigorated the field of HIV-1 vaccine research. Nonetheless, the way forward is formidable with the realization that a successful vaccine may have to overcome multiple immunoregulatory controls of BNAbs, and focus the immune response on subdominant epitopes. Our ultimate success will likely require an in depth knowledge of host-pathogen immunobiology unlike that required for any prior vaccine. Taking nature’s lead, the development of a vaccine that elicits potent and cross-reactive NAbs is possible and such a vaccine has the prospect of effective prevention of HIV-1 infection.
Acknowledgements
We thank Tony Moody and Rebecca Lynch for their review and suggestions for this manuscript. Supported by the intramural research program of the Vaccine Research Center, NIAID, NIH and by the NIH, NIAID Center for HIV/AIDS Vaccine immunology-Immunogen Design UM1 AI100645 and by Collaboration for AIDS Vaccine Discovery grants to BFH. The authors have no conflicts of interest to declare.
References
- 1.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 2.Barre-Sinoussi F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 3.Gallo RC, et al. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220:865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- 4.Gallo RC, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 5.Starcich BR, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 6.Robey WG, et al. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science. 1985;228:593–595. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- 7.Veronese FD, DeVico AL, Copeland TD, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science. 1985;229:1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- 8.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 9.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 10.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Deng H, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 14.Salzwedel K, Berger EA. Cooperative subunit interactions within the oligomeric envelope glycoprotein of HIV-1: functional complementation of specific defects in gp120 and gp41. Proc Natl Acad Sci USA. 2000;97:12794–12799. doi: 10.1073/pnas.230438497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss RA, Clapham PR, Weber JN, Dalgleish AG, Lasky LA, Berman PW. Variable and conserved neutralization antigens of human immunodeficiency virus. Nature. 1986;324:572–575. doi: 10.1038/324572a0. [DOI] [PubMed] [Google Scholar]
- 16.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 17.Douek DC, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 18.Folks T, Powell DM, Lightfoote MM, Benn S, Martin MA, Fauci AS. Induction of HTLV-III/LAV from a nonvirus-producing T-cell line: implications for latency. Science. 1986;231:600–602. doi: 10.1126/science.3003906. [DOI] [PubMed] [Google Scholar]
- 19.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 20.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang J, et al. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J Virol. 2002;76:11273–11282. doi: 10.1128/JVI.76.22.11273-11282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 24.Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 25.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 26.Overbaugh J, Morris L. The Antibody Response against HIV-1. Cold Spring Harbor Perp Med. 2012;2:a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong L, Sattentau QJ. Antigenicity and immunogenicity in HIV-1 antibody-based vaccine design. J AIDS Clinic Res. 2012;S8:003. doi: 10.4172/2155-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melbye M, Biggar RJ, Chermann JC, Montagnier L, Stenbjerg S, Ebbesen P. High prevalence of lymphadenopathy virus (LAV) in European haemophiliacs. Lancet. 1984;2:40–41. doi: 10.1016/s0140-6736(84)92023-3. [DOI] [PubMed] [Google Scholar]
- 30.Cheng-Mayer C, Seto D, Tateno M, Levy JA. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 31.Robert-Guroff M, Brown M, Gallo RC. HTLV-III-neutralizing antibodies in patients with AIDS and AIDS-related complex. Nature. 1985;316:72–74. doi: 10.1038/316072a0. [DOI] [PubMed] [Google Scholar]
- 32.Weiss RA, et al. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature. 1985;316:69–72. doi: 10.1038/316069a0. [DOI] [PubMed] [Google Scholar]
- 33.Steimer KS, Scandella CJ, Skiles PV, Haigwood NL. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to Gp120. Science. 1991;254:105–108. doi: 10.1126/science.1718036. [DOI] [PubMed] [Google Scholar]
- 34.Chakrabarti S, Robert-Guroff M, Wong-Staal F, Gallo RC, Moss B. Expression of the HTLV-III envelope gene by a recombinant vaccinia virus. Nature. 1986;320:535–537. doi: 10.1038/320535a0. [DOI] [PubMed] [Google Scholar]
- 35.Graham BS, et al. Vaccination of vaccinia-naive adults with human immunodeficiency virus type 1 gp160 recombinant vaccinia virus in a blinded, controlled, randomized clinical trial. The AIDS Vaccine Clinical Trials Network. J Infect Dis. 1992;166:244–252. doi: 10.1093/infdis/166.2.244. [DOI] [PubMed] [Google Scholar]
- 36.Dolin R, et al. The safety and immunogenicity of a human immunodeficiency virus type 1 (HIV-1) recombinant gp160 candidate vaccine in humans. NIAID AIDS Vaccine Clinical Trials Network. Ann Intern Med. 1991;114:119–127. doi: 10.7326/0003-4819-114-2-119. [DOI] [PubMed] [Google Scholar]
- 37.Kovacs JA, et al. Induction of humoral and cell-mediated anti-human immunodeficiency virus (HIV) responses in HIV sero-negative volunteers by immunization with recombinant gp160. J Clin Invest. 1993;92:919–928. doi: 10.1172/JCI116667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz DH, et al. Induction of HIV-1-neutralising and syncytium-inhibiting antibodies in uninfected recipients of HIV-1IIIB rgp120 subunit vaccine. Lancet. 1993;342:69–73. doi: 10.1016/0140-6736(93)91283-r. [DOI] [PubMed] [Google Scholar]
- 39.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 40.Mascola JR, et al. Two antigenically distinct subtypes of human immunodeficiency virus type 1: viral genotype predicts neutralization serotype. J Infect Dis. 1994;169:48–54. doi: 10.1093/infdis/169.1.48. [DOI] [PubMed] [Google Scholar]
- 41.Sawyer LS, et al. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrin T, Loh TP, Vennari JC, Schuitemaker H, Nunberg JH. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daar ES, Li XL, Moudgil T, Ho DD. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrin T, Nunberg JH. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS. 1994;8:1622–1623. doi: 10.1097/00002030-199411000-00017. [DOI] [PubMed] [Google Scholar]
- 45.Mascola JR, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 46.Palker TJ, et al. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Javaherian K, et al. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci USA. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goudsmit J, et al. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc Natl Acad Sci USA. 1988;85:4478–4482. doi: 10.1073/pnas.85.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VanCott TC, et al. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary human immunodeficiency virus type 1 envelope. J Virol. 1999;73:4640–4650. doi: 10.1128/jvi.73.6.4640-4650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanCott TC, et al. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J Virol. 1997;71:4319–4330. doi: 10.1128/jvi.71.6.4319-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matthews TJ. Dilemma of neutralization resistance of HIV-1 field isolates and vaccine development. AIDS Res Hum Retroviruses. 1994;10:631–632. doi: 10.1089/aid.1994.10.631. [DOI] [PubMed] [Google Scholar]
- 52.Wu X, et al. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology. 2008;380:285–295. doi: 10.1016/j.virol.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binley JM, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vancott TC, Polonis VR, Loomis LD, Michael NL, Nara PL, Birx DL. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res Hum Retroviruses. 1995;11:1379–1391. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- 55.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 56.Gilbert PB, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 57.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 58.McCoy LE, Weiss RA. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med. 2013;210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graham BS, Mascola JR. Lessons from failure--preparing for future HIV-1 vaccine efficacy trials. J Infect Dis. 2005;191:647–649. doi: 10.1086/428406. [DOI] [PubMed] [Google Scholar]
- 60.Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5:579–595. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- 61.Pilgrim AK, et al. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J Infect Dis. 1997;176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- 62.Deeks SG, et al. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol. 2006;80:6155–6164. doi: 10.1128/JVI.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatos NM, et al. Neutralizing antibodies from the sera of human immunodeficiency virus type 1-infected individuals bind to monomeric gp120 and oligomeric gp140. J Virol. 1998;72:9656–9667. doi: 10.1128/jvi.72.12.9656-9667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 65.Trkola A, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 69.Mascola JR, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89. 6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 71.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathogens. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wyatt R, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 75.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 76.Cardoso RM, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 77.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 80.Alam SM, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies. 2005;14:59–67. [PMC free article] [PubMed] [Google Scholar]
- 82.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zwick MB, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burton DR, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 87.Johnston MI, Fauci AS. An HIV vaccine--challenges and prospects. N Engl J Med. 2008;359:888–890. doi: 10.1056/NEJMp0806162. [DOI] [PubMed] [Google Scholar]
- 88.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Walker LM, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathogens. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gray ES, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Albert J, et al. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 94.Montefiori DC, et al. Homotypic antibody responses to fresh clinical isolates of human immunodeficiency virus. Virology. 1991;182:635–643. doi: 10.1016/0042-6822(91)90604-a. [DOI] [PubMed] [Google Scholar]
- 95.Montefiori DC, et al. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J Infect Dis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- 96.Darden JM, et al. A flow cytometric method for measuring neutralization of HIV-1 subtype B and E primary isolates. Cytometry. 2000;40:141–150. [PubMed] [Google Scholar]
- 97.Mascola JR, et al. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J Virol. 2002;76:4810–4821. doi: 10.1128/JVI.76.10.4810-4821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mascola JR, et al. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79:10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li M, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Doria-Rose NA, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathogens. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol. 2008;82:7932–7941. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 105.Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS. 2007;21:693–702. doi: 10.1097/QAD.0b013e32805e8727. [DOI] [PubMed] [Google Scholar]
- 106.Lynch RM, et al. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J Virol. 2011;85:905–915. doi: 10.1128/JVI.02006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 109.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 113.Scheid JF, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morris L, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PloS One. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PloS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013 doi: 10.1038/nature12053. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou T, et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu Z, et al. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J Virol. 2011;85:11401–11408. doi: 10.1128/JVI.05312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 124.Wu X, Zhou T, O’Dell S, Wyatt RT, Kwong PD, Mascola JR. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J Virol. 2009;83:10892–10907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.West AP, Jr., Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hoot S, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathogens. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiao X, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochemi Biophys Res Comm. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moir S, Malaspina A, Fauci AS. Prospects for an HIV vaccine: leading B cells down the right path. Nat Struct Mol Biol. 2011;18:1317–1321. doi: 10.1038/nsmb.2194. [DOI] [PubMed] [Google Scholar]
- 131.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Doria-Rose NA, et al. A short segment of the HIV-1 gp120 V1/V2 region Is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol. 2012;86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pancera M, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Douagi I, et al. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol. 2010;84:1683–1695. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sundling C, et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Trans Med. 2012;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sundling C, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Beddows S, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360:329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 139.Kwong PD, Mascola JR, Nabel GJ. Rational Design of Vaccines to Elicit Broadly Neutralizing Antibodies to HIV-1. Cold Spring Harbor Persp Med. 2011;1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Burton DR, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tomaras GD, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Parren PW, Burton DR, Sattentau QJ. HIV-1 antibody--debris or virion? Nat Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- 145.Sun ZY, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 146.Ofek G, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Julien JP, et al. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol. 2010;84:4136–4147. doi: 10.1128/JVI.02357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pietzsch J, Scheid JF, Mouquet H, Seaman MS, Broder CC, Nussenzweig MC. Anti-gp41 antibodies cloned from HIV-infected patients with broadly neutralizing serologic activity. J Virol. 2010;84:5032–5042. doi: 10.1128/JVI.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Song L, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci USA. 2009;106:9057–9062. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mehandru S, et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J Virol. 2007;81:11016–11031. doi: 10.1128/JVI.01340-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vcelar B, et al. Reassessment of autoreactivity of the broadly neutralizing HIV antibodies 4E10 and 2F5 and retrospective analysis of clinical safety data. AIDS. 2007;21:2161–2170. doi: 10.1097/QAD.0b013e328285da15. [DOI] [PubMed] [Google Scholar]
- 153.Li Y, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Verkoczy L, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci USA. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Verkoczy L, et al. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH x VL knockin mice reveals multiple tolerance controls. J Immunol. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PloS One. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liao HX, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr. Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. J Immunol. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 160.Meffre E, Milili M, Blanco-Betancourt C, Antunes H, Nussenzweig MC, Schiff C. Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. J Clin Invest. 2001;108:879–886. doi: 10.1172/JCI13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Briney BS, Willis JR, Crowe JE., Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PloS One. 2012;7:e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Larimore K, McCormick MW, Robins HS, Greenberg PD. Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol. 2012;189:3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 163.Briney BS, Willis JR, Hicar MD, Thomas JW, 2nd, Crowe JE., Jr. Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology. 2012;137:56–64. doi: 10.1111/j.1365-2567.2012.03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bonsignori M, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moody MA, et al. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. J Virol. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liao HX, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dal Porto JM, Haberman AM, Shlomchik MJ, Kelsoe G. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol. 1998;161:5373–5381. [PubMed] [Google Scholar]
- 168.Dal Porto JM, Haberman AM, Kelsoe G, Shlomchik MJ. Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med. 2002;195:1215–1221. doi: 10.1084/jem.20011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shih TA, Meffre E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- 170.Bonsignori M, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pancera M, et al. N332-directed broadly veutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PloS One. 2013;8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu J, et al. Somatic populations of PGT135-137 HIV-1-neutralizing antibodies identified by 454 pyrosequencing and bioinformatics. Frontiers Microbiol. 2012;3:315. doi: 10.3389/fmicb.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Briney BS, Willis JR, McKinney BA, Crowe JE., Jr. High-throughput antibody sequencing reveals genetic evidence of global regulation of the naive and memory repertoires that extends across individuals. Genes Immun. 2012;13:469–473. doi: 10.1038/gene.2012.20. [DOI] [PubMed] [Google Scholar]
- 174.Moore PL, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathogens. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Moore PL, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tobin GJ, et al. Deceptive imprinting and immune refocusing in vaccine design. Vaccine. 2008;26:6189–6199. doi: 10.1016/j.vaccine.2008.09.080. [DOI] [PubMed] [Google Scholar]
- 177.Srivastava IK, VanDorsten K, Vojtech L, Barnett SW, Stamatatos L. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J Virol. 2003;77:2310–2320. doi: 10.1128/JVI.77.4.2310-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hu SL, Stamatatos L. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr HIV Res. 2007;5:507–513. doi: 10.2174/157016207782418542. [DOI] [PubMed] [Google Scholar]
- 179.Dey B, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathogens. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dennison SM, et al. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PloS One. 2011;6:e27824. doi: 10.1371/journal.pone.0027824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Srivastava IK, et al. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol. 2003;77:11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xu R, et al. Immunization with HIV-1 SF162-derived Envelope gp140 proteins does not protect macaques from heterologous simian-human immunodeficiency virus SHIV89.6P infection. Virology. 2006;349:276–289. doi: 10.1016/j.virol.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 183.Barnett SW, et al. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J Virol. 2001;75:5526–5540. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pantophlet R, Wilson IA, Burton DR. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J Virol. 2003;77:5889–5901. doi: 10.1128/JVI.77.10.5889-5901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ma BJ, et al. Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathogens. 2011;7:e1002200. doi: 10.1371/journal.ppat.1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Morner A, et al. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J Virol. 2009;83:540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guenaga J, et al. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PloS One. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci USA. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Azoitei ML, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 190.Correia BE, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 191.Tong T, Crooks ET, Osawa K, Binley JM. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol. 2012;86:3574–3587. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci USA. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Leaman DP, Zwick MB. Increased Functional Stability and Homogeneity of Viral Envelope Spikes through Directed Evolution. PLoS Pathogens. 2013;9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Montefiori DC, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alam SM, et al. Differential Reactivity of Germ Line Allelic Variants of a Broadly Neutralizing HIV-1 Antibody to a gp41 Fusion Intermediate Conformation. J Virol. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cho MW, et al. Polyvalent envelope glycoprotein vaccine elicits a broader neutralizing antibody response but is unable to provide sterilizing protection against heterologous simian/human immunodeficiency virus infection in pigtailed macaques. J Virol. 2001;75:2224–2234. doi: 10.1128/JVI.75.5.2224-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pal R, et al. Immunization of rhesus macaques with a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine elicits protective antibody response against simian human immunodeficiency virus of R5 phenotype. Virology. 2006;348:341–353. doi: 10.1016/j.virol.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 198.Wang S, et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine. 2008;26:3947–3957. doi: 10.1016/j.vaccine.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Xiao X, Chen W, Feng Y, Dimitrov DS. Maturation Pathways of Cross-Reactive HIV-1 Neutralizing Antibodies. Viruses. 2009;1:802–817. doi: 10.3390/v1030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dey B, et al. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol. 2007;81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang X, et al. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol. 2004;78:12975–12986. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Joyce MG, et al. Outer domain of HIV-1 gp120: antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. J Virol. 2013;87:2294–2306. doi: 10.1128/JVI.02717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu L, et al. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol. 2009;83:5077–5086. doi: 10.1128/JVI.02600-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jardine J, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013 doi: 10.1126/science.1234150. DOI:10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lonberg N. Human antibodies from transgenic animals. Nat Biotechnol. 2005;23:1117–1125. doi: 10.1038/nbt1135. [DOI] [PubMed] [Google Scholar]
- 206.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 208.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rolland M, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tomaras G, et al. HIV-1 vaccine-induced envelope gp120 C1-region IgA blocks binding and effector function of gp120 IgG. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1301456110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zolla-Pazner S, et al. Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PloS One. 2013;8:e53629. doi: 10.1371/journal.pone.0053629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hladik F, Hope TJ. HIV infection of the genital mucosa in women. Current HIV/AIDS Rep. 2009;6:20–28. doi: 10.1007/s11904-009-0004-1. [DOI] [PubMed] [Google Scholar]