Abstract
Advances in imaging and reductionist approaches provide a high-resolution understanding of nuclear pore complex structure and transport, revealing unexpected mechanistic complexities based on nucleoporin functions and specialized import and export pathways.
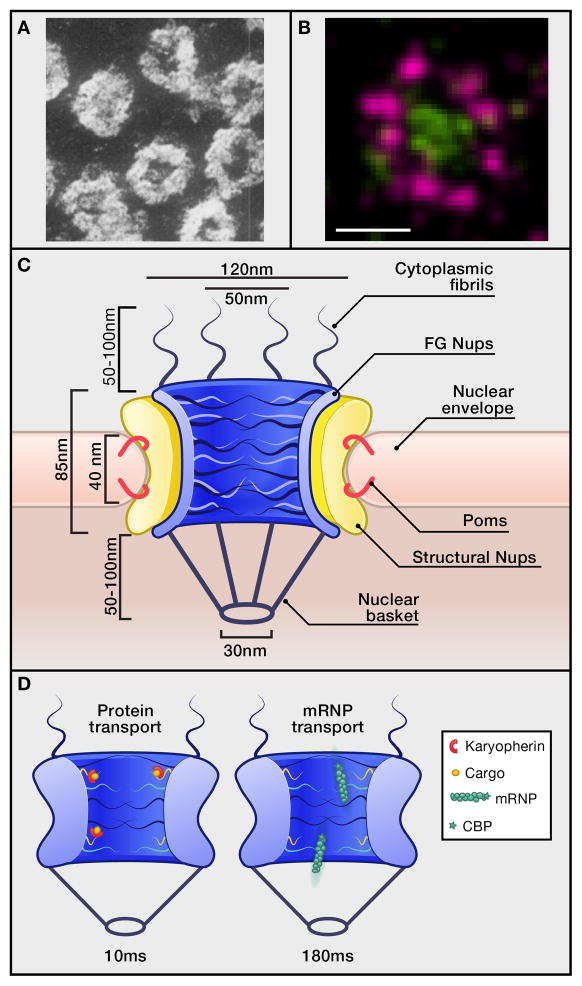
First impressions can be misleading. Pioneering transmission electron microscopy (EM) approaches 60 years ago first revealed a structure within the eukaryotic nuclear envelope (NE): the nuclear pore complex (NPC) (Gall, 1954) (Figure 1A). The original view is striking yet deceptively simple, with the ~100 MDa proteineacous NPC assembly spanning the NE to provide a passageway between the nucleus and cytoplasm. Over time, insights into NPC structure and function have revealed unexpected complexities.
Figure 1. NPC structure and transport.
A. Early EM image of the NPC cytoplasmic face in a salamander oocyte NE. Reprinted with permission from Gall, 1954. Scale bar, 100nm. B. 8-fold symmetry of the NPC in the NE plane resolved by dSTORM microscopy. Lumenal domain of the transmembrane Nup gp210 (magenta) and the FG Nups (green) in a Xenopus laevis oocyte NE. Reprinted with permission from Löschberger, et al, 2012. Scale bar, 100nm. C. Schematic of NPC architecture. Measurements indicate dimensions for the human NPC from cryoET (Maimon et al., 2012). D. Transport pathways through the NPC, with distinct FG Nup requirements for karyopherin transport versus mRNA export (Terry and Wente, 2009). Protein transport occurs in ~10ms (Yang and Musser, 2006), whereas mRNA export takes 180ms (Grünwald and Singer, 2010). Transport cargo sizes are to scale with NPC: protein cargo as ~80kDa globular shape, and mRNP size potentially proportional to the transcript length (including the 5′ Cap-binding protein complex (CBP) (star) and other RNA binding proteins (circles).
NPC pathways for nucleocytoplasmic transport are based on the type of cargo. Diffusion through NPCs is inhibited for molecules > ~40kDa, larger macromolecules and/or accumulation against a concentration gradient requires facilitated transport (Aitchison and Rout, 2012). Nuclear RNAs are actively exported for function in the cytoplasm while nuclear import being required for proteins made in the cytoplasm during interphase. Increased The eukaryotic proteome and RNA repertoires have expanded range and bulk of macromolecules that require facilitated transport through NPCs.
Do all NPCs in a given cell and all transport pathways in a given NPC function the same? Recent work uncovers unanticipated layers of complexity in NPC structure and function. High-resolution imaging has allowed dynamic visualization of NPC transport events while reductionist approaches pinpoint how both complex and simple components contribute to transport pathway specialization. How such specialization might contribute to the transport mechanism and high cargo load capacity is intriguing. This also sets the stage for future studies taking into account possible heterogeneity between within NPCs.
Insights gained from high-resolution NPC structural analysis
The original EM views of the NPC documented a simple structure with 8-fold rotational symmetry in the plane of the NE. Details of cytoplasmic filaments and a nuclear basket structure were defined by scanning EM (Aitchison and Rout, 2012) (Figure 1C). Leaps in structural resolution come from a combination of both high-resolution cryo-electron tomography (cryo-ET) of NPCs in intact NEs, x-ray crystallography studies (Bilokapic and Schwartz, 2012) and cryo-ET work yielding a 6.6nm resolution image of the human NPC (Maimon et al., 2012). Coupling these with strategies to individually pinpoint different Nups may allow crystal structures of components to be modeled into the entire NPC. Tour de force analysis of most yeast S. cerevisiae Nups (“NPC-wide”) by parallel structural and biochemical approaches enabled in silico computational modeling, generating new insights into NPC molecular architecture (Alber et al., 2007).
Importantly, while previous low-resolution studies show conservation of structure between humans and other eukaryotes, high-resolution cryo-ET unravel subtle differences in divergent NPCs. Variations in the cavities near the periphery of the central transport channel suggest functional divergence in this part of the NPC (Maimon et al., 2012). These may arise from the protein composition differences across species and innovations in super-resolution light microscopy should allow Nup localization to be examined at an EM-level resolution. These methods have already permitted visualization of the 8-fold symmetry of Nups in fixed cells (Löschberger et al., 2012) (Figure 1B), and direct live cell observations of the asymmetric nuclear-cytoplasmic distribution of Nups in NPCs (Hayakawa et al., 2012). Further studies employed to map Nups to particular NPCs could establish how specific Nup subcomplexes are oriented in NPCs.
Functional complexity revealed by NPC-wide analysis
S. cerevisiae and human NPC-constituting proteins were identified a decade ago. The ~30 proteins are grouped into three functional classes (Terry and Wente, 2009): transmembrane Nups that anchor the NPC in the NE, also called pore membrane proteins (Poms); structural Nups that stabilize the NE curvature at nuclear pores and provide scaffolding for assembling other peripheral Nups; or FG Nups that contribute to the permeability barrier for nonspecific transport and facilitate movement as direct binding sites for transport receptors. Nups adopt a limited variety of structural folds such as beta-propeller, alpha solenoid, or FG domains (Aitchison and Rout, 2012; Bilokapic and Schwartz, 2012). Parts of this simple structural assembly reflect the Nups’ ancestral relationship with proteins in vesicle coat complexes. Thus, this complex machine derives its function through surprisingly simple structural elements.
The complexity in NPC function comes from several elements. First, different Nups are associated with NPCs for different time periods. Recent structural Nups are among the most stable proteins in a cell, persisting for months or years in a non-dividing cell (Savas et al., 2012); these remain stably NPC-associated once assembled into the NPC (Rabut et al., 2004). In contrast, FG Nups are highly dynamic (Rabut et al., 2004), with seconds-minutes residence times in the NPC. How this dichotomy in association times for different components might affect transport unknown. Second, NPC cargo load can alter the transport mechanism. Single-molecule microscopy studies show increasing concentrations of the import in beta transport receptor altering transport time of both its cargo and molecules that passively diffuse (Yang and Musser, 2006). It is intriguing to consider that the environment of a given transport channel might be temporally impacted due to either cargo load or the specific associated FG Nups.
Third, diversity in function among the FG Nups is illuminated by several key NPC-wide studies. FG Nups have been considered interchangeable and of uniform function due to their common attributes. FG Nups contain motifs enriched in phenylalanine (F) and glycine (G) repeats, such as FXFG and GLFG (L, leucine; X, any amino acid); the spacer sequences between FG repeats consist of ~5–30 residues typically enriched in polar amino acids. Analyses to date indicate that FG domains are unstructured and occupy the central NPC channel (Terry and Wente, 2009; Yamada et al., 2010; Aitchison and Rout, 2012). Although these FG domains constitute ~12% of the NPC, they are not resolved in high-resolution structures due. Immuno-EM analysis indicate that a single FG domain type occupies multiple topologies within an NPC (Fahrenkrog et al., 2002). Thus, all FG Nups may share an unexpected structural flexibility as a defining feature.
Several notable distinctions are also defined amongst the FG domains. NPC-wide analysis of biochemical and biophysical properties of individual FG domains or subdomains show differences in cohesive properties in terms of self- and inter-FG interactions, and in levels of compaction (collapsed versus random coil) (Yamada et al., 2010). In vivo evidence reveals distinct functions for FG domains. In an analysis of FG domain deletion mutants, S. cerevisiae viability required only specific combinations of FG domains: individual ones were dispensable with few required in double or triple mutant combinations (Terry and Wente, 2009). Importantly, FG domain deletion mutants were defective in specific nuclear transport pathways. For example, a FG deletion mutant defective in Kap121 import was competent for mRNA export, and vice versa (Terry and Wente, 2009). Recently, the Nup98 was shown to be necessary for generation of the permeability barrier that inhibits diffusion of macromolecules (Hülsmann et al., 2012). Without the Nup98 FG domain, only substitution with another cohesive FG domain restored the barrier. That the permeability barrier function could be attributed to one specific FG Nup further provides evidence that all FG Nups are neither the same nor interchangable.
A final layer of complexity stems from Nup post-translational modifications. It is known that vertebrate FG Nups are O-linked glycosylated, and this may regulate the vertebrate NPC permeability barrier (Labokha et al., 2012). Nup98 phosphorylation is an initial step in the breakdown of the NPC during open mitosis (Laurell et al., 2011). Phosphorylation increases permeability of the NPC, either through altering the conformation of the Nup98 GLFG domain or inducing its dissociation from NPCs (Hülsmann et al., 2012). In a NPC-wide analysis of ubiquitylation carried out in S. cerevisiae (Hayakawa et al., 2012), this modification was discovered on almost all Nups. Interestingly, proper nuclear migration during mitosis requires Nup159 ubiquitylation. Future work should reveal how these layers of complexity impact nuclear transport function.
Dynamic and diverse transport pathways uncovered within NPCs
NPC translocation is defined by docking, translocation and release steps for cargo complexes (Aitchison and Rout, 2012). A protein must display a nuclear localization sequence (NLS) for entry or nuclear export sequence (NES) for exit. These motifs provide binding sites for transport receptors (karyopherins, importins, exportins, transportins). RNA transport receptors either recognize the RNA directly (tRNA, miRNA) or interact with an RNA-binding adaptor protein (in the mRNA ribonucleoprotein (mRNP) complex). Besides cargo interactions, transport receptors also contain hydrophobic pockets that bind the phenylalanine residues of FG domains (Terry and Wente, 2009).
Alternative models for how transport receptor-FG interactions mediate NPC translocation are under investigation. However, the understanding of how transport directionality is dictated has been agreed upon. For karyopherins, accumulation of cargo against its concentration gradient and recycling of the transport receptor is based on localized control of Ran GTPase activity (GTP state in the nucleus, GDP in the cytoplasm). Specifically, the importin-cargo complex binding to Ran-GTP in the nucleus causes cargo release. In contrast, a RanGTP-exportin-cargo complex disassembles in the cytoplasm with GTP hydrolysis (Aitchison and Rout, 2012). An analogous non-RanGTP mechanism exists for mRNA export by the NXF1 receptors (S. cerevisiae Mex67), wherein ATP/ADP cycling of a RNA-dependent DEAD-box ATPase (Dbp or DDX) localized on the NPC cytoplasmic filaments drives directional transport (Folkmann et al., 2011). Overall, directional facilitated translocation is dictated by spatially controlled, nucleotide-dependent switches at exit sites.
The requirements of different FG Nups for specific transport receptors underscores the potential for multiple preferential pathways existing in an NPC (Figure 1D) (Terry and Wente, 2009). Whether the active and passive transport pathways are both functionally and spatially distinct in the NPC central channel has been debated. Recent microscopy technologies have documented real-time single translocation events (Yang and Musser, 2006; Grünwald and Singer, 2010; Lowe et al., 2010; Mor et al., 2010; Ma et al., 2012 based on both high spatial resolution and temporal parameters, coupled with single-molecule innovations for specific protein cargo labeling (Yang and Musser, 2006) and large quantum dots (Lowe et al., 2010). NPC interaction times during facilitated protein transport were measured as ~10ms, with a reported range of 2–34ms (Yang and Musser, 2006), with RanGTP driving release of large cargo from the NPC (Lowe et al., 2010). The three-dimensional pathways for importin beta cargo may be more peripheral to that for diffusive cargo (Figure 1D) (Ma et al., 2012).
Single mRNAs have also been observed moving across the NPC by engineering sequence specific RNA stem-loops into endogenous or inducible transcripts and co-expressing fluorescently-tagged MS2 RNA stem-loop binding proteins (Grünwald and Singer, 2010; Mor et al., 2010). Here, the observed time frame for mRNA transport through the pore is 180ms (Grünwald and Singer, 2010) - 500ms (Mor et al., 2010), with nuclear and a cytosolic rate-limiting steps (Grünwald and Singer, 2010). The rate-limiting interval at the cytoplasmic face is likely due to mRNP remodeling to promote directionality. Although both fast and slow (>800ms) transport rates are observed for a single mRNA type (Grünwald and Singer, 2010), mRNP translocation through the NPC occurred 15-fold faster than diffusion through the nucleus (Mor et al., 2010),
Comparing the transport of protein and mRNA reveal differences with a longer duration for mRNA transport across the NPC, likely due to the size differences in the respective protein versus mRNP cargos (Figure 1D). mRNA export also has a rate-limiting step at the NPC entry site that might be attributed to the mRNA quality control and surveillance mechanisms prior to export. For protein and mRNA transport single-molecule experiments, a striking common conclusion is that cargo enters the NPC and explores the channel in a diffusive/sub diffusive manner with observed back and forth movements, suggesting he lack of a straight path through the NPC and that movement itself is not inherently directional It is remarkable that the transport events are most often unsuccessful (Grünwald and Singer, 2010; Yang and Musser, 2006). raising the question of how the NPC accommodates not only a large amount of successful transport events but an even larger number of unsuccessful events.
Models impacted by nuclear pore complexity and heterogeneity
The NPC’s inherent complexity has favored reductionist approaches to gain molecular insights into transport mechanisms. Innovations include the development of in vitro nanopores and hydrogels for testing the selective barrier properties with transport receptors and cargo. In a nanopore approach, recombinant FG domains were coupled to a small nanopore (30nm holes) (Jovanovic-Talisman et al., 2009). In contrast, the hydrogels self-formed under experimentally determined conditions with recombinant FG domains (Labokha et al., 2012). These have demonstrated that FG domains are sufficient for allowing selective passage of transport receptors. A recent hydrogel study characterized individual FG domains of Xenopus laevis on an NPC-wide level finding that resulting hydrogels had different capacities for selective transport (Labokha et al., 2012). To effectively mimic the heterogeneous and dynamic NPC environment, these systems will require constructing single nanopores and hydrogels with multiple different FG domains included. Because of the now known complexity, one FG domain type cannot be considered in isolation, nor are all FG domains the same.
Several different models have been proposed for the mechanism of NPC translocation, especially the entropic barrier and selective phase models. These differ in how the intermolecular interactions between FG domains contribute to facilitated transport and a selective barrier (Terry and Wente, 2009; Aitchison and Rout, 2012; Hülsmann et al., 2012). The entropic barrier model suggests that the unstructured FG domains function to exclude non-interacting molecules. Alternatively, the selective phase model proposes that inter-domain hydrophobic interactions form a gel-like meshwork locally “dissolved” by transport receptor interactions. For both models, work is needed to account for the heterogeneity of FG domains in vivo and in vitro. A hybrid model is also quite appealing wherein functions for cohesive (for permeability barrier) and noncohesive (for entropic bristles) interactions are considered (Yamada et al., 2010). These complexities provide an exciting challenge for further investigations.
Perspective
Currently, a single mechanism of nuclear transport across the NPC likely does not exist; rather, layers of complexity lead to multiple specialized pathways in a given NPC. Whether different transport pathways allow multiple transport events to take place within a single NPC is still unresolved. Classic immuno-EM experiments demonstrated that an individual NPC is capable of carrying out both import and export (Feldherr, et al, 1984); however, it has not been tested directly whether such can be simultaneous. Tracking single mRNA transcripts reveal transient association with multiple NPCs before exit (Grünwald and Singer, 2010), due to the inherent properties of stochastic cargo movement with the NPC. It is possible this may reflect a full cargo load for a given NPC inhibiting entry and new translocation events. This may also involve the absence of specific factors/Nups at a given NPC or quality control mechanisms detecting incomplete processing of the transcript. To directly address simultaneous transport, a future challenge will be to monitor single-molecule facilitated transport of different cargoes at the same time within one cell/NPC.
While specialized transport pathways exist within the heterogenous environment of the NPC, it is unclear whether different NPCs in a single cell are specialized for distinct types of transport. Distinctions might exist in each NPC as a result of dynamic Nup associations, post-translational or conformational changes or temporal changes in expression. There is evidence for differential NPC function in specific animal tissues at specific times in cellular differentiation. A recent study found that a transmembrane Nup (gp210) was absent in proliferating myoblasts, but was required for differentiation into neuroprogenitors (D’Angelo et al., 2012). Using genome-wide RNA-sequencing, gp210 expression caused differential regulation of a subset of transcripts without globally affecting NPC transport. How a transmembrane Nup has these effects is unclear; however, NPC function is evidently altered by differential Nup association. Advances in imaging and NPC-wide, or genome wide, approaches will be needed to further analyze NPC mechanisms of specialization on cellular and organism levels.
Finally, the complexity of Nups extends beyond the NPC, as independent functions have been uncovered for some Nups (Raices and D’Angelo, 2012). Thus, a full understanding of nuclear pore complexity is needed to position the field in evaluating the molecular mechanisms underlying nup mutants linked to human developmental diseases (Raices and D’Angelo, 2012). The wealth of innovations has unveiled NPC structure and function as much more complex than anticipated.
Acknowledgments
We thank Joe Gall (Carnegie Institution for Science) and Markus Sauer (Julius-Maximilians-University Wurzburg) for permission to reprint the images in Figure 1A, B,; Wente laboratory members and Elizabeth Bowman for discussion. Due to space constraints, we regret not being able to cite all primary references. The authors were supported by grants from the National Institute of Health (R37GM051219 (S.R.W.) and T32HD007502 (R.L.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitchison JD, Rout MP. Genetics. 2012;190:855–883. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. Curr Opin Cell Biol. 2012;24:86–91. doi: 10.1016/j.ceb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B, Maco B, Fager AM, Köser J, Sauder U, Ullman KS, Aebi U. J Struct Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Kallenbach E, Schultz N. J Cell Biol. 1984;99:2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Noble KN, Cole CN, Wente SR. Nucleus. 2011;2:540–548. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Exp Cell Res. 1954;7:197–200. doi: 10.1016/0014-4827(54)90054-3. [DOI] [PubMed] [Google Scholar]
- Grünwald D, Singer RH. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A, Babour A, Sengmanivong L, Dargemont C. J Cell Biol. 2012;196:19–27. doi: 10.1083/jcb.201108124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülsmann BB, Labokha AA, Görlich D. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. Nature. 2009;457:1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. EMBO J. 2012;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Löschberger A, van de Linde S, Dabauvalle MC, Rieger B, Heilemann M, Krohne G, Sauer M. J Cell Sci. 2012;125:570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- Lowe AR, Siegel JJ, Kalab P, Siu M, Weis K, Liphardt JT. Nature. 2010;467:600–603. doi: 10.1038/nature09285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Goryaynov A, Sarma A, Yang W. Proc Natl Acad Sci USA. 2012;109:7326–7331. doi: 10.1073/pnas.1201724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Nat, Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Raices M, D’Angelo MA. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. Eukaryot Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, Gopinathan A, Lau EY, Colvin ME, Uversky VN, Rexach MF. Mol Cell Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Musser SM. J Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]