Primary sclerosing cholangitis (PSC) is a severe liver disease of unknown etiology leading to fibrotic destruction of the bile ducts and ultimately the need for liver transplantation1-3. We compared 3,789 European ancestry PSC cases to 25,079 population controls across 130,422 single-nucleotide polymorphisms (SNPs) genotyped using Immunochip4. We identified 12 genome-wide significant associations outside the human leukocyte antigen (HLA) complex, nine of which were novel, thereby increasing the number of known PSC risk loci to 16. Despite comorbidity with inflammatory bowel disease (IBD) in 72% of the patients, six of the 12 loci showed significantly stronger association with PSC than IBD, suggesting an overlapping yet distinct genetic architecture. We incorporated pleiotropy with seven diseases clinically co-occurring with PSC and found suggestive evidence for 33 additional PSC risk loci. Together with network analyses, these findings further complete the genetic risk map of PSC and considerably expand on the relationship between PSC and other immune-mediated diseases.
The pathogenesis of PSC is poorly understood, and due to lack of effective medical therapy, PSC remains a leading indication for liver transplantation in Northern Europe and the US5, despite the relatively low prevalence (1/10,000). Affected individuals are diagnosed at a median age of 30-40 years and suffer from an increased frequency of IBD (60-80%)5,6 and autoimmune diseases (25%)7. Conversely, approximately only 5% of patients with IBD develop PSC5,6. A 9-39-fold sibling relative risk indicates a strong genetic component to PSC risk8. In addition to multiple strong associations within the HLA complex, recent association studies have identified genome-wide significant loci at 1p36 (MMEL1/TNFRSF14), 2q13 (BCL2L11), 2q37 (GPR35), 3p21 (MST1), 10p15 (IL2RA) and 18q21 (TCF4)9-13.
Several theories have been proposed to explain the development of PSC5. The strong HLA associations and the clinical co-occurrence of immune-mediated diseases suggest that autoimmunity plays a role. To further characterize the genetic etiology of PSC, we recruited PSC patients throughout Europe and North America, more than doubling the number of ascertained cases included in previous genetic studies11. We genotyped 196,524 SNPs in 4,228 PSC cases and 27,077 population controls (see Online Methods and Supplementary Note) using the Immunochip4,14, a targeted genotyping array with dense marker coverage across 186 known disease loci from 12 immune-mediated diseases. Outside these 186 loci, Immunochip also assays thousands of SNPs of intermediate significance from multiple meta-analyses of immune-mediated diseases.
Following quality control (QC; see Online Methods), 130,422 SNPs from 3,789 PSC cases and 25,079 population controls were available for analysis (Supplementary Tables 1 and 2, Supplementary Figures 1 and 2). We imputed a further 80,183 SNPs located in the Immunochip fine mapping regions using the 1000 Genomes reference panel (Online Methods). We performed case-control association tests using a linear mixed model as implemented in MMM15 to minimize the effect of population stratification and sample relatedness (λGC = 1.02, estimated using 2,544 “null” SNPs, see Online Methods).
We identified twelve non-HLA genome-wide significant (P<5×10−8) susceptibility loci (Table 1), nine of which were novel (Fig. 1). The most associated SNP within each locus was a common variant (all risk allele frequencies >0.18) of moderate effect (odds ratios (ORs) between 1.15 and 1.4) (Table 1). Genotype imputation and stepwise conditional regressions16 within each locus did not identify additional independent genome-wide significant signals, nor did genotype-genotype or gender-genotype interaction analyses (Online Methods).
Table 1. Association results of twelve non-HLA genome-wide significant risk loci for primary sclerosing cholangitis (PSC).
| Chr | SNPa | RA | RAF cases |
RAF controls |
P-value | OR (95%CI) |
LD regionb (Kb) |
RefSeq genes in LD region |
Notable nearby gene(s)c |
Functional annotationd |
|---|---|---|---|---|---|---|---|---|---|---|
| 1p36 | rs3748816 | A | 0.698 | 0.656 | 7.41×10−12 | 1.21 (1.14-1.27) |
2,398-2,775 | 9 |
MMEL1, TNFRSF14 |
eQTL,MS, OC, PB, HM |
| 2q33 | rs7426056 | A | 0.277 | 0.229 | 1.89×10−20 | 1.3 (1.23-1.37) | 204,155-204,397 | 1 | CD28 | HM, OC |
| 3p21 | rs3197999 | A | 0.352 | 0.285 | 2.45×10−26 | 1.33 (1.26-1.4) |
48,388-51,358 | 89 | MST1 | eQTL,MS, OC, PB HM |
| 4q27 | rs13140464 | C | 0.871 | 0.836 | 8.87×10−13 | 1.3 (1.21-1.4) |
123,204-123,784 | 4 | IL2, IL21 | OC, PB |
| 6q15 | rs56258221 | G | 0.213 | 0.183 | 8.36×10−12 | 1.23 (1.16-1.31) |
90,967-91,150 | 1 | BACH2 | OC, PB |
| 10p15 | rs4147359 | A | 0.401 | 0.349 | 8.19×10−17 | 1.24 (1.18-1.3) |
6,070-6,206 | 2 | IL2RA | PB |
| 11q23 | rs7937682 | G | 0.298 | 0.265 | 3.17×10−09 | 1.17 (1.11-1.24) |
110,824-111,492 | 19 | SIK2 | OC, PB, HM |
| 12q13 | rs11168249 | G | 0.506 | 0.466 | 5.49×10−09 | 1.15 (1.1-1.21) |
46,442-46,534 | 3 | HDAC7 | OC, PB, HM |
| 12q24 | rs3184504 | A | 0.527 | 0.488 | 5.91×10−11 | 1.18 (1.12-1.24) |
110,186-111,512 | 16 |
SH2B3, ATXN2 |
MS, OC, HM |
| 18q22 | rs1788097 | A | 0.518 | 0.483 | 3.06×10−08 | 1.15 (1.1-1.21) |
65,633-65,721 | 2 | CD226 | MS, OC, PB, HM |
| 19q13 | rs60652743 | A | 0.864 | 0.836 | 6.51×10−10 | 1.25 (1.16-1.34) |
51,850-51,998 | 6 |
PRKD2, STRN4 |
OC, PB, HM |
| 21q22 | rs2836883 | G | 0.777 | 0.728 | 3.19×10−17 | 1.28 (1.21-1.36) |
39,374-39,404 | - | PSMG1 | OC, PB, HM |
Chr: chromosome; CI: confidence interval; eQTL: expression quantitative trait locus, HM: overlaps a region of histone modification; Kb: kilobasepairs; LD: linkage disequilibrium; MS: missense mutation; OC: overlaps known region of open chromatin; OR: odds ratio; PB: overlaps a region of protein binding; RA: risk allele; RAF: risk allele frequency
SNPs from novel PSC-associated loci are shown in bold.
LD regions around lead SNPs were calculated by extending in both directions a distance of 0.1 centimorgans as defined by the HapMap recombination map.
Candidate gene(s) within same LD region as the associated SNPs.
Denotes if there are SNPs with r2>0.8 with the hit SNP that have functional annotations (Supplementary Tables 4-7).
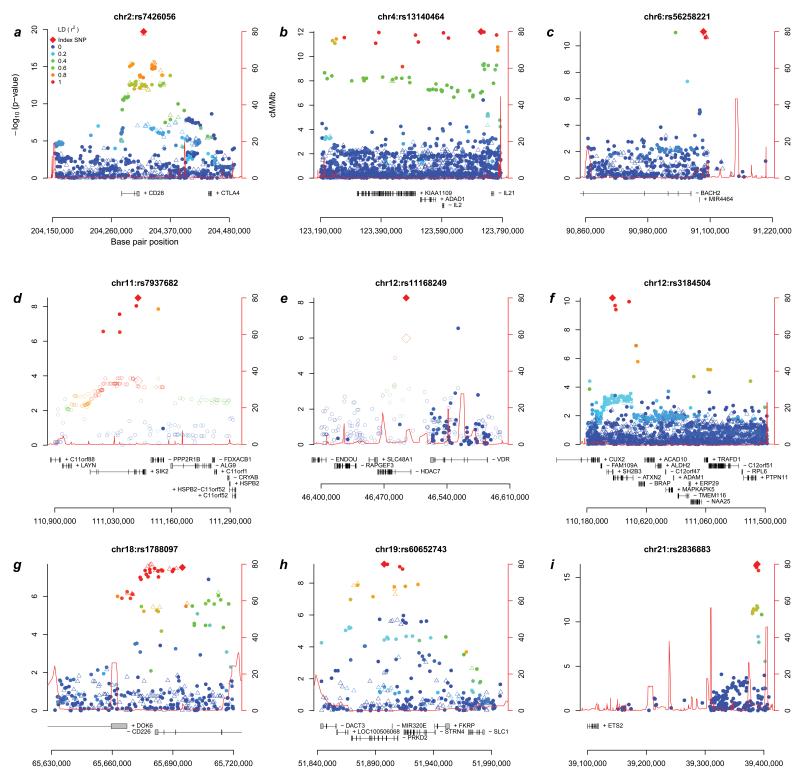
Figure 1. Association results across the nine newly associated primary sclerosing cholangitis (PSC) loci.
Regional association plots of the 9 loci newly associated with PSC at genome-wide significance (P<5×10−8). Filled-in circles are directly genotyped and hollow triangles are imputed (see Online Methods) single-nucleotide polymorphisms (SNPs). The color of the marker (see legend in panel a) illustrates linkage disequilibrium with the most associated SNP. Since the most associated SNPs in panels d) and e) are located outside Immunochip fine mapping regions, association results from the discovery panel of the largest PSC genome-wide association study to date12 are shown as hollow circles and the most associated SNP is a hollow diamond (genotyped and imputed HapMap release 22 SNPs, cases overlap with the current study).
For seven of the nine novel loci, the most significantly associated SNP in the locus was the same SNP or was in strong linkage disequilibrium (LD; r2>0.8) with the original association reports for another disease (Supplementary Table 3). The two exceptions were 11q23, where only independent disease associations (r2<0.01) have so far been reported17, and 6q15, where the most significantly associated PSC variant, rs56258221 (OR=1.23, P=8.36×10−12), is in low-to-moderate LD with the previously reported BACH2 variants in Crohn’s disease (r2=0.23) and type 1 diabetes (r2=0.12). Three out of four known non-HLA PSC risk loci present on the Immunochip passed genotyping QC and were confirmed in our analysis (1p36, 3p21 and 10p15; see Supplementary Note and Supplementary Fig. 3).
To prioritize candidate genes within the non-HLA genome-wide significant loci, we searched for functional consequences of the most associated SNPs or SNPs in high LD (r2>0.8), i.e. missense SNPs (Supplementary Table 4 and Supplementary Fig. 4) and expression quantitative trait loci (eQTLs) (Supplementary Table 5), and we functionally annotated risk loci using data from the ENCODE project (Supplementary Table 6 and Supplementary Note)18. We also constructed networks based on functional similarity measures (Supplementary Fig. 5 and Online Methods), known protein-protein interactions (DAPPLE19, Supplementary Table 7 and Supplementary Note), and the published literature (GRAIL20, Supplementary Fig. 6 and Supplementary Note) to identify important disease-relevant genes. For six of the 12 genome-wide significant loci, the same gene (MMEL1, CD28, MST1, SH2B3, CD226 and SIK2) was annotated by more than one method (Supplementary Table 7), suggesting these as candidates for further investigation at these loci.
Two newly associated loci are located outside of the Immunochip fine mapping regions (Figures 1d and 1e). At 11q23, the most strongly associated SNP, rs7937682 (OR=1.17, P=3.18×10−9), is located in an intron of salt-inducible kinase 2 (SIK2), which both influences the expression of interleukin-10 in macrophages and Nur77, an important transcription factor in leukocytes21. The association at 12q13 is with an intronic SNP (rs11168249, OR=1.15, P=5.49×10−9) within the histone deacetylase 7 (HDAC7) gene, which has also been associated with IBD22. HDAC7 has been implicated in negative selection of T cells in the thymus23, a key factor in the development of immune tolerance. A role for HDAC7 in PSC etiology is supported by the novel association at 19q13, where the most associated SNP, rs60652743 (OR=1.25, P=6.51×10−10) is located within an intron of serine-threonine protein kinase D2 (PRKD2). When T cell receptors of thymocytes are engaged, PRKD2 phosphorylates HDAC7, leading to nuclear exclusion of HDAC7 and loss of its gene regulatory functions, ultimately resulting in apoptosis and negative selection of immature T cells24,25. Interestingly, this negative selection takes place due to a loss of HDAC7-mediated repression of Nur77 (regulated by SIK2)26, linking three novel PSC loci to this pathway.
The associations at the HLA complex at 6p21 were refined by imputing alleles at HLA-A HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, HLA-DQA and HLA-DPB1 (see Supplementary Note)27. The top associated SNP (rs4143332) was in almost perfect LD (r2=0.996) with HLA-B*08:01 (Supplementary Note). In a stepwise conditional analysis including both SNP and HLA allele genotypes, rs4143332 (tagging HLA-B*08:01) and a complex HLA class II association signal determined by HLA-DQA1*01:03 and SNPs rs532098, rs1794282 and rs9263964 (Supplementary Fig. 7) explain most of the HLA association signal in PSC.
When performing a stepwise regression of the HLA alleles only, the class II associations are coherent with previous reports, apart from a novel association with HLA-DQA1*01:01 (see Supplementary Note and Supplementary Tables 8, 9, 10)9,28,29. The HLA-DRB1*15:01 association overlaps with that of ulcerative colitis (risk increasing) and Crohn’s disease (risk decreasing)30,31. Since imputed genotypes at the class II region were only available for four (HLA-DRB1, HLA-DQB1, HLA-DQA1 and HLA-DPB1) out of 20 loci32, further studies involving direct sequencing of all HLA class II loci along with assessments of their protein structure and peptide binding are required to causally resolve the link between this HLA subregion and PSC development33,34.
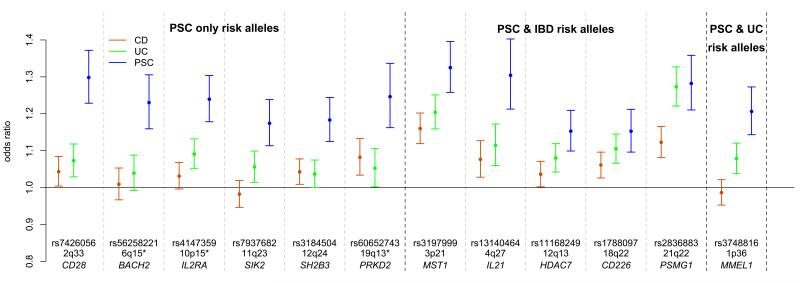
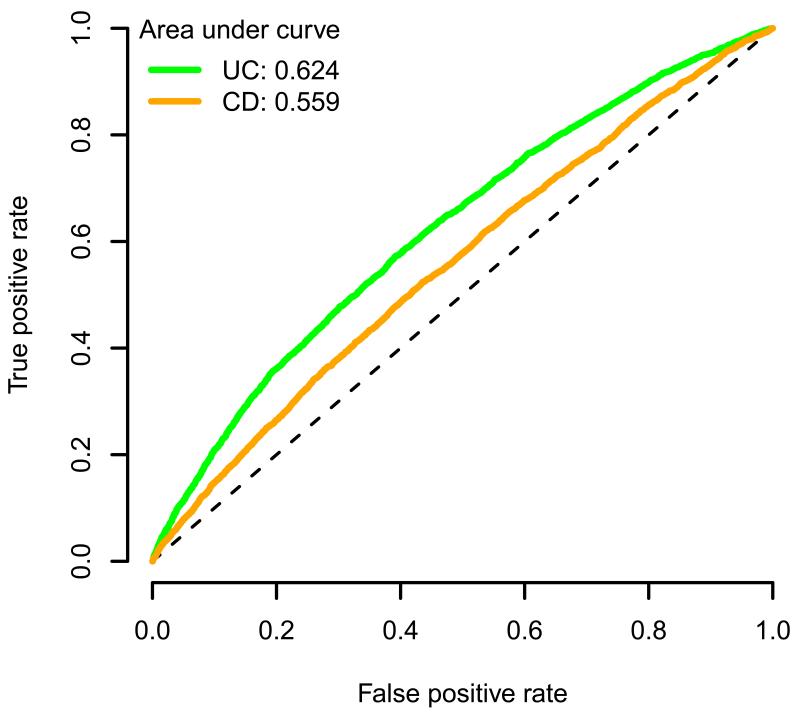
Although 72% of the PSC patients in this study have a diagnosis of concomitant IBD (Supplementary Table 11), only half of our genome-wide significant loci were associated with IBD in the recent International IBD Genetics Consortium (IIBDGC) Immunochip analysis (Fig. 2a, Supplementary Table 3 and Supplementary Fig. 8)22, despite the greater sample size of that study (25,683 cases and 15,977 controls). Across the 12 non-HLA PSC loci we observed greater similarity between the OR estimates for PSC and ulcerative colitis than for PSC and Crohn’s disease. We used the Crohn’s disease and ulcerative colitis OR estimates for the 163 IBD-associated loci to predict PSC case/control status in our sample (Online Methods)22, and found a significantly greater area under the receiver operating characteristic curve (AUC) when applying ulcerative colitis ORs compared to Crohn’s disease ORs (ulcerative colitis AUC=0.62, Crohn’s disease AUC=0.56, P=1.2x10−57, Fig. 2b). This suggests that PSC is genetically more similar to ulcerative colitis than Crohn’s disease and is consistent with clinical observations of greater comorbidity between PSC with ulcerative colitis than Crohn’s disease35. To further compare the genetic profile of PSC and IBD, we combined our genome-wide significant PSC loci with the 163 confirmed IBD loci22 in a functional similarity network (Supplementary Fig. 9 and Supplementary Table 12). The figure shows that the PSC loci are distributed throughout the IBD loci, suggesting that there is no particular functional subcluster of IBD susceptibility genes associated with PSC and vice versa.
Figure 2. Genetic similarity of primary sclerosing cholangitis (PSC) and inflammatory bowel disease (IBD) associated loci.
a) Comparison of odds ratios (ORs) between the most associated risk allele in PSC and the same allele in Crohn’s disease and ulcerative colitis across the 12 genome-wide significant PSC-associated loci. Ulcerative colitis and Crohn’s disease ORs, and the denomination of IBD loci as ulcerative colitis, Crohn’s disease or both (IBD), were obtained from Jostins et al.22. The error bars represent 95% confidence intervals around the ORs. *The PSC associated alleles at 6q15 (BACH2), 10p15 (IL2RA) and 19q13 (PRKD2) are independent of the reported IBD associations (r2<0.3), but locate to the same broad genetic region; for this reason these loci are defined as PSC-IBD loci in Supplementary Figure 9, but as PSC only allelic associations here.
b) Predicting PSC using OR estimates across 163 IBD-associated loci. The green and orange lines represent the receiver operating characteristic (ROC) curves for discriminating PSC cases from population controls using ulcerative colitis or Crohn’s disease ORs from 163 loci associated with IBD, respectively22. The dashed diagonal line is y=x, and specifies the ROC curve of a random predictor.
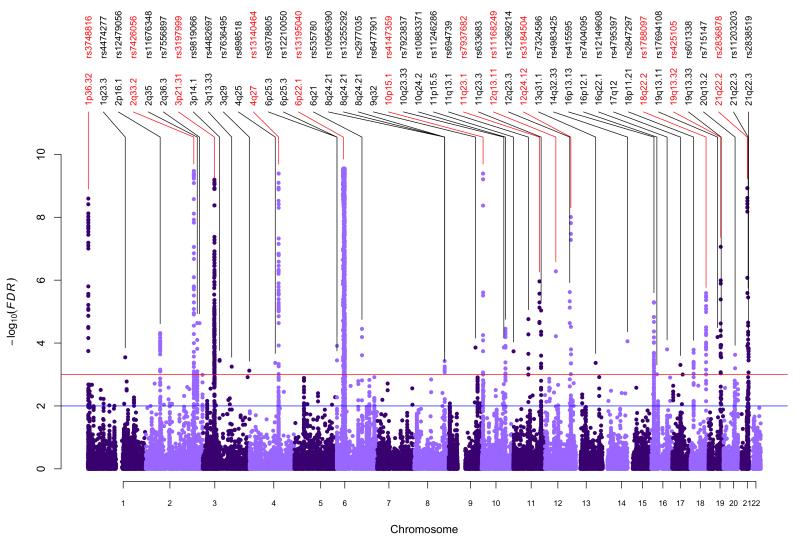
While we consider only those loci reaching a stringent significance threshold (P<5×10−8) to be conclusively associated to PSC, it is likely that additional true associations lie among SNPs with weaker associations. An alternative approach for controlling for multiple hypothesis testing is false discovery rate (FDR) control, which regulates the expected proportion of incorrectly rejected null hypotheses. FDR is well suited to focused genotyping platforms such as Metabochip36 and Immunochip because it implicitly accounts for the expected enrichment of association. To further increase this enrichment, we exploited the known pleiotropy between related immune-mediated traits37, and calculated the FDR38-40 for association with PSC conditional on previously published summary statistics from each of the related phenotypes (yielding a per SNP conditional FDR)41 (Online Methods). We identified 33 non-HLA loci with a conditional FDR<0.001 in this analysis (Fig. 3), all of which showed suggestive significance (5×10−5≤P<5×10−8) in the standard association analysis (Supplementary Table 13 and Supplementary Figures 10-12). These loci were integrated in the functional similarity network analysis (Supplementary Fig. 13), highlighting potential candidate susceptibility genes.
Figure 3. Pleiotropic primary sclerosing cholangitis (PSC) loci.
Manhattan plot of conditional associations in PSC calculated as stratified false discovery rates (FDRs) based on the results of the present PSC analysis and genetic associations previously reported in seven immune-mediated diseases (Crohn’s disease, celiac disease, psoriasis, rheumatoid arthritis, sarcoidosis; type 1 diabetes and ulcerative colitis) (see Online Methods and Supplementary Figures 10-12). Single-nucleotide polymorphisms (SNPs) in red represent genome-wide significant findings from the regular association analysis, while SNPs listed in black are significantly associated with PSC conditional on their pleiotropic effects across the related immune-mediated diseases. The horizontal red line represents a threshold of FDR<0.001 (Supplementary Table 13), while the blue line represents a threshold of FDR<0.01 (see Supplementary Table 14 for full listing).
In conclusion, the present study increases the number of genome-wide significant loci in PSC from seven to 16 (including the HLA complex). The nine novel variants together explain 0.9% of variance in PSC liability, increasing the total amount of variance explained by the 16 known loci to 7.3% (Online Methods). The data convincingly show that genetic susceptibility to PSC extends considerably beyond the risk factors involved in the closely related IBD phenotype and into autoimmune pathophysiology. Furthermore, analysis of pleiotropic immune-related genetic variants highlights 33 additional suggestive loci in PSC, overall representing major new avenues for research into disease pathogenesis.
Online Methods
Study Subjects
The study participants are described in the Supplementary Note and Supplementary Table 15.
Ethical approval
The patient recruitment was approved by the ethics committees or institutional review boards of all participating centers. Written informed consent was obtained from all participants.
Quality control
SNPs with a call rate <80% were removed prior to commencing sample QC (n=235). Per individual genotype call rate and heterozygosity rate were calculated using PLINK42 and outlying samples were identified using Aberrant43, which automatically identifies outliers from otherwise Gaussian distributions (Supplementary Fig. 1). A set of 20,837 LD-pruned (r2<0.1) SNPs with MAF>10% present on both the Immunochip and the Illumina Omni2.5-8 array used in the 1000 Genomes project (see URLs) were used to estimate identity by descent and ancestry. For each pair of individuals with estimated identity by descent≥0.9, the sample with lower call rate was removed (unless case/control status was discordant between the pair, in which case both samples were removed, n=92). Related individuals (0.1875<identity by descent<0.9) remained in the analysis to maximize power because the mixed model association analysis can correctly account for the relatedness. Principal components analysis, implemented in SMARTPCA (Eigenstrat)44, was used to identify samples of non-European ancestry. Principal components were defined using population samples from the 1000 Genomes project45 genotyped using the Illumina Omni2.5-8 genotyping array (see URLs) and then projected into cases and controls (Supplementary Fig. 2)14,22,46. Following sample QC, 3,789 PSC cases and 25,079 controls remained. SNPs with a minor allele frequency less than 0.1%, Hardy-Weinberg equilibrium P<10−5, call rate lower than 98%, or failing the PLINK v1.07 non-random differential missing data rate test between cases and controls (P<10−5) were excluded. After completion of marker QC (Supplementary Table 2), 131,220 SNPs were available for analysis, further reduced to 130,422 after cluster plot inspection (see below).
Statistical methods
Genomic inflation factor
The Immunochip contains 3,120 SNPs that were part of a bipolar disease replication effort and other non-immune-related studies. After QC, 2,544 of these were used as null markers to estimate the overall inflation of the distribution of association test statistics.
Imputation
Using 85,747 post-QC SNPs located in the Immunochip fine mapping regions, additional genotypes were imputed using IMPUTE2 with the 1000 Genomes Phase 1 (March, 2012) reference panel of 1,092 individuals47 and 744,740 SNPs. Imputation was performed separately in ten batches, with the case:control and country of origin ratios constant across batches. SNPs with a posterior probability less than 0.9 and those with differential missingness (P<10−5) between the 10 batches were removed, as were SNPs failing the exclusion thresholds used for genotyped SNP QC. After imputation, a total of 163,379 SNPs in the Immunochip fine mapping regions, including 153,857 SNPs from the reference panel, were available for analysis.
Association analysis
Case-control association tests were performed using a linear mixed model as implemented in MMM15. A covariance matrix, R, of a random effects component was included in the model to explicitly account for confounding due to population stratification and cryptic relatedness between individuals. This method has been shown to better control for population stratification than correction for principal components or meta-analyses of matched subgroups of cases and controls48-50. R is a symmetric n×n matrix with each entry representing the relative sharing of alleles between two individuals compared to the average in the sample, and is typically estimated using genome-wide SNP data15. To avoid biases in the estimation of R due to the design of the Immunochip, SNPs were first pruned for LD (r2<0.1). Of the remaining SNPs, we then removed those that lie in the HLA region or have a minor allele frequency<10%. Finally, we excluded SNPs that showed modest association (P<0.005) with PSC in a linear regression model fitting the first 10 principal components as covariates. A total of 17,260 SNPs were used to estimate R.
Due to computational limitations, we estimated the R matrix and performed all association analyses applying R separately for UK (n=9,696) and non-UK (n=19,172) samples, and then combined the results using a fixed-effects (inverse-variance weighting) meta-analysis, as done previously48. This reduced the λGC, estimated using the 2,544 “null” SNPS, from 1.24 to 1.02 (Supplementary Fig. 14), showing excellent control for population stratification. Stepwise conditional regression was used to identify possible independent associations at genome-wide significant loci. SNP×SNP interactions between all pairs of genome-wide significant SNPs were tested using the PLINK --epistasis command. Signal intensity plots of all non-HLA loci with association P-value<5×10−6 were visually inspected using Evoker51. SNPs that clustered poorly were removed (n=798).
Prediction of PSC using IBD SNPs
ORs for Crohn’s disease and ulcerative colitis in 163 IBD-associated SNPs were obtained from Jostins et al.22. We used the R package Mangrove (see URLs) to estimate each individual’s probability of developing PSC among our 3,789 PSC cases and 25,079 controls assuming additive risk (log-additive OR). The performance of our predictor using either Crohn’s disease or ulcerative colitis ORs, was assessed by constructing a ROC curve, showing the proportion of true and false positives at each probability threshold. The AUC was calculated to compare the predictive power of the ulcerative colitis and Crohn’s disease ORs. The DeLong method was used to test if the AUC was significantly greater using ulcerative colitis than Crohn’s disease ORs52.
Functional similarity networks
In functional similarity networks, each edge represents strong functional similarity of two genes based on annotated Gene Ontology terms as determined by the functional similarity measure rfunSim53. The rfunSim similarity values above the recommended cutoff 0.8 were retrieved using the FunSimMat web service54. The resulting networks were visualized and analyzed using Cytoscape55.
To construct PSC-specific networks from functional similarity networks that contained more than one gene per locus (Supplementary Figures 5 and 13), the connectivity of each gene was assessed by computing different topology measures for the corresponding node: (1) degree (number of direct edges to other nodes), (2) shortest path closeness (inverted average shortest path distance to other nodes) and (3) shortest path betweenness (fraction of shortest paths passing through the node). Similarity edges between genes in the same locus and gene nodes that were not contained in the resulting largest connected subnetworks were ignored. The genes were first ranked according to each measure and then assigned the best of the three ranks. The PSC-specific network was generated from the top ranked genes in their respective locus.
Pleiotropy analysis
We included summary statistics from genome-wide association studies in seven PSC-associated diseases (Crohn’s disease, celiac disease, psoriasis, rheumatoid arthritis, sarcoidosis, type 1 diabetes, ulcerative colitis, see Supplementary Table 16). For all diseases we constructed conditional stratified Q-Q plots of empirical quantiles of nominal −log10(P) values for SNP association with PSC for all SNPs (see Supplementary Fig. 10), and for different overlapping subsets of SNPs determined by the significance of their association with the PSC-associated autoimmune disorder (SNP subsets defined as P<1, P<0.1, P<0.01 and P<0.001 in the pleiotropic phenotype, respectively). For a given PSC associated phenotype, ‘enrichment’ for pleiotropic signals in PSC can be observed as an increasing leftward deflection from the expected null line with lower P-value thresholds in the second phenotype (Supplementary Note). The ‘enrichment’ in the stratified Q-Q plots is directly interpretable in terms of the true discovery rate (TDR), equivalent to one minus the FDR56. Specifically, it can be shown that a conservative estimate of FDR can be calculated from the horizontal shift of the Q-Q curve from the expected line x=y, with a larger shift corresponding to a smaller FDR for a given nominal P-value (Supplementary Note). We calculated the conditional TDR as a function of P-value in PSC across a series of P-value thresholds in the pleiotropic trait (Supplementary Fig. 10).
In order to assess significance of the association with PSC, we assigned a pleiotropic (conditional) FDR value for PSC per SNP. The pleiotropic FDR value for each SNP is based on the P-value of the SNP in PSC relative to the P-value distribution of other SNPs in the same conditioning subset, where subsets are defined by the pleiotropic association (lowest P-value among associated diseases) of the SNP. Importantly, the conditioning procedure is blind to the P-value of the SNP with respect to PSC. The pleiotropic FDR is then interpolated from conditional FDR curves using established stratified FDR methods41,57 (see Supplementary Note). The increase in power from using pleiotropic FDR is demonstrated by dividing the total sample in half and observing that empirical replication rates between the training and test halves increase with decreasing P-value in the pleiotropic disease (Supplementary Fig. 15). The SNP with lowest FDR within each LD block (as defined by 1000 Genomes) was considered the lead SNP of a new pleiotropic PSC locus, if below a 0.001 threshold (loci defined by FDR<0.001 and FDR<0.01 shown in Supplementary Tables 13 and 14). All test statistics were adjusted for population stratification by genomic control (see Supplementary Note and Supplementary Fig. 16).
Variance explained and heritability
The proportion of variance explained by the genome-wide significant loci and HLA alleles was calculated using a liability threshold model58 assuming a disease prevalence of 10/100,000 and multiplicative risk.
Supplementary Material
Acknowledgements
We thank all PSC patients and healthy controls for their participation, and we are indebted to all physicians and nursing staff who recruited patients. We thank Tanja Wesse, Tanja Henke, Sanaz Sedghpour Sabet, Rainer Vogler, Gunnar Jacobs, Ilona Urbach, Wolfgang Albrecht, Virpi Pelkonen, Kristian Holm, Hege Dahlen Sollid, Bente Woldseth, Jarl Andreas Anmarkrud and Liv Wenche Torbjørnsen for expert help. Prof. Ulrich Beuers, Dr. Felix Braun, Dr. Wolfgang Kreisel, Dr. Thomas Berg and Dr. Rainer Günther are acknowledged for contributing German PSC patients. Benedicte A. Lie and The Norwegian Bone Marrow Donor Registry at Oslo University Hospital, Rikshospitalet in Oslo and the Nord-Trøndelag Health Study (HUNT) are acknowledged for sharing the healthy Norwegian controls..Banco Nacional de ADN, Salamanca, Spain is acknowledged for providing Spanish control samples. This study makes use of genotyping data generated by DILGOM (see URLs), the Cooperative Research in the Region of Augsburg (KORA) study and by the Heinz Nixdorf Recall (Risk Factors, Evaluation of Coronary Calcification, and Lifestyle) study. We acknowledge the members of the International PSC Study Group, the NIDDK Inflammatory Bowel Disease Genetics Consortium (IBDGC), the UK-PSC Consortium and the Alberta IBD Consortium for their participation. Individuals who have been sharing summary statistics and statistical software are acknowledged in the Supplementary Note.
The study was supported by The Norwegian PSC Research Center (see URLs), by the German Ministry of Education and Research through the National Genome Research Network (01GS0809-GP7), by the Deutsche Forschungsgemeinschaft (FR 2821/2-1), by the EU Seventh Framework Programme FP7/2007-2013 (262055) ESGI, by the Integrated Research and Treatment Center - Transplantation (01EO0802) and the PopGen biobank (see URLs).
J.Z.L., T.S., C.A.A. are supported by a grant from the Wellcome Trust (098051). Additional financial support to the study and the co-authors is listed in the Supplementary Note.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
URLs
1000 Genomes Omni2.5M genotype data: (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20110921_phase2_omni_genoty pes/Omni25_genotypes_1856_samples.b36.20110921.vcf.gz) Mangrove: (http://cran.r-project.org/web/packages/Mangrove/) DILGOM: (http://www.aka.fi/en-GB/A/Programmes-and-Cooperation/Research-programmes/Ongoing/ELVIRA/Projects/DILGOM/) The Norwegian PSC Research Center: (http://ous-research.no/nopsc/) PopGen biobank: (http://www.popgen.de)
References
- 1.Aadland E, et al. Primary sclerosing cholangitis: a long-term follow-up study. Scand J Gastroenterol. 1987;22:655–64. doi: 10.3109/00365528709011139. [DOI] [PubMed] [Google Scholar]
- 2.Broome U, et al. Natural history and prognostic factors in 305 Swedish patients with primary sclerosing cholangitis. Gut. 1996;38:610–5. doi: 10.1136/gut.38.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrant JM, et al. Natural history and prognostic variables in primary sclerosing cholangitis. Gastroenterology. 1991;100:1710–7. doi: 10.1016/0016-5085(91)90673-9. [DOI] [PubMed] [Google Scholar]
- 4.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsen TH, Schrumpf E, Boberg KM. Update on primary sclerosing cholangitis. Dig Liver Dis. 2010;42:390–400. doi: 10.1016/j.dld.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Karlsen TH, Kaser A. Deciphering the genetic predisposition to primary sclerosing cholangitis. Semin Liver Dis. 2011;31:188–207. doi: 10.1055/s-0031-1276647. [DOI] [PubMed] [Google Scholar]
- 7.Saarinen S, Olerup O, Broome U. Increased frequency of autoimmune diseases in patients with primary sclerosing cholangitis. Am J Gastroenterol. 2000;95:3195–9. doi: 10.1111/j.1572-0241.2000.03292.x. [DOI] [PubMed] [Google Scholar]
- 8.Bergquist A, et al. Increased risk of primary sclerosing cholangitis and ulcerative colitis in first-degree relatives of patients with primary sclerosing cholangitis. Clin Gastroenterol Hepatol. 2008;6:939–43. doi: 10.1016/j.cgh.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Karlsen TH, et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 2010;138:1102–11. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava B, et al. Fine mapping and replication of genetic risk loci in primary sclerosing cholangitis. Scand J Gastroenterol. 2012;47:820–6. doi: 10.3109/00365521.2012.682090. [DOI] [PubMed] [Google Scholar]
- 11.Folseraas T, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012;57:366–75. doi: 10.1016/j.jhep.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melum E, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 2011;43:17–9. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinghaus D, et al. Genome-wide association analysis in sclerosing cholangitis and ulcerative colitis identifies risk loci at GPR35 and TCF4. Hepatology. 2012 doi: 10.1002/hep.25977. [DOI] [PubMed] [Google Scholar]
- 14.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirinen M, Donnelly P, Spencer C. Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann Appl Stat. 2012 In press. [Google Scholar]
- 16>.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70:124–41. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters U, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131:217–34. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna RN, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol. 2011;12:778–85. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasler HG, et al. Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. J Immunol. 2011;186:4782–93. doi: 10.4049/jimmunol.1001179. [DOI] [PubMed] [Google Scholar]
- 24.Dequiedt F, et al. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity. 2003;18:687–98. doi: 10.1016/s1074-7613(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 25.Dequiedt F, et al. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J Exp Med. 2005;201:793–804. doi: 10.1084/jem.20042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark K, et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1215450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrumpf E, et al. HLA antigens and immunoregulatory T cells in ulcerative colitis associated with hepatobiliary disease. Scand J Gastroenterol. 1982;17:187–91. doi: 10.3109/00365528209182038. [DOI] [PubMed] [Google Scholar]
- 29.Spurkland A, et al. HLA class II haplotypes in primary sclerosing cholangitis patients from five European populations. Tissue Antigens. 1999;53:459–69. doi: 10.1034/j.1399-0039.1999.530502.x. [DOI] [PubMed] [Google Scholar]
- 30.Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395–401. doi: 10.1136/gut.45.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada Y, et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn’s disease. Gastroenterology. 2011;141:864–871. e1–5. doi: 10.1053/j.gastro.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 32.Horton R, et al. Gene map of the extended human MHC. Nature. 2004;5:889–99. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 33.Hov JR, et al. Electrostatic modifications of the human leukocyte antigen-DR P9 peptide-binding pocket and susceptibility to primary sclerosing cholangitis. Hepatology. 2011;53:1967–76. doi: 10.1002/hep.24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hovhannisyan Z, et al. The role of HLA-DQ8 beta57 polymorphism in the anti-gluten T-cell response in coeliac disease. Nature. 2008;456:534–8. doi: 10.1038/nature07524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broome U, Bergquist A. Primary sclerosing cholangitis, inflammatory bowel disease, and colon cancer. Semin Liver Dis. 2006;26:31–41. doi: 10.1055/s-2006-933561. [DOI] [PubMed] [Google Scholar]
- 36.The CDC, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2012 doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 39.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Statist. 2003;31:2013–2035. [Google Scholar]
- 40.Efron B. Simultaneous Inference: When Should Hypothesis Testing Problems Be Combined? Ann Appl Statist. 2008;2:197–223. [Google Scholar]
- 41.Sun L, Craiu RV, Paterson AD, Bull SB. Stratified false discovery control for large-scale hypothesis testing with application to genome-wide association studies. Genet Epidemiol. 2006;30:519–30. doi: 10.1002/gepi.20164. [DOI] [PubMed] [Google Scholar]
- 42.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellenguez C, Strange A, Freeman C, Donnelly P, Spencer CC. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2012;28:134–5. doi: 10.1093/bioinformatics/btr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu JZ, et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2012 doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korte A, et al. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet. 2012;44:1066–71. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris JA, Randall JC, Maller JB, Barrett JC. Evoker: a visualization tool for genotype intensity data. Bioinformatics. 2010;26:1786–7. doi: 10.1093/bioinformatics/btq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 53.Schlicker A, Domingues FS, Rahnenfuhrer J, Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlicker A, Albrecht M. FunSimMat update: new features for exploring functional similarity. Nucleic Acids Res. 2010;38:D244–8. doi: 10.1093/nar/gkp979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Efron B. Size, power and false discovery rates. Ann Statist. 2007;35:1351–77. [Google Scholar]
- 57.Yoo YJ, Pinnaduwage D, Waggott D, Bull SB, Sun L. Genome-wide association analyses of North American Rheumatoid Arthritis Consortium and Framingham Heart Study data utilizing genome-wide linkage results. BMC Proc. 2009;3(Suppl 7):S103. doi: 10.1186/1753-6561-3-s7-s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–7. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.