Abstract
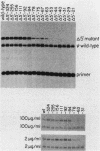
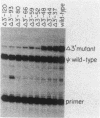
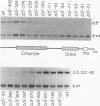
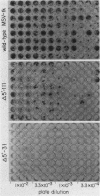
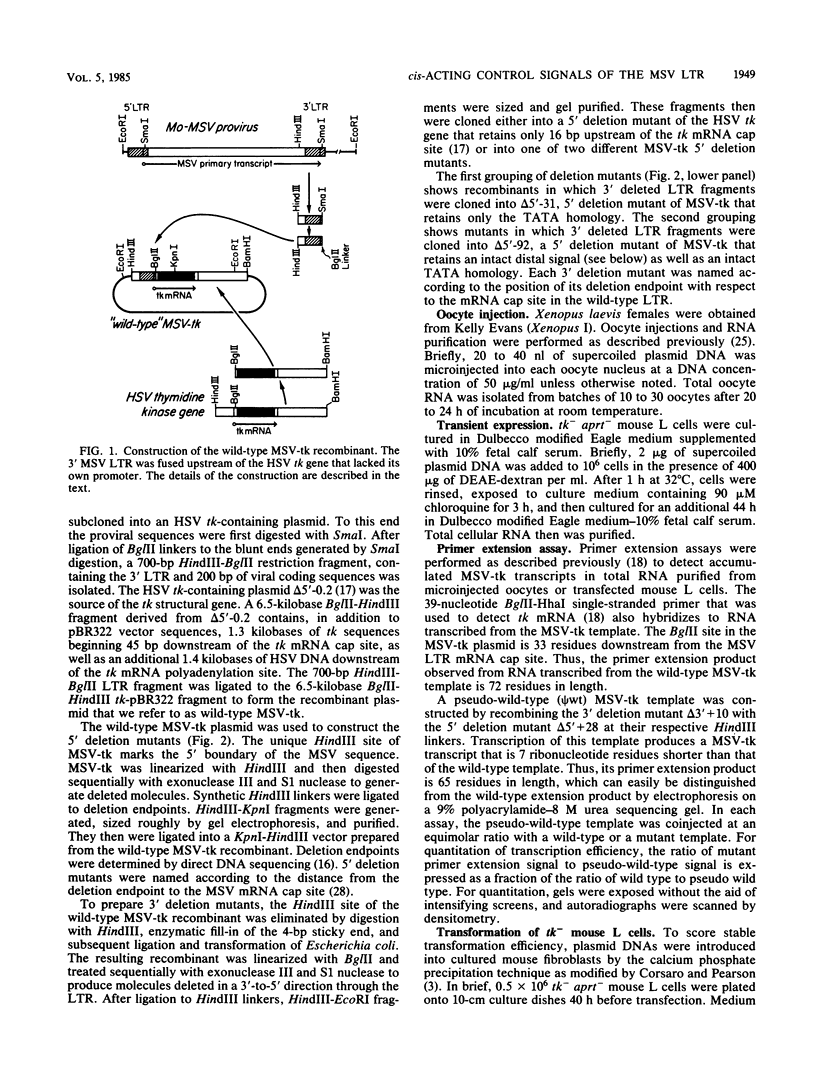
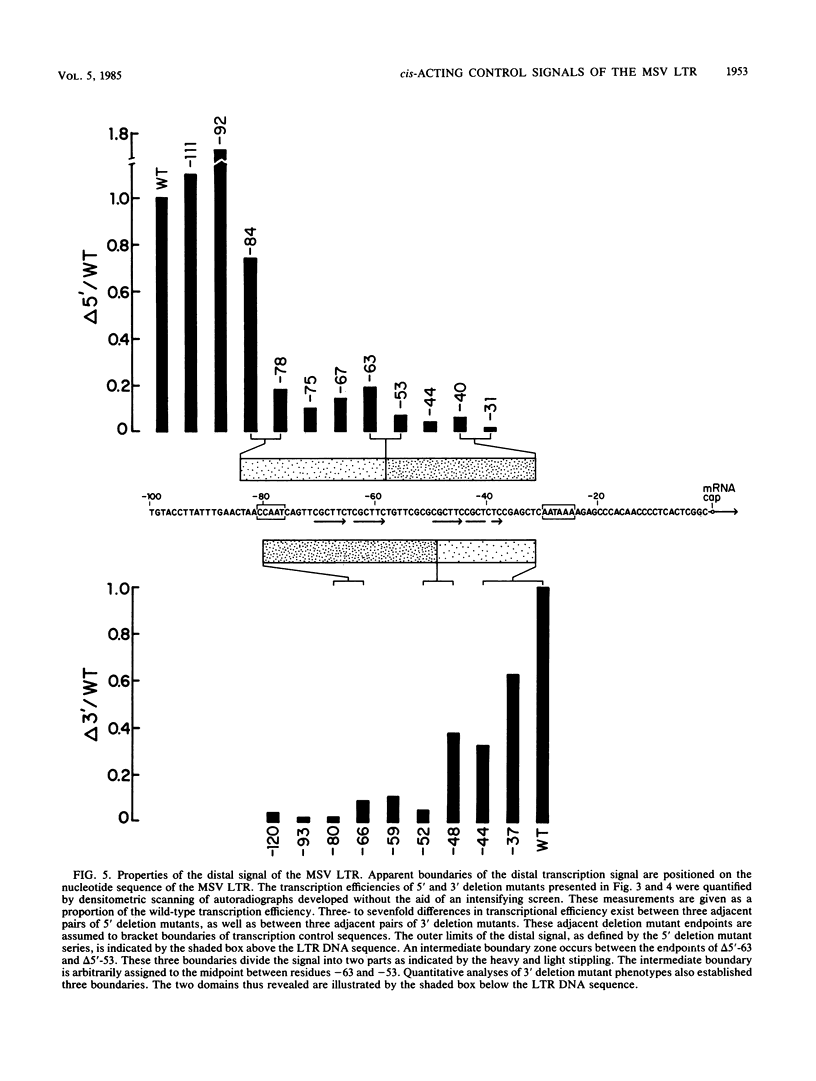
We identified three distinct elements within the Moloney murine sarcoma virus long terminal repeat that control transcription. The phenotypes of unidirectional deletion mutants of the long terminal repeat were assayed in microinjected frog oocytes and in transfected mouse fibroblasts. Steady-state levels of RNA bearing the same 5' terminus as the authentic Moloney murine sarcoma viral transcripts were measured by primer extension in assays that included a pseudo-wild-type internal reference. Mutant phenotypes define the boundaries of three functional elements. A region between 21 and 31 base pairs upstream from the mRNA cap site contains AT-rich sequences that function to establish the transcription start site. A second control element, termed the distal signal, lies between 31 and 84 base pairs upstream of the mRNA cap site. A CAT box consensus sequence is located at the 5' boundary of the distal signal. Additional components of the distal signal include a hexanucleotide sequence that is repeated four times. The distal signal augments transcription efficiency in oocytes but contributes only weakly to long terminal repeat-mediated expression in mouse fibroblasts. A third transcriptional control element lies between 156 and 364 base pairs upstream of the mRNA cap site. This element includes the 75-base-pair repeats previously identified as the Moloney murine sarcoma virus enhancer. In contrast to the distal signal, the Moloney murine sarcoma virus enhancer is crucial for significant expression in mouse fibroblasts but does not contribute to transcriptional expression in frog oocytes.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blair D. G., Oskarsson M., Wood T. G., McClements W. L., Fischinger P. J., Vande Woude G. G. Activation of the transforming potential of a normal cell sequence: a molecular model for oncogenesis. Science. 1981 May 22;212(4497):941–943. doi: 10.1126/science.7233190. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Corsaro C. M., Pearson M. L. Enhancing the efficiency of DNA-mediated gene transfer in mammalian cells. Somatic Cell Genet. 1981 Sep;7(5):603–616. doi: 10.1007/BF01549662. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Van Beveren C., Verma I. M. Identification of a RNA polymerase II initiation site in the long terminal repeat of Moloney murine leukemia viral DNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5411–5415. doi: 10.1073/pnas.78.9.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Eisenberg S. P., Coen D. M., McKnight S. L. Alternate utilization of two regulatory domains within the Moloney murine sarcoma virus long terminal repeat. Mol Cell Biol. 1985 Aug;5(8):1959–1968. doi: 10.1128/mcb.5.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Jolly D. J., Esty A. C., Subramani S., Friedmann T., Verma I. M. Elements in the long terminal repeat of murine retroviruses enhance stable transformation by thymidine kinase gene. Nucleic Acids Res. 1983 Mar 25;11(6):1855–1872. doi: 10.1093/nar/11.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R. Expression of the herpes thymidine kinase gene in Xenopus laevis oocytes: an assay for the study of deletion mutants constructed in vitro. Nucleic Acids Res. 1980 Dec 20;8(24):5931–5948. doi: 10.1093/nar/8.24.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. C., Spence A., Smith M. The distal transcription signals of the herpesvirus tk gene share a common hexanucleotide control sequence. Cell. 1984 May;37(1):253–262. doi: 10.1016/0092-8674(84)90321-0. [DOI] [PubMed] [Google Scholar]
- Ostrowski M. C., Berard D., Hager G. L. Specific transcriptional initiation in vitro on murine type C retrovirus promoters. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4485–4489. doi: 10.1073/pnas.78.7.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Probst E., Kressmann A., Birnstiel M. L. Expression of sea urchin histone genes in the oocyte of Xenopus laevis. J Mol Biol. 1979 Dec 15;135(3):709–732. doi: 10.1016/0022-2836(79)90173-6. [DOI] [PubMed] [Google Scholar]
- Srinivasan A., Reddy E. P., Dunn C. Y., Aaronson S. A. Molecular dissection of transcriptional control elements within the long terminal repeat of the retrovirus. Science. 1984 Jan 20;223(4633):286–289. doi: 10.1126/science.6322296. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Function of the retrovirus long terminal repeat. Cell. 1982 Jan;28(1):3–5. doi: 10.1016/0092-8674(82)90367-1. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]