Abstract
Despite countermeasures against influenza virus that prevent (vaccines) and treat (antivirals) infection, this upper respiratory tract human pathogen remains a global health burden, causing both seasonal epidemics and occasional pandemics. More potent and safe new vaccine technologies would contribute significantly to the battle against influenza and other respiratory infections. Using plasmid-based reverse genetics techniques, we have developed a single-cycle infectious influenza virus (sciIV) with immunoprotective potential. In our sciIV approach, the fourth viral segment, which codes for the receptor-binding and fusion protein hemagglutinin (HA), has been removed. Thus, upon infection of normal cells, although no infectious progeny are produced, the expression of other viral proteins occurs and is immunogenic. Consequently, sciIV is protective against influenza homologous and heterologous viral challenges in a mouse model. Vaccination with sciIV protects in a dose- and replication-dependent manner, which is attributed to both humoral responses and T cells. Safety, immunogenicity, and protection conferred by sciIV vaccination were also demonstrated in ferrets, where this immunization additionally blocked direct and aerosol transmission events. All together, our studies suggest that sciIV may have potential as a broadly protective vaccine against influenza virus.
INTRODUCTION
For centuries, the global community has been waging a war with the influenza A virus. Pandemics such as the 1918 “Spanish influenza” (H1N1), 1957 “Asian influenza” (H2N2), 1968 “Hong Kong influenza” (H3N2), and 2009 “swine influenza” (H1N1) ranged unpredictably in disease severity, but they all managed to become widespread in a matter of weeks (1, 2). With the exception of “Spanish influenza,” the remaining pandemics occurred despite vaccine technology that was developed in the early 1940s and that we still employ today (3). Thus, perhaps it was not surprising that recent meta-analysis of the two Food and Drug Administration (FDA)-approved influenza vaccines (trivalent inactivated virus, TIV; live-attenuated influenza virus, LAIV) used to protect against influenza disease revealed a mere 35% effectiveness (4).
Influenza virus belongs to the family Orthomyxoviridae of negative-sense, single-stranded RNA viruses, and that family is further divided into three types, A, B, and C, the first two of which cause significant respiratory disease in humans (5). Influenza A virus causes seasonal epidemics and occasional pandemics that account for an average of 250,000 to 500,000 deaths worldwide each year (6). Influenza A virus possesses two well-characterized mechanisms to escape host immunity: antigenic drift, where the viral glycoproteins gradually accumulate mutations to evolve a viral coat unrecognizable to preexisting antibodies (Abs), and antigenic shift, where two or more influenza A virus strains reassort in a coinfected individual to generate a novel virus whose glycoproteins have not been previously found in humans (2). To fight emerging antigenic variants, extensive surveillance programs have been aimed at identifying novel circulating strains. The World Health Organization (WHO) uses surveillance data from the Global Influenza Surveillance Network (GISN) to select biyearly vaccine formulations based on the predominant strain (6).
Both TIV and LAIV historically consist of three strains of influenza virus, two subtypes of influenza A virus (H1N1 and H3N2) and one influenza B virus. However, recent quadrivalent formulations of TIV and LAIV have been approved by the FDA that contain an additional strain of influenza B virus (7, 8). The subtypes of influenza A virus are classified by the hemagglutinin (HA; 17 subtypes) and neuraminidase (NA; 9 subtypes) they possess, as these two envelope glycoproteins are the major antigenic determinants of the virus and are both necessary and intricately related (5, 9). HA possesses receptor-binding and fusion functions, and NA possesses sialidase activity (also referred to as receptor-destroying activity), which is required for virus release (5). Influenza can be inhibited by neutralizing antibodies (NAbs) that bind HA and block receptor attachment, and vaccine responses are most routinely measured by hemagglutination inhibition (HAI) assay, with a 4-fold increase in Ab titers after vaccination indicative of seroconversion (10).
Both safety and protective efficacy are desirable when evaluating vaccine candidates. TIV is noninfectious and safe and is recommended for all people above 6 months of age (11), but this vaccine relies on the generation of NAbs to the highly mutable viral glycoproteins, and immunogenicity is poor in patients above 65 years of age (4). In contrast, LAIV mimics a natural infection and elicits robust protection in patients aged 6 months to 7 years (4), but due to possible complications in the very young and in patients with preexisting conditions, only healthy people aged 2 to 49 are recommended to receive it.
To overcome the shortcomings of current influenza vaccines, new technologies must be developed that safely produce broadly cross-reactive, protective immune responses. To create a robust immune response that is safe, single-cycle infectious influenza viruses (sciIV) offer a wider potential patient range than LAIV since sciIV is self-limiting and therefore likely to cause fewer complications. We have previously generated and characterized a sciIV that lacks the open reading frame (ORF) of HA within the fourth genomic segment and instead carries between the 3′ and 5′ noncoding regions and packaging signals the ORF for green fluorescent protein (GFP) (12). SciIV is trans-complemented in vitro by HA-expressing cells and, within this context, behaves similarly to wild-type (WT) influenza virus. We used sciIV to infect mice intranasally and hypothesized that the virus would not cause severe pathology and, by following the route of natural infection, would provide protective mucosal immunity. We observed that sciIV is safe and immunogenic, suggesting that sciIV is a promising candidate to protect against future influenza virus infection. Heterosubtypic immunity was not conferred by transfer of immune sera alone and was retained only partially in the absence of T cells. SciIV provides homologous and heterologous protection that is partially dependent upon CD4 and CD8 T cells. Our vaccine approach might provide a safer alternative with a wider breadth of protection than that gained by LAIV or TIV.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney (293T) cells (ATCC CRL-11268) and Madin-Darby canine kidney (MDCK) cells (ATCC CCL-34) were maintained in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Inc.) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and 1% P-S-G (penicillin, 100 units/ml; streptomycin, 100 μg/ml; l-glutamine, 2 mM; Mediatech, Inc.). Cells were grown at 37°C in a 5% CO2 atmosphere. MDCK cells constitutively expressing influenza A/WSN/33 H1N1 virus have been previously described (12). MDCK cells stably expressing HA from A/California/4_NYICE_E3/09 (H1N1) (13) (pH1N1/E3-HA) were generated by cotransfecting pCAGGS pH1N1/E3-HA and pCB7 (1:3 ratio) for eukaryotic expression of HA and hygromycin B resistance, respectively (12). HA-expressing cells were grown in DMEM–10% FBS–1% P-S-G supplemented with 200 μg/ml hygromycin B (12). After viral infections, cells were maintained in DMEM containing 0.3% bovine serum albumin (BSA), 1% P-S-G, and 0.3 μg/ml tosyl-sulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma).
Influenza A/Puerto Rico/8/1934 virus (PR8; H1N1) (14), a recombinant virus containing the HA and NA segments of A/Hong Kong/1/1968 (H3N2) in the background of PR8 (X31) (15), and the prototypic pandemic influenza A/California/04_NYICE_E3/2009 virus (pH1N1/E3; H1N1) (13) were prepared in eggs as described previously (13, 16, 17). PH1N1/E3 is a spontaneous laboratory adaptation of A/California/04/2009 (pH1N1) that contains 1 mutation (D107G) in the nucleoprotein (NP) and 3 mutations (K136N, S200P, and D239G) in the receptor-binding domain of HA (13). Virus titers for immunization and challenge experiments were determined by immunofocus assay (with results quantified in fluorescent focus-forming units [FFU]) as described below or by standard plaque assay (with results quantified in PFU) in MDCK cells as described previously (18). WSN-sciIV was generated as previously published (12) and has the ORF of GFP substituted into the fourth segment. WSN-sciIV was amplified on MDCK-HA cells, and virus titers were determined by plaque assay as described above but in MDCK-HA cells (12). For virus titration in multicycle analyses and lung or nasal titrations, FFU/ml were enumerated using a fluorescence microscope with fluorescent viruses or secondary Ab detection with a monoclonal Ab (MAb) against influenza virus NP (HT103), as described below.
Construction of plasmids.
The reverse genetics plasmids used to generate pH1N1/E3-sciIV (19) and the plasmid pPolI HA(45)GFP(80) have been previously described (20). Plasmid pCAGGS pH1N1 HA was generated by reverse transcription-PCR (RT-PCR) from RNA isolated from pH1N1-infected MDCK cells and cloned into pCAGGS using EcoRI and XhoI (New England BioLabs). Construction of pCAGGS pH1N1/E3 HA was performed by isolating RNA from pH1N1/E3-infected MDCK cells and using RT-PCR to amplify the viral cDNA of HA. Restriction enzymes SacI and NsiI (New England BioLabs) were used to subclone the fragment of pH1N1/E3 that contained 3 mutations (K136N, S200P, and D239G) into pCAGGS pH1N1 HA using a rapid DNA ligation kit (Promega). All plasmids were generated using standard cloning techniques and purified using a Wizard SV kit (Promega). Primers for the generation of the described plasmid constructs are available upon request. All plasmid constructs were verified by DNA sequencing.
Transient HA complementation of sciIV.
Parental MDCK cells were transfected with 1 μg of pCAGGS HA-protein expression plasmids and Lipofectamine 2000 (1:1 ratio) and, 16 h posttransfection, infected with WSN-sciIV (12). Transient complementation was monitored by GFP expression 24 and 48 h postinfection using a fluorescence microscope (DM IRB; Leica). Empty pCAGGS plasmid was used as a negative control.
Rescue of pH1N1/E3-sciIV.
Rescue of pH1N1/E3-sciIV from plasmid DNA was performed in a manner similar to that previously described for WSN-sciIV (12). Briefly, cocultures (1:1) of 293T/MDCK WSN-HA (6-well plate format, 106 cells) were cotransfected, in suspension, using Lipofectamine 2000 (Invitrogen) with 1 μg each of the seven pH1N1 ambisense plasmids (pDZ PB2, PB1, PA, NP, NA, M, and NS) (19), pPolI HA(45)GFP(80) (20), and pCAGGS pH1N1/E3-HA to facilitate viral rescue (12). Sixteen hours posttransfection, the media were replaced with DMEM containing 0.3% BSA, 1% P-S-G, and 0.3 μg/ml TPCK-treated trypsin (Sigma). At 48 h posttransfection, tissue culture supernatant (TCS) was collected, clarified, and used to infect fresh WSN HA-expressing MDCK cells (6-well plate format, 106 cells). Three days postinfection, virus was plaque purified and scaled up in MDCK pH1N1/E3-HA cells at 34°C and 5% CO2 for 3 days (19). Virus stocks were generated by infecting confluent 10-cm-diameter dishes of MDCK pH1N1/E3 at a low multiplicity of infection (MOI) (0.001) for 3 days as described above. Stocks were titrated by plaque assay on MDCK WSN-HA cells as previously described (12).
Indirect immunofluorescence assay (IFA).
For cell line characterization, monolayers of parental or HA-expressing MDCK cells (105 cells, 48-well plate format) were fixed with 4% paraformaldehyde for 15 min at room temperature and blocked overnight with 2.5% BSA at 4°C. Cells were then incubated with 1 μg/ml of MAbs (2G9, anti-WSN HA [12]; 31C2, anti-pH1N1 HA [21]) for 1 h at 37°C. After washes with 1× phosphate-buffered saline (1× PBS), cells were incubated with secondary anti-mouse fluorescein isothiocyanate (FITC) (Dako) (1:150) and 4′,6′-diamidino-2-phenylindole (DAPI; Research Organics) (1:1,000) for 30 min at 37°C. Cells were visualized using a fluorescence microscope, photographed (Cooke Sensicam QE) at ×40 magnification, and colored and merged using Adobe Photoshop CS4 (v11.0) software. Fields representative of at least three independent fields are shown.
For virus characterization, monolayers of MDCK cells (2.5 × 105 cells, 24-well plate format) were infected at an MOI of 5. Ten hours postinfection, cells were fixed as described above and permeabilized with 0.2% Triton X-100. Viral NP was detected with HT103 primary and anti-mouse Alexa-594 (Jackson ImmunoResearch Laboratories, Inc.) (1:1,000) secondary Abs.
Growth kinetics of recombinant viruses.
To evaluate the replicative capabilities of sciIV, confluent monolayers of MDCK-HA stable cell lines (6-well plate format, 106 cells) were infected at an MOI of 0.001 in triplicate and incubated at 34°C for pH1N1 viruses or 37°C for WSN viruses for 3 days (12). At indicated times postinfection (12, 24, 48, and 72 h), GFP expression was assessed by fluorescence microscopy, and viral titers in the TCSs were measured by evaluating FFU/ml with a fluorescence microscope in an immunofocus assay. Briefly, triplicate wells of MDCK WSN-HA cells (96-well format, 5 × 104 cells) were infected with 10-fold serial dilutions of TCS. Eighteen hours postinfection, cells were washed with 1× PBS and foci were visualized and enumerated using a fluorescence microscope. Titrations of WT viruses were performed in a manner similar to that described above, but individual infected cells were detected 10 h postinfection by immunofluorescence of the influenza virus NP using MAb HT103 to determine FFU/ml. Mean values and standard deviations were calculated using Microsoft Excel software.
To evaluate the complementation of MDCK-HA cells or the growth phenotype of sciIV, plaque phenotype analysis was performed in parental, WSN-HA, or pH1N1/E3-HA MDCK cells. Briefly, confluent monolayers (106 cells, 6-well plate format) were infected and incubated for 3 days in agar-containing postinfection media at 37°C or 34°C for WSN or pH1N1 backbone viruses, respectively. Monolayers were stained with crystal violet.
Assays to determine NAbs.
Virus neutralization (VN) assays were performed against pH1N1 virus as previously described (22) with ferret sera. Briefly, sera were treated with receptor-destroying enzyme (RDE) following the instructions of the manufacturer (Denka Seiken) prior to heat inactivation and serially diluted 2-fold in a 96-well plate. Two hundred PFU of pH1N1 was then added to Ab dilutions at a 1:1 ratio and incubated for 1 h at room temperature. Confluent monolayers of MDCK cells (96-well plate format, 5 × 104 cells) were then infected for 1 h at room temperature with the virus-Ab mixture and incubated for 3 days at 34°C. Neutralization was determined by hemagglutination assay (HA) using a 96-well plate format and TCS with 0.5% turkey red blood cells (RBCs) for 30 min on ice as previously described (13). Triplicate wells were used to calculate the geometric mean titer (GMT).
Hemagglutination inhibition (HAI) assays were used to determine the HA-neutralizing capabilities of immune mice or ferret sera. Mouse or ferret sera were RDE treated (Sigma or Denka Seiken, respectively) prior to heat inactivation for 30 min at 56°C and mixed 1:1 with 4 hemagglutinating units (HAU) of pH1N1/E3 or pH1N1 for mice or ferret sera, respectively, for 30 min at room temperature. HAI was visualized by adding 0.5% turkey RBCs to virus-Ab mixtures for 30 min on ice as previously described (13).
Animal experiments.
For mouse immunization and challenge experiments, female 6-to-8-week-old C57BL/6 mice were purchased from The Jackson Laboratory and maintained at the University of Rochester under specific pathogen-free conditions. All mouse experiments were approved by the University Committee of Animal Resources and complied with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (28a). For vaccinations and infections, mice were anesthetized intraperitoneally (i.p.) with 2,2,2 tribromoethanol (Avertin; 240 mg/kg of body weight) and then inoculated intranasally (i.n.) with 30 μl of virus preparations as indicated. For immunizations, mice received one or two doses at 2-week intervals of non- or UV-inactivated pH1N1/E3-sciIV (102 to 105 FFU/mouse, as indicated) or 30 PFU pH1N1/E3 (for generation of convalescent-phase sera or evaluation of CD8+ T cells in lungs). For the challenge, mice were inoculated with 1.5 × 103 PFU pH1N1/E3, 103 PFU PR8, or 104 PFU X31.
Morbidity was monitored by body weight loss after immunization or vaccination, as well as by clinical signs of infection (hunching, ruffling of fur, malaise, or respiratory distress). Each day, percent body weight was determined relative to starting weight prior to manipulation. Mice losing greater than 30% body weight were euthanized humanely. At 3 and 6 days postchallenge, lungs were surgically extracted, placed into an Eppendorf tube, and frozen on dry ice. Virus titers were determined from homogenized lung preparations. T cells from the lungs were collected by placing lungs in DMEM and were purified as previously described (18) using Histopaque 1083 purification (Sigma).
For ferret immunization and challenge and transmission experiments, 5-month-old male Fitch ferrets (Triple F Farms) seronegative for pH1N1 were used. Animals were maintained in modified transmission cages as previously described (23) in accordance with the Icahn School of Medicine at Mount Sinai Institutional Biosafety Program and Institutional Animal Use and Care Committee guidelines. Ferrets were anesthetized through intramuscular (i.m.) injection of a ketamine-xylazine mixture prior to i.n. virus inoculation (0.5 ml) or nasal wash treatment (1 ml). For immunization, ferrets received two doses of 106 FFU pH1N1/E3-sciIV at 4-week intervals prior to challenge with 106 PFU of pH1N1.
Virus titers in homogenized mouse lungs or ferret nasal washes were determined by infecting triplicate wells of confluent MDCK cells (96-well plate format, 5 × 104 cells) with 10-fold serial dilutions of virus. Ten hours postinfection, cells were fixed and permeabilized (4% formaldehyde–0.5% Triton X-100–1× PBS) for 15 min at room temperature, stained with primary anti-NP MAb HT103 (24) for 1 h at 37°C, washed with 1× PBS, and then stained with secondary anti-mouse FITC (Dako, 1:150) for 1 h at 37°C. NP-expressing cells were enumerated to determine the virus titer (FFU/ml). Geometric mean titer and statistical analyses (Mann-Whitney test) were performed using GraphPad Prism software.
Viral UV inactivation.
For UV inactivation, sciIV was placed in uncovered 12-well plates and exposed to shortwave UV radiation at 254 nm (Mineralight UV lamp, UV S-68; Ultra-Violet Products) for 20 min on ice at a distance of 6 cm (25). Following UV irradiation, infectivity was measured as the 50% tissue culture infectious dose (TCID50) in MDCK WSN-HA cells. Briefly, confluent MDCK WSN-HA cells (96-well plate format, 5 × 104 cells) were infected with 10-fold serial dilutions of mock- or UV-treated virus for 3 days at 34°C before crystal violet staining.
Passive immune transfer.
Two weeks following prime and boost vaccination with 105 FFU pH1N1/E3-sciIV, mouse sera were collected and individually verified for anti-influenza virus Ab concentrations comparable to those of convalescent-phase sera by enzyme-linked immunosorbent assay (ELISA; data not shown). Pooled sera were diluted 1:1 in 1× PBS, and 200 μl was transferred intraperitoneally (i.p.) into naive mice 24 h prior to challenge with the indicated viruses.
ELISA.
Mouse sera were collected 24 h prior to challenge by submandibular bleeding and stored at −20°C. ELISAs were performed by coating plates for 18 h at 4°C with lysates from mock- or pH1N1/E3-infected MDCK cells (MOI of 0.005, 24 h postinfection; total anti-influenza virus Abs) or with 2 μg/ml of recombinant protein (PR8 NP, a gift from Troy D. Randall; pH1N1 HA, NR-13691, BEI Resources). After blocking with 1% BSA was performed, plates were incubated with 2-fold dilutions of sera for 1 h at 37°C, washed with H2O, and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Southern Biotech) (1:2,000) for 30 min at 37°C. Reactions were then developed with TMB substrate (BioLegend) for 5 min at room temperature, quenched with 2N H2SO4, and read at 450 nm (Vmax kinetic microplate reader; Molecular Devices).
Flow cytometry.
To determine the presence of T cells in the lungs of mice, flow cytometry was performed on isolated cells 10 days postimmunization or postinfection with 30 PFU pH1N1. Briefly, cells were washed with staining buffer (1× PBS–1% FBS), blocked with anti-CD16/32 for 20 min, and then stained with the following Abs for 30 min at 4°C: CD3ε allophycocyanin (APC) (145-2C11), CD4 phycoerythrin (PE)-Cy5 (RM4-5), CD8 APC-Cy7 (53-6.7) (purchased from BD Pharmingen), and Live/Dead violet (Invitrogen). Cells specific for influenza were identified using PE-conjugated H2-Db/influenza virus pH1N1/E3 NP366–374 prepared by the NIH Tetramer Core Facility at Emory University (26). Samples were analyzed by the use of an LSR II system (BD Biosciences) and data analyzed using FlowJo software (Tree Star) and statistical analyses performed using GraphPad Prism software (student's one-tailed t test).
T cell depletion.
Immunized mice were depleted of T cells by three i.p. injections (challenge days −2, 0, and 2) consisting of 100 μg per dose as previously described (13) with anti-CD4 (GK1.5) or anti-CD8 (2.43) from BioXCell or isotype (rat IgG2bκ; eBioscience).
RESULTS
Generation and characterization of pH1N1/E3-sciIV.
Our previous studies (12) have demonstrated that sciIV based on the genetic backbone of A/WSN/33 (WSN) could be generated where the coding region of HA that is not required for segment packaging was replaced with that of GFP. The HA-deficient influenza virus obtained was efficiently complemented in HA-expressing MDCK cell lines (12). Using a similar strategy, we intended to establish a sciIV based on the genetic backbone of the 2009 pandemic H1N1, using the prototypic A/California/04/2009 (pH1N1) strain.
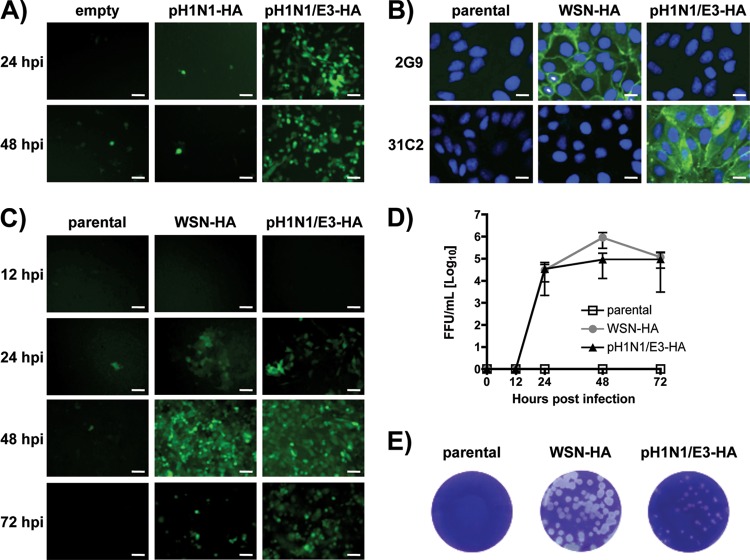
To generate an MDCK-HA cell line that pseudotypes sciIV with HA from pH1N1 (Fig. 1), we initially cloned the HA of the WT pH1N1 into the pCAGGS vector (pH1N1-HA) and transfected MDCK cells before infection with WSN-sciIV. However, upon several trials, we could not achieve successful complementation of sciIV, as indicated by the absence of green fluorescence (Fig. 1A), although HA was indeed present and recognized by an anti-pH1N1 MAb (data not shown). The original isolate of the pH1N1 virus does not replicate well in MDCK cells but can be made to propagate well in vitro if critical mutations are incorporated into the receptor-binding pocket of HA (13, 27, 28). An example is the pH1N1/E3 virus generated in our laboratory by selection of a high-growth clone of pH1N1 in 10-day-old embryonated chicken eggs (13). Therefore, we cloned the HA of pH1N1/E3 into pCAGGS (pH1N1/E3-HA) and tested it in transient complementation with the WSN-sciIV. Successful complementation of sciIV using pCAGGS pH1N1/E3-HA was achieved as indicated by green fluorescence 24 and 48 h postinfection (Fig. 1A). We consequently generated MDCK stable cell lines expressing pH1N1/E3-HA as previously described (12) (Fig. 1B) that could be used to complement a WSN-sciIV to levels comparable to those of WSN HA-expressing stable cell lines, as shown by GFP expression (Fig. 1C), multicycle growth kinetics (Fig. 1D), and plaque phenotype (Fig. 1E). Interestingly, the plaque size of WSN-sciIV in the pH1N1/E3-HA cell line was slightly less than that seen with WSN-HA stable cells. We performed a GFP-based microneutralization assay (12) using specific neutralizing antibodies that confirmed the identity of the HA-pseudotyped viruses with either WSN or pH1N1/E3 HA (data not shown).
Fig 1.
Three mutations in the receptor-binding domain of pH1N1 HA allow for complementation of sciIV. (A) Transient HA complementation in MDCK cells. MDCK cells were transfected with pCAGGS protein expression plasmids for HA from pH1N1 (pH1N1-HA) or pH1N1/E3 (pH1N1/E3-HA) or empty plasmid and, 24 h posttransfection, infected with WSN-sciIV (12) (MOI, 0.01) to evaluate trans-complementation by GFP focus formation. GFP-expressing cells were visualized by fluorescence microscopy 24 and 48 h postinfection. Representative images at ×10 magnification are illustrated. Scale bars, 40 μM. (B) HA protein expression by MDCK stable cell lines. Monolayers of MDCK cells stably transfected with WSN-HA or pH1N1/E3-HA or parental cells were fixed and incubated with MAbs specific for WSN (2G9) or pH1N1 (31C2) HA and with DAPI for nuclear staining. Representative images are shown at ×40 magnification. Scale bars, 10 μM. (C and D) Multicycle growth of sciIV. Parental, WSN-HA, or pH1N1/E3-HA MDCK cells were infected with WSN-sciIV at an MOI of 0.001. At various times postinfection, GFP expression was visualized by fluorescence microscopy (×10 representative images; scale bars, 40 μM) (C), and TCSs were collected for titration in MDCK WSN-HA cells (D). Data represent means ± SD of the results determined for triplicates. (E) Plaque morphology of sciIV in HA-expressing cells. WSN-HA, pH1N1/E3-HA, or parental MDCK cells were infected with WSN-sciIV. Three days postinfection, monolayers were stained with crystal violet.
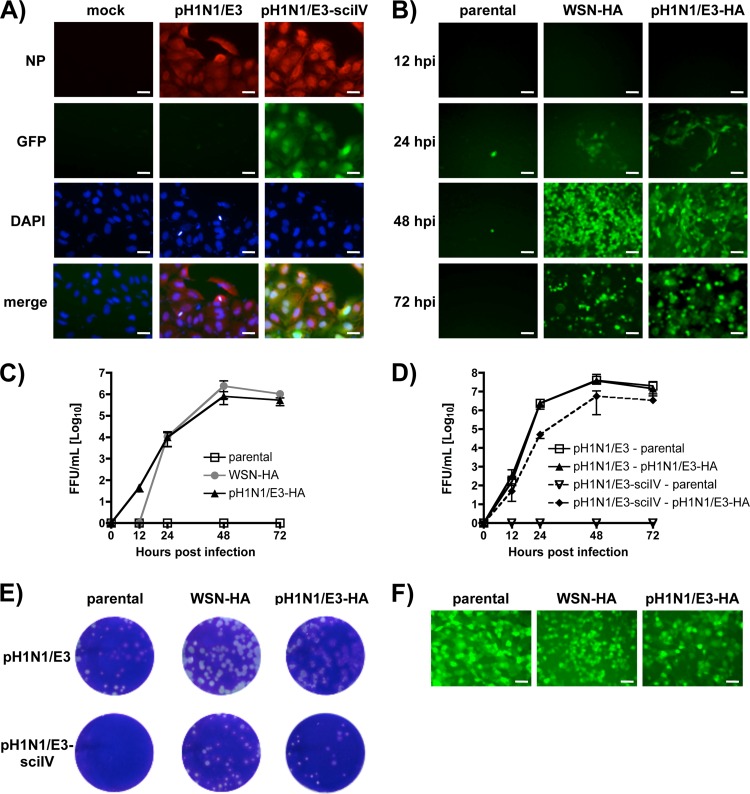
With the pH1N1/E3-HA cell lines established, we sought to generate a pH1N1 backbone sciIV (pH1N1/E3-sciIV) using pH1N1 plasmid-based reverse genetics as previously described (19). To characterize the pH1N1/E3-sciIV (Fig. 2), we first verified that infected cells expressed GFP and nucleoprotein (NP) simultaneously (Fig. 2A). We next demonstrated that only HA-expressing cells could complement the virus by GFP propagation (Fig. 2B) and that similar levels of FFU were generated in the WSN- and pH1N1/E3-HA cell lines (Fig. 2C). In comparisons of pH1N1/E3-sciIV to pH1N1/E3 WT, pH1N1/E3 WT replicates more efficiently, as determined by multicycle growth (Fig. 2D) and plaque formation (Fig. 2E). The efficiency might perhaps be due to the relative abundance of HA generated by RNA polymerase II (in the case of the stable cell line) versus the viral promoter as previously observed (12). Because we believed that pH1N1/E3-sciIV would lead to a single round of infection and gene expression in normal cells or in vivo, we evaluated the ability of pH1N1/E3-sciIV to infect both parental and HA-expressing cells at a high MOI by GFP expression (Fig. 2F). As previously shown (12), infection of non HA-expressing MDCK cells with a high MOI of the pH1N1/E3-sciIV resulted in 100% GFP expression. These results demonstrate that, similarly to WSN-sciIV (12), pH1N1/E3-sciIV grows with kinetics comparable to those of pH1N1 in MDCK-HA cells but can replicate for only one cycle in cells lacking HA.
Fig 2.
Characterization of pH1N1/E3-sciIV. Plasmid-based reverse genetics techniques were used to rescue a sciIV with genes derived from pH1N1, except the fourth segment which codes for HA(45)GFP(80), as previously described (12). (A) GFP is expressed by cells infected with pH1N1/E3-sciIV. MDCK cells were mock infected or infected with pH1N1 or pH1N1/E3-sciIV at an MOI of 5. Ten hours postinfection, monolayers were fixed, permeabilized, and incubated with a MAb against NP (HT103) and subjected to nuclear counterstaining with DAPI. Representative ×20 images (NP, GFP, DAPI, and merged [bottom row]) are shown. Scale bars, 20 μM. (B and C) Multicycle growth of pH1N1/E3-sciIV. WSN-HA, pH1N1/E3-HA, or parental MDCK cells were infected with pH1N1/E3-sciIV at an MOI of 0.001. At various times postinfection, GFP expression was visualized by fluorescence microscopy (B) (×10 representative images shown; scale bars, 40 μM), and TCSs were collected for titration in MDCK WSN-HA cells (C). Data represent means ± SD of the results determined for triplicates. (D) Multicycle growth of pH1N1/E3-sciIV and pH1N1/E3. Parental or pH1N1/E3-HA-expressing MDCK cells were infected with pH1N1/E3 or pH1N1/E3-sciIV at an MOI of 0.001. At various times postinfection, TCSs were collected for titration on MDCK cells by IFA using the NP MAb HT103. Data represent means ± SD of the results determined for triplicates. (E) Plaque morphology of pH1N1/E3-sciIV. WSN-HA, pH1N1/E3-HA, or parental MDCK cells were infected with pH1N1 or pH1N1/E3-sciIV for plaque assay. Three days postinfection, monolayers were stained with crystal violet. (F) pH1N1/E3-sciIV can infect cells that do not express HA. Parental, WSN-HA, or pH1N1/E3-HA MDCK cells were infected at an MOI of 5 with pH1N1/E3-sciIV. Ten hours postinfection, monolayers were analyzed by fluorescence microscopy for GFP expression. Representative images (×10 magnification) are shown. Scale bars, 40 μM.
pH1N1/E3-sciIV induces an Ab response.
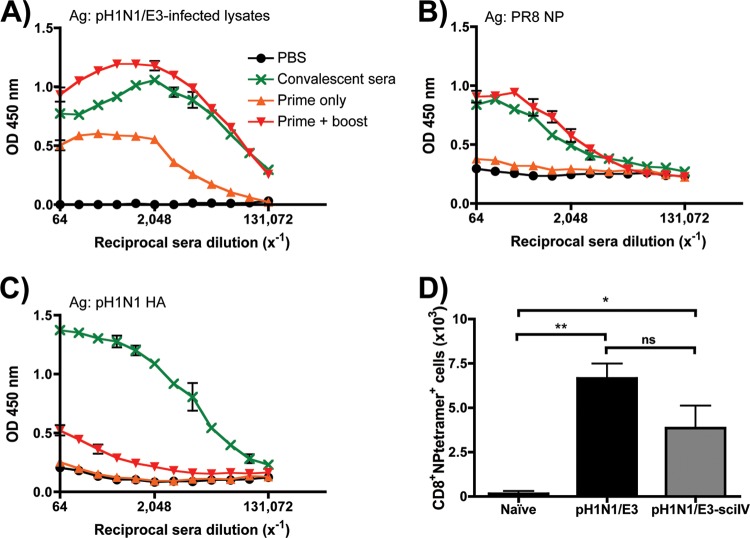
Although pH1N1/E3-sciIV can replicate only once within the airway target cells that lack influenza HA expression, the replication and transcription of other viral proteins after infection suggest that Abs could be elicited for some, if not all, of the remaining components of pH1N1/E3-sciIV. To test this, groups of mice were i.n. inoculated once or twice at 2-week intervals. Two weeks postprime (one dose) or postboost (two doses), sera were collected to measure Abs against total pH1N1/E3 virus, NP, or HA. Additionally, HAI was used to examine the antiviral activity of the sera from immunized mice (Table 1). As seen in Fig. 3, Abs against total pH1N1/E3 virus were detectable after prime and were enhanced significantly after boost to a level that is comparable to that seen with WT pH1N1/E3 convalescent mice (Fig. 3A). To test if Abs were generated against an influenza virus protein that was expressed upon viral replication in situ, we determined anti-NP IgG Abs (Fig. 3B) in inoculated mice. Anti-NP IgG Abs were produced to levels comparable to those seen with convalescent mice. In contrast, and as expected, anti-HA Abs were detectable only slightly and the levels from pH1N1/E3-sciIV prime-boost mice were significantly lower than those seen with convalescent mice (Fig. 3C). All together, these results indicate that although the Ab immune response is activated with a single dose of pH1N1/E3-sciIV, two doses achieve humoral responses to the whole virus at comparable levels of convalescence. As expected, infection with sciIV does not induce high levels of anti-HA Ab titers, because the virus lacks the ability to replicate and express the HA protein upon infection.
Table 1.
Immunogenicity of sciIV in mice
| Treatment | Collection timea | Serum GMTb |
|
|---|---|---|---|
| HAI | ± SD | ||
| PBS | Prime + boost | <20 | ND |
| pH1N1/E3-sciIV | Prime only | 29 | 56 |
| Prime + boost | 660 | 750 | |
| pH1N1/E3-sciIV, UV inactivated | Prime + boost | <20 | ND |
| Convalescent-phase serac | 2,100 | 780 | |
Vaccinations were administered at 2 week intervals; samples were collected 2 weeks after the indicated dose.
Data were calculated from 10 immunized or 2 convalescent individual animals. ND, not determined.
Mice were inoculated with a nonlethal dose of pH1N1/E3; sera were collected 2 weeks later.
Fig 3.
Immunogenicity of pH1N1/E3-sciIV in mice. (A to C) Humoral immune response to vaccination. Female 6-to-8-week-old C56BL/6 mice (n = 5) were inoculated at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV once (orange triangles, prime only) or twice (red triangles, prime plus boost) or with PBS (black circles) as a negative control. Two weeks postvaccination, sera were collected and evaluated for IgG Abs against total influenza virus protein (A), recombinant NP from PR8 (B), or recombinant HA from pH1N1 (C) by ELISA. Convalescent-phase sera (green) collected 2 weeks after naive mice were infected with 0.02 MLD50 pH1N1/E3 were used as a positive control (n = 3). Data represent means ± standard errors of the means (SEM) of the results from 4 independent experiments. Ag, antigen; OD, optical density. (D) CD8+ T cells present in the lungs after immunization with pH1N1/E3-sciIV. Female 6-to-8-week-old C56BL/6 mice were inoculated twice at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV or PBS (Naive) or were infected with 0.02 MLD50 pH1N1/E3 (n = 3). Ten days after the final vaccination or infection, lungs were extracted and resident cells were pooled and prepared for flow cytometry. Live, CD3+ CD4− CD8+ cells specific for pH1N1 NP366-374 tetramer were counted. Data represent means ± SEM of the results from 4 independent experiments. Statistical analysis was performed using Student's one-tailed paired t test. * = P ≤ 0.05; ** = P ≤ 0.01; ns = not significant.
pH1N1/E3-sciIV elicits a CD8 T cell response.
To further examine the immunogenicity of pH1N1/E3-sciIV, we evaluated if the virus can induce a localized CD8 T cell response. Mice were given a prime and boost with 105 FFU pH1N1/E3-sciIV or a 0.02 50% mouse lethal dose (MLD50) of WT pH1N1/E3 virus. Ten days postinfection, lungs samples were collected and single-cell preparations made for flow cytometric analysis. Live, CD8 T cells were further gated for a pH1N1/E3-specific population by using a tetramer specific for H2-Db-restricted NP366 peptide derived from pH1N1/E3 NP. As shown in Fig. 3D, the i.n. prime-boost strategy with pH1N1/E3-sciIV led to elevated levels of NP366-specific lung CD8 cells similar to those induced by WT pH1N1/E3 and both results were individually statistically significant compared to the results seen with mock-immunized mice. The data presented indicate that protein delivered and replicated by pH1N1/E3-sciIV can elicit a specific and tissue-homing acute CD8 T cell response.
Vaccination with pH1N1/E3-sciIV confers full protection against homologous virus challenge.
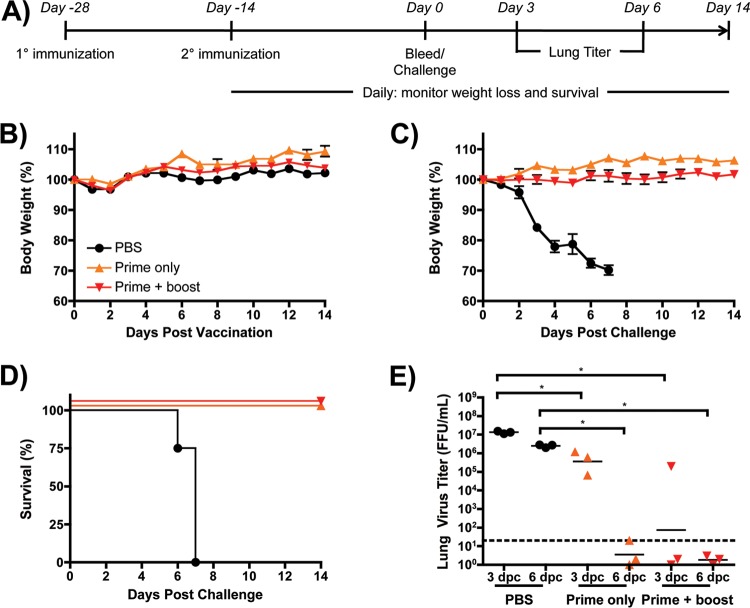
After observing that immunization with pH1N1/E3-sciIV can generate both Ab and CD8 T cell responses, we sought to evaluate if a single cycle of replication by pH1N1/E3-sciIV elicited a measurable level of immunity and provided protection against influenza virus challenge (Fig. 4). To that end, mice were primed once or twice with 105 FFU of pH1N1/E3-sciIV and then challenged with 10 MLD50 of pH1N1/E3 (Fig. 4A). To determine any gross morbidity induced by sciIV vaccination, we followed the body weight loss of mice for 2 weeks following prime or boost (Fig. 4B). As expected, mice did not lose any body weight after vaccination, because sciIV is not complemented within mouse airways. Moreover, sciIV-immunized mice were protected from the challenge, whereas PBS immunized mice rapidly lost weight and all succumbed to infection (Fig. 4C and Fig. 4D). To quantify the benefit of prime versus prime-boost with sciIV, the amount of challenge virus in the lungs was measured at day 3 and day 6 postchallenge (Fig. 4E). The prime-boost regimen resulted in a lower geometric mean virus titer at day 3 than the prime regimen, though one animal had comparable titer levels, likely due to an inefficient vaccination. At day 6 postchallenge, both prime and prime-boost animals did not have detectable levels of challenge virus in the lungs. Taken together, these data suggest that one or two doses of pH1N1/E3-sciIV can afford mice full protection against a lethal influenza virus challenge.
Fig 4.
pH1N1/E3-sciIV vaccination protects mice from lethal homologous challenge. (A) Representation of the immunization and challenge schedule. (B) Tolerability of pH1N1/E3-sciIV. Female 6-to-8-week-old C56BL/6 mice were inoculated at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV once or twice or with PBS as a negative control (n = 5). Weight loss was monitored daily for 2 weeks after the PBS or first (or ange triangles, prime only) or second (red triangles, prime plus boost) sciIV inoculation. Plotted data represent means ± SEM. (C and D) Protection from lethal challenge conferred by pH1N1/E3-sciIV vaccination. Immunized mice from the experiment described for panel B were challenged with 10 MLD50 pH1N1/E3, and weight loss (plotted data represent means ± SEM) (C) and survival (D) were monitored daily for 2 weeks postchallenge. (E) Virus lung titers. Mice immunized and challenged as described for panels B and D were sacrificed at 3 and 6 days postchallenge (dpc) to evaluate levels of replicating challenge virus in the lungs by IFA as described for Fig. 2D (n = 3). Symbols represent data from individual mice; bars represent geometric mean lung virus titers, and the dotted line denotes the limit of detection (20 FFU/ml). Statistical analysis was performed using the Mann-Whitney test; * = P ≤ 0.05.
Protection conferred by pH1N1/E3-sciIV is replication and dose dependent.
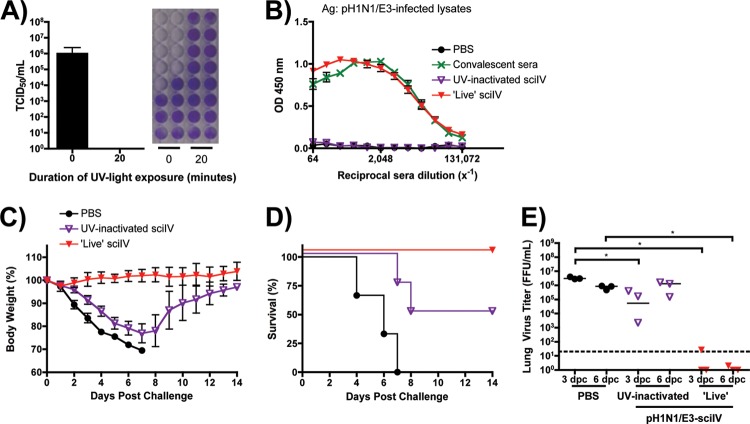
To evaluate if the sciIV inoculum must be infectious in order to induce protection, we inactivated the pH1N1/E3-sciIV preparation prior to inoculation using UV light exposure (Fig. 5). UV-inactivated sciIV did not produce a measurable TCID50 titer (Fig. 5A), and priming did not lead to the generation of detectable HAI titers (Table 1) or total anti-influenza IgG Abs (Fig. 5B). When mice were challenged after receiving UV-inactivated sciIV, they were not fully protected from pH1N1/E3 challenge, as indicated by rapid weight loss (Fig. 5C), death (Fig. 5D), and viral titers comparable to those seen with mock-immunized mice at 6 days postchallenge (Fig. 5E). Interestingly, half of the mice died from infection whereas the other half eventually recovered, which could be reflective of the antigen dose, suggesting that infection and replication of pH1N1/E3-sciIV are required for complete protection against a lethal challenge with the pH1N1/E3 WT virus.
Fig 5.
UV-inactivated pH1N1-sciIV does not fully protect mice from lethal homologous challenge. (A) UV light treatment renders sciIV noninfectious. SciIV was exposed to UV light for 20 min on ice, and infectivity was determined by TCID50 assay on MDCK WSN-HA cells. (B) Immunogenicity of UV-inactivated sciIV. Female 6-to-8-week-old C56BL/6 mice were inoculated twice at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV that was (purple triangles; UV-inactivated sciIV) or was not (red triangles; ‘Live’ sciIV) exposed to UV light on ice for 20 min or with PBS (black circles) as a negative control (n = 5). Two weeks postvaccination, sera were collected and evaluated for IgG Abs against total influenza virus protein by ELISA. Convalescent-phase sera (green) were used as a positive control (n = 3); plotted data represent means ± SEM. (C and D) Protection from lethal challenge by pH1N1/E3-sciIV vaccination. Immunized mice from the experiment described for panel B were challenged with 10 MLD50 pH1N1/E3, and weight loss (C) (plotted data represent meana ± SEM) and survival (D) were monitored daily for 2 weeks postchallenge. (E) Virus lung titers. Mice immunized and challenged as described for panels C and D were sacrificed at 3 and 6 days postchallenge to evaluate levels of replicating challenge virus in the lungs by IFA (n = 3). Symbols represent individual mice; bars represent geometric mean lung virus titers, and the dotted line denotes the limit of detection (20 FFU/ml). Statistical analysis was performed using the Mann-Whitney test; * = P ≤ 0.05.
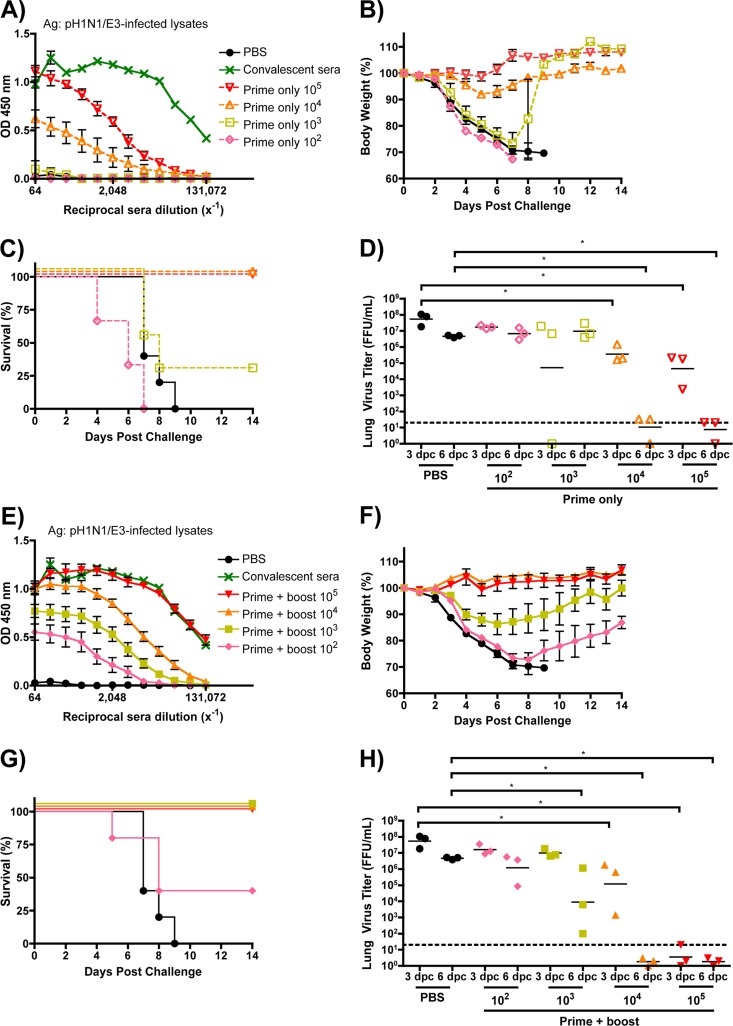
To examine whether the observed protection afforded by vaccination with pH1N1/E3-sciIV is dose dependent (Fig. 6), we inoculated mice once or twice with 10-fold dilutions of pH1N1/E3-sciIV (102 to 105 FFU per mouse). We first evaluated the total Ab responses from immunized mice (Fig. 6A and E). As expected, total levels of anti-influenza virus Abs directly correlated with the amount of pH1N1/E3-sciIV. After challenge with 10 MLD50 of pH1N1/E3, mice that received one dose of 104 FFU of pH1N1/E3-sciIV demonstrated a slight weight loss (Fig. 6B) but significantly less viral lung replication compared to mock-immunized mice detected at day 3 and day 6 (Fig. 6D), and all animals quickly recovered and survived (Fig. 6C). Boosting with this same dose provided full protection against the challenge, as shown by the absence of weight loss (Fig. 6F), significantly diminished viral titers in the lungs (Fig. 6H), and no animal death (Fig. 6C and G). When mice were primed once with 103 FFU of pH1N1/E3-sciIV, weight loss (Fig. 6B) and virus replication in the lungs (Fig. 6D) after the challenge were similar to those seen with the group with no priming and the majority died within 8 days (Fig. 6C). However, of the group receiving the 103 FFU prime only, one mouse lost only 15% body weight and recovered (Fig. 6C) and another had no evidence of virus in the lungs (Fig. 6D). However, the mice receiving both a prime and boost with 103 FFU of pH1N1/E3-sciIV all survived (Fig. 6G), lost less weight (Fig. 6F), and exhibited hastened virus clearance at day 6 postchallenge (Fig. 6H). The mice given a prime of 102 FFU of pH1N1/E3-sciIV lost weight quickly after the challenge (Fig. 6B) and had high levels of viruses in the lungs (Fig. 6D), and none survived (Fig. 6C). When given two doses of 102 FFU pH1N1/E3-sciIV, mice still suffered rapid weight loss (Fig. 6F) and had high viral titers in the lungs (Fig. 6H) and more than half died (Fig. 6G), although a small portion of animals recovered. Taken together, the data indicate that protective immunity induced by the pH1N1/E3-sciIV is dose dependent and that the prime-boost strategy gives better protection.
Fig 6.
Dose-dependent immunogenicity and protective efficacy conferred by sciIV vaccination. Female 6-to-8-week-old C56BL/6 mice were inoculated once (A to D, dotted lines) or twice (E to H, solid lines) at 2-week intervals i.n. with 10-fold dilutions of pH1N1/E3-sciIV (red, 105; orange, 104; yellow, 103; pink, 102) or with PBS (black) as a negative control (n = 5). Two weeks postvaccination, sera were collected and evaluated for IgG Abs against total influenza virus protein by ELISA (A and E; plotted data represent means ± SEM). Convalescent-phase sera (green) were used as a positive control (n = 3). Immunized mice from the experiment described for panels A and E were challenged with 10 MLD50 pH1N1/E3, and weight loss (B and F; plotted data represent means ± SEM) and survival (C and G) were monitored daily for 2 weeks postchallenge. (D and H) Virus lung titers. Mice immunized and challenged as described above were sacrificed at 3 and 6 days postchallenge to evaluate levels of replicating challenge virus in the lungs by IFA (n = 3). The experiments represented in panels D and H were performed simultaneously; thus, the same PBS-vaccinated controls were plotted in both graphs. Symbols represent individual mice; bars represent geometric mean lung virus titers, and the dotted line denotes the limit of detection (20 FFU/ml). Statistical analysis was performed using the Mann-Whitney test; * = P ≤ 0.05.
Immunization with pH1N1/E3-sciIV provides protection against heterologous virus challenge.
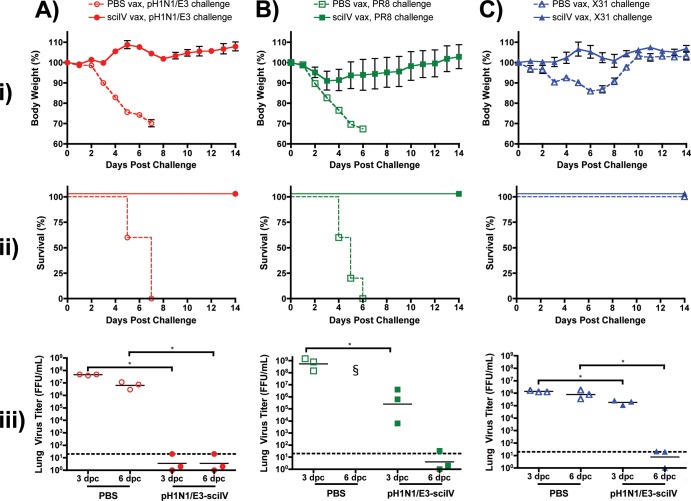
After we observed that pH1N1/E3-sciIV vaccination could provide protection against homologous pH1N1/E3 virus challenge, we further investigated the breadth of immunity conferred by sciIV vaccination (Fig. 7). To this end, we used the prime-boost strategy to inoculate mice with 105 FFU of pH1N1/E3-sciIV, as this dose confers optimal protection against homologous virus challenge. For heterologous viral challenge, we used laboratory-adapted influenza A/Puerto Rico/8/1934 virus (H1N1; PR8) (Fig. 7B) and a reassortant X31 H3N2 virus, which carries the same six internal segments as PR8 virus and the HA and NA segments from the A/Hong Kong/1/1968 (H3N2) strain of influenza virus (Fig. 7C). As an internal control, we challenged mice with the pH1N1/E3 virus (Fig. 7A), as in previous experiments. Vaccinated mice challenged with a lethal dose of PR8 experienced a slight loss of weight (Fig. 7Bi) and regained the weight after day 3, whereas PBS-treated mice drastically lost weight and succumbed to viral infection by day 6 (Fig. 7Bii). In addition, virus replication was, significantly, 3 logs less in the lungs of immunized mice at day 3 compared to those of mice that were mock immunized (Fig. 7Biii), and the virus was cleared by day 6, whereas mock-immunized animals succumbed to infection by that time. When challenged with X31, nonvaccinated mice experienced mild morbidity with quick recovery as shown by a moderate loss of body weight (Fig. 7Ci). In contrast, with pH1N1/E3-sciIV vaccination, such weight loss was not observed. Furthermore, the viral lung titers were also significantly lower in the primed animals at 3 and 6 days postchallenge (Fig. 7Ciii). All vaccinated and nonvaccinated mice survived X31 challenge due to the limited lethality of 104 PFU in C57BL/6 mice (Fig. 7Cii). As previously shown and as expected, all vaccinated mice that received homologous challenge did not lose weight (Fig. 7Ai) and survived (Fig. 7Aii) with undetectable levels of pH1N1/E3 virus in the lungs at days 3 and 6 postinfection (Fig. 7Aiii). These results indicate that pH1N1/E3-sciIV vaccination induces broad immunity to both homologous and heterologous virus infections.
Fig 7.
Homosubtypic and heterosubtypic immunity conferred by sciIV vaccination. (i and ii) Female 6-to-8-week-old C56BL/6 mice were inoculated (vax) twice at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV (filled symbols) or with PBS (open symbols) as a negative control (n = 5). Two weeks postvaccination, mice were challenged with 10 MLD50 pH1N1/E3 (A, red circles) or PR8 (B, green squares) or with 104 PFU X31 (C, blue triangles) and weight loss (panels i; plotted data represent means ± SEM) and survival (panels ii) were monitored daily for 2 weeks postchallenge. (iii) Virus lung titers. Mice immunized and challenged as described above were sacrificed at 3 and 6 days postchallenge to evaluate levels of replicating challenge virus in the lungs by IFA (n = 3). Symbols represent individual mice; bars represent geometric mean lung virus titers, and the dotted line denotes the limit of detection (20 FFU/ml). Statistical analysis was performed using the Mann-Whitney test; * = P ≤ 0.05. §, no mice surviving at time of lung extraction.
Immune mechanisms for pH1N1/E3-sciIV mediated protection.
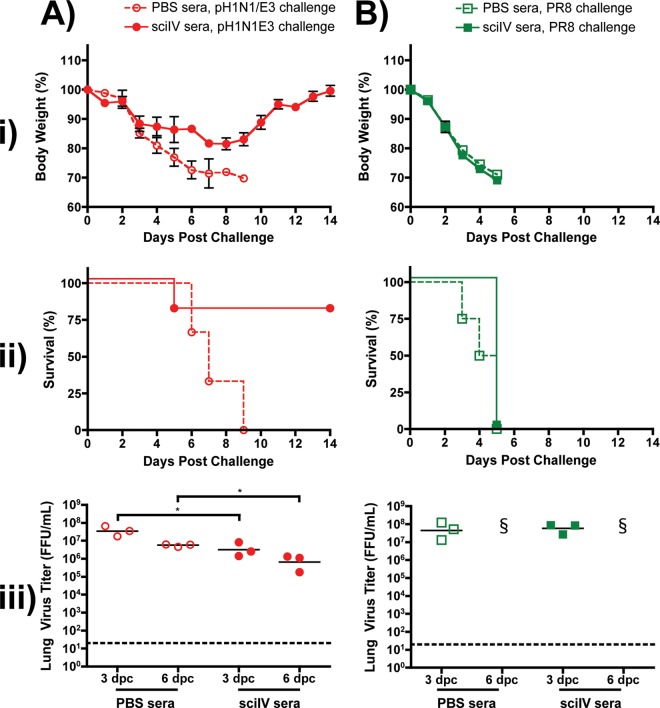
We have observed that inoculation with pH1N1/E3-sciIV could induce protective immunity from both homologous and heterologous influenza viruses, suggesting that both Abs and CD8 T cell immunity may mediate the protection (13). To confirm the potential involvement of Abs in protection, we transferred immune sera to naive mice (Fig. 8). Sera collected from mock-immunized mice or from mice receiving a prime-boost with 105 FFU pH1N1/E3-sciIV were pooled, and 100 μl was transferred i.p. into individual naive mice before challenge 1 day later with pH1N1/E3 (Fig. 8A) or PR8 (Fig. 8B) viruses (n = 11). When naive mice receiving the control sera collected from mock-immunized mice were challenged with a lethal dose of pH1N1/E3, rapid weight loss occurred (Fig. 8Ai) and all died within 9 days postchallenge (Fig. 8Aii). In contrast, naive mice receiving sera from pH1N1/E3-sciIV-immunized animals displayed modest weight loss, four of five animals recovered from challenge, and the level of virus detected in the lungs at 3 and 6 days postchallenge was significantly lower than that detected in the lungs of the group that received control sera (Fig. 8Aiii). However, upon lethal challenge with PR8, mice receiving sera from either PBS- or pH1N1/E3-sciIV-immunized mice showed dramatic weight loss (Fig. 8Bi) and all died (Fig. 8Bii). These data demonstrate that antigenically well-matched Abs induced by sciIV can be highly protective, even against lethal challenge. However, challenge with a virus that is a poor antigenic match requires more than just cross-reactive Abs and may also rely on the T cell response induced by sciIV.
Fig 8.
Passive immune transfer does not protect from lethal heterosubtypic challenge. (i and ii) Female 6-to-8-week-old C56BL/6 mice were inoculated twice at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV or with PBS as a negative control (n = 5). Two weeks after the final vaccination, sera were collected over the course of 1 week, checked for seroconversion by ELISA against total influenza virus protein (data not shown), and pooled. Naive mice received 100 μl of immune (sciIV sera, filled symbols) or nonimmune (PBS sera, open symbols) sera (n = 11) i.p. 1 day prior to challenge with 10 MLD50 pH1N1/E3 (A, red circles) or PR8 (B, green squares). Weight loss (panels i; plotted data represent means ± SEM) and survival (panels ii) were monitored daily for 2 weeks postchallenge. (iii) Virus lung titers. Mice passively immunized and challenged as described above were sacrificed at 3 and 6 days postchallenge to evaluate levels of replicating challenge virus in the lungs by IFA (n = 3). Symbols represent individual mice; bars represent geometric mean lung virus titers, and the dotted line denotes the limit of detection (20 FFU/ml). Statistical analysis was performed using the Mann-Whitney test; * = P ≤ 0.05. §, no mice surviving at time of lung extraction.
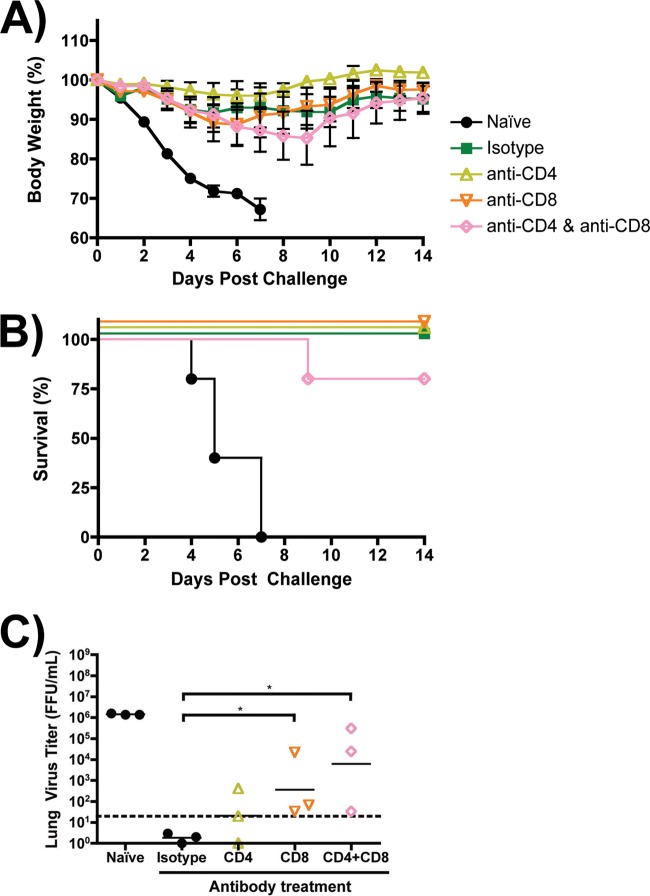
To address the importance of T cells in protection conferred by sciIV immunization, immune mice were depleted of CD4 or CD8 or both CD4 and CD8 T cells at the time of challenge by injecting subset-specific Abs prior to challenge (Fig. 9) (13). To focus on T cell responses, mice were challenged with PR8 (n = 11), which we demonstrated to not be controlled by passive immune transfer of Abs generated through pH1N1/E3-sciIV vaccination (Fig. 8B). At 3 days postchallenge, lungs were harvested and we found by flow cytometry that the Ab depletion of T cells was successful (data not shown). The depleted groups showed no significant increase in weight loss compared with the positive control (Isotype Ab) (Fig. 9A). The most profound effect of depletion was seen in mice that were depleted of both CD4 and CD8 T cells, where one out of five mice died (Fig. 9B). However, both the group that received anti-CD8 alone and the group that received anti-CD8 and anti-CD4 had significantly higher geometric mean viral lung titers than the control isotype group at day 6 postchallenge (Fig. 9C), though no significant difference was observed at day 3 (data not shown). Additionally, all immunized groups had significantly less detectable virus in the lungs than mock-immunized animals at day 6 (statistics not represented). Taken together, the data suggest that both CD4 and CD8 T cells induced by sciIV can contribute to protection from lethal challenge, as suggested by the higher viral titers in both the CD8- and the CD4-plus-CD8-depleted groups. The results do not rule out a contribution of mucosal Abs, and we cannot exclude the possibility of an additive or synergistic role for both the cell-mediated and humoral responses (29).
Fig 9.
T cells in sciIV-vaccinated mice enhance clearance of heterologous challenge. (A and B) Female 6-to-8-week-old C56BL/6 mice were inoculated twice at 2-week intervals i.n. with 105 FFU of pH1N1/E3-sciIV or with PBS (naïve; black circles) as a negative control (n = 11). Two weeks postvaccination, immunized mice were depleted of CD4 (yellow open triangles) or CD8 (orange open triangles) or both CD4 and CD8 (pink open diamonds) T cells by i.p. injection with 3 doses of blocking Abs (−2, 0, and 2 days postchallenge) or were given an IgG2b isotype control (green filled squares). All mice were challenged with 10 MLD50 PR8, and weight loss (A; plotted data represent means ± SEM) and survival (B) were monitored daily for 2 weeks postchallenge. (C) Virus lung titers. Mice immunized, depleted of T cells, and challenged as described above were sacrificed at 3 and 6 days postchallenge to evaluate levels of replicating challenge virus in the lungs by IFA (n = 3). Symbols represent individual mice; bars represent geometric mean lung virus titers, and the dotted line denotes the limit of detection (20 FFU/ml). Statistical analysis was performed using the Mann-Whitney test; * = P ≤ 0.05. Statistical significance (P ≤ 0.05) was also achieved in comparisons of each treatment group with the naive group (not shown).
Ferrets immunized with pH1N1/E3-sciIV are protected from influenza virus challenge and do not transmit virus.
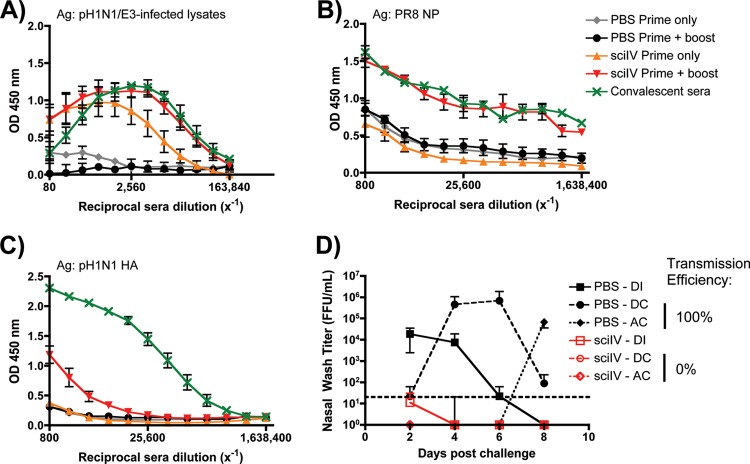
We sought to evaluate if the mechanisms of protection conferred by pH1N1/E3-sciIV immunization in mice translated to another animal model (Fig. 10). Ferrets are invaluable for influenza research, because, like humans, their sialic acids contain α2,6 linkages in the upper respiratory tract (30), and their respiratory tract anatomy and clinical symptoms are similar to those in humans (reviewed in reference 31). Additionally, contact- and aerosol-dependent transmission events occur in ferrets challenged with human H1N1 and H3N2 influenza A viruses (32–35). In ferrets receiving one or two doses of pH1N1/E3-sciIV, Abs were induced to levels similar to those induced within mice (Fig. 10A and 3A, respectively). Compared to ferrets convalescing from influenza virus infection, those receiving the prime-boost of sciIV showed similar levels of both total anti-influenza IgG Abs (Fig. 10A) and Abs against the in situ-replicated NP (Fig. 10B), whereas one dose elicited lower levels of both total and NP-specific Abs. As expected, and mirroring the response in mice, sciIV-immunized ferrets did not generate substantial anti-HA Abs (Fig. 10C and 3C, respectively), although HAI results indicated low levels of seroconversion (Table 2). In pH1N1/E3-sciIV-immunized ferrets, the challenge virus did not replicate in the nasal passages (Fig. 10D), and this sterilizing immunity also prevented transmission through direct and aerosol contacts. In contrast, PBS-immunized ferrets all replicated the challenge virus in nasal passages and subsequently transmitted the virus through both direct and aerosol contacts. Taking the results together, animals vaccinated with pH1N1/E3-sciIV produced a quantifiable Ab response that directly correlates with sterilizing immunity to a matched challenge that prevents transmission in ferrets.
Fig 10.
SciIV are immunogenic, induce protection from homologous infection, and prevent transmission in ferrets. (A to C) Five-month-old male ferrets were inoculated twice at 4-week intervals i.n. with 106 FFU of pH1N1/E3-sciIV or with PBS as a negative control (n = 3). Sera were collected 4 weeks postprime and 4 weeks postboost and evaluated for IgG Abs against total influenza virus protein (A; plotted data represent means ± SEM), recombinant NP from PR8 (B; plotted data represent means ± SEM), or recombinant HA from pH1N1 (C; plotted data represent means ± SEM) by ELISA. Sera from ferrets convalescing from A/California/07/2009 influenza virus infection were used as a positive control (n = 3). (D) Protection from challenge and transmission conferred by pH1N1/E3-sciIV vaccination. Immunized ferrets from the experiments represented in panels A to C were then divided among 6 total housing units and challenged with 106 pH1N1 FFU (direct infection [DI]; squares) 1 day prior to being cohoused with a direct contact (DC; circles) and adjacent to an aerosol contact (AC; diamonds; n = 3/group). Starting 2 days postchallenge and every other day following, nasal washes were collected and used to evaluate levels of replicating challenge virus by IFA. Symbols represent mean virus titers ± SD; the dotted line denotes the limit of detection (20 FFU/ml).
Table 2.
Immunogenicity of sciIV in ferrets
| Treatment | Collection timea | Sera GMTb |
|
|---|---|---|---|
| HAI | VNA | ||
| PBS | Prebleed | <10 | <10 |
| Prime only | <10 | <10 | |
| Prime + boost | <10 | <10 | |
| pH1N1/E3-sciIV | Prebleed | <10 | <10 |
| Prime only | 40 | 64 | |
| Prime + boost | 64 | 160 | |
| Convalescent-phase serac | 1,800 | 3,600 | |
Vaccinations were administered at 4 week intervals; samples were collected 4 weeks after the indicated dose.
Data were calculated from 3 immunized or 2 convalescent individual animals. VNA, virus neutralization assay.
Ferrets were inoculated with a nonlethal dose of A/California/07/2009; sera were collected 4 weeks later.
DISCUSSION
Using reverse genetics techniques, we have engineered a recombinant influenza A virus (sciIV) that cannot undergo multicycle replication due to the lack of the HA gene. However, sciIV can be amplified in HA-expressing stable cell lines and appears to be a safe and protective vaccine against influenza virus infections in mice and ferrets. It is difficult to directly compare the immune response generated by sciIV with that of current vaccines (TIV and LAIV) because of their differences in replication, but the marked protection and HAI titer > 1:40 we observed are consistent with seroconversion and successful vaccination in human trials (10). Vaccination with sciIV drives robust protection that is mediated by both T and B cells, as demonstrated by homo- and heterosubtypic challenge experiments.
The HA-deficient, reporter-expressing sciIV was first generated to evaluate NAbs using single-cycle HA-pseudotyped viruses. Because this virus still retains all the essential components for replication and transcription (PB1, PB2, PA, and NP), we sought to inoculate mice i.n. to mimic a single round of bona fide influenza A virus infection in vivo. Though the contribution of preexisting immunity to sciIV uptake has yet to be assessed, any hindrance of uptake can presumably be nullified in patients by pseudotyping with an HA that humans are not typically exposed to.
In addition to the use of sciIVs, alternative methods have been shown to be effective in overcoming the moderate efficacy of TIV while maintaining its tolerability profile. The elderly are a group particularly sensitive to influenza illness, and unfortunately, TIV has not been very effective in this population (36). However, marked increases in HAI titers have been obtained when elderly patients aged 65 years and older received a high-dose formulation of TIV (37), which is now recommended by the Advisory Committee on Immunization Practices (ACIP) (38). Additionally, technologies that have proven successful in protecting against other pathogens have been applied to influenza virus with great success, albeit with a number of caveats. Vaccination with recombinant HA produced in insect cells induces humoral immunity (39). The use of virus-like particles (VLPs) that contain the four influenza virus proteins HA, NA, M1, and M2 (40) has been shown to induce protection through the induction of protective Abs that either neutralize the virus or lead to antibody-dependent cell cytotoxicity (41, 42). Lastly, VLPs that express tandem repeats of the highly conserved M2 ectodomain (M2e) (43) induce cross-reactive Ab-mediated protection (41). The drawbacks of these approaches lie in the reliance upon systemic, humoral protection and the lack of replication-induced immunostimulation. Also, all of the vaccine candidates, besides the M2e-based vaccines, generate an anti-HA response, which is ineffective when reassortant pandemic viruses emerge (44, 45) or when the vaccine is a poor match to the circulating seasonal strains (46).
In this study, we also sought to identify the mechanism by which sciIV protects. Several of our observations have led us to believe that both T and B cells are playing a role in protection. First, there appear to be three distinct phases of morbidity observed from mice that are protected by low doses of replicating sciIV. Over the first 2 to 3 days postchallenge, mice uniformly maintain body weight, suggesting the presence of an established protective environment, likely due to a primed innate environment or the induction of Abs. Mice then lose weight on days 4 to 7 but subsequently recover simultaneously with T cell peak lung accumulation (47). Second, that we observed protection from homologous and heterologous virus challenge suggests induction of T cells that recognize internal, conserved epitopes (47, 48). Our data suggest that sciIV-induced Abs alone can confer protection against only homologous virus challenge and not heterologous virus challenge. However, when addressing whether T cells alone were driving heterologous protection, we observed that CD4 and CD8 T cells contribute to protection.
Here, we have demonstrated that a recombinant single-cycle influenza virus stimulated robust immunity to influenza in both mice and ferrets. Due to current limitations of FDA-approved influenza vaccines in the United States, alternative methodologies are needed to help overcome the hurdle presented by this rapidly evolving human pathogen. We believe that sciIV holds promise not only to perform as a standalone vaccine prototype but also to enhance the current TIV approach or other protein-based approaches if the strains are delivered simultaneously. Administration of LAIV is limited to immunocompetent patients between 2 and 49 years of age, but like sciIV, LAIV vaccination mimics a natural infection and immunogenicity is strong in unexposed individuals (4). However, since sciIV is self-limiting, the patient range can potentially be broadened due to lower safety concerns. Recent effort has been redoubled for adapting existing egg-based vaccine technologies to mammalian cells (including recent FDA approval of MDCK-derived influenza vaccines [49]) to limit the requirement of large quantities of eggs, reactogenicity issues, problems with maintaining antigenicity, and the time required from surveillance to vaccine distribution. By inclusion of the protein-coding region for HA into MDCK cells, as we have demonstrated, large quantities of sciIV vaccine stocks could be produced, though we have yet to test the adaptability of our system to growth in suspension. Indeed, seroconversion of patients by HI would not be applicable with our vaccination strategy, but a rapid ELISA could be employed that measures Abs against total influenza virus proteins. Beyond influenza, we hope that sciIV can be adapted for use as a vaccine vector for immunization against both influenza virus and other respiratory pathogens by replacing GFP with the ORF of a viral or bacterial immunogenic protein.
ACKNOWLEDGMENTS
We thank members of the laboratory of L.M.-S. for their discussion and Snezhana Dimitrova for technical support. We thank Troy D. Randall (currently at UAB, Birmingham, AL) for providing the PR8 NP. The H1 hemagglutinin (HA) protein from influenza virus A/California/04/09 (H1N1) virus was provided by the NIH/NIAID Biodefense and Emerging Infections Research Resources Repository. The HT103 monoclonal antibody was generated at the Center for Therapeutic Antibody Discovery at Icahn School of Medicine at Mount Sinai. Class I tetramers were kindly provided by the NIH Tetramer Facility.
S.F.B. is a recipient of an intercollaborative training grant award from the Centers for Excellence in Influenza Research & Surveillance (CEIRS). Research in the laboratory of L.M.-S. is funded by the NIH (grants RO1 AI077719, R21NS075611-01, and R03AI099681-01A) and The University of Rochester Center for Biodefense Immune Modeling (CBIM), contract HHSN272201000055C. Research in the laboratory of L.M.-S. and D.J.T. is supported by the NIH/NIAID network of Centers of Excellence in Influenza Research and Surveillance (CEIRS), contract HHSN266200700008C. Research in the laboratory of A.G.-S. is partly supported by the NIH/NIAID Centers of Excellence for Influenza Research and Surveillance (HHSN266200700010C) and by NIAID grant P01AI097092.
Footnotes
Published ahead of print 29 May 2013
REFERENCES
- 1. Lemey P, Suchard M, Rambaut A. 2009. Reconstructing the initial global spread of a human influenza pandemic: a Bayesian spatial-temporal model for the global spread of H1N1pdm. PLoS Curr. 1:RRN1031. 10.1371/currents.RRN1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wright PF, Neumann G, Kawaoka Y. 2007. Orthomyxoviruses, p 1691–1740 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Commission on Influenza, Board for the Investigation and Control of Influenza and Other Epidemic Diseases in the Army, Preventive Medicine Service, Office of the Surgeon General, United States Army 1944. A clinical evaluation of vaccination against influenza. Joint Report with Members of the Commission on Influenza, Board for the Investigation and Control of Influenza and Other Epidemic Diseases in the Army, Preventive Medicine Service, Office of the Surgeon General, United States Army. JAMA 124:982–985 [Google Scholar]
- 4. Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 12:36–44 [DOI] [PubMed] [Google Scholar]
- 5.Palese PS, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 6. WHO 2009. Influenza (seasonal) fact sheet no. 211. WHO, Geneva, Switzerland [Google Scholar]
- 7. Sun W. 2012. Approval letter—Fluarix Quadrivalent (BL 125127/513). U.S. Food and Drug Administration (FDA), Silver Spring, MD [Google Scholar]
- 8. Sun W. 2012. Approval letter—FluMist® Quadrivalent (BL 125020/1668). U.S. Food and Drug Administration (FDA), Silver Spring, MD [Google Scholar]
- 9. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dowdle WN, Kendal AP, Noble GR. 1979. Influenza viruses, p 585–609 In Lennette EH, Schmidt NJ. (ed), Diagnostic procedures for viral, rickettsial, and chlamydial infections, 5th ed American Public Health Association, Washington, DC [Google Scholar]
- 11. Centers for Disease Control and Prevention (CDC) 2012. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP)—United States, 2012–13 influenza season. MMWR Morb. Mortal. Wkly. Rep. 61:613–618 [PubMed] [Google Scholar]
- 12. Martínez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, Garcia-Sastre A. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. 2011. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 85:448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schickli JH, Flandorfer A, Nakaya T, Martinez-Sobrido L, Garcia-Sastre A, Palese P. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1965–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kilbourne ED. 1969. Future influenza vaccines and the use of genetic recombinants. Bull. World Health Organ. 41:643–645 [PMC free article] [PubMed] [Google Scholar]
- 16. Polakos NK, Klein I, Richter MV, Zaiss DM, Giannandrea M, Crispe IN, Topham DJ. 2007. Early intrahepatic accumulation of CD8+ T cells provides a source of effectors for nonhepatic immune responses. J. Immunol. 179:201–210 [DOI] [PubMed] [Google Scholar]
- 17. Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. 2007. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J. Virol. 81:7608–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chapman TJ, Castrucci MR, Padrick RC, Bradley LM, Topham DJ. 2005. Antigen-specific and non-specific CD4+ T cell recruitment and proliferation during influenza infection. Virology 340:296–306 [DOI] [PubMed] [Google Scholar]
- 19. Hai R, Schmolke M, Varga ZT, Manicassamy B, Wang TT, Belser JA, Pearce MB, Garcia-Sastre A, Tumpey TM, Palese P. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsh GA, Hatami R, Palese P. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 81:9727–9736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medina RA, Manicassamy B, Stertz S, Seibert CW, Hai R, Belshe RB, Frey SE, Basler CF, Palese P, Garcia-Sastre A. 2010. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nat. Commun. 1:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steel J, Lowen AC, Pena L, Angel M, Solorzano A, Albrecht R, Perez DR, Garcia-Sastre A, Palese P. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seibert CW, Kaminski M, Philipp J, Rubbenstroth D, Albrecht RA, Schwalm F, Stertz S, Medina RA, Kochs G, Garcia-Sastre A, Staeheli P, Palese P. 2010. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J. Virol. 84:11219–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindenmann J, Burke DC, Isaacs A. 1957. Studies on the production, mode of action and properties of interferon. Br. J. Exp. Pathol. 38:551–562 [PMC free article] [PubMed] [Google Scholar]
- 26. Guo H, Topham DJ. 2012. Multiple distinct forms of CD8+ T cell cross-reactivity and specificities revealed after 2009 H1N1 influenza A virus infection in mice. PLoS One 7:e46166. 10.1371/journal.pone.0046166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Z, Wang W, Zhou H, Suguitan AL, Jr, Shambaugh C, Kim L, Zhao J, Kemble G, Jin H. 2010. Generation of live attenuated novel influenza virus A/California/7/09 (H1N1) vaccines with high yield in embryonated chicken eggs. J. Virol. 84:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suphaphiphat P, Franti M, Hekele A, Lilja A, Spencer T, Settembre E, Palmer G, Crotta S, Tuccino AB, Keiner B, Trusheim H, Balabanis K, Sackal M, Rothfeder M, Mandl CW, Dormitzer PR, Mason PW. 2010. Mutations at positions 186 and 194 in the HA gene of the 2009 H1N1 pandemic influenza virus improve replication in cell culture and eggs. Virol. J. 7:157. 10.1186/1743-422X-7-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang S, Mozdzanowska K, Palladino G, Gerhard W. 1994. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152:1653–1661 [PubMed] [Google Scholar]
- 30. van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maher JA, DeStefano J. 2004. The ferret: an animal model to study influenza virus. Lab. Anim. (NY) 33:50–53 [DOI] [PubMed] [Google Scholar]
- 32. Herlocher ML, Carr J, Ives J, Elias S, Truscon R, Roberts N, Monto AS. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 54:99–111 [DOI] [PubMed] [Google Scholar]
- 33. Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. 2009. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc. Natl. Acad. Sci. U. S. A. 106:3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vu T, Farish S, Jenkins M, Kelly H. 2002. A meta-analysis of effectiveness of influenza vaccine in persons aged 65 years and over living in the community. Vaccine 20:1831–1836 [DOI] [PubMed] [Google Scholar]
- 37. Falsey AR, Treanor JJ, Tornieporth N, Capellan J, Gorse GJ. 2009. Randomized, double-blind controlled phase 3 trial comparing the immunogenicity of high-dose and standard-dose influenza vaccine in adults 65 years of age and older. J. Infect. Dis. 200:172–180 [DOI] [PubMed] [Google Scholar]
- 38. Centers for Disease Control and Prevention (CDC) 2010. Licensure of a high-dose inactivated influenza vaccine for persons aged ≥ 65 years (Fluzone High-Dose) and guidance for use—United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:485–486 [PubMed] [Google Scholar]
- 39. Treanor JJ, Wilkinson BE, Masseoud F, Hu-Primmer J, Battaglia R, O'Brien D, Wolff M, Rabinovich G, Blackwelder W, Katz JM. 2001. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 19:1732–1737 [DOI] [PubMed] [Google Scholar]
- 40. Latham T, Galarza JM. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, Van Rooijen N, Verbeek S, Fiers W, Saelens X. 2011. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 186:1022–1031 [DOI] [PubMed] [Google Scholar]
- 42. Pushko P, Tumpey TM, Bu F, Knell J, Robinson R, Smith G. 2005. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine 23:5751–5759 [DOI] [PubMed] [Google Scholar]
- 43. Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. 1999. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 5:1157–1163 [DOI] [PubMed] [Google Scholar]
- 44. Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361:1945–1952 [DOI] [PubMed] [Google Scholar]
- 46. Belongia EA, Kieke BA, Donahue JG, Greenlee RT, Balish A, Foust A, Lindstrom S, Shay DK. 2009. Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004–2005 season to the 2006–2007 season. J. Infect. Dis. 199:159–167 [DOI] [PubMed] [Google Scholar]
- 47. Kohlmeier JE, Woodland DL. 2009. Immunity to respiratory viruses. Annu. Rev. Immunol. 27:61–82 [DOI] [PubMed] [Google Scholar]
- 48. LaMere MW, Lam HT, Moquin A, Haynes L, Lund FE, Randall TD, Kaminski DA. 2011. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J. Immunol. 186:4331–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gruber M. 2012. Approval letter—Flucelvax (BL 125408/0). U.S. Food and Drug Administration (FDA), Silver Spring, MD [Google Scholar]