Spatial control of EB3 phosphorylation by Aurora B during mitotic exit regulates microtubule dynamics necessary for the stabilization of focal adhesions and coordinated daughter cell spreading, as well as for midbody stabilization and efficient cytokinesis.
Abstract
During mitosis, human cells round up, decreasing their adhesion to extracellular substrates. This must be quickly reestablished by poorly understood cytoskeleton remodeling mechanisms that prevent detachment from epithelia, while ensuring the successful completion of cytokinesis. Here we show that the microtubule end-binding (EB) proteins EB1 and EB3 play temporally distinct roles throughout cell division. Whereas EB1 was involved in spindle orientation before anaphase, EB3 was required for stabilization of focal adhesions and coordinated daughter cell spreading during mitotic exit. Additionally, EB3 promoted midbody microtubule stability and, consequently, midbody stabilization necessary for efficient cytokinesis. Importantly, daughter cell adhesion and cytokinesis completion were spatially regulated by distinct states of EB3 phosphorylation on serine 176 by Aurora B. This EB3 phosphorylation was enriched at the midbody and shown to control cortical microtubule growth. These findings uncover differential roles of EB proteins and explain the importance of an Aurora B phosphorylation gradient for the spatiotemporal regulation of microtubule function during mitotic exit and cytokinesis.
Introduction
Human cells round up during mitosis as a result of increased hydrostatic pressure and actomyosin cortex contraction, which counteracts adhesion to extracellular substrates (Stewart et al., 2011). Thus, mitosis represents a short period in the cell cycle where loss of substrate adhesion is maximal. If cell-substrate adhesion is not rapidly reestablished upon completion of mitosis, cells may detach from epithelia, which has been proposed as a mechanism for cancer cell dissemination and metastasis (Vasiliev et al., 2004).
Upon mitotic entry, adhesion complexes are disassembled in a process that involves the phosphorylation of FAK and its release from other adhesion components such as paxillin and p130/Cas (Yamakita et al., 1999). Interaction of mitotic cells with the extracellular matrix is achieved through actin-rich structures called retraction fibers (Mitchison, 1992). These not only provide attachment of the cell to the substrate but also play an active role during mitosis by providing spatial cues for spindle positioning (Théry et al., 2005). However, how the adhesion machinery cross-talks with spindle microtubules (MTs) and their respective reorganization throughout cell division remains largely unknown.
End-binding (EB) proteins are a conserved family of MT plus-end tracking proteins (+TIPs; for review see Akhmanova and Steinmetz, 2008). In humans, they include three related members: EB1, EB2, and EB3. EB1 has been the most studied due to its interaction with the C terminus of adenomatosis polyposis coli (APC), which is often disrupted in colon cancers (Su et al., 1995). During early mitosis, EB1 is involved in spindle orientation in yeast, Drosophila melanogaster, and human cells (Lee et al., 2000; Rogers et al., 2002; Green et al., 2005; Toyoshima and Nishida, 2007). This role of EB1 involves stabilization of astral MTs and interaction with APC (Green et al., 2005; Toyoshima and Nishida, 2007). Interaction between EB1 and APC was also shown to promote MT stabilization in interphase cells (Mimori-Kiyosue et al., 2000; Nakamura et al., 2001; Wen et al., 2004). Loss-of-function studies of EB1 have reported somewhat contradictory requirements for mitosis. In fixed cells, it has been proposed that EB1 plays a role in chromosome congression and segregation (Green et al., 2005; Toyoshima and Nishida, 2007). However, live-cell studies revealed that cells depleted of EB1 by RNAi or upon microinjection of function-blocking antibodies progress normally through mitosis (Draviam et al., 2006; Brüning-Richardson et al., 2011), which supports timely satisfaction of the spindle-assembly checkpoint.
The roles of EB2 and EB3 are less well studied. Similar to EB1, EB3 is ubiquitously expressed in cultured cells and localizes to centrosomes and the mitotic spindle throughout mitosis (Bu and Su, 2001; Su and Qi, 2001). EB3 depletion was reported to cause a prometaphase accumulation, resulting in a slight increase in the mitotic index in fixed material (Ban et al., 2009). Curiously, this was not observed upon EB1/EB3 co-depletion. EB3 is also highly expressed in differentiated cells from the central nervous system and skeletal muscle, and has been shown to specifically regulate MT dynamics near the cell cortex, while being essential for proper myoblast differentiation (Nakagawa et al., 2000; Straube and Merdes, 2007). EB2, however, appears to be the most divergent member of the family, with variable expression levels in different cell types (Su and Qi, 2001).
EB proteins are currently viewed as major regulators of MT dynamics that facilitate MT growth by promoting MT dynamicity and inhibiting catastrophes (Rogers et al., 2002; Tirnauer et al., 2002; Komarova et al., 2009). In addition, EB proteins control MT growth by regulating association of other factors with the plus ends (Gouveia and Akhmanova, 2010).
EB proteins are similar in structure and can adopt a homo- or heterodimeric conformation. EB1 and EB3 readily form heterodimers in vitro and in cells, whereas EB2 does not appear to participate in the formation of heterotypic complexes (De Groot et al., 2010). The MT-binding domain of EBs, which is necessary and sufficient to recognize MT plus ends, is defined by a calponin homology domain in the N terminus. In contrast, the dimerization domain of EBs is in the C terminal and has been implicated in the interaction with EB binding partners, including APC, CLIPs, CLASPs, p150Glued/dynactin, spectraplakins, STIM1, and mitotic centromere-associated kinesin (MCAK; Gouveia and Akhmanova, 2010; Jiang et al., 2012). Finally, EB1 and EB3 have been shown to interact with Aurora kinases, but only EB3 was found to be phosphorylated on serine 176 (S176) during mitosis (Sun et al., 2008; Ban et al., 2009). These data suggest differential regulation of EB protein function and raise important questions regarding their individual roles during cell division.
In this study we focused on understanding the respective roles and regulatory mechanism of human EB1 and EB3 in MT function throughout mitosis and cytokinesis. We uncovered spatially and temporally distinct roles of EB1 and EB3 in spindle orientation, postmitotic cell-substrate adhesion, and cytokinesis. We provide functional evidence that the roles of EB3 during late mitosis and cytokinesis rely on distinct Aurora B–mediated phosphorylation states of S176 and discuss the implications for the spatiotemporal regulation of MT function in light of the recently discovered Aurora B phosphorylation gradient at the spindle midzone.
Results
EB1 and EB3 play temporally distinct roles in spindle orientation and postmitotic cell adhesion to the substrate
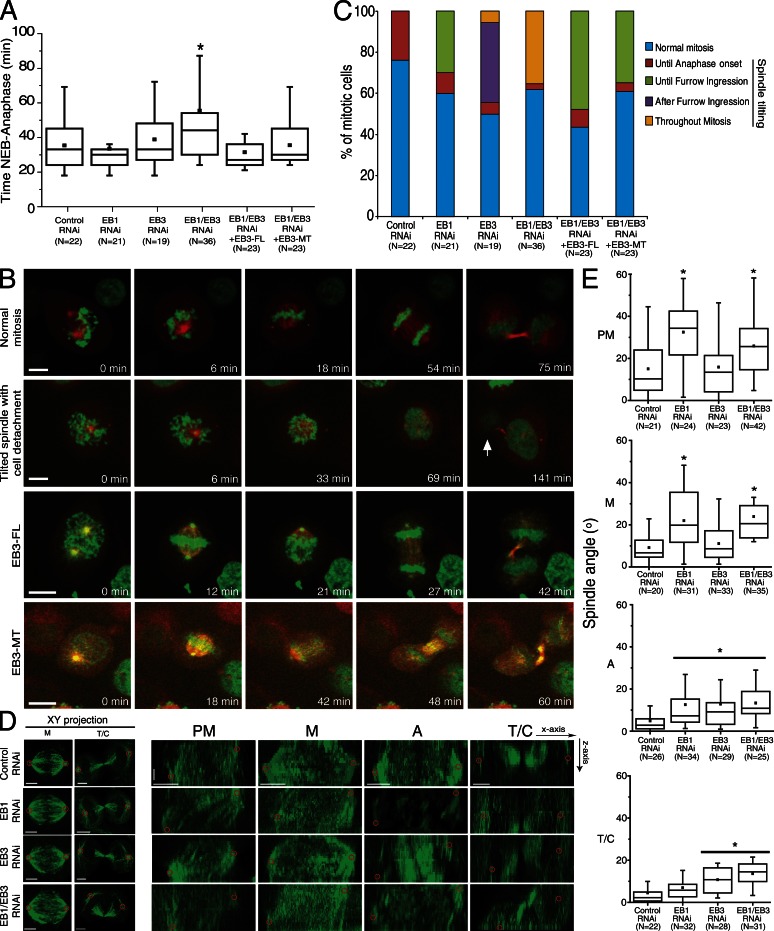
Here we were interested in investigating whether EB proteins play distinct or overlapping roles throughout mitosis. For this purpose, we performed individual or simultaneous depletions of EB1 and/or EB3 in HeLa cells, followed by thorough fixed- and live-cell microscopy analyses of phenotypes. Immunofluorescence and Western blot analyses confirmed that EB1 and EB3 are expressed in HeLa cells, colocalizing to MT plus ends in interphase and throughout mitosis/cytokinesis, while decorating mitotic spindles, spindle poles, and midbodies (Fig. S1 A). The specific localization of EB1 and/or EB3 was sensitive to RNAi-mediated depletion of each respective protein (Fig. S1, B and C). As opposed to individual depletions, only co-depletion of EB1 and EB3 caused a slight but significant increase in the mitotic index of HeLa cells (Fig. S1 D). This likely reflected the enrichment of double-depleted cells in prometaphase (Fig. S1 D). Spinning-disk confocal microscopy of live RNAi-depleted HeLa cells stably coexpressing H2B-Histone-GFP/α-tubulin–mRFP to monitor both chromosome and mitotic spindle behavior revealed that the observed prometaphase accumulation caused a delay in anaphase onset (Fig. 1 A). Accordingly, HeLa cells upon control (luciferase) RNAi spent 35 ± 13 min from nuclear envelope breakdown (NEB) to anaphase onset (Video 1). When EB1 or EB3 were individually depleted, we found a marginal but not significant increase in time between NEB to anaphase onset (33 ± 14 min and 38 ± 16 min, respectively). The only significant difference relative to controls, but not relative to EB3 depletion, was found in EB1/EB3 co-depleted cells, which went from NEB to anaphase in 44 ± 25 min (P < 0.05; Fig. 1 A and Video 2). Curiously, we noticed that the longest observed delay in anaphase onset (54.5 ± 25 min) upon EB1/EB3 co-depletion in a subpopulation of cells correlated with the presence of spindles with excessive rotation. Overall, these data suggest that EB1 and EB3 are largely dispensable for mitotic progression.
Figure 1.
Depletion of EB proteins leads to different phenotypes during mitosis. HeLa cells expressing H2B-GFP/α-tubulin–mRFP were depleted of EB1 and/or EB3 and imaged by spinning disk microscopy. (A) Quantification of NEB-to-anaphase duration (*, P < 0.05 using nonparametric ANOVA followed by a post-hoc Dunn’s test). (B) Frames from time-lapse movies showing the phenotypes quantified. The arrow indicates a daughter cell that failed to attach to the substrate. Bars, 10 µm. (C) Cumulative quantification of mitotic phenotypes in all experimental groups. The spindle was considered “tilted” when the two poles were not on the same focal plane for >5 min. (D) HeLa cells were immunostained with an α-tubulin antibody, and top (xy) and lateral (yz) projections of the spindle were used to identify spindle poles and quantify spindle angle in relation to the substrate. Red circles highlight the spindle poles. Horizontal bar, 5 µm; vertical bar, 1.5 µm. (E) Quantification of spindle angles relative to the substrate in prometaphase (PM), metaphase (M), anaphase (A), and telophase/cytokinesis (T/C) for all treatment groups (*, P < 0.05 using parametric ANOVA followed by a post-hoc Student-Newman-Keuls test). Experiments were done in triplicate and N represents the number of cells quantified in each condition.
We further observed that EB1 depletion caused spindle tilting (regarded as such when the two spindle poles were not in the same focal plane for >5 min) in ∼40% of the cells. Most of these spindles did not align with the substrate until furrow ingression (Fig. 1, B and C; and Video 3). In the same period, individual EB3 depletion did not cause significant problems in spindle alignment (Fig. 1 C and Video 4). However, after furrow ingression, spindle tilting was observed in 35% of EB3-depleted cells (Fig. 1 C and Video 4). Finally, EB1/EB3 co-depleted cells showed spindle tilting throughout mitosis in ∼40% of the cells (Fig. 1, B and C; and Video 2). Overall, these observations suggest that EB1 and EB3 are important for spindle alignment relative to the substrate at distinct mitotic stages.
EB proteins intervene in several MT-dependent cellular functions, either directly by suppression of catastrophes (and thus promotion of MT growth), or indirectly by loading other factors onto growing MT plus ends (Honnappa et al., 2009; Jiang et al., 2012). To distinguish between these possibilities we investigated whether the observed spindle tilting in double-depleted cells could be rescued either by the expression of full-length EB3 fused with GFP (EB3-FL) or an EB3-NL-LZ-GFP construct (from here on referred to as EB3-MT for simplicity) comprising the EB3 N-terminal portion with the MT-binding domain and a part of the linker region, but lacking the EB partner-binding domain at the C terminus (Komarova et al., 2009). This construct allows EB dimerization due to the presence of the leucine zipper domain of GCN4 and is sufficient to suppress MT catastrophes in interphase cells (Komarova et al., 2009). When EB1/EB3 co-depleted cells expressed any of the EB3-GFP rescue constructs at near endogenous levels, the duration from NEB to anaphase was indistinguishable from control or individual EB depletions. Additionally, the spindle tilting occurred mostly until furrow ingression, which is equivalent to EB1 depletion only (Fig. 1, B and C). This demonstrates that exogenous EB3 expression can rescue the EB3-specific but not the EB1-specific phenotype. These data further indicate that the role of EB3 in preventing spindle tilting in late mitosis is independent of its C-terminal binding partners.
To quantify the observed spindle alignment defects, we determined the spindle angle relative to the substrate throughout mitosis in fixed cells after depleting EB1, EB3, or both (Fig. 1, D and E). During prometaphase in control cells, the spindle is still aligning with the substrate and, therefore, the measured angle was between 5° and 24° in 75% of the cells. Spindle angle then gradually decreased to ∼8° as cells reached metaphase, with minimal values from anaphase onwards, indicating nearly complete alignment relative to the substrate. EB1-depleted cells, however, were unable to align their spindles properly from prometaphase until anaphase onset. For comparison, the spindle angle in metaphase upon EB1 depletion was higher than in prometaphase in control cells. This may be caused by the fact that EB1 depletion induced an ∼50% decrease in astral MT area and respective fluorescence intensity (Fig. S2), which might compromise the capacity of spindles to interact normally with the cell cortex, which is in agreement with previous reports (Théry et al., 2005; Toyoshima and Nishida, 2007). Nonetheless, EB1-depleted cells were still able to maintain a low spindle angle in telophase, which suggests that they were able to establish proper attachment to the substrate upon mitotic exit. EB3 depletion, however, did not induce changes in spindle angle in early mitosis compared with controls, nor did it decrease astral MT area or respective fluorescence intensity (Fig. 1, D and E; and Fig. S2). However, anaphase and telophase spindles showed increased angles relative to the substrate, comparable to those observed for control cells in prometaphase (Fig. 1, D and E). In agreement with our live-cell observations, EB1/EB3 co-depletion resulted in an increased spindle angle throughout mitosis, which is consistent with a requirement for individual EB proteins in spindle alignment with the substrate at different mitotic stages (Fig. 1, C–E).
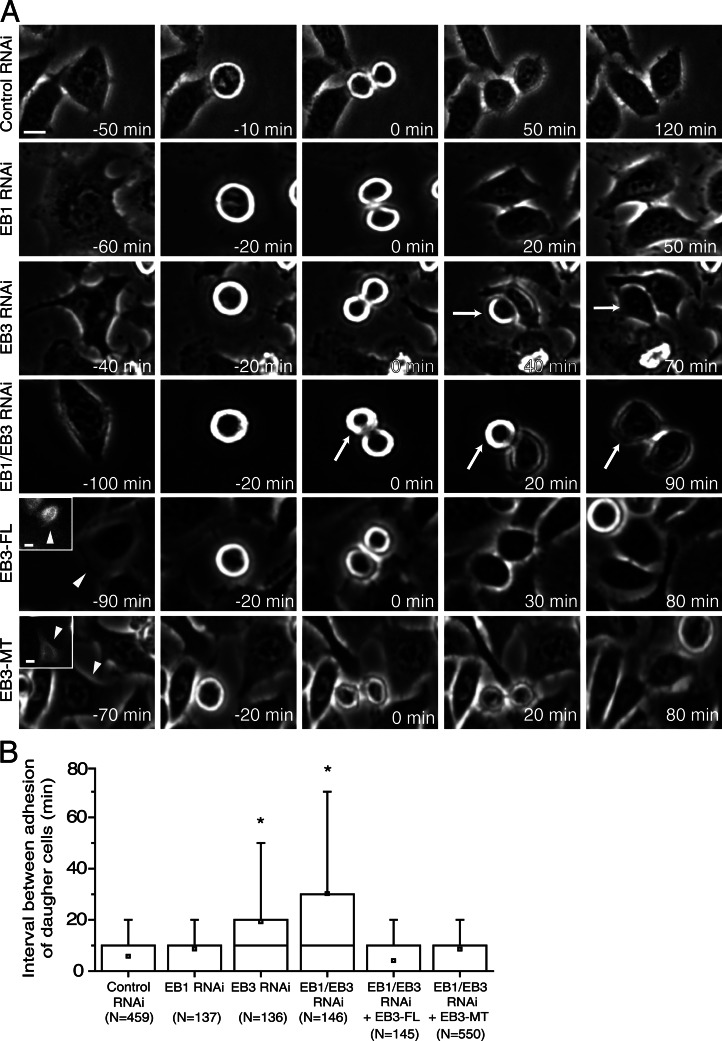
From our live-cell confocal microscopy analysis we noticed that the increased spindle tilting observed in late mitosis upon EB3 or EB1/EB3 depletion normally correlated with a defect in substrate adhesion of one of the daughter cells as they exit from mitosis (Fig. 1 B and Video 2). A more extensive live-cell analysis by phase-contrast microscopy revealed that EB3 or EB1/EB3, but not control or EB1 RNAi, resulted in a significant delay in adhesion and spreading between the two daughter cells (Fig. 2, A and B). Expression of any of the EB3-GFP rescue constructs in EB1/EB3 co-depleted cells also reduced the observed delay in adhesion and spreading between the two daughter cells to levels similar to those observed in control or EB1 RNAi cells (Fig. 2 B). Overall, these results suggest that the observed requirement for EB3 function in postanaphase spindle position/orientation and postmitotic daughter cell spreading is independent of its C-terminal binding partners.
Figure 2.
Characterization of postmitotic cell adhesion to the substrate. (A) HeLa cells were filmed using phase-contrast microscopy. 0 min corresponds to the first frame in anaphase. Arrows indicate cells with uncoordinated adhesion. Arrowheads and insets indicate cells expressing either the full-length EB3-GFP (EB3-FL) or the MT binding domain of EB3 (EB3-MT) that were tracked in this particular example. Bars, 10 µm. (B) Quantification of the adhesion delay between the first and the second daughter cells (*, P < 0.001 using nonparametric ANOVA followed by a post-hoc Dunn’s test). Experiments were done in triplicate and N represents the number of cells quantified in each condition.
EB3 regulates the stabilization of focal adhesions (FAs) during mitotic exit
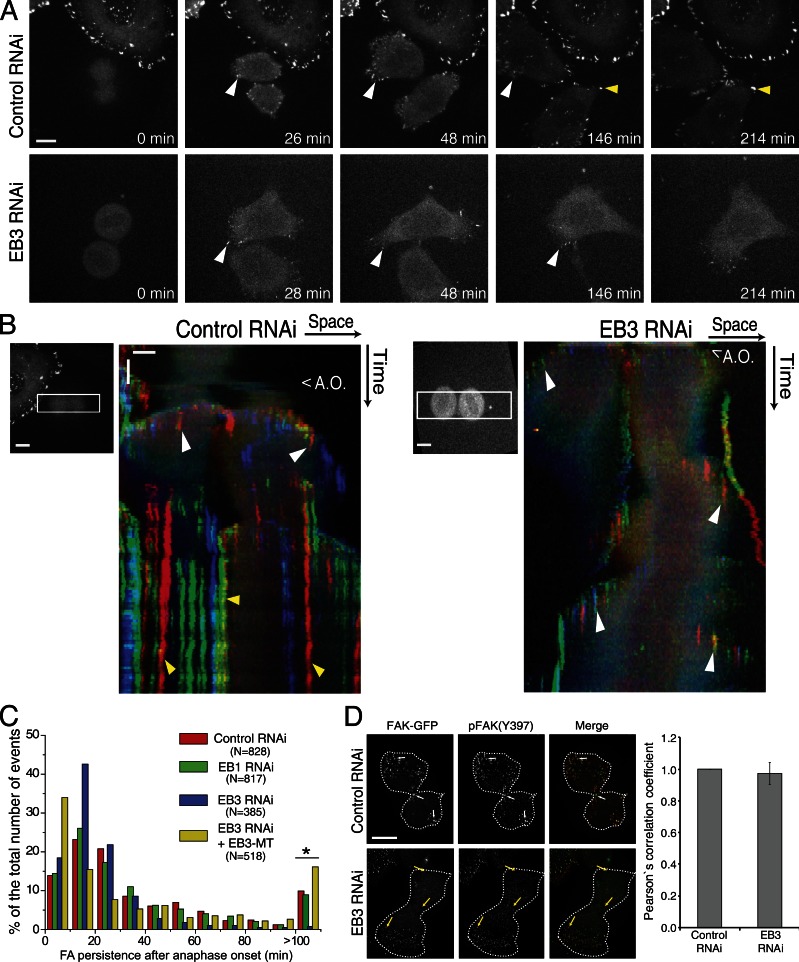
The reestablishment of cell-substrate adhesion at the end of mitosis relies on the recruitment of FAK to FAs (Yamakita et al., 1999; Pugacheva et al., 2006). By using spinning-disk live-cell microscopy analysis of HeLa cells expressing FAK-GFP during mitotic exit, we determined that FAK normally accumulated in focal complexes near the substrate shortly after anaphase onset (Fig. 3, A and B; and Video 5). These structures showed high turnover (Fig. 3 C) and accumulated at the periphery as cells changed shape (Fig. 3, B–D). Notably, as postmitotic daughter cells spread, initial focal complexes disappeared and gave way to longer-lived FAs that allowed for stabilization of the cell shape (Fig. 3, B and C). Moreover, all the FAs identified by FAK-GFP were also positive upon immunostaining with a pFAK(Y397) antibody, which indicates the presence of catalytically active FAK (Fig. 3 D). Upon depletion of EB1 alone we did not find any alteration in the occurrence or persistency of FAs during mitotic exit (Fig. 3 C and Video 6).
Figure 3.
Stabilization of FAs upon mitotic exit. (A) HeLa cells expressing FAK-GFP were imaged by spinning disk microscopy during mitotic exit. (B) Chromo-kymographs of control and EB3-depleted cells expressing FAK-GFP during mitotic exit. To generate the chromo-kymographs, an ROI was selected that was aligned with the long axis of the cells in anaphase, and RGB components were attributed so that a smoothly varying color was assigned to objects at different y positions. This allows objects that colocalize in the x axis to be differentiated by color. White arrowheads highlight the nascent FAs formed after anaphase onset during cell spreading. Yellow arrowheads highlight the more stable FAs that appear later on as cells stabilized their shape. A.O., anaphase onset. Horizontal bar, 10 µm; vertical bar, 20 min. (C) Quantification of FA persistency after anaphase onset, as measured by the stability of FAK-positive structures. Depletion of EB3 leads to the disappearance of the more stable FAs, which can be rescued by expression of the EB3-MT (*, P < 0.001 using nonparametric ANOVA followed by a post-hoc Dunn’s test). N represents the number of FAs measured in three independent experiments. (D) Immunostaining showing the colocalization of FAK-GFP with the active form of FAK (pFAK-Y397). Note the extensive colocalization of both forms of the protein as determined by the Pearson’s correlation coefficient. Broken lines indicate the outline of the cells. White arrows show FAs in control cells appearing in the equator and the polar regions of both daughter cells. Bar, 10 µm. Yellow arrows show FAs in an EB3-depleted cell appearing in one of the daughter cells but not in the equator or the polar region of the other cell. Error bars indicate mean ± SEM.
Depletion of EB3 did not impair the formation of the short-lived active focal complexes that appeared upon mitotic exit similar to control cells (Fig. 3, A–D; and Video 7). However, in EB3-depleted daughter cells, the formation of adhesion complexes was delayed, leading to uncoordinated spreading and attachment problems (Fig. 3 B). Importantly, lack of EB3 prevented the formation of long-lived FAs, and daughter cells kept moving constantly upon spreading (Fig. 3, B and C; and Video 7), which suggested that the increased cell mobility resulted from incapacity to stabilize FAs. Indeed, quantification of cell migration parameters revealed that EB3-depleted HeLa cells moved faster and covered longer distances (unpublished data). Finally, we found that the observed reestablishment of coordinated daughter cell spreading at the end of mitosis upon expression of EB3-MT in an EB3 RNAi background correlated with the accumulation of long-lived FAs (Fig. 3 C). These results support the finding that EB3-mediated regulation of MT dynamics controls postmitotic daughter cell mobility and adhesion required for the stabilization of FAs.
EB3 dephosphorylation on S176 upon mitotic exit is necessary for coordinated daughter cell spreading
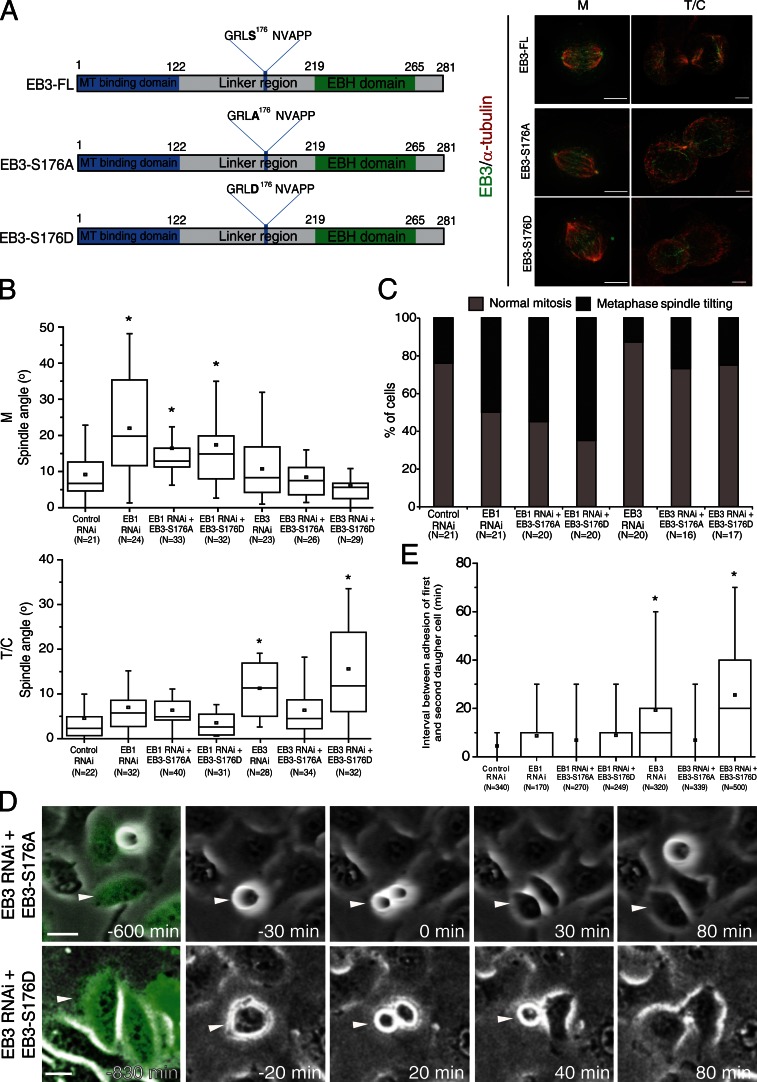
It has been reported that phosphorylation of EB3 at S176 by Aurora kinases during mitosis decreases as cells enter into G1 (Ban et al., 2009). We hypothesized that dephosphorylation of EB3 upon mitotic exit could act as a functional switch, which is required for coordinated daughter cell spreading and adhesion to the substrate. To test this we generated EB3 mutants that carried S-A or S-D mutations at S176 to mimic either constitutively absent or persistent phosphorylation, respectively (Fig. 4 A). Both EB3 mutants localized normally to the plus ends of MTs throughout mitosis (Fig. 4 A), and their expression on an EB1-depleted background did not affect the metaphase spindle angle typically observed upon EB1 depletion (Fig. 4, B and C). This reinforces the idea that spindle positioning parallel to the substrate in metaphase depends specifically on EB1. Notably, expression of the nonphosphorylatable, but not the phosphomimetic mutant in cells depleted of endogenous EB3 successfully restored telophase spindle angle and coordinated daughter cell adhesion to control values (Fig. 4, B and E). In agreement, cells depleted of endogenous EB3, which expressed EB3-S176D, showed problems in late spindle alignment leading to uncoordinated daughter cell spreading and delayed adhesion (Fig. 4, B, D, and E). Overall, these data indicate that EB3 dephosphorylation on S176 is required for postmitotic daughter cell adhesion to the substrate.
Figure 4.
Cortical dephosphorylation of EB3 during mitotic exit is required for proper attachment to the substrate. (A) Diagram of full-length EB3 and the EB3-S176A and EB3-S176D mutants (left). Immunolocalization of α-tubulin and the EB3-FL, EB3-S176A, or EB3-S176D constructs in metaphase (M) and telophase (T/C) cells that were depleted of endogenous EB3 (right). Note the plus end localization of the mutants. (B) Quantification of spindle angles in metaphase (M) and telophase/cytokinesis (T/C) using fixed cells. (C) Quantification of mitotic phenotypes observed using spinning disk live-cell imaging. Note that neither EB3 mutant is able to rescue the EB1 phenotypes observed in metaphase cells. (D) Selected frames from time-lapse movies of HeLa cells depleted of endogenous EB3 and expressing EB3-S176A or EB3-S176D. Arrowheads highlight a cell that was tracked in each of the groups. Note the simultaneous spreading of both daughter cells in the EB3-S176A treatment, in contrast with the uncoordinated spreading observed in the EB3-S176D group. (E) Quantification of the delay in adhesion of the two daughter cells. The EB3-S176A mutant is able to induce coordinated attachment of both daughter cells to the substrate, whereas the EB3-S176D mutant fails to do so (*, P < 0.001 using nonparametric ANOVA [Kruskal-Wallis] followed by a post-hoc Dunn’s test). Experiments were done in triplicate and n represents the number of cells quantified for each condition. Bars: (A) 5 µm; (D) 20 µm.
EB3 phosphorylation status on S176 controls cortical MT growth
Our previous results showed that expression of the EB3-MT (which includes S176) or the EB3-S176A mutants significantly rescued coordinated daughter cell spreading and adhesion, which suggests a direct role of EB3 phosphorylation in the control of MT dynamics required for this process. To test this, we determined the impact of EB3 depletion on cortical MT dynamics during mitotic exit and the respective rescue potential of GFP-tagged EB3-FL, EB3-MT, EB3-S176A, or EB3-S176D mutants. For this purpose we tracked the different GFP-tagged EB3-decorated comets (or EB1-GFP in the case of EB3 RNAi alone) upon depletion of endogenous EB3 in adherent telophase cells and measured the comet lifetime, traveled distance, and growth velocity near the cortex (Table 1). Note that all the EB3 constructs were expressed at equivalent levels, but because they were all overexpressed relative to endogenous EB3 (Fig. S1 C), we focused our analysis on cells expressing the lowest tractable EB3-GFP (or EB1-GFP in specified cases) levels on MT plus ends. We found that EB3 depletion leads to longer MT growth episodes and that this effect can be reversed by expression of EB-FL, EB3-MT, and EB3 S176A, but not S176D mutant (Table 1), which suggests that S176 phosphorylation inhibits EB3 function related with cortical MT dynamics.
Table 1.
Quantification of comet tracking parameters
| Parameters | Telophase (cortical MTs) | ||
| Comet lifetime | Growth distance | Mean velocity | |
| s | µm | µm/s | |
| Control RNAi | |||
| EB1-GFP (359/21)a | 8.0 ± 3.7 | 2.9 ± 1.5 | 0.406 ± 0.12 |
| EB3 RNAi | |||
| EB1-GFP (467/30) | 11.0 ± 5.5c | 3.5 ± 1.9c | 0.349 ± 0.11c |
| EB3-FL (275/18) | 6.7 ± 2.4 | 2.2 ± 0.9 | 0.335 ± 0.11 |
| EB3-MT (276/16) | 6.3 ± 3.0 | 2.0 ± 1.1 | 0.318 ± 0.16 |
| EB3-S176A (282/14) | 6.6 ± 3.7 | 2.7 ± 1.7 | 0.407 ± 0.13b |
| EB3-S176D (197/12) | 12.2 ± 6.0b | 3.9 ± 1.9b | 0.330 ± 0.11 |
Numbers within parenthesis represent the number of tracked comets/number of cells in three independent experiments.
P < 0.001 when compared to EB3-FL (nonparametric ANOVA followed by a post-hoc Dunn’s test).
P < 0.001 when compared to EB1-GFP (nonparametric ANOVA followed by a post-hoc Dunn’s test).
EB3 promotes midbody MT stability necessary for efficient cytokinesis
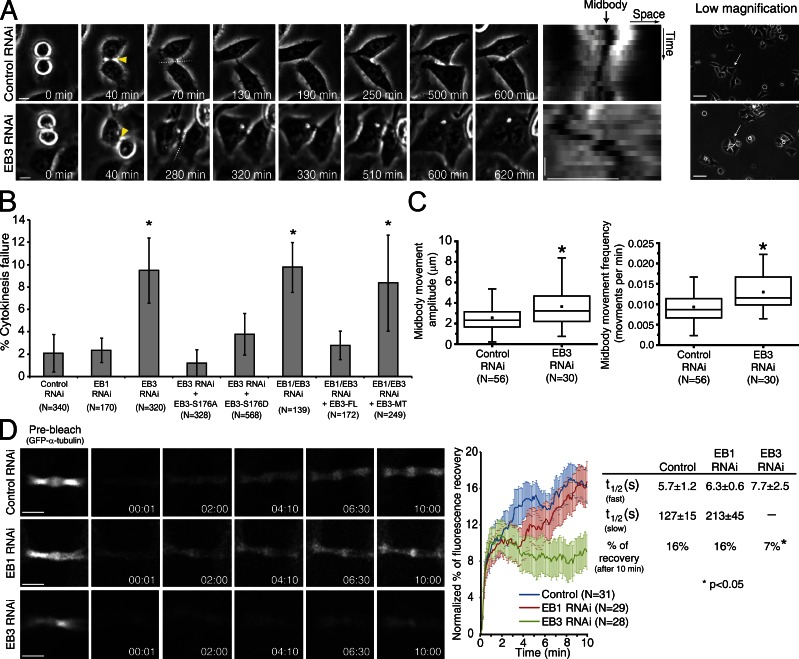
MTs are essential for the completion of cytokinesis (Wheatley and Wang, 1996), and EB-like proteins have been previously implicated in this process in yeast, Dictyostelium discoideum, and sea urchin (Muhua et al., 1998; Strickland et al., 2005; King et al., 2010). We therefore investigated the respective requirements of human EB1 and/or EB3 during cytokinesis upon RNAi and live-cell microscopy in HeLa cells. We found that EB3 and EB1/EB3 depletion induced cytokinesis failure in ∼10% of attempts, whereas EB1 depletion alone had no significant impact in this process (Fig. 5, A and B; see also King et al., 2010). Interestingly, ectopic expression of EB3-FL, but not EB3-MT, rescued the cytokinesis defects associated with EB3 depletion (Fig. 5 B), which suggests that this role of EB3 requires binding with other proteins through its C-terminal domain. Moreover, and in contrast to control cells, we found that the midbodies of ∼76% of EB3-depleted cells that failed cytokinesis were unable to establish a stable position between the two daughter cells and showed increased amplitude and frequency of motion (Fig. 5, A and C), which suggests that stabilization of midbody position is important for cytokinesis. Importantly, these differences in midbody stability were not caused by different cell densities in control or EB3-depleted cells (Fig. 5 A).
Figure 5.
EB3 is required for stabilization of the midbody upon mitotic exit. (A) HeLa cells were filmed in phase contrast to determine the number of failed cytokinesis events. Yellow arrowheads indicate the first time-frame where the midbody could be observed. The broken lines indicate the region that was used to generate the kymograph of the midbody position. Horizontal bar, 10 µm; vertical bar, 50 min. Note the formation of a binucleated cell in an EB3-depleted cell, where the midbody fails to stabilize. In the low-magnification images, white arrows indicate the cells that were tracked in this particular example. 0 min corresponds to the first frame in anaphase. (B) Quantification of the number of failed cytokinesis per total divisions observed upon EB3 and/or EB1 RNAi and respective rescue efficiencies with ectopic constructs. Error bars indicate mean ± SEM. *, P < 0.05 when compared to control RNAi using nonparametric ANOVA followed by a post-hoc Dunn’s test. (C) Quantification of midbody stability in control RNAi and EB3-depleted cells. Midbody movement amplitude and frequency were extracted from kymographs generated in the midbody region. Movement amplitude corresponds to the lateral deviation of the midbody throughout time. Movement frequency was defined as the number of midbody oscillations per unit of time. (*, P < 0.001 using nonparametric ANOVA followed by a post-hoc Dunn’s test). All experiments were done in triplicate, and n represents the number of cells quantified in each condition. (D) FRAP analysis of the midbody region in cells expressing α-tubulin–GFP. The percentages of fluorescence recovery and half-time recovery were determined by applying a double (control and EB1 RNAi) or single (EB3 RNAi) exponential fitting to the recovery curve. Time is given in minutes:seconds. Horizontal bar, 10 µm; vertical bars, 3 µm. *, P < 0.05 when compared to control RNAi using nonparametric ANOVA followed by a post-hoc Dunn’s test.
Next, we investigated whether EB proteins play a role in midbody MT stability. To do so, we determined midbody MT half-life and renewal capacity by quantifying FRAP of GFP–α-tubulin in control, EB1-depleted, or EB3-depleted cells. In agreement with previous work (Saxton and McIntosh, 1987), we found that midbody MTs in control cells turn over very slowly, with FRAP on the order of 16% over a 10-min period. Interestingly, whereas the FRAP curves for EB3 RNAi were best fit by a single exponential equation, control and EB1 RNAi were best fit by double exponential equations, which indicates biphasic kinetics caused by fast and slow recovering MT populations (Fig. 5 D). Quantification of the respective MT half-life in different experimental conditions revealed that although EB1-depleted cells were indistinguishable from controls, EB3 depletion specifically abolished the slow recovery component corresponding to more stable MTs, restricting fluorescence recovery to only 7% after 10 min (Fig. 5 D). These data indicate that EB3, but not EB1, promotes midbody MT stability necessary for efficient cytokinesis. Moreover, our data suggest that this effect over midbody MT stabilization is not exclusively caused by the impact of EB3 on MT dynamics and likely involves interaction with other proteins binding through its C-terminal domain.
EB3 phosphorylation on S176 is spatially regulated by Aurora B kinase to coordinate cell adhesion with cytokinesis
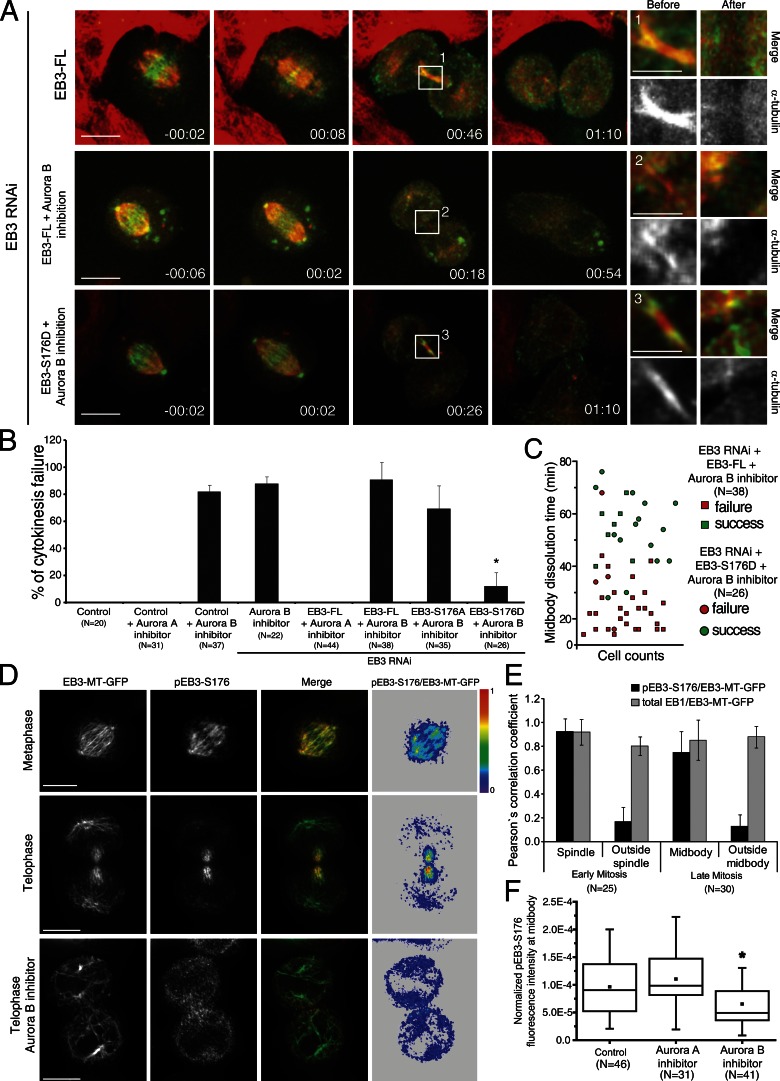
Aurora B kinase is a major regulator of cytokinesis (Ruchaud et al., 2007) that interacts with and phosphorylates EB3 on S176 (Ban et al., 2009). To investigate whether this specific phosphorylation of EB3 is required/sufficient for cytokinesis, we depleted cells of endogenous EB3 by RNAi and expressed RNAi-resistant GFP-tagged EB3-FL, EB3-S176A, or EB3-S176D mutants, followed by Aurora A or B inhibition at the metaphase-to-anaphase transition. As negative and positive controls, we used cells treated with only Aurora A or B inhibitors, respectively. Accordingly, we found that treatment of live anaphase cells stably expressing α-tubulin–mRFP with the more specific Aurora A inhibitor MLN8054 (Manfredi et al., 2007) did not lead to any detectable cytokinesis defects in control or EB3-depleted cells rescued with EB3-FL (Fig. 6, A and B). However, inhibition of Aurora B with ZM447439 (Ditchfield et al., 2003) in control, EB3-depleted, or EB3-depleted cells rescued with EB3-FL induced cytokinesis failure in ∼80% of the cells (Fig. 6, A and B; and Video 8). Remarkably, when EB3-depleted cells expressing EB3-S176D but not the EB3-S176A mutant were treated with ZM447439, there was a significant rescue of the observed cytokinesis defects associated with Aurora B inhibition, which correlated with the prevention of premature midbody disassembly (Fig. 6, A–C; and Video 9). These data suggest that EB3 phosphorylation on S176 by Aurora B promotes midbody MT stability. Interestingly, expression of either EB3-S176A or EB3-S176D was able to rescue the cytokinesis defects induced by EB3 depletion (Fig. 5 B), which indicates that EB3 phosphorylation on S176 by Aurora B is sufficient but not required to ensure the successful completion of cytokinesis. One possible redundant mechanism might have to do with the capacity of the EB3-S176A construct to rescue normal spindle orientation and daughter cell adhesion during mitotic exit (Fig. 4, B and E), indirectly leading to the stabilization of the midbody. Indeed, we observed a strong correlation between the incapacity to orient the spindle during late mitosis and the probability (83%) to fail cytokinesis upon EB3 depletion (Fig. 5 A).
Figure 6.
Phosphorylation of EB3 by spatially regulated Aurora B. (A) Frames from time-lapse movies of cells expressing α-tubulin–mRFP and either EB3-FL or EB3-S176D tagged with GFP in the presence or absence of the Aurora B inhibitor ZM447439 upon EB3 RNAi. Higher magnification images highlight the midbodies before and after dissolution. Time is given in hours:minutes. Bars: (main panels) 10 µm; (insets) 3 µm. (B) Quantification of the percentage of cells that fail cytokinesis after inhibition of Aurora A (MLN8054) or Aurora B (ZM447439), and the respective dependence on EB3 phosphorylation. Error bars indicate mean ± SEM. (C) Correlation between the midbody dissolution time and cytokinesis outcome in cells expressing either EB3-FL or EB3-S176D and treated with the Aurora B inhibitor upon EB3 RNAi. Note the positive correlation between the midbody dissolution time and cytokinesis success. (D) Immunolocalization of endogenous pEB3-S176. Cells expressing the EB3-MT-GFP construct were used to facilitate detection of MT plus ends. The ratio between pEB3-S176 and EB3-MT-GFP is shown on the right. Bars, 10 µm. (E) Quantification of the subcellular colocalization of endogenous pEB3-S176. Note that phosphorylation of EB3 at S176 occurs throughout the spindle in early mitosis but is concentrated in the midbody in telophase. Error bars indicate mean ± SEM. (F) Inhibition of Aurora B leads to a significant decrease in the levels of pEB3-S176 in the midbody when normalized relative to EB3-MT-GFP and tubulin. (*, P < 0.001 using nonparametric ANOVA followed by a post-hoc Dunn’s test). Experiments were done in triplicate and n represents the number of cells quantified in each condition.
It has recently been demonstrated that an Aurora B activity gradient is established from the spindle midzone during anaphase/telophase (Fuller et al., 2008), but its biological significance remained unclear. Therefore, we sought to investigate whether EB3 phosphorylation on S176 was spatially regulated by Aurora B during mitotic exit/cytokinesis. For this purpose we used a phospho-specific antibody against S176 (Ban et al., 2009) and determined the respective fluorescence ratio relative to endogenous EB3, as well as ectopic EB3-MT-GFP or EB3-FL-GFP, which we used to enhance MT plus-end labeling. As a control, we determined the fluorescence ratio of endogenous EB1 relative to EB3-MT-GFP. We found that EB3 phosphorylation on S176 is enriched on midbody MTs and is low near the cortex (Fig. 6, D and E; and Fig. S3). Finally, Aurora B inhibition with ZM447439 or expression of EB3-S176A mutant in EB3-depleted cells significantly reduced the epitope recognized by the phospho-specific antibody against S176 (Figs. 6 F and S3). Collectively, these data support the finding that EB3 phosphorylation on S176 is spatially regulated by an Aurora B activity gradient and is important to coordinate daughter cell adhesion with cytokinesis.
Discussion
Here we investigated the respective roles and regulatory mechanisms behind EB1 and EB3 during mitosis, postmitotic cell adhesion, and cytokinesis. Given the central role played by EB proteins in regulating MT dynamics during interphase (Akhmanova and Steinmetz, 2008; Honnappa et al., 2009; Komarova et al., 2009), we were surprised that individual or simultaneous depletion of EB1 and EB3 gave rise to relatively mild spindle MT phenotypes during early mitosis in HeLa cells. Nevertheless, and consistent with previous findings, our experiments showed that EB1 controls spindle position relative to the substrate, but it does so specifically during early mitosis. This is thought to occur through the interaction between the cell cortex and astral MTs (Green et al., 2005; Toyoshima and Nishida, 2007), which are functionally compromised upon EB1 depletion. However, EB3 was not required for this purpose and, unlike EB1 knockdown, cells depleted of EB3 still established a visible astral MT array. Curiously, EB3, but not EB1, was required for efficient spindle alignment with the substrate only after anaphase onset, which compromised daughter cell adhesion as they spread during mitotic exit. Because both EB1 and EB3 colocalize at spindle MT plus ends throughout mitosis, our findings raise the question, Why is EB3 dispensable for spindle orientation during early mitosis but important during later stages, as opposed to EB1? One possibility might have to do with the relative abundance and half-life of EB1 and EB3 proteins in HeLa cells throughout mitosis. In fact, although EB1 is expressed constitutively throughout the cell cycle, EB3 has a shorter half-life, and its levels peak with high cyclin B expression as cells enter and progress through mitosis (Ban et al., 2009). Another nonexclusive possibility might be related with the relative affinity of EB1 and EB3 to MT plus ends. Interestingly, recent in vitro studies on the dimerization properties of EB1 and EB3 suggested that these proteins are part of a common pool, and depletion of one will have implications on the homo- and heterotypic complex formation of EB1 and/or EB3 and their subsequent association to the MT plus ends, ultimately determining the existence of different functional species (De Groot et al., 2010). Finally, the finding that only EB3, but not EB1, is phosphorylated on S176 throughout mitosis by Aurora A and B (Ban et al., 2009) might regulate the functional state of EB3 and/or its affinity to MT plus ends, causing a direct impact on MT dynamics or control EB3 association with other proteins. In the latter case, there are two interesting candidates: the MT depolymerizing protein MCAK and the tumor suppressor protein APC. MCAK is known to associate with both EB1 and EB3 at MT plus ends to promote the rapid switching between MT growth and shortening (Moore et al., 2005; Lee et al., 2008; Montenegro Gouveia et al., 2010). Interestingly, MCAK localization and activity in mitosis is spatially and temporally regulated by Aurora B and Cdk1 (Lan et al., 2004; Ohi et al., 2004; Moore et al., 2005; Sanhaji et al., 2010; Tanenbaum and Medema, 2011), which could account for spatial differences in MT plus-end dynamics in the spindle context throughout mitosis, consistent with the presence of an Aurora B activity gradient in metaphase as well (Tan and Kapoor, 2011). However, this scenario is unlikely, given that expression of a mutant EB3 construct lacking the C-terminal domain necessary to bind MCAK (Lee et al., 2008) was sufficient to restore normal MT dynamics (see also Komarova et al., 2009). APC is also a well-established EB-interacting protein, and APC truncating mutations associated with the development of colon cancer were shown to disrupt the interaction with EB proteins and compromise early spindle orientation (Su et al., 1995; Su and Qi, 2001; Green et al., 2005). More recently, it was suggested that early spindle orientation in yeast relies on the interaction between EB and APC homologues (Bim1p and Kar9p, respectively) through the linker region of Bim1p and is negatively regulated by Aurora B/Ipl1p–mediated phosphorylation (Hüls et al., 2012). Interestingly, EB3 depletion or its phosphorylation on S176 has a similar effect over cortical MT dynamics in HeLa cells, which suggests that this specific phosphorylation renders EB3 to become “inactive” during early mitosis, thus explaining its dispensability at this particular stage.
At anaphase onset, the reorganization of the MT cytoskeleton is coordinated with the reattachment of postmitotic cells to the substrate, which involves the reassembly of adhesion complexes that were previously inactivated as cells entered mitosis. However, how MTs influence the reformation of adhesion complexes as cells remodel their cytoskeleton at the exit from mitosis remained unclear. Here we show that, during mitotic exit, there is a sudden enrichment of FAK at the FA sites and that EB3 is required for FA stabilization independently of its C-terminal cargo domain, resulting in abnormal daughter cell adhesion and migration. Surprisingly, we provide evidence that EB3 depletion or its phosphorylation on S176 promotes cortical MT growth during mitotic exit, whereas dephosphorylation of EB3 on the same residue ensures normal daughter cell adhesion to the substrate. These results reveal unexpected features behind EB3 functional and spatial regulation throughout the cell cycle and show that the respective effects of EB1 and EB3 on MT growth processivity are more complex than previously thought (Komarova et al., 2009). This possibly translates the fact that EB proteins can cooperate with both positive and negative regulators of MT polymerization, and their balance might be tipped differently in different phases of the cell cycle.
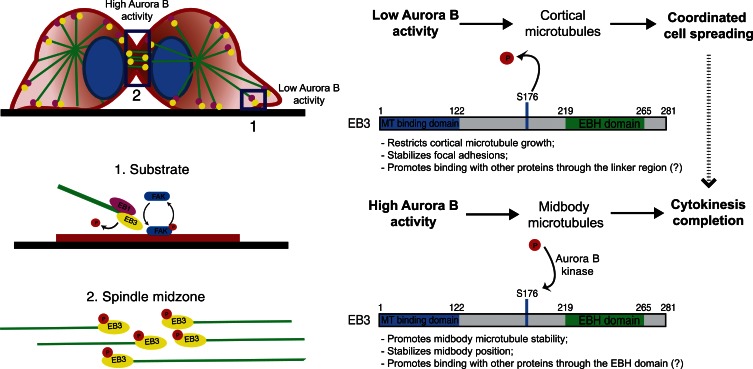
Overall, our data support a model (Fig. 7) where EB3, and more specifically its dephosphorylation on S176, is required to restrict cortical MT growth necessary for the stabilization of FAs (Kaverina et al., 1999; Ezratty et al., 2005). Nevertheless, the possibility that EB3 dephosphorylation on S176 might promote interaction with other proteins (e.g., APC) through the linker region cannot be formally excluded (Hüls et al., 2012). Interestingly, induction of cell protrusions was shown to specifically require growing MTs (Waterman-Storer et al., 1999), suggesting that simply tipping the balance in favor of MT growth is sufficient to induce destabilization of postmitotic cell adhesion factors, which might be the basis of cancer cell dissemination and metastasis. While this paper was in revision, another work reported that EB3 phosphorylation, now on S162, suppressed MT growth and promoted the formation of adherens junctions in response to VE-cadherin signaling in human endothelial cells during interphase (Komarova et al., 2012). The role of EB3 phosphorylation in the regulation of MT dynamics to mediate cell–cell and cell–substrate interactions certainly waits for further development.
Figure 7.
Proposed model for the phosphoregulation of EB3 function during mitotic exit and cytokinesis. In late mitosis, an Aurora B phosphorylation gradient ensures that only MTs in the furrow region remain phosphorylated, whereas MTs in the vicinity of the substrate contain dephosphorylated EB3. Phosphorylation of EB3 at S176 by Aurora B ensures successful cytokinesis completion by promoting midbody MT stability and midbody stabilization. Dephosphorylation of EB3 occurs near the substrate and restricts MT growth, allowing for coordinated daughter cell spreading, which, by itself, indirectly potentiates the successful completion of cytokinesis.
In humans, Aurora B relocalizes from the chromosomes to the spindle midzone at the end of mitosis, generating a phosphorylation gradient that diffuses toward the cell periphery (Fuller et al., 2008; Tan and Kapoor, 2011). The functional significance of this phosphorylation gradient in MT reorganization at the exit from mitosis remained unknown. Recently it was shown that the single EB-like orthologue in yeast is specifically phosphorylated during anaphase by Aurora B/Ipl1p, which was required for normal spindle elongation and disassembly (Zimniak et al., 2009). However, our results support the finding that during mitotic exit in human cells, there is a shift in EB “dominance” required for spindle orientation, with EB3 becoming the key player in coordinating the re-adhesion and spreading of daughter cells to the substrate and completion of cytokinesis. Interestingly, coordinated daughter cell spreading depended on dephosphorylation of S176 on EB3, whereas completion of cytokinesis could be achieved in two possible ways: one that involves EB3 phosphorylation on S176, which directly promotes midbody MT stability; and another that is indirectly potentiated by promoting daughter cell adhesion to the substrate (e.g., due to EB3 dephosphorylation on S176). In agreement, we found that phosphorylated EB3 on S176 was enriched at, and followed a gradient pattern from, the spindle midzone/midbody during late anaphase and cytokinesis in an Aurora B–dependent manner. EB3 phosphorylation on S176 may prevent MT catastrophe events required for the stabilization of a specific MT pool at the midbody. Altogether, our data highlights the fact that systems that normally rely on a single EB-like protein must centralize different functions, which are mediated by distinct EB proteins in human cells. Moreover, our findings imply that EB3 exists in distinct phosphorylation states associated with different MT subpopulations that coordinate daughter cell adhesion with the successful completion of cytokinesis and are consistent with the presence of an Aurora B phosphorylation gradient diffusing from the spindle midzone/midbody regions toward the cell periphery (Fig. 7). Interestingly, it has long been proposed that an EB-dependent cytokinesis checkpoint might be sensitive to mitotic spindle orientation in yeast (Muhua et al., 1998), which could explain how proper spindle orientation throughout mitosis leading to coordinated cell adhesion also contributes to the successful completion of cytokinesis. In line with this hypothesis, integrin-mediated adhesion to the substrate has been shown to be important for spindle orientation parallel to the substrate in an EB1-dependent manner (Toyoshima and Nishida, 2007) and to ensure proper midbody anchoring required for efficient cytokinesis (Pellinen et al., 2008).
Finally, it is interesting that although EB3 depletion results in only ∼10% failure of cytokinesis due to disruption of a highly stable MT pool at the midbody, Aurora B inhibition results in ∼80% of unsuccessful cytokinesis attempts. This suggests that, normally, cytokinesis requires additional Aurora B targets at the spindle midzone/midbody, such as the centralspindlin complex (Glotzer, 2009). Intriguingly, mimicking constitutive EB3 phosphorylation on S176, but not EB3 depletion, extensively rescued the cytokinesis defects caused by Aurora B inhibition, which implies that during cytokinesis, unlike daughter cell adhesion, EB3 RNAi and S176 phosphorylation do not act in a similar way, at least in conditions of Aurora B inhibition. This could mean that either EB3 phosphorylation on S176 brings in a specific partner or competes with EB1, displacing some partners away. This is supported by our observation that an EB3 construct lacking the C-terminal partner binding region, which is sufficient to rescue daughter cell adhesion, failed to rescue the cytokinesis defects associated with EB3 depletion. In this context, it will be important in the future to investigate the importance of the turnover of EB3 phosphorylation on S176 for the spatial regulation of MT stability and interaction with other EB partners.
Materials and methods
Cell culture
All cell lines used were cultured in DMEM with 10% FBS and grown in a 5% CO2 atmosphere at 37°C. HeLa H2B-GFP/α-tubulin–mRFP and HeLa α-tubulin–mRFP cell lines were a gift from P. Meraldi (University of Geneva, Geneva, Switzerland). Cells were seeded on fibronectin (FBN; 50 µg/ml; Sigma-Aldrich)-treated coverslips for the indicated time points and fixed for immunofluorescence or used for live-cell imaging. The Aurora A kinase inhibitor MLN8054 (Selleckchem) was used at a final concentration of 250 nM. The Aurora B kinase inhibitor ZM447439 (Tocris Bioscience) was used at a final concentration of 2–4 µM.
shRNA and transfection experiments
Cells were transfected using FUGENE HD or X-tremeGENE HP (Roche) 24 h after seeding. pSUPER-based vectors were used for RNAi experiments with EB1, EB3, or EB1/EB3 in tandem (sequences were previously described in Komarova et al., 2005). All shRNA-based depletion experiments were performed after 120 h. For the rescue experiments, cells were transfected simultaneously with pSUPER-EB1/EB3 and the full-length EB3-GFP rescue construct (EB3-FL) or the EB3-GFP MT-binding domain (EB3-MT). The EB3-MT construct is comprised of the MT-binding domain of EB3 (aa 1–122) followed by the linker region and artificially dimerized by the addition of the leucine zipper (LZ) domain of GCN4. pEGFP-FAK was a gift from P. Wang (University of California, San Diego, La Jolla, CA). mRFP-FAK was a gift from G. Gundersen (Columbia University, New York, NY). To generate the EB3-S176A-GFP and EB3-S176D-GFP constructs, a synthesized sequence with the respective mutation was introduced in pEGFP-N1 using SalI and BamHI. Rescue experiments with the EB3 mutants were performed by transfecting cells simultaneously with pSUPER-EB3 and either EB3-S176A-GFP or EB3-S176D-GFP constructs.
Antibodies and immunofluorescence
Cells were grown on FBN-coated coverslips and processed for immunofluorescence as described previously (Maiato et al., 2006). In brief, cells were fixed in cold methanol followed by 4% paraformaldehyde, blocked with 10% FBS in PBS-T, and incubated with the specified primary antibodies: mouse anti–α-tubulin clone B512 (1:2,000; Sigma-Aldrich); rat anti–α-tubulin YL1/2 1:10 (AbD Serotec); rat anti-EB1 and anti-EB3 1:20 and 1:10, respectively; rabbit anti-EB3 1:300; mouse anti-EB1 1:200 (Abcam); rabbit anti-pFAK(Y397) (1:400, Santa Cruz Biotechnology, Inc.); and mouse anti–pEB3-S176 (1:1; a gift from T. Urano, Shimane University School of Medicine, Izumo, Japan). Secondary antibodies used were Alexa Fluor 488, 568, and 647 (1:2,000; Invitrogen), and 1 µg/ml DAPI. Images were acquired with a microscope (Axio Imager Z1; Carl Zeiss) equipped with an imagine camera (Axiocam MR; Carl Zeiss) using an immersion oil 63× 1.4 NA Plan-Apochromatic objective lens, controlled by AxioVision software. Immunofluorescence images were deconvolved with AutoQuant software (Media Cybernetics) and contrast was adjusted with Photoshop software (Adobe).
Time-lapse microscopy
For live-cell imaging experiments, cell lines were grown with L-15 medium with 10% FBS on FBN-treated coverslips or plated in 35-mm glass bottom microwell (14 mm, no 1.5 coverglass) dishes (MatTek Corporation). Four-dimensional datasets were acquired with a spinning disc confocal system (Revolution; Andor Technology) equipped with an electron-multiplying charge-coupled device camera (iXonEM+; Andor Technology) and a CSU-22 unit (Yokogawa Corporation of America) based on an inverted microscope (IX81; Olympus). Two laser lines (488 and 561 nm) were used for near-simultaneous excitation of GFP and mRFP, and the system was driven by iQ software (Andor Technology). Time-lapse imaging of z stacks with 0.7-µm steps covering the entire volume of the mitotic apparatus were collected every 2 or 3 min with a Plan-Apochromatic 1.40 NA 60× immersion oil objective lens. Phase-contrast microscopy was performed on a microscope (Eclipse TE2000-U; Nikon) driven by NIS Elements 3.0 software with a 0.40 NA 20× objective lens. Time-lapse photography was performed every 10 min.
Quantitative analysis of spindle angle and length
Quantification of spindle angles and length was performed using Volocity LE (PerkinElmer). Images were collected by taking z stacks with a step of 0.3 µm covering the entire volume of the mitotic spindle. Distances tangent (ΔR) and perpendicular (ΔZ) to the substrate were measured after defining the position of the two poles and correcting for projection errors. The angle between the spindle axis and the substrate was defined by the inverse trigonometric function α = arctg (ΔZ/ΔR). Spindle length is defined as L = √[(ΔR)2+(ΔZ)2].
Quantification of astral MT area and fluorescence intensity
To determine astral MT area, maximal projections of z stacks covering the entire mitotic apparatus were generated and a freehand selection tool in ImageJ software (National Institutes of Health) was used to outline the spindle pole and astral MTs. Astral MT fluorescence intensity was performed by selecting the pole regions using the freehand tool of ImageJ and measuring the total fluorescence intensity in the entire z stacks. The resulting value was normalized to the corresponding area.
Quantitative analysis of adhesion of the daughter cells to the substrate after mitosis
Individual cells were manually tracked until they exited mitosis and respread out in the substrate, and the lag time between adhesion of the first and last daughter was quantified.
Quantification of cytokinesis failure
To quantify cytokinesis failure events, we counted the number of mitoses that failed to segregate properly, leading to bi-nucleated cells. These were then divided by the total number of mitoses observed, thus defining the percentage of cytokinesis failures per total mitoses.
Chromo-kymographic analysis of FAs
To determine FA formation and persistency, cells expressing either GFP-FAK or mRFP-FAK were followed with a 2-min time lapse as they exited mitosis. Subsequent analysis of FA formation and persistence were done using chromo-kymographic methods that have been previously described (Pereira and Maiato, 2010). To prepare a kymograph for analysis of FA assembly, we used a custom routine written in MATLAB (MathWorks). A region of interest (ROI) was selected that was parallel to the anaphase axis. Chromo-kymographs are generalized kymographs that retain information contained along the small axis (y) of the ROI by attributing a particular RGB combination to thin ROI subslices. RGB components are attributed so that a smoothly varying color is assigned to objects at different y positions. This allows objects that colocalize in the x axis to be differentiated by color. A conventional kymograph is a particular case of a chromo-kymograph in which the subslices are attributed the same color (or none).
FRAP of midbodies
To perform FRAP analysis, an ROI containing the entire midbody structure was identified and bleached using a 488-nm laser at a power of 100% for 2 s. Images were then collected with a time lapse of 5 s in a z stack of 1.5 µm. Quantification of fluorescence recovery was obtained using the FRAP profiler plugin of ImageJ, which accounts for correction of overall bleaching. Recovery rates for control and EB1 RNAi were determined after fitting a double exponential curve using the formula y = y0 + A1e−x/t1 + A2e−x/t2, where A1 represents the less stable MT population and A2 the more stable MT population with decay rates of k1 and k2, respectively. For the EB3 RNAi experimental group, recovery rates were determined after fitting a single exponential curve using the formula y = y0 + Ae−x/t. Half-time recovery was determined as ln2/k.
Inhibition of Aurora kinases during mitotic exit
To inhibit Aurora kinases during mitotic exit, cells were filmed during mitosis and either Aurora A (MLN8054) or Aurora B (ZM447439) inhibitors were immediately added before anaphase onset. Cells were then filmed until the midbody structure disappeared or they failed cytokinesis.
Phospho-EB3 fluorescence intensity quantifications
Fluorescence signal quantification in the spindle and midbody was done using a MATLAB script. A sum-projection image of the z stack is generated upon which the user manually chooses several points along the selected path. A closed cubic spline curve is generated to smoothly connect the points. The spline curve is then convolved with a disc-shaped kernel of variable diameter, which allows control of the ROI curve thickness. Fluorescence intensity is then calculated as the integrated signal inside the inner boundary of the ROI.
Comet tracking
Individual comets were imaged by using the indicated EB3 constructs tagged with GFP. Movies with a time lapse of 2 s were acquired on a spinning disk confocal microscope, and individual comets were manually tracked using the “Manual Tracking” plugin of ImageJ. Comets were followed continuously until the fluorescent signal disappeared. The comet tracking time is defined as the continuous time a single comet track is followed (in seconds). The mean growth distance is defined as the distance the comet traveled until the signal disappeared (in micrometers). The mean velocity is defined as the instantaneous velocity between consecutive time frames as given by the formula v = mean distance (micrometers)/time (seconds).
Statistical analysis and data presentation
When data are represented as box-whisker plots, the box size represents 75% of the population and the line inside the box represents the median of the sample. The size of the bars (whiskers) represents the maximum (in the upper quartile) and the minimum (in the lower quartile) values. Statistical analysis for multiple group comparison was performed using a parametric one-way analysis of variance (ANOVA) when the samples had a normal distribution. When the sample did not have a normal distribution, multiple group comparison was done using a nonparametric ANOVA (Kruskal-Wallis). All pairwise multiple comparisons were subsequently analyzed using either post-hoc Student-Newman-Keuls (parametric) or Dunn’s (nonparametric) tests. When comparing only two experimental groups, a parametric t test was used when the sample had a normal distribution, or a nonparametric Mann-Whitney test was used for samples without normal distribution. All statistical analyses were performed using SigmaStat 3.5 (Systat Software, Inc.).
Online supplemental material
In Fig. S1 we describe the localization of EB1 and EB3 throughout mitosis and determine the efficiency of depletion of EB proteins by shRNA. Furthermore, we show that individual depletions do not affect mitotic progression. In Fig. S2 we show the effect of EB depletion on astral MT area and respective fluorescence intensity. Fig. S3 gives representative immunofluorescence images of mitotic HeLa cells immunostained for endogenous EB3 or expressing EB3-FL or EB3-S176A that were also immunoreacted with a pEB3-S176 antibody, demonstrating the presence of a phosphorylation gradient in late mitosis with endogenous EB3 and EB3-FL, but not with EB3-S176A. Video 1 illustrates a control cell progressing through mitosis. Video 2 shows the mitotic progression of a cell depleted of EB1/EB3 with spindle tilting and daughter cell detachment. Video 3 shows a cell depleted of EB1 that has a tilted spindle followed by normal daughter cell attachment. In Video 4 we show a cell depleted of EB3 that aligns the spindle in metaphase but shows defects in daughter cell attachment. In Video 5 we show the mitotic exit of a control cell expressing FAK-GFP. In Video 6 we show the mitotic exit of a cell depleted of EB1 and expressing FAK-GFP. In Video 7 we show the mitotic exit of a cell depleted of EB3 and expressing FAK-GFP, demonstrating the lack of stability of FAs. In Video 8 we show the mitotic exit of a cell expressing EB3-FL-GFP and α-tubulin–mRFP that was treated with Aurora B inhibitor and fails cytokinesis due to premature midbody disassembly. In Video 9 we show the mitotic exit of a cell expressing EB3-S176D-GFP and α-tubulin–mRFP that was treated with Aurora B inhibitor and completes cytokinesis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201301131/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201301131.dv.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Patrick Meraldi, Peter Wang, Gregg Gundersen, and Takeshi Urano for the kind gift of reagents and João Relvas for critical reading of the manuscript.
A. Akhmanova is supported by the Netherlands Organization for Scientific Research NWO-ALW VICI grant. H. Maiato is funded by grants PTDC/SAU-GMG/099704/2008 and PTDC/SAU-ONC/112917/2009 from Fundação para a Ciência e a Tecnologia (FCT; COMPETE-FEDER), the Human Frontier Science Program, and the seventh framework program grant PRECISE from the European Research Council.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- APC
- adenomatosis polyposis coli
- EB
- end binding
- FA
- focal adhesion
- FBN
- fibronectin
- MCAK
- mitotic centromere-associated kinesin
- MT
- microtubule
- NEB
- nuclear envelope breakdown
- ROI
- region of interest
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Ban R., Matsuzaki H., Akashi T., Sakashita G., Taniguchi H., Park S.Y., Tanaka H., Furukawa K., Urano T. 2009. Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J. Biol. Chem. 284:28367–28381 10.1074/jbc.M109.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüning-Richardson A., Langford K.J., Ruane P., Lee T., Askham J.M., Morrison E.E. 2011. EB1 is required for spindle symmetry in mammalian mitosis. PLoS ONE. 6:e28884 10.1371/journal.pone.0028884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W., Su L.K. 2001. Regulation of microtubule assembly by human EB1 family proteins. Oncogene. 20:3185–3192 10.1038/sj.onc.1204429 [DOI] [PubMed] [Google Scholar]
- De Groot C.O., Jelesarov I., Damberger F.F., Bjelić S., Schärer M.A., Bhavesh N.S., Grigoriev I., Buey R.M., Wüthrich K., Capitani G., et al. 2010. Molecular insights into mammalian end-binding protein heterodimerization. J. Biol. Chem. 285:5802–5814 10.1074/jbc.M109.068130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V.M., Shapiro I., Aldridge B., Sorger P.K. 2006. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 25:2814–2827 10.1038/sj.emboj.7601168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty E.J., Partridge M.A., Gundersen G.G. 2005. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7:581–590 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia S.M., Akhmanova A. 2010. Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int. Rev. Cell Mol. Biol. 285:1–74 10.1016/B978-0-12-381047-2.00001-3 [DOI] [PubMed] [Google Scholar]
- Green R.A., Wollman R., Kaplan K.B. 2005. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell. 16:4609–4622 10.1091/mbc.E05-03-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Hüls D., Storchova Z., Niessing D. 2012. Post-translational modifications regulate assembly of early spindle orientation complex in yeast. J. Biol. Chem. 287:16238–16245 10.1074/jbc.M112.347872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Toedt G., Montenegro Gouveia S., Davey N.E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L.B., Bezstarosti K., et al. 2012. A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22:1800–1807 10.1016/j.cub.2012.07.047 [DOI] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J.V. 1999. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 146:1033–1044 10.1083/jcb.146.5.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.S., Veltman D.M., Georgiou M., Baum B., Insall R.H. 2010. SCAR/WAVE is activated at mitosis and drives myosin-independent cytokinesis. J. Cell Sci. 123:2246–2255 10.1242/jcs.063735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G.G., Akhmanova A. 2005. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell. 16:5334–5345 10.1091/mbc.E05-07-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., De Groot C.O., Grigoriev I., Gouveia S.M., Munteanu E.L., Schober J.M., Honnappa S., Buey R.M., Hoogenraad C.C., Dogterom M., et al. 2009. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184:691–706 10.1083/jcb.200807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y.A., Huang F., Geyer M., Daneshjou N., Garcia A., Idalino L., Kreutz B., Mehta D., Malik A.B. 2012. VE-cadherin signaling induces EB3 phosphorylation to suppress microtubule growth and assemble adherens junctions. Mol. Cell. 48:914–925 10.1016/j.molcel.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Lee L., Tirnauer J.S., Li J., Schuyler S.C., Liu J.Y., Pellman D. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 287:2260–2262 10.1126/science.287.5461.2260 [DOI] [PubMed] [Google Scholar]
- Lee T., Langford K.J., Askham J.M., Brüning-Richardson A., Morrison E.E. 2008. MCAK associates with EB1. Oncogene. 27:2494–2500 10.1038/sj.onc.1210867 [DOI] [PubMed] [Google Scholar]
- Maiato H., Hergert P.J., Moutinho-Pereira S., Dong Y., Vandenbeldt K.J., Rieder C.L., McEwen B.F. 2006. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 115:469–480 10.1007/s00412-006-0076-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi M.G., Ecsedy J.A., Meetze K.A., Balani S.K., Burenkova O., Chen W., Galvin K.M., Hoar K.M., Huck J.J., LeRoy P.J., et al. 2007. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc. Natl. Acad. Sci. USA. 104:4106–4111 10.1073/pnas.0608798104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., Tsukita S. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868 10.1016/S0960-9822(00)00600-X [DOI] [PubMed] [Google Scholar]
- Mitchison T.J. 1992. Actin based motility on retraction fibers in mitotic PtK2 cells. Cell Motil. Cytoskeleton. 22:135–151 10.1002/cm.970220207 [DOI] [PubMed] [Google Scholar]
- Montenegro Gouveia S., Leslie K., Kapitein L.C., Buey R.M., Grigoriev I., Wagenbach M., Smal I., Meijering E., Hoogenraad C.C., Wordeman L., et al. 2010. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 20:1717–1722 10.1016/j.cub.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Moore A.T., Rankin K.E., von Dassow G., Peris L., Wagenbach M., Ovechkina Y., Andrieux A., Job D., Wordeman L. 2005. MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 169:391–397 10.1083/jcb.200411089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L., Adames N.R., Murphy M.D., Shields C.R., Cooper J.A. 1998. A cytokinesis checkpoint requiring the yeast homologue of an APC-binding protein. Nature. 393:487–491 10.1038/31014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Koyama K., Murata Y., Morito M., Akiyama T., Nakamura Y. 2000. EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene. 19:210–216 10.1038/sj.onc.1203308 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Zhou X.Z., Lu K.P. 2001. Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr. Biol. 11:1062–1067 10.1016/S0960-9822(01)00297-4 [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T.J. 2004. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 15:2895–2906 10.1091/mbc.E04-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellinen T., Tuomi S., Arjonen A., Wolf M., Edgren H., Meyer H., Grosse R., Kitzing T., Rantala J.K., Kallioniemi O., et al. 2008. Integrin trafficking regulated by Rab21 is necessary for cytokinesis. Dev. Cell. 15:371–385 10.1016/j.devcel.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Pereira A.J., Maiato H. 2010. Improved kymography tools and its applications to mitosis. Methods. 51:214–219 10.1016/j.ymeth.2010.01.016 [DOI] [PubMed] [Google Scholar]
- Pugacheva E.N., Roegiers F., Golemis E.A. 2006. Interdependence of cell attachment and cell cycle signaling. Curr. Opin. Cell Biol. 18:507–515 10.1016/j.ceb.2006.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Rogers G.C., Sharp D.J., Vale R.D. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884 10.1083/jcb.200202032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Sanhaji M., Friel C.T., Kreis N.N., Krämer A., Martin C., Howard J., Strebhardt K., Yuan J. 2010. Functional and spatial regulation of mitotic centromere-associated kinesin by cyclin-dependent kinase 1. Mol. Cell. Biol. 30:2594–2607 10.1128/MCB.00098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.M., McIntosh J.R. 1987. Interzone microtubule behavior in late anaphase and telophase spindles. J. Cell Biol. 105:875–886 10.1083/jcb.105.2.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M.P., Helenius J., Toyoda Y., Ramanathan S.P., Muller D.J., Hyman A.A. 2011. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 469:226–230 10.1038/nature09642 [DOI] [PubMed] [Google Scholar]
- Straube A., Merdes A. 2007. EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr. Biol. 17:1318–1325 10.1016/j.cub.2007.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland L.I., Wen Y., Gundersen G.G., Burgess D.R. 2005. Interaction between EB1 and p150glued is required for anaphase astral microtubule elongation and stimulation of cytokinesis. Curr. Biol. 15:2249–2255 10.1016/j.cub.2005.10.073 [DOI] [PubMed] [Google Scholar]
- Su L.K., Qi Y. 2001. Characterization of human MAPRE genes and their proteins. Genomics. 71:142–149 10.1006/geno.2000.6428 [DOI] [PubMed] [Google Scholar]
- Su L.K., Burrell M., Hill D.E., Gyuris J., Brent R., Wiltshire R., Trent J., Vogelstein B., Kinzler K.W. 1995. APC binds to the novel protein EB1. Cancer Res. 55:2972–2977 [PubMed] [Google Scholar]
- Sun L., Gao J., Dong X., Liu M., Li D., Shi X., Dong J.T., Lu X., Liu C., Zhou J. 2008. EB1 promotes Aurora-B kinase activity through blocking its inactivation by protein phosphatase 2A. Proc. Natl. Acad. Sci. USA. 105:7153–7158 10.1073/pnas.0710018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Kapoor T.M. 2011. Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proc. Natl. Acad. Sci. USA. 108:16675–16680 10.1073/pnas.1106748108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Medema R.H. 2011. Localized Aurora B activity spatially controls non-kinetochore microtubules during spindle assembly. Chromosoma. 120:599–607 10.1007/s00412-011-0334-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Racine V., Pépin A., Piel M., Chen Y., Sibarita J.B., Bornens M. 2005. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7:947–953 10.1038/ncb1307 [DOI] [PubMed] [Google Scholar]
- Tirnauer J.S., Grego S., Salmon E.D., Mitchison T.J. 2002. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626 10.1091/mbc.02-04-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26:1487–1498 10.1038/sj.emboj.7601599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliev J.M., Omelchenko T., Gelfand I.M., Feder H.H., Bonder E.M. 2004. Rho overexpression leads to mitosis-associated detachment of cells from epithelial sheets: a link to the mechanism of cancer dissemination. Proc. Natl. Acad. Sci. USA. 101:12526–12530 10.1073/pnas.0404723101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Worthylake R.A., Liu B.P., Burridge K., Salmon E.D. 1999. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat. Cell Biol. 1:45–50 10.1038/9018 [DOI] [PubMed] [Google Scholar]
- Wen Y., Eng C.H., Schmoranzer J., Cabrera-Poch N., Morris E.J., Chen M., Wallar B.J., Alberts A.S., Gundersen G.G. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6:820–830 10.1038/ncb1160 [DOI] [PubMed] [Google Scholar]
- Wheatley S.P., Wang Y. 1996. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135:981–989 10.1083/jcb.135.4.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakita Y., Totsukawa G., Yamashiro S., Fry D., Zhang X., Hanks S.K., Matsumura F. 1999. Dissociation of FAK/p130(CAS)/c-Src complex during mitosis: role of mitosis-specific serine phosphorylation of FAK. J. Cell Biol. 144:315–324 10.1083/jcb.144.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.