The MSL (male-specific lethal) complex is required for increased transcription of the X chromosome in males during Drosophila dosage compensation. Soruco et al. now show that the conserved non-sex-specific CLAMP (chromatin-linked adaptor for MSL proteins) zinc finger protein functions as the previously unknown link between the MSL complex and the male X chromosome. The data support a novel mechanism for X identification that involves integration of the functions of X-linked long noncoding RNAs and a sequence-specific DNA-binding protein.
Keywords: dosage compensation, chromatin, zinc finger protein, Drosophila
Abstract
The Drosophila male-specific lethal (MSL) dosage compensation complex increases transcript levels on the single male X chromosome to equal the transcript levels in XX females. However, it is not known how the MSL complex is linked to its DNA recognition elements, the critical first step in dosage compensation. Here, we demonstrate that a previously uncharacterized zinc finger protein, CLAMP (chromatin-linked adaptor for MSL proteins), functions as the first link between the MSL complex and the X chromosome. CLAMP directly binds to the MSL complex DNA recognition elements and is required for the recruitment of the MSL complex. The discovery of CLAMP identifies a key factor required for the chromosome-specific targeting of dosage compensation, providing new insights into how subnuclear domains of coordinate gene regulation are formed within metazoan genomes.
Diverse mechanisms have evolved to correct for dosage imbalances between genes on the X chromosomes and autosomes in mammals, Caenorhabditis elegans, and Drosophila (Gelbart and Kuroda 2009). In mammals, dosage compensation involves inactivation of one of the two X chromosomes in females (Nguyen and Disteche 2006; Payer and Lee 2008; Meyer 2010). In C. elegans, gene expression along each of the two hermaphroditic X chromosomes is reduced (Meyer 2010). In Drosophila, dosage compensation increases the transcript levels from most active genes along the length of the single male X chromosome by at most twofold to equal expression from the two female X chromosomes (Belote and Lucchesi 1980; Hamada et al. 2005; Larschan et al. 2007). A unifying feature of these dosage compensation mechanisms is that they all involve distinguishing the X chromosome from autosomes to coordinately regulate a large number of genes—a process that remains poorly understood.
The key regulator of dosage compensation in Drosophila is the male-specific lethal (MSL) complex, which is expressed specifically in males and distinguishes the X chromosome from the autosomes (Belote and Lucchesi 1980; Gelbart and Kuroda 2009). It has been hypothesized that the MSL complex identifies its X-chromosome target genes in a three-stage process. First, the MSL complex recognizes the sites of transcription of its roX1 and roX2 long noncoding RNA components, which are encoded on the X chromosome, and cotranscriptionally incorporates them (Kelley et al. 1999, 2008). Second, the MSL complex recognizes 150–300 additional genomic loci distributed across the X chromosome, called “chromatin entry sites” (CESs). CESs were defined by their high levels of MSL complex occupancy and retention of the core MSL components in the absence of MSL3 (Kelley et al. 1999; Alekseyenko et al. 2008). Most CESs contain one or multiple 21-base-pair (bp) GA-rich cis-acting DNA sequences called MSL recognition elements (MREs) that are required for MSL complex recruitment, but there are no known sequence-specific contacts between the MSL complex and MREs (Alekseyenko et al. 2008; Straub et al. 2008; Fauth et al. 2010). Third, the MSL complex spreads from CESs to the bodies of active genes on the X chromosome by recognizing histone modifications associated with transcriptional activation, including trimethylation of H3K36 (H3K36me3) (Larschan et al. 2007; Bell et al. 2008). Once active genes are recognized, their transcript levels are increased by the MSL complex (Hamada et al. 2005; Larschan et al. 2011).
While both the roX noncoding RNAs and MREs contribute to the recognition of the male X chromosome, they are individually insufficient to generate X-chromosome specificity (Meller and Rattner 2002; Park et al. 2002; Alekseyenko et al. 2008; Straub et al. 2008). Therefore, we hypothesized that additional factors are required to tether and enrich the MSL complex at its target sites on the male X chromosome. Thus, we performed a cell-based genome-wide RNAi screen, allowing for the possibility that MSL regulators might have genome-wide roles in both males and females in addition to X-specific roles in males (Larschan et al. 2012) that would not have been recovered from the powerful MSL screens that identified all of the MSL complex components (Belote and Lucchesi 1980). Such non-sex-specific regulators provide a key opportunity to understand the first steps in X identification, including recognition of highly conserved MREs within CESs (Alekseyenko et al. 2013).
To define new regulators of MSL complex targeting or function, our genome-wide RNAi screen identified proteins that specifically modulate the activity of a luciferase reporter gene fused to the CESs adjacent to roX2 (Larschan et al. 2012). Among many other candidate-positive regulators, we identified the conserved non-sex-specific CG1832 protein that has a glutamine-rich N terminus and a C-terminal domain containing seven C2H2 zinc fingers (Larschan et al. 2012). We found that CG1832 is enriched at CESs and that CG1832 RNAi strongly reduces MSL complex recruitment at five CESs tested in male tissue culture cells (Larschan et al. 2012). However, key questions remained: (1) Does CG1832 provide a direct link between the MSL complex and the X chromosome? (2) Does CG1832 regulate MSL complex recruitment to the entire X chromosome in flies? (3) Is CG1832 enriched on the X chromosome independent of the MSL complex to facilitate the discrimination of the X chromosome from autosomes?
Here, we named CG1832 CLAMP (chromatin-linked adaptor for MSL proteins). We identify CLAMP as the previously unknown link between the MSL complex and the X chromosome. First, we show that CLAMP directly recognizes MREs and is required for recruitment of the MSL complex to the entire X chromosome. Second, we demonstrate that CLAMP is enriched at key CESs independent of the MSL complex. Third, we show that the MSL complex and CLAMP have a synergistic occupancy relationship that increases the X enrichment of both factors at critical CESs such as those at the roX loci. Our results support a mechanism whereby the MSL complex uses the X-enriched CLAMP protein at CESs followed by further enrichment from synergistic interactions between the two factors.
Results and Discussion
CLAMP directly recognizes MREs to tether the MSL complex to the X chromosome
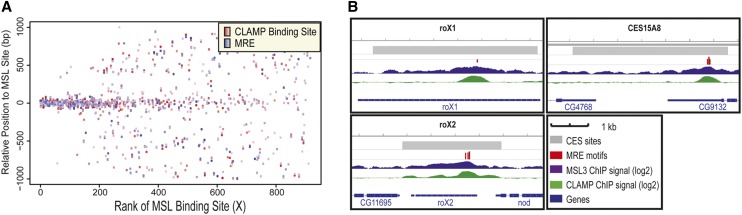
To determine whether CLAMP and the MSL complex colocalize at CESs in vivo, we performed CLAMP chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) experiments in male SL2 cells and compared CLAMP occupancy profiles with available MSL complex occupancy profiles (Fig. 1; Alekseyenko et al. 2008; Larschan et al. 2012). We found CLAMP occupancy at many loci throughout the genome (X: 2695; autosomes: 10,009) and strong colocalization with MRE sequences (Fig. 1A,B; Supplemental Fig. S1). Moreover, CLAMP and the MSL complex exhibit their highest occupancy levels when colocalized (Fig. 1A; Supplemental Fig. S2A,B). Nonoverlapping CLAMP occupancy sites occur at 5′ ends of active genes on all chromosomes (Supplemental Fig. S2C), and nonoverlapping MSL-binding sites are localized over the bodies of active X-linked genes (Supplemental Fig. S2B). Furthermore, CLAMP sites occur precisely over MREs within CESs compared with the broader domains of MSL complex occupancy (Figs. 1A,B, 3A [below]). Together, these patterns are consistent with a role for CLAMP in recruiting the MSL complex to CESs, followed by spreading of the MSL complex to the bodies of active genes and additional non-sex-specific roles for CLAMP at 5′ ends of active genes.
Figure 1.
CLAMP localizes precisely at MREs at high levels when colocalized with the MSL complex. (A) Occupancy relationships calculated from input-normalized CLAMP ChIP-seq from male SL2 cells (Larschan et al. 2012) and previously defined CESs (Alekseyenko et al. 2008) (two biological replicates). MSL complex-binding sites ranked based on MSL complex occupancy from ChIP-seq read counts (X-axis; strongest on the left and weakest on the right) (Alekseyenko et al. 2008). The Y-axis indicates the position relative to the center of the MSL complex sites. Each red dot represents the position of a CLAMP-binding site that is within 1 kb of an MSL complex-binding site. The intensity of the red color is proportional to CLAMP occupancy (see the Materials and Methods). The intensity of the blue color is proportional to similarity of the individual MRE sequence to the consensus sequence. (B) Examples of CLAMP and MSL complex occupancy at three CESs are shown from SL2 cells. Input-normalized read counts are shown on a log scale for CLAMP and the MSL complex.
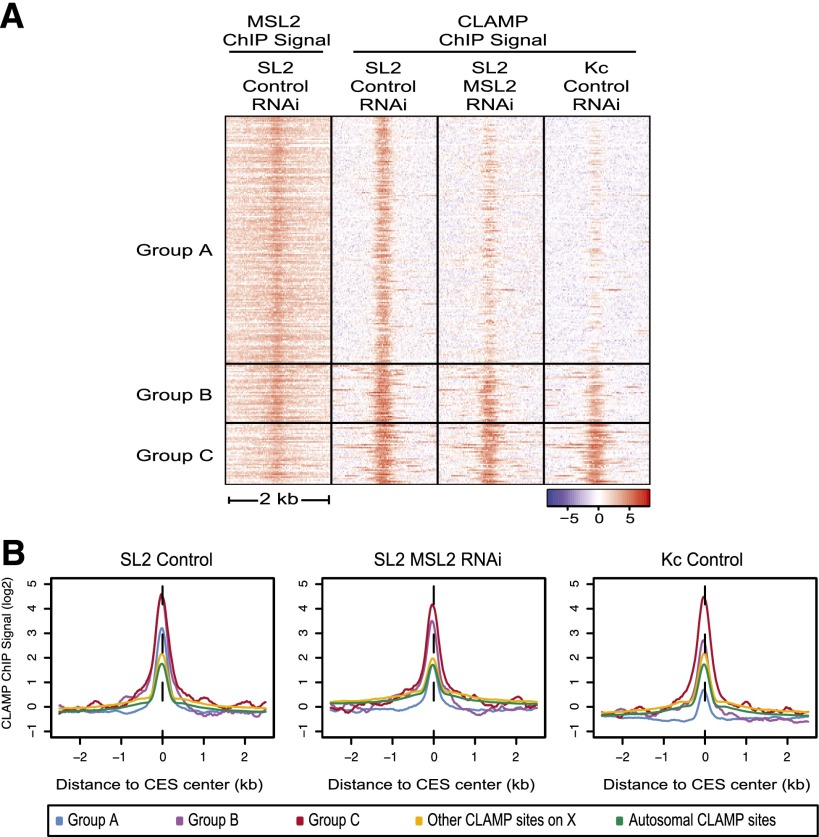
Figure 3.
CESs have three classes of CLAMP occupancy patterns. (A) CLAMP or MSL2 occupancy patterns (reads per kilobase per million mapped reads [RPKM] normalized to input) are shown within a 2-kb window centered at the MRE for each member of each subclass of CESs (265 total CESs). (1) Group A sites are highly MSL-dependent. (2) Group B sites are partially MSL-dependent. (3) Group C sites are independent of the MSL complex. (B) Average profiles of CLAMP occupancy (RPKM) normalized to input are centered at group A, group B, or group C CES peaks and are shown compared with average occupancy at all CLAMP sites on the X or autosomes (log2 scale).
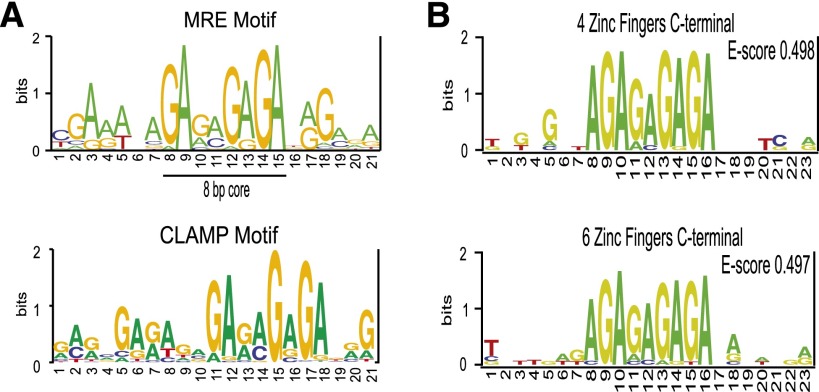
Next, we used the MEME software package (Bailey et al. 2009) to generate a position weight matrix (PWM) representing the most enriched sequence within 500 regions surrounding CLAMP occupancy sites (2 kb each) at the 5′ and 3′ ends of genes genome-wide. All CLAMP localization motifs on the X chromosome and on autosomes shared an 8-bp GA-rich core region that is very similar to the previously identified MRE consensus sequence (Fig. 2A). Therefore, we hypothesized that CLAMP functions as an MRE recognition factor.
Figure 2.
CLAMP directly binds to MRE sequences. (A, top) PWM for the original MRE motif (Alekseyenko et al. 2008). (Bottom) PWMs were derived from CLAMP ChIP-seq data from SL2 cells. MEME was used to analyze 200-bp regions flanking 500 CLAMP occupancy sites that were located at the 3′ ends of active genes on the X chromosome. (B) PBM analysis resulted in direct in vitro binding specificities of the C-terminal CLAMP four (top) and six (bottom) zinc finger fragments.
To test this hypothesis, we characterized the in vitro DNA-binding specificity of CLAMP. We used protein-binding microarrays (PBMs) to assay CLAMP binding to all possible 10-bp sequences (Berger et al. 2006). Each PBM is a dsDNA array that contains all possible 10-bp sequences embedded within variable flanking sequences. After testing the complete seven-finger CLAMP construct and many additional combinations of zinc fingers, we were able to express soluble protein for the C-terminal six-finger and four-finger zinc finger regions of CLAMP (see the Materials and Methods). The in vitro PBM analysis for these proteins yielded statistically significant 8-bp motifs with high PBM enrichment (E-score = 0.497; E-score = 0.498), in excellent agreement with the CLAMP in vivo binding-site motif and the previously defined MRE consensus (Fig. 2B). Therefore, CLAMP binds directly to the MRE motif in vitro.
Next, we examined whether MREs are required for the recruitment of CLAMP in vivo in male and female larvae using available stocks with three intact or mutated MREs embedded within a minimal 150-bp fragment of CES5C2 inserted site specifically into an autosomal location (Alekseyenko et al. 2008; Supplemental Fig. S2D). Occupancy of both the MSL complex and CLAMP is greater on the X chromosome compared with the autosomal insertion (Supplemental Fig. S2D). The autosomal insertion with three intact MREs shows greater occupancy of CLAMP than that in which the 8-bp core of each of the three MREs is mutated. Therefore, MREs are required for CLAMP recruitment in both males and females, as expected for a non-sex-specific factor.
To determine whether enrichment at CESs is specific to CLAMP or is a common feature shared with other factors that can recognize MRE-like sequences, we examined a well-characterized Drosophila DNA-binding protein, the GAGA factor (GAF) (Lee et al. 2008). In contrast to CLAMP, which is strongly enriched at CESs, GAF occupancy was lower at CESs compared with other GAF-binding sites throughout the genome (Supplemental Fig. S2E,F). These data suggest that CLAMP, but not GAF, directly recognizes MRE elements within CESs. In addition, CLAMP was recently shown to physically associate with the MSL complex on chromatin using an approach that combined ChIP with mass spectrometry (Wang et al. 2013). Therefore, CLAMP directly recognizes MREs and physically associates with the MSL complex, providing the first link between the MSL complex and the X chromosome.
CLAMP is required for MSL complex recruitment to the male X chromosome in vivo
We previously demonstrated that CLAMP is required for recruitment of the MSL complex to five CESs in SL2 male tissue culture cells (Larschan et al. 2012). To determine whether CLAMP is required for MSL complex recruitment to the entire X chromosome in flies, we used a publicly available RNAi transgenic Drosophila line to reduce CLAMP mRNA levels (http://www.flyrnai.org; see the Materials and Methods). Because CLAMP is highly expressed in all tissues, we expressed the RNAi construct ubiquitously (http://www.flybase.org; see the Materials and Methods). Although the CLAMP RNAi construct did not completely eliminate CLAMP mRNA levels (Supplemental Fig. S3A), immunostaining of polytene chromosomes for CLAMP (Supplemental Fig. S3A,C) and CLAMP ChIP occupancy (Supplemental Fig. S3D,E) were strongly reduced. CLAMP RNAi causes a pupal lethal phenotype in both males and females (Larschan et al. 2012).
Next, we used ChIP for the MSL2 core component to examine MSL complex recruitment to five CESs in the presence and absence of CLAMP RNAi treatment (Supplemental Fig. S3F). Our results showed that CLAMP RNAi strongly reduced the occupancy of the MSL complex in male larvae (Supplemental Fig. S3F). We measured H3 occupancy by ChIP to determine whether CLAMP RNAi globally disrupts chromatin and found only modest changes after CLAMP RNAi (Supplemental Fig. S3G). Also, we performed polytene staining for the MSL3 protein after CLAMP RNAi and observed no detectable MSL3 staining, revealing that CLAMP is required for MSL complex recruitment to the entire X chromosome (Supplemental Fig. S3B). Thus, CLAMP promotes MSL complex localization to the entire X chromosome in flies.
It was previously shown that loss of targeting of the MSL complex reduces complex stability, likely due to ubiquitylation of complex members by MSL2 (Copps et al. 1998; Villa et al. 2012). Therefore, measuring MSL protein levels after CLAMP RNAi would not distinguish between effects on complex stability and targeting to the X chromosome. However, we previously demonstrated that CLAMP RNAi does not affect mRNA levels of MSL complex components (Larschan et al. 2012), and CLAMP is strongly enriched at 92% of CESs (Fig. 1; Supplemental Fig. S1), suggesting that indirect effects on MSL complex stability are unlikely (Larschan et al. 2012).
The MSL complex is highly enriched on the X chromosome compared with autosomal controls (Supplemental Fig. S3B; Alekseyenko et al. 2008) but mediates at most a twofold increase in transcription. Furthermore, only a 1.4-fold decrease in expression of X-linked genes is seen after MSL2 RNAi in male SL2 cells (Hamada et al. 2005). To determine how CLAMP regulates gene expression globally, we conducted mRNA sequencing (mRNA-seq) experiments in male SL2 cells before and after CLAMP RNAi treatment. As expected for a non-sex-specific factor present on both the X chromosome and autosome (Supplemental Figs. S2, S3B), CLAMP RNAi caused changes in gene expression genome-wide. However, CLAMP RNAi caused a greater decrease in the gene expression of X-linked genes compared with autosomal genes (P < 9.1 ×10−5), consistent with a function in MSL complex targeting. Consistent with its localization throughout the genome and enrichment on the X chromosome (Supplemental Fig. S1), CLAMP RNAi affects gene expression on both the X chromosome and autosomes with a bias toward X-linked genes.
In contrast to average X-linked genes, the genes that encode the roX noncoding RNA components are highly dependent on the MSL complex (Bai et al. 2004). Therefore, we measured the levels of roX RNAs before and after CLAMP RNAi treatment by quantitative RT–PCR (qRT–PCR) in male and female third instar larvae (Supplemental Fig. S3A). In males, we found that roX2 levels were reduced fourfold after CLAMP RNAi, indicating that CLAMP serves as an activator of roX2 expression. In contrast, roX1 levels were largely unchanged in males after CLAMP RNAi. In females, roX1 was significantly increased after CLAMP RNAi treatment (6.5-fold), and roX2 levels remained largely unchanged. These patterns of CLAMP-mediated regulation agree with the established differential regulation of roX1 and roX2 and recent genetic evidence implicating roX2 as the first site of X identification (Meller and Rattner 2002; Bai et al. 2004; Lim and Kelley 2012).
CLAMP is enriched on the X chromosome independent of the MSL complex
Identifying CLAMP as the protein that directly recognizes MREs provides a key opportunity to reveal new insight into the process of X identification. We hypothesized that X enrichment of CLAMP independent of the MSL complex would indicate that CLAMP could function as a beacon on the X chromosome to distinguish it from autosomes.
To determine how the MSL complex regulates CLAMP occupancy, we compared CLAMP occupancy at MREs on the X chromosome and on autosomes in male cells before and after MSL2 RNAi and in female cells that lack the MSL complex. Overall, we found that the percentage of MREs occupied by CLAMP was greater on the X chromosome when compared with autosomes in both male cells (50% on X vs. 32% on autosomes) and female cells (62% on X vs. 38% on autosomes) (Supplemental Fig. S4A–C). Therefore, CLAMP is enriched at X-chromosome MREs compared with autosomal MREs, independent of the MSL complex.
Next, we determined how CLAMP occupancy varies in its dependence on the MSL complex at MREs within CESs. To define the total number of likely CESs for subsequent analysis, we determined how the largest class of previously characterized CESs (306 loci) (Sural et al. 2008) overlapped with CLAMP sites present in male cells and found 265 common sites (87% overlap). Next, we clustered these sites based on their overall binding pattern and occupancy level of CLAMP in male cells, male cells after hypomorphic MSL2 RNAi (Hamada et al. 2005), and female cells (Fig. 3A; see the Supplemental Material). We also plotted the average occupancy at each of these classes of CESs compared with the average CLAMP site on the X chromosome and the autosome (Fig. 3B). In this way, three distinct classes of CESs were defined (Fig. 3A,B). Group A: Largely MSL-dependent CESs exhibit a strongly decreased CLAMP occupancy in the absence of the MSL complex, and their occupancy in female cells is much below that of the average CLAMP site (177 sites). Group B: Partially MSL-dependent CESs, including both roX loci, exhibit higher than average levels of CLAMP occupancy in the absence of the MSL complex and further increase in their occupancy and breadth in the presence of the MSL complex (43 sites). Group C: Largely MSL-independent CESs have higher than average CLAMP occupancy in both male and female cell lines (45 sites). ChIP-qPCR was used to validate these classes in both cell culture and larvae (Supplemental Fig. S4D).
Thus, group B and group C CESs, including both roX loci, have higher than average levels of MSL-independent CLAMP occupancy compared with group A sites, which have lower than average MSL-independent CLAMP occupancy. These data suggest that group B and group C CESs may function as beacons to promote X identification because they are occupied by CLAMP independent of the MSL complex. In addition, CLAMP associates with chromatin interdependently at group A and group B sites (Fig. 3; Supplemental Fig. S3B,F). Therefore, synergistic interactions between CLAMP and the MSL complex at group A and group B sites likely increase the occupancy of both factors.
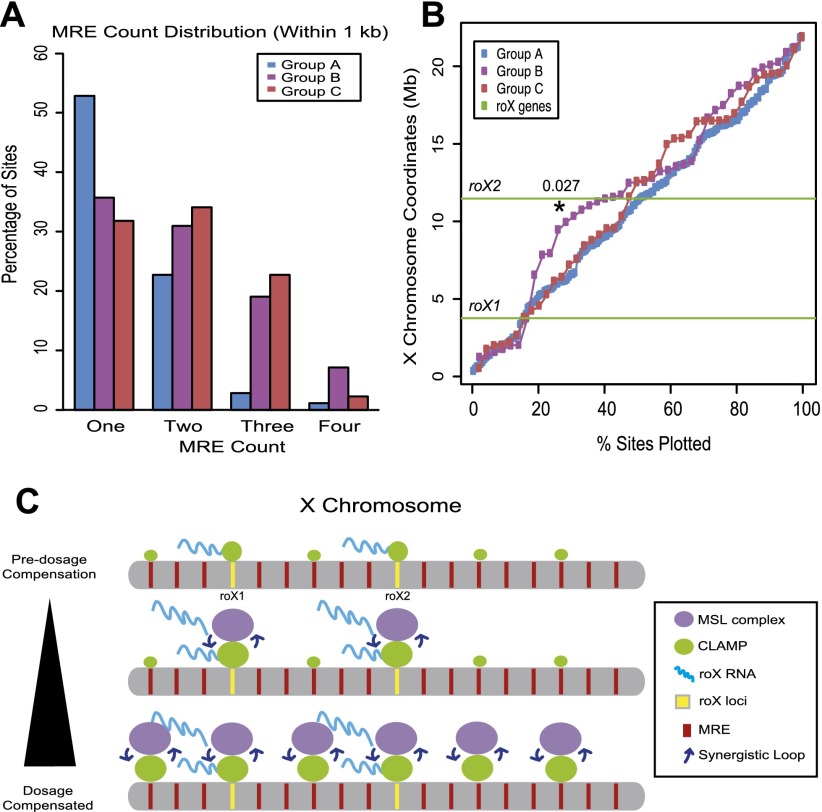
To define additional common features that distinguish each subclass of CESs, we compared the following properties: the number of tandem MREs within each CES, the two-dimensional clustering of CESs along the X chromosome, and the average nucleosome occupancy in the absence of the MSL complex (Fig. 4; Supplemental Fig. S4). First, we found a strong increase in the percentage of group B and group C sites with three or more tandem MREs present within each CES locus compared with group A sites (Fig. 4A). Second, group B sites are clustered in two dimensions along the length of the X within a 5-Mb region surrounding roX2 (P < 0.027), with the majority of sites between the roX genes (Fig. 4B). In contrast, group A sites are uniformly distributed, and group C sites exhibit minimal clustering distal to the group B sites. Third, we examined chromatin organization at each subclass of sites in the absence of the MSL complex using publicly available female cell data sets (Supplemental Fig. S4E; Henikoff et al. 2009; http://www.modencode.org). Under high-salt extraction conditions, which are likely to provide the most accurate nucleosome occupancy profiles over CESs, the decreased nucleosome density at group B and group C loci in female cells resembles that previously reported for male cells (Supplemental Fig. S4E; Alekseyenko et al. 2012).
Figure 4.
Three subclasses of CESs differ in their number of tandem MREs and two-dimensional clustering along the X chromosome. (A) Analysis of the number of tandem MREs within each subclass of CESs. (B) The locations of CESs from each subclass of sites are plotted to allow their relative positions along the length of the X chromosome to be determined. The Y-axis is the position along the length of the X chromosome, and the X-axis shows the percentage of sites plotted. The positions of the roX loci are shown as green lines. (C) Model for CLAMP and roX RNAs functioning together during X identification: (1) CLAMP binds to MREs within group B and group C independent of the MSL complex, and roX RNAs are transcribed independent of MSL complex recruitment. (2) The MSL complex is likely to be cotranscriptionally assembled at roX loci. CLAMP tethers the MSL complex to CESs, including roX loci. The MSL complex catalyzes H4K16ac, thereby opening chromatin and increasing the accessibility of MRE sequences for CLAMP recognition. Next, increased levels of CLAMP associate with CESs and thereby recruit additional MSL complexes via synergistic interactions. (3) The MSL complex spreads to additional CESs, potentially stabilized by CLAMP, thereby initiating additional synergistic interactions.
In summary, the number of tandem MREs within each CES locus (Fig. 4A), two-dimensional clustering along the length of the X chromosome (Fig. 4B), and MSL-independent nucleosome occupancy (Supplemental Fig. S4E) are features that distinguish subclasses of CESs and were not previously identified by analysis of the average properties of all CESs (Alekseyenko et al. 2008, 2012). By defining three distinct subclasses of CESs, we provide a new tool for examining how the MSL complex identifies the X chromosome.
Our identification of CLAMP as the previously unknown link that tethers the MSL complex to the X chromosome supports the following mechanism for X identification (Fig. 4C): (1) Prior to MSL complex targeting, maternally loaded CLAMP binds directly to MREs within group B and group C CESs, including roX loci, due to clustering of tandem MREs within these loci (Fig. 4A). (2) During early embryogenesis, MSL-independent transcription of roX genes is stimulated (Kelley et al. 1997; Meller and Rattner 2002) and is likely to facilitate cotranscriptional MSL complex assembly (Lim and Kelley 2012). CLAMP is enriched at the CESs adjacent to roX loci independent of the MSL complex and tethers the MSL complex to the X chromosome (Figs. 1–3). (3) The MSL complex catalyzes acetylation of H4K16 (H4K16ac) (Smith et al. 2001), thereby opening chromatin (Shogren-Knaak et al. 2006) and increasing the accessibility of MRE sequences for recognition by CLAMP. Synergistic interactions between the MSL complex and CLAMP (Fig. 3A; Supplemental Fig. S3B,C,F) are likely to contribute to the high occupancy of both factors at the roX loci, followed by spreading to additional CESs (Fig. 3A; Supplemental Figs. S3F, S4). Through this proposed mechanism, we suggest that CLAMP functions together with roX RNAs to target the MSL complex to the X chromosome. The high degree of conservation of both CLAMP and MREs across Drosophila species (Alekseyenko et al. 2013) suggests that they are key components of a conserved mechanism for establishing an active chromatin domain within a metazoan genome.
Materials and methods
Flies and crosses
The CLAMP RNAi fly line was obtained from the Transgenic RNAi Project (TRiP) (http://www.flyrnai.org). See the Supplemental Material for details.
ChIP from third instar larvae
For ChIP from third instar larvae, a protocol was adapted from Larschan et al. (2007). See the Supplemental Material for details.
Cell culture and RNAi
Standard cell culture approaches were used to maintain Drosophila SL2, and Kc cells, and RNAi treatment conditions are described at http://www.flyrnai.org. See the Supplemental Material for details.
mRNA sample preparation
For mRNA sample preparation, a protocol was adapted from Gelbart et al. (2009). See the Supplemental Material for details.
Preparation of Illumina sequencing libraries for ChIP-seq and mRNA-seq
For preparation of Illumina sequencing libraries for ChIP-seq and mRNA-seq, a protocol was adapted from Alekseyenko et al. (2008). See the Supplemental Material for details.
Bioinformatics
See the Supplemental Material for details on bioinformatics.
In vitro protein expression for PBMs and PBM
For in vitro protein expression for PBMs and PBM, protocol was adapted from Berger et al. (2006). See the Supplemental Material for details.
Immunostaining of polytene chromosomes
For immunostaining of polytene chromosomes, protocol was adapted from Kelley et al. (1999). See the Supplemental Material for details.
Acknowledgments
We thank Mitzi Kuroda, Fred Winston, and Judith Bender for critical reading of the manuscript. We are grateful to Mitzi Kuroda for the gift of the MSL3 and MSL2 antibodies. We thank Timothy Hughes for helpful advice. This work was supported by the following grants to E.L.: NIH grant GM098461-01, a Pew Biomedical Scholars Program grant, a NSF-ADVANCE pilot project grant, a RI-INBRE pilot project grant, a Rhode Island Foundation grant, and the Brown University Salomon grant. M.M.L.S. is supported by NIH grant F31 GM099399-01. M.L.B. is supported by NIH grant R01 HG005287, and P.J.P. is supported by NIH grant U01HG004258. All of the Illumina sequencing in this study was conducted at the Brown University Center for Genomics and Proteomics and was supported by grant number P30RR031153 from the National Center for Research Resources. M.M.L.S., J.C, J.F., P.X., K.G., A.R.L., A.U.S., E.L., and A.V. performed experiments. E.P.B., M.Y.T., and T.S. performed computational analysis. E.L., M.L.B., and P.J.P. designed and consulted on experiments. E.L. and M.M.L.S. prepared the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.214585.113.
References
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Ho JW, Peng S, Gelbart M, Tolstorukov MY, Plachetka A, Kharchenko PV, Jung YL, Gorchakov AA, Larschan E, et al. 2012. Sequence-specific targeting of dosage compensation in Drosophila favors an active chromatin context. PLoS Genet 8: e1002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Ellison CE, Gorchakov AA, Zhou Q, Kaiser VB, Toda N, Walton Z, Peng S, Park PJ, Bachtrog D, et al. 2013. Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev 27: 853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Alekseyenko AA, Kuroda MI 2004. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J 23: 2853–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schubeler D 2008. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol 28: 3401–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote JM, Lucchesi JC 1980. Male-specific lethal mutations of Drosophila melanogaster. Genetics 96: 165–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW 3rd, Bulyk ML 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copps K, Richman R, Lyman LM, Chang KA, Rampersad-Ammons J, Kuroda MI 1998. Complex formation by the Drosophila MSL proteins: Role of the MSL2 RING finger in protein complex assembly. EMBO J 17: 5409–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauth T, Muller-Planitz F, Konig C, Straub T, Becker PB 2010. The DNA binding CXC domain of MSL2 is required for faithful targeting the dosage compensation complex to the X chromosome. Nucleic Acids Res 38: 3209–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI 2009. Drosophila dosage compensation: A complex voyage to the X chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, Kuroda MI 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev 19: 2289–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K 2009. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Wang J, Bell L, Kuroda MI 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Lee OK, Shim YK 2008. Transcription rate of noncoding roX1 RNA controls local spreading of the Drosophila MSL chromatin remodeling complex. Mech Dev 125: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI 2007. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28: 121–133 [DOI] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI 2011. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471: 115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Soruco MM, Lee OK, Peng S, Bishop E, Chery J, Goebel K, Feng J, Park PJ, Kuroda MI 2012. Identification of chromatin-associated regulators of MSL complex targeting in Drosophila dosage compensation. PLoS Genet 8: e1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CK, Kelley RL 2012. Autoregulation of the Drosophila noncoding roX1 RNA gene. PLoS Genet 8: e1002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Rattner BP 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ 2010. Targeting X chromosomes for repression. Curr Opin Genet Dev 20: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet 38: 47–53 [DOI] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH 2002. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Smith ER, Allis CD, Lucchesi JC 2001. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem 276: 31483–31486 [DOI] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB 2008. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4: e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI 2008. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol 15: 1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R, Forne I, Muller M, Imhof A, Straub T, Becker PB 2012. MSL2 combines sensor and effector functions in homeostatic control of the Drosophila dosage compensation machinery. Mol Cell 48: 647–654 [DOI] [PubMed] [Google Scholar]
- Wang CI, Alekseyenko AA, Leroy G, Elia AE, Gorchakov AA, Britton LM, Elledge SJ, Kharchenko PV, Garcia BA, Kuroda MI 2013. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol 20: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]