Abstract
Mitochondria continually change shape through the combined actions of fission, fusion, and movement along cytoskeletal tracks. The lengths of mitochondria and the degree to which they form closed networks are determined by the balance between fission and fusion rates. These rates are influenced by metabolic and pathogenic conditions inside mitochondria and by their cellular environment. Fission and fusion are important for growth, for mitochondrial redistribution, and for maintenance of a healthy mitochondrial network. In addition, mitochondrial fission and fusion play prominent roles in disease-related processes such as apoptosis and mitophagy. Three members of the Dynamin family are key components of the fission and fusion machineries. Their functions are controlled by different sets of adaptor proteins on the surface of mitochondria and by a range of regulatory processes. Here, we review what is known about these proteins and the processes that regulate their actions.
The main mitochondrial fission and fusion proteins are members of the Dynamin family. Their functions are controlled by adaptor proteins and by a range of regulatory processes (e.g., protein modification and proteolysis).
Mitochondrial movement and fission were first observed with light microscopy almost 100 years ago (Lewis and Lewis 1914). For a long time, these observations remained something of a curiosity and they were all but forgotten when electron microscopy popularized the idea that mitochondria exist as isolated sausage-shaped organelles floating in a sea of cytoplasm. Renewed appreciation for mitochondrial dynamics emerged some 20 or 30 years ago when technological advances made it much easier to track mitochondria in live cells. Careful observations, first with phase contrast microscopy, then with vital dyes and finally with targeted fluorescent proteins, showed that mitochondria continually divide and fuse, even in resting cells (Johnson et al. 1981; Bereiter-Hahn and Voth 1994; Rizzuto et al. 1996). Their lengths are determined by the balance between fission and fusion. Mitochondrial morphologies can change dramatically by shifting this balance. In some cells they fuse together, forming a single closed network, whereas in other cells or under different circumstances mitochondria convert into large numbers of small fragments. Because of these morphological changes mitochondria are now known to be very dynamic.
The importance of frequent mitochondrial fission and fusion events for cell survival was also not fully appreciated until fairly recently. Obvious reasons, such as accommodating cell growth, cell division, and the redistribution of mitochondria during differentiation, did not fully explain why mitochondria fuse nor did they explain the high frequencies of these occurrences. However, in more recent years, the biological relevance of these phenomena has become clear with the discovery of human diseases that are caused by mutations in fission and fusion proteins and the discovery of numerous connections with apoptosis and mitophagy (Westermann 2010; Chan 2012; Nunnari and Suomalainen 2012; Youle and van der Bliek 2012). Mitochondrial fission and fusion are now considered cornerstones for cell survival because of their contributions to health and disease.
DYNAMIN FAMILY MEMBERS THAT MEDIATE MITOCHONDRIAL FISSION AND FUSION
The main mitochondrial fission and fusion proteins are members of the Dynamin family (Fig. 1). The first mitochondrial Dynamin was named Mgm1, because mutations in the yeast gene cause a mitochondrial genome maintenance defect (Jones and Fangman 1992). This mitochondrial DNA (mtDNA) distribution defect was later shown to be a secondary consequence of defects in mitochondrial inner membrane fusion (Wong et al. 2000). Localization studies also showed that Mgm1 is anchored in the mitochondrial inner membrane with the bulk of the protein facing the mitochondrial inner membrane space (Wong et al. 2000). A human homolog of Mgm1 was discovered through sequence homologies and through genetic mapping of a late onset neurodegenerative eye disease, named Dominant Optic Atrophy (DOA). The human homolog was named Opa1 for the optic atrophy defects (Alexander et al. 2000; Delettre et al. 2000).
Figure 1.
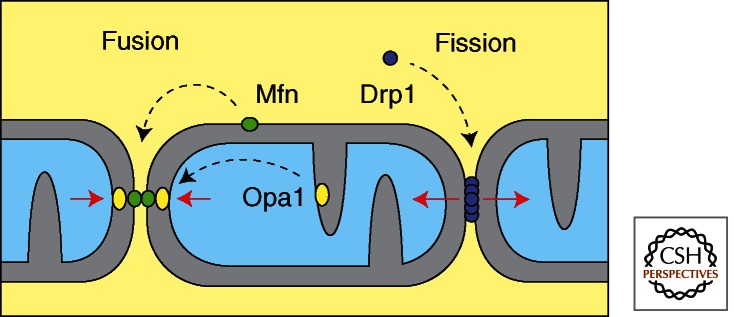
Functions of the mitochondrial Dynamin family members. Mitofusins mediate mitochondrial outer membrane fusion in mammals. Opa1 mediates mitochondrial inner membrane fusion. Drp1, which cycles between the cytosol and the mitochondrial outer membrane, mediates mitochondrial fission.
Fusion of mitochondrial outer membranes is mediated by a different set of Dynamin family members. The first of these was discovered in Drosophila sperm cells, where it was named fuzzy onions for the onion-like and fuzzy appearance of unfused mitochondria in electron micrographs of these mutants (Hales and Fuller 1997). A second mitochondrial outer membrane protein, named Marf, was later shown to mediate fusion in other cell types (Dorn et al. 2011). The sequences of Marf and fuzzy onions are similar, but their expression patterns are different so the mutants have different phenotypes: fuzzy onions mutants are sterile whereas Marf mutants are lethal (Dorn et al. 2011). Yeast and Caenorhabditis elegans each have only one fuzzy onions or Marf homolog (Fzo1p and FZO-1, respectively) (Hermann et al. 1998; Rapaport et al. 1998). Mammals do have two homologs (the Mitofusins Mfn1 and Mfn2), but those are often expressed in the same cells (Santel and Fuller 2001; Chen et al. 2003). Although some functional differences between Mfn1 and Mfn2 have been observed, both of these proteins are also able to support mitochondrial fusion by themselves, suggesting that they fulfill partially redundant functions in this process (Chen et al. 2003).
Mitochondrial outer membrane fusion is almost always coordinated with inner membrane fusion. There are, however, some instances in which outer membrane fusion can occur without concomitant inner membrane fusion as, for example, when mutations or loss of membrane potential block inner membrane fusion whereas outer membrane fusion can still occur (Olichon et al. 2003). Localized loss of membrane potential and its effects on inner membrane fusion may explain why the matrix compartments in larger mitochondrial networks sometimes have breaks, whereas their outer membranes appear to be fully fused (Twig et al. 2006). The physiological relevance of these observations has not yet been explored.
Unlike the fusion machinery, which has both outer and inner membrane components, it would appear that most higher eukaryotes and yeast use a single Dynamin family member for mitochondrial fission. This Dynamin-like protein is largely cytosolic, but can cycle on and off of the mitochondrial outer membrane. Homologs of this protein were discovered with genetic screens for mitochondrial morphology defects in yeast (yeast Dnm1) and through functional analyses of different Dynamin family members in C. elegans and mammalian cells (Bleazard et al. 1999; Labrousse et al. 1999; Smirnova et al. 2001). The mammalian protein was alternatively named Drp1, Dnm1L, DLP1, Dymple, and DVLP1 (Shin et al. 1997; Imoto et al. 1998; Kamimoto et al. 1998; Smirnova et al. 1998; Yoon et al. 1998). Here we refer to it as Drp1, because this is the most commonly used name.
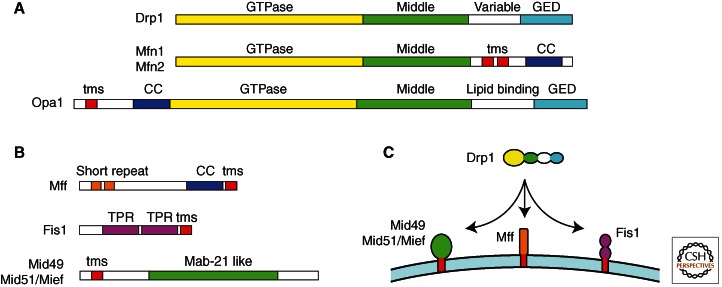
The Drp1 homologs have a domain structure that is characteristic of the Dynamin family (Fig. 2). Each one has a GTPase domain followed by a middle domain, a variable domain, and a GTPase effector domain (GED) (Praefcke and McMahon 2004). The variable domain, also referred to as insert B, is generally required for binding to the target membrane. In the fission Dynamins, this domain binds to adaptor proteins on the mitochondrial surface. The lipid-binding domain in the fusion proteins, Mgm1 and Opa1, is specific for cardiolipin (Meglei and McQuibban 2009), whereas the outer membrane fusion proteins, Fzo1 and Mitofusins, each have two trans-membrane segments that serve as membrane anchors (Hoppins et al. 2007).
Figure 2.
Schematic diagrams of fission and fusion proteins. (A) The main Dynamin family members in mammals, Drp1, Mfn1, Mfn2, and Opa1 are shown with their different protein domains. Each one has a GTPase domain, a middle domain, and a GED, which together form the canonical Dynamin structure. The GED in Mitofusins is not as well conserved, but it still has coiled-coil segments, suggesting that it can participate in multimeric assembly. The variable domain in Drp1, which most likely serves as a mitochondrial targeting sequence, is replaced by trans-membrane segments (tms) in the Mitofusins and by a cardiolipin binding domain in Opa1. (B) Adaptor proteins that can bind to drp1 on the surface of mitochondria. Each one has a membrane anchor (tms), but they have different protein interaction domains, suggesting that they are functionally distinct. (C) Illustration of the multiplicity of Drp1 receptors on the surface of mammalian mitochondria.
The conserved middle domain and the GED of Dynamin family members fold back on each other to form a stalk (van der Bliek and Payne 2010). As a result, the GTPase domain is directed away from the membrane. Biophysical, cryo-electron microscopy, and X-ray crystallography studies showed that Dynamin family members assemble into multimeric spirals through a series of binding interactions between the stalks of different subunits (Chappie et al. 2010; Gao et al. 2010). Once a spiral completes a turn, contacts between the GTPase domains of adjacent rungs trigger GTP hydrolysis (van der Bliek and Payne 2010). This enzymatic activity drives conformational changes within the multimers causing constriction of the spiral (Mears et al. 2011). Constriction driven by the mitochondrial fission Dynamins severs both mitochondrial inner and outer membranes.
Electron microscopy (EM) studies have provided insight into the mechanisms of mitochondrial fission. Like Dynamin, the yeast Drp1 homolog Dnm1 can tubulate membranes by assembling into large multimeric spirals that wrap around those tubules (Ingerman et al. 2005). There are, however, two striking differences with the Dynamin spirals. First, the diameter is much larger. Instead of the 50 nm for Dynamin, these spirals have an average diameter of 120 nm (Ingerman et al. 2005; Mears et al. 2011). This large diameter is enough to wrap around a double membrane structure, which would be formed by constrictions in mitochondria when both inner and outer membranes are still intact. How might the spiral generate the additional force needed to sever both membranes? Cryo-EM data show that Dnm1 forms “two-start” helices, perhaps accommodating the larger initial diameter, but it might also provide more force because the two intertwined spirals would have more contact points (Mears et al. 2011).
Although less is known about the mechanisms of fusion, genetic data show that the fusion Dynamins are needed on both of the opposing membranes. Current models suggest that the fusion Dynamins from opposing membranes dimerize and then pull the membranes together with a SNARE-like mechanism (Koshiba et al. 2004). Alternative mechanisms, however, have not yet been ruled out. It remains possible that the fusion Dynamins assemble into spirals on membranes like the other Dynamin family members. Protruding membrane tubules formed by this process might then be mated by contacts between opposing Dynamin spirals (Devay et al. 2009). Forced contacts between the convex lipid surfaces at the tips of the Dynamin spirals could be fusogenic, similar to the forced contacts between opposing convex membranes during SNARE-mediated fusion. Time will tell whether fusion Dynamins use either of these mechanisms or perhaps use a totally different mechanism.
Isolated mitochondria have been used to study biophysical requirements for mitochondrial inner and outer membrane fusion processes (Meeusen et al. 2006). As expected, both processes require GTP, but only inner membrane fusion requires membrane potential. The reasons for this last requirement are not yet clear. More importantly, it was shown that fusion Dynamins are required on both opposing membranes and that these Dynamins can form large protein complexes during the fusion process (Meeusen et al. 2006). There are, as of yet, no good in vitro assays to study mitochondrial fission, but the analogies with classical Dynamin, as well as EM studies of Drp1 homologs on membranes and studies with selective inhibitors have been remarkably informative. Several drugs, like Dynasore, inhibit both Dynamin and Drp1 (Macia et al. 2006), but there are also more selective drugs, like Mdivi-1, which only inhibit Drp1 homologs (Cassidy-Stone et al. 2008). Mdivi-1 has been used to study the role of Drp1 in cytochrome c release during apoptosis and it may represent a new class of drugs with therapeutic potential (Tanaka and Youle 2008).
ADAPTOR PROTEINS
Genetic screens for mutations that suppress fusion defects led to the discovery of additional fission proteins in yeast. One of the fission proteins that came out of these screens is Fis1, which has two tetratrico peptide repeat (TPR) domains and a carboxy-terminal hydrophobic segment that anchors this protein in the mitochondrial outer membrane (Mozdy et al. 2000). The second protein discovered with the genetic screens was Mdv1, which is a cytosolic protein with a poorly defined amino-terminal segment followed by a series of withdrawal (protein motif of approximately 40 amino acids ending with trypophan (W) and aspartic acid (D) [WD]) motifs (Tieu et al. 2002; Cerveny and Jensen 2003). The amino terminus of Mdv1 binds to Fis1 on the surface of mitochondria, whereas the WD repeats form a β propeller structure that binds to and recruits the yeast Drp1 homolog Dnm1 to the surface of mitochondria (Koirala et al. 2010). Yeast has a second Mdv1-like protein, Caf4, which is partially redundant with Mdv1 (Griffin et al. 2005). These proteins are the only known adaptors for Dnm1 on mitochondria in yeast.
The situation in higher eukaryotes is more confusing. Higher eukaryotes have no obvious Mdv1 homologs, but they do have Fis1 homologs. The functions of Fis1 in higher eukaryotes were, however, brought into question by genetic data showing that Fis1 is not required for fission (Otera et al. 2010). A number of alternative receptors for Drp1 have been proposed. Among these are Mff, Mid49, Mid51/Mief, and GDAP1 (Fig. 2). Mff was first isolated with a small interfering RNA (siRNA) screen for mitochondrial morphology defects (Gandre-Babbe and van der Bliek 2008). It is a relatively small protein with a short amino-terminal repeat, a coiled-coil segment, and a carboxy-terminal hydrophobic segment that anchors the protein in the mitochondrial outer membrane, similar to the tail anchor of Fis1. Subsequent work with a mistargeted protein in mammalian cells showed that Mff is sufficient to act as a Drp1 receptor (Otera et al. 2010). Knockdown and overexpression studies of Mid49, Mid51/Mief, and GDAP1 have independently shown effects on mitochondrial fission, suggesting that there are multiple ways to control fission (Niemann et al. 2005; Palmer et al. 2011; Zhao et al. 2011). This was confirmed in a recent paper showing that Fis1, Mff, Mid49, and Mid51/Mief act in a partially redundant way to promote Drp1-dependent fission (Loson et al. 2013). Interestingly, Fis1 and Mff are also present on peroxisomes, consistent with results showing that peroxisome fission is mediated by Drp1 in higher eukaryotes (Gandre-Babbe and van der Bliek 2008). It seems likely that this part of the fission machinery can act independently. It is not yet clear whether the other fission adaptors or receptors take part in redundant pathways or act at different steps in the same pathway.
No similar collection of adaptors or receptors has been found for mitochondrial fusion proteins with the possible exception of Ugo1 in yeast. Ugo1 is an integral mitochondrial outer membrane protein that coIPs with the inner and outer membrane fusion proteins (Sesaki and Jensen 2004). It may help coordinate the two membrane fusion events, but higher eukaryotes have no obvious Ugo1 homologs. The proapoptotic BH3 protein Bax is required to maintain Mitofusin tetramers in mammalian cells, but it is unclear whether Bax affects Mitofusin assembly or is directly involved in the fusion process (Karbowski et al. 2006). In addition, a cytosolic medium-chain dehydrogenase/reductase named MIB for Mitofusin binding protein was shown to selectively inhibit the functions of Mfn1 causing mitochondrial elongation with knockdown and fragmentation when it was overexpressed in mammalian cells (Eura et al. 2006). Last, it was found that the lipid metabolizing enzyme mitoPLD functions downstream from Mitofusins during mitochondrial fusion, potentially providing a link between fission and fusion (Choi et al. 2006). The role of this protein is discussed in more detail in the section on the regulation of fission. It remains possible that other, as yet undiscovered, proteins control fusion, but so far the data suggest that the control of fusion is much simpler than the control of fission.
REGULATION OF FISSION BY PHOSPHORYLATION AND UBIQUITINATION
Mitochondrial fission proteins are regulated by a range of protein modifications, including phosphorylation, ubiquitination, sumoylation, and nitrosylation. Different kinases control the activities of Drp1 by phosphorylating this protein at three different sites. These sites include Ser616, which is phosphorylated by protein kinase C (PKC) δ, Rock kinase, CDK1/Cyclin B or CAMK-Ia (Taguchi et al. 2007; Han et al. 2008; Qi et al. 2011; Wang et al. 2012), and Ser637, which is phosphorylated by protein kinase A (PKA) (Chang and Blackstone 2007; Cribbs and Strack 2007). Phosphorylation of Ser616 is likely to activate fission, because it promotes binding to other fission proteins, whereas phosphorylation of Ser637 could be an inactivating step (Chang and Blackstone 2010). The third site in Drp1 is Ser693, which is phosphorylated by GSK3β to inhibit mitochondrial fission during apoptosis (Chou et al. 2012). This residue is in the GED, where it most likely affects Drp1 oligomerization or GTP hydrolysis.
Ser616 and Ser637 are both toward the end of the variable domain. It is likely that the variable domain is in close contact with mitochondrial outer membrane proteins, so phosphorylation at these sites may regulate the association of Drp1 with mitochondria by changing the contacts with mitochondrial outer membrane proteins or with the membrane itself. This concept is supported by the enhanced binding of Drp1 to Fis1 when Ser616 is phosphorylated by CAMK-I (Han et al. 2008), followed by the putative inactivation step by PKA, which is localized to mitochondria through the A-kinase activating protein (AKAP) Rab32 in mammals (Alto et al. 2002; Merrill et al. 2011). The phosphate on Ser637 is removed by Calcineurin (PP2A), which is a calcium-activated cytosolic phosphatase (Cribbs and Strack 2007; Cereghetti et al. 2008). These observations suggest that phosphorylation regulates the cycling of Drp1 between cytosol and the mitochondrial outer membrane.
After Drp1 is primed for another round of fission by phosphatases, it needs a trigger to initiate the fission event, for example, by the phosphorylation of Ser616. Because PKC is one of the kinases that can phosphorylate Ser616 and different PKC isoforms are generally activated by calcium and diacylglycerol, this trigger may react to localized signals. Diacylglycerol is produced on mitochondria by Lipin1, which can be activated by phosphatidic acid on mitochondria (Huang et al. 2011). Phosphatidic acid in turn is produced by mitoPLD during mitochondrial fusion (Choi et al. 2006). These different steps suggest a cycle in which fusion is quickly followed by fission through the sequential actions of mitoPLD, Lipin1, PKC, and Drp1. This sequence fits remarkably well with live cell imaging studies in which it was shown that fission often follows fusion, more or less at the same place and within 20 min (Twig et al. 2008). It is tempting to speculate that these live images show results of the regulatory pathway outlined in this paragraph.
Alternative fission-inducing pathways may also exist because the conditions that activate some of the other kinases that phosphorylate Drp1 do not necessarily come from fusion. There is, for example, massive fragmentation when cells are treated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or with apoptosis-inducing drugs (Frank et al. 2001; Ishihara et al. 2003). This fragmentation results from all but complete mobilization of Drp1 and a large number of fission events without corresponding fusion events. It has been suggested that calcium release from depolarized mitochondria may activate Calcineurin (Cribbs and Strack 2007; Cereghetti et al. 2008), whereas apoptosis-induced fission may rely on calcium release from the ER (Scorrano et al. 2003). Other pathways may exist, but they all seem to lead to Drp1 at the center of the mitochondrial fission apparatus.
Several other forms of protein modification may regulate mitochondrial fission. The membrane-bound E3 ligase March5 may regulate Drp1 through ubiquitination (Karbowski et al. 2007). Another membrane-bound E3 ligase, Mulan or Mapl, was proposed to regulate Drp1 through sumoylation (Braschi et al. 2009), and NO-dependent nitrosylation of a Drp1 has been proposed to activate fission in a way that promotes neurodegeneration in Alzheimer’s patients (Cho et al. 2009). These alternative forms of regulation are still somewhat controversial. March5 and Mulan were both reported to have other targets, whereas the effects of nitrosylation were brought into question by independent investigators (Nakamura et al. 2006; Bossy et al. 2010). Further investigations may help determine which of these mechanisms are physiologically relevant regulators of Drp1.
COORDINATION OF MITOCHONDRIAL FISSION WITH THE CYTOSKELETON, ER, AND ITS ROLE IN APOPTOSIS
Frequent close contacts between mitochondria and ER in mitochondrion-associated membranes (MAMs) have been studied for years (Rowland and Voeltz 2012). They are known to contribute to lipid and calcium transfer between these organelles, but their relevance for mitochondrial fission was only recently discovered through high-resolution light microscopy of live cells (Friedman et al. 2011). These images showed that ER tubules often cross paths with mitochondria at points of impending fission or they wrap around mitochondria at those points (Friedman et al. 2011). In this report, it was also shown that the contacts between ER and mitochondria are made and that mitochondria are still constricted at those sights when the recruitment of Drp1 to mitochondria is prevented by siRNA for Drp1 or for Mff. These results suggested that ER might play an active role during the earliest stages of fission, well before Drp1-mediated severing of the membrane. This suggestion was nicely confirmed by the recent discovery that the ER protein INF-2 can promote mitochondrial fission by inducing constriction before Drp1 is recruited to mitochondria (Korobova et al. 2013). INF-2 is a Formin-like protein that triggers actin filament assembly. Actin was previously shown to be important for mitochondrial fission through studies with inhibitors (De Vos et al. 2005), but the connections with the fission apparatus were only now coming into focus.
Evidence for contributions from ER to mitochondrial fission also comes from earlier work on apoptosis. Cytochrome c is released through Bax-lined pores at sites of Drp1-mediated mitochondrial fission (Montessuit et al. 2010). Interestingly, Bax also affects calcium release from the ER, which might then affect Drp1-mediated fission, and it induces cleavage of the ER protein Bap31 (Iwasawa et al. 2011). Bap31 has been shown to be enriched in MAMs and it can co-immunoprecipitate with Fis1, suggesting that it is part of a larger fission complex (Simmen et al. 2005; Iwasawa et al. 2011). Although the molecular details have not yet been worked out, these studies suggest that the MAM is somehow involved in the regulation of cytochrome c release during apoptosis and that Drp1 plays a central role in this process. Because apoptosis is an end point that does not require functional organelles, this process may have co-opted certain parts of the MAM in abnormal ways.
There may be other connections between the MAM and mitochondrial fission or fusion. The yeast Miro homolog Gem1p was shown to regulate contacts between the ER and mitochondria (Kornmann et al. 2011); but in higher eukaryotes, Miro regulates mitochondrial transport along microtubules through interactions with dynein and the adaptor Milton (Glater et al. 2006). It is not yet clear to what extent these functions overlap. The small GTP-binding protein Rab32, which recruits PKA for inactivation of Drp1, was also shown to regulate the MAM, perhaps through its ability to recruit PKA, but the targets of these proteins on the ER are not yet known (Bui et al. 2010). Although this area is still in its infancy, it is already clear that the MAM plays a central role in mitochondrial fission.
REGULATION OF MITOCHONDRIAL FUSION BY PROTEOLYTIC CLEAVAGE AND UBIQUITINATION
Mitochondrial fusion proteins are regulated by proteolysis and ubiquitination. The predominant form of regulation for mitochondrial outer membrane fusion Dynamins is inactivation through ubiquitin-mediated degradation, whereas the functions of inner membrane fusion Dynamins are altered by proteolytic cleavage at a select few sites. The ubiquitin-mediated degradation of yeast Fzo1 is triggered by the actions of two mitochondrial outer membrane F-box proteins (Fritz et al. 2003; Neutzner and Youle 2005; Durr et al. 2006). Fzo1 accumulates when either one of these F-box proteins is mutated, suggesting that they promote Fzo1 turnover. Fzo1 is degraded by the proteasome as part of the yeast mating response, suggesting that regulation through turnover is physiologically relevant (Neutzner and Youle 2005). In higher eukaryotes, ubiquitin-mediated degradation of the mitochondrial fusion machinery is induced by stress. When mitochondria lose membrane potential, Mitofusins are degraded by PTEN-induced putative kinase 1 (Pink1)- and Parkin-mediated ubiquitination during mitophagy (Youle and Narendra 2011). After ubiquitination, these proteins are extracted by the AAA protein p97 and then degraded by the proteasome (Tanaka et al. 2010), as discussed in more detail elsewhere in this issue. The degradation of Mfn2 can also be induced by other kinds of stress, such as Doxorubicin, which triggers ubiquitination by the E3 ligase Huwe1 (Leboucher et al. 2012), or during muscle wasting with increased Mfn2 ubiquitination by the E3 ligase Mulan (Lokireddy et al. 2012). These results show tight temporal control of Mitofusin levels through different stress-induced pathways.
The mitochondrial inner membrane fusion dynamins, Mgm1 in yeast, EAT-3 in C. elegans, and Opa1 in mammals are also proteolytically cleaved but the effects are very different. These proteins each have a mitochondrial targeting sequence that is cleaved on import, whereas the bulk of the protein remains exposed to the intermembrane space (Wong et al. 2000; Olichon et al. 2002). In yeast, approximately half of the protein is further cleaved by the mitochondrial inner membrane rhomboid protease Pcp1 to generate a short and a long form (Esser et al. 2002; Herlan et al. 2003; Sesaki et al. 2003). The long form retains a membrane anchor at the amino terminus, whereas the short form loses this membrane anchor. The ratio of short to long forms is determined by the energy status of mitochondria (Herlan et al. 2004). Although the functional consequences of this form of regulation are not yet known, both short and long forms of Mgm1 are needed for fusion between mitochondria. C. elegans and Drosophila homologs of Mgm1 are also cleaved by a rhomboid protease (van der Bliek AM, unpubl.) (McQuibban et al. 2006). It was initially reported that mammalian Opa1 is also cleaved by a rhomboid protease (Cipolat et al. 2006), but subsequent experiments with knockout cells for this rhomboid did not show an effect on Opa1 (Griparic et al. 2007). Instead, Opa1 is cleaved by a different set of proteases in a complex pattern governed by alternative splicing of the Opa1 mRNA.
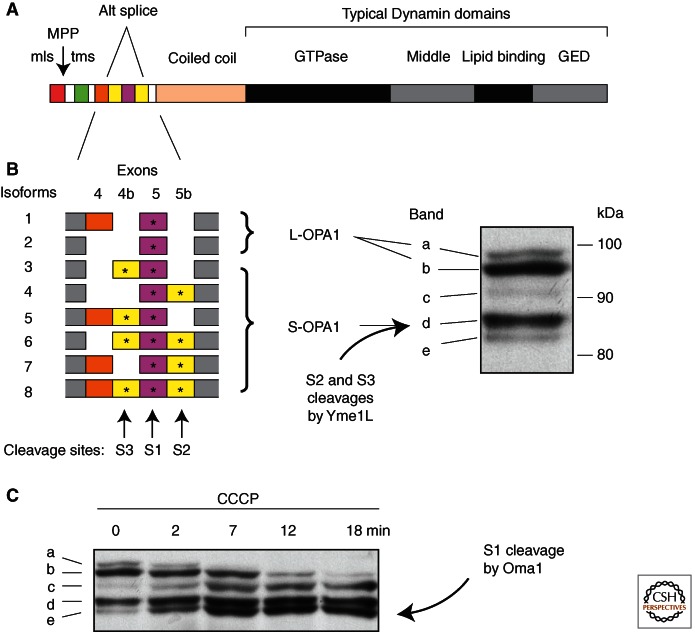
Mammals have eight different Opa1 isoforms generated by alternative splicing (Olichon et al. 2007). Differences between the isoforms result from the presence or absence of several short exons near the amino terminus. These exons replace the single rhomboid cleavage site in lower eukaryotes with three different proteolytic cleavage sites in mammals. The S1 cleavage site is present in all isoforms, whereas S2 and S3 cleavage sites are present in approximately half of the isoforms (Fig. 3). Those with S2 or S3 sites are constitutively cleaved by the large intermembrane space AAA-protease Yme1L (Griparic et al. 2007; Song et al. 2007). As a result, mammalian cells typically have a 50/50 ratio of short (Yme1L cleavage products) and long (uncleaved) forms of Opa1, similar to the distribution in lower eukaryotes. However, mammalian cells also have an S1 site, which is present in all isoforms and is only cleaved when mitochondria lose membrane potential, have low levels of ATP, or lose some of the other quality-control mechanisms (mutations in the AFG3L2 protease or the Prohibitin chaperone) (Baricault et al. 2007; Merkwirth et al. 2008; Ehses et al. 2009). This inducible cleavage is mediated by the small inner membrane Zn-protease Oma1 (Ehses et al. 2009; Head et al. 2009). Oma1 cleaves all remaining long forms of Opa1 within minutes after treatment with CCCP or other drugs that affect mitochondrial ATP production (Fig. 3). Oma1-mediated cleavage of Opa1 prevents inner membrane fusion and it may cause some changes in cristae architecture, well before other stress-management systems, such as the Pink1 Parkin pathway, are activated. The reasons for first shutting down the inner membrane fusion machinery are not yet clear.
Figure 3.
Proteolytic cleavage sites in mammalian Opa1. (A) Protein domains in Opa1. The amino terminus has a mitochondrial leader sequence, followed by a mitochondrial processing peptidase cleavage site (MPP), a trans-membrane segment (tms) and several alternatively spliced exons. (B) Alternatively spliced exons have three more cleavage sites. S2 and S3 sites are constitutively cleaved by Yme1L, giving rise to S-Opa1 (band d). Isoforms lacking exons 4b and 5b are not normally cleaved, giving rise to L-Opa1 (bands a and b). (C) Oma1 cleaves the S1 site in isoforms that were not already cleaved by Yme1L, but only when mitochondria lose membrane potential. Bands a and b disappear, giving rise to the intermediate form band c and the final product band e. (The blots were from Griparic et al. 2007; reprinted, with permission, from the authors.)
ADDITIONAL ROLES OF FUSION PROTEINS
The phenotypic effects of mutations in mitochondrial inner and outer membrane fusion proteins are strikingly different. This was noted for yeast Fzo1 and Mgm1, where Mgm1 appears to have additional functions in helping to preserve mtDNA and cristae morphology (Guan et al. 1993; Meeusen et al. 2006). Differences in viability and tissue-specific effects have also been noted with mutations in the analogous C. elegans and Drosophila proteins (Kanazawa et al. 2008; Yarosh et al. 2008). Whereas homozygous mutations in mice are lethal, there are differences in the most severely affected stages and in heterozygous animals (Chen et al. 2003; White et al. 2009). Humans with disease-causing mutations in Mitofusins or Opa1 are generally heterozygous, but the diseases caused by these mutations are different: mutations in Opa1 cause optic atrophy through progressive loss of retinal ganglion cells, whereas mutations in Mfn2 cause a form of peripheral neuropathy (Charcot-Marie-Tooth disease CMT2A) (Chan 2012).
Mammalian cell culture experiments and work with model organisms, such as C. elegans, Drosophila, and mice, shed light on the functional differences between mitochondrial inner and outer membrane fusion proteins. In mammals, loss of both inner and outer membrane fusion proteins promotes cytochrome c release during apoptosis, but the effects of losing the inner membrane fusion protein Opa1 are much more severe (Olichon et al. 2003; Arnoult et al. 2005). Mutations in C. elegans and Drosophila Opa1 homologs also severely impact growth and survival, much more so than mutations in the outer membrane fusion proteins, but not through apoptosis. These organisms do not release cytochrome c release as part of their apoptotic program. Instead, the C. elegans and Drosophila mutants become very sensitive to reactive oxygen species (ROS) producing agents such as Paraquat (Kanazawa et al. 2008; Yarosh et al. 2008). This effect is distinct from the fusion defect, because complete loss of the C. elegans Opa1 homolog is not suppressed by fission defects (Kanazawa et al. 2008). Similar dual functions were proposed for mammalian Opa1 and yeast Mgm1 (Cipolat et al. 2006; Meeusen et al. 2006).
Clues to this second function of Opa1 come from mammalian cell culture experiments. Immunogold-labeled EM shows that only 8% of Opa1 localizes to the rim, where fusion is likely to take place (Griparic et al. 2004). The remaining 92% of the Opa1 protein is inside cristae. Partial loss of Opa1 at early time points after transfection with siRNA causes some changes in mitochondrial morphology (alternate swelling and constriction of mitochondrial tubules and swollen cristae), consistent with defects in cristae maintenance (Griparic et al. 2004). It has been suggested that Opa1 may reside at the necks of cristae (cristae junctions), because those have the same diameter as Dynamin rings (Frezza et al. 2006), but this is not supported by the even distribution of Opa1 distributed throughout cristae as detected with Immuno-EM (Griparic et al. 2004). Instead, it seems likely that Opa1 somehow helps maintain cristae morphology, similar to the proposed functions of Mitofilin and ATP synthase (Rabl et al. 2009). Swelling of cristae morphologies could promote ROS production, for example, by dislodging different parts of the respiratory supercomplex (Schafer et al. 2006). Increased ROS production might also promote apoptosis in mammalian cells by oxidizing cardiolipin, which then promotes the release of cytochrome c from the inner membrane (Ott et al. 2007).
PATHOGENIC EFFECTS OF MUTATIONS IN FISSION AND FUSION PROTEINS
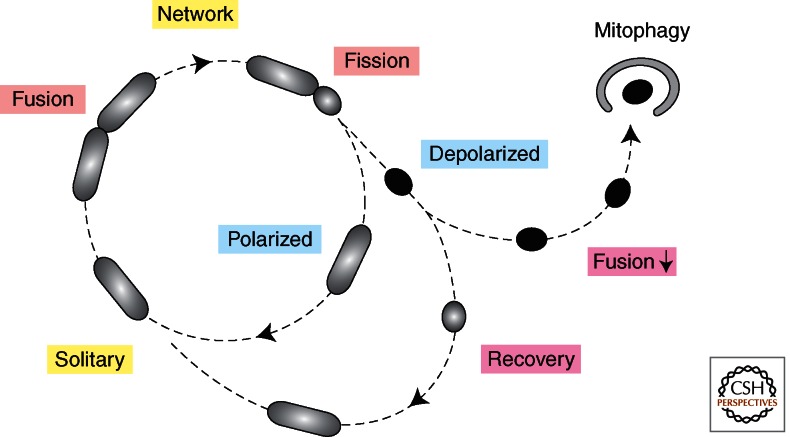
With live cell microscopy of mammalian cells, it was shown that mitochondria primarily exist as solitary units that periodically fuse with other mitochondria (Twig et al. 2008) (Fig. 4). These fusion events are often rapidly followed by fission, which can be asymmetric, because the daughter mitochondria often have different membrane potentials. This could to be a sorting event, giving rise to some mitochondria with a heavy load of defective components (Youle and van der Bliek 2012). Mitochondria that cannot recover membrane potential because of these defects become targets for Pink1- and Parkin-mediated degradation (Narendra et al. 2010). As discussed in more detail elsewhere in this collection and also touched on in this paper, a range of human diseases has been connected with mutations in these pathways. Many of these diseases are neurodegenerative (Parkinson’s, Charcot-Marie-Tooth, Dominant Optic Atrophy, and Alzheimer’s), but effects on other tissues have also been observed (diabetes, ischemia reperfusion, and liver disease) (Liesa et al. 2009; Chan 2012; Nunnari and Suomalainen 2012).
Figure 4.
Mitochondrial life cycle. Live cell imaging showed that mitochondria often exist as solitary units. They do occasionally fuse to other mitochondria but fusion is often followed within 20 min by fission at the same place. Fission can give rise to daughter mitochondria with different membrane potentials, suggesting that fission is preceded by a sorting event. Daughter mitochondria with lower membrane potential often recover, allowing them to rejoin the mitochondrial network by fusion, but persistently low membrane potential will inhibit fusion and cause elimination through mitophagy. (The figure was created from data adapted from Twig et al. 2008.)
Two opposing trends in fission and fusion rates are used to counter different levels of stress: increased fusion and/or decreased fission helps overcome low levels of stress, whereas decreased fusion and/or increased fission occurs with high levels of stress. Low levels of stress are seen with starvation, during which mitochondrial fission is inhibited to protect cells from excessive autophagosomal degradation of mitochondria (Gomes et al. 2011; Rambold et al. 2011). Similar effects are achieved by increasing the rates of fusion, which occurs when cells are treated with low levels of toxins. This phenomenon is called stress-induced mitochondrial hyperfusion (SIMH) (Tondera et al. 2009). SIMH is distinct from other forms of fusion because it relies on a specific subset of fusion proteins (Mfn1, L-Opa1, SLP2). Interestingly, knockdown of Yme1L, which increases the proportion of L-Opa1 in a cell, also increases the numbers of fused mitochondria, suggesting that this treatment leads to SIMH as well (Griparic et al. 2007). Increased fusion and decreased fission are likely beneficial because the functions of partially defective mitochondrial can be restored by mixing the contents as a form of complementation.
As discussed before, fission rates are drastically increased during apoptosis or when mitochondria lose membrane potential. In mammalian cells, fusion is then also blocked by Oma1-mediated cleavage of Opa1 (Ehses et al. 2009; Head et al. 2009). Similar steps may occur, but much less frequently, in normal cells to eliminate unsalvageable mitochondria through mitophagy (Frank et al. 2012). In these defective mitochondria, the Mitofusins are eventually eliminated by Pink1- and Parkin-mediated degradation (Youle and Narendra 2011). Parkinson’s disease can be caused by mutations in Pink1 or Parkin, which most likely lead to the accumulation of damaged mitochondria. These damaged mitochondria may eventually kill the cell through ROS or other toxic agents in the dopaminergic neurons of Parkinson’s patients (Chinta and Andersen 2008). Paradoxically, however, another neurodegenerative disease, the peripheral neuropathy Charcot-Marie-Tooth disease CMT2A, is caused by mutations in a Mitofusin (Bradbury 2004). So the Mitofusins that inappropriately remain active in Parkinson’s disease are actually defective in patients with Charcot-Marie-Tooth disease. Perhaps the initial fusogenic stress response is more important for cell survival in peripheral neurons, whereas the elimination of malfunctioning parts through mitophagy is more important for the survival of dopaminergic neurons.
SOME EVOLUTIONARY CONSIDERATIONS
Lower eukaryotes, such as Dictyostelium and Trypanosomes, have Dynamin homologs that affect mitochondrial fission, but there is no obvious distinction in these organisms between Dynamins involved in organelle fission and those involved in membrane traffic, unlike the yeast Dynamins Vps1 and Dnm1 or Dynamin and Drp1 in higher eukaryotes (Wienke et al. 1999; Chanez et al. 2006). This lack of specialization may reflect an early stage in eukaryotic evolution. Bacteria also have Dynamin-like proteins. The closest Dynamin relatives appear to be a bacterial Dynamin-like protein (BDLP) in Nostoc punctiforme and dynA in Bacillus subtilus. Structural studies with BDLP suggest that it can form spirals around membrane tubules, similar to the Dynamin spirals in higher eukaryotes (Low et al. 2009), whereas dynA is able to fuse membranes in vitro (Burmann et al. 2011). Although bacteria are not known to have internal organelles or membrane traffic and these proteins are on the wrong side of the membrane for Drp1-like fission, membrane fusion may be needed at a late stage of cytokineses. Moreover, it was recently shown that dynA affects ftsZ-mediated bacterial fission, but this was only detected with additional mutations in bacterial Flotillin homologs (Dempwolff et al. 2012). These new data suggest that bacterial Dynamins play a nonessential role during cytokinesis. It will be interesting to see whether these proteins are directly involved in bacterial cell division and whether this might help us better understand mitochondrial fission and fusion.
The apparent lack of a mitochondrial inner membrane fission apparatus in higher eukaryotes raises some evolutionary questions as well. The endosymbiont that gave rise to mitochondria most likely had an elaborate inner membrane fission machinery, similar to the ftsZ-based fission apparatus of present-day α proteobacteria. Indeed, some lower eukaryotes like Dictyostelium and primitive algae still have mitochondrial ftsZ homologs, along with a Dynamin homolog for outer membrane fission (Takahara et al. 1999; Gilson et al. 2003; Kiefel et al. 2004). The presence of ftsZ in the mitochondria of certain algae, but not in the mitochondria of higher plants or animals, suggests that ftsZ was independently lost in plants and animals after these kingdoms diverged, but well before the emergence of the most modern phyla. It is tempting to speculate that the loss of inner membrane fission proteins was facilitated by a change in the environment, such as the dramatic increase in oxygen levels that more or less coincides with this time (Canfield et al. 2007), but other environmental factors have not been ruled out.
CONCLUDING REMARKS
The main components of the mitochondrial fission and fusion machinery are Dynamin family members. Keeping these machineries fully functional is important for cell survival. The plethora of regulatory mechanisms that control their actions are just now being pieced together. Disruptions in these processes lead to a range of neurodegenerative and other diseases.
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet 26: 211–215 [DOI] [PubMed] [Google Scholar]
- Alto NM, Soderling J, Scott JD 2002. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J Cell Biol 158: 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D, Grodet A, Lee YJ, Estaquier J, Blackstone C 2005. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J Biol Chem 280: 35742–35750 [DOI] [PubMed] [Google Scholar]
- Baricault L, Segui B, Guegand L, Olichon A, Valette A, Larminat F, Lenaers G 2007. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res 313: 3800–3808 [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M 1994. Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech 27: 198–219 [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1: 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy B, Petrilli A, Klinglmayr E, Chen J, Lutz-Meindl U, Knott AB, Masliah E, Schwarzenbacher R, Bossy-Wetzel E 2010. S-nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer’s disease. J Alzheimers Dis 20: S513–S526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury J 2004. Mitochondrial fusion protein mutated in CMT2A. Lancet Neurol 3: 326. [DOI] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride HM 2009. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep 10: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Gilady SY, Fitzsimmons RE, Benson MD, Lynes EM, Gesson K, Alto NM, Strack S, Scott JD, Simmen T 2010. Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties. J Biol Chem 285: 31590–31602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann F, Ebert N, van Baarle S, Bramkamp M 2011. A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol Microbiol 79: 1294–1304 [DOI] [PubMed] [Google Scholar]
- Canfield DE, Poulton SW, Narbonne GM 2007. Late-Neoproterozoic deep-ocean oxygenation and the rise of animal life. Science 315: 92–95 [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, et al. 2008. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 14: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L 2008. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci 105: 15803–15808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny KL, Jensen RE 2003. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell 14: 4126–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC 2012. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu Rev Genet 46: 265–287 [DOI] [PubMed] [Google Scholar]
- Chanez AL, Hehl AB, Engstler M, Schneider A 2006. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J Cell Sci 119: 2968–2974 [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C 2007. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 282: 21583–21587 [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C 2010. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann NY Acad Sci 1201: 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F 2010. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 465: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinta SJ, Andersen JK 2008. Redox imbalance in Parkinson’s disease. Biochim Biophys Acta 1780: 1362–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA 2009. S-nitrosylation of Drp1 mediates β-amyloid-related mitochondrial fission and neuronal injury. Science 324: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8: 1255–1262 [DOI] [PubMed] [Google Scholar]
- Chou CH, Lin CC, Yang MC, Wei CC, Liao HD, Lin RC, Tu WY, Kao TC, Hsu CM, Cheng JT, et al. 2012. GSK3β-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PloS ONE 7: e49112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126: 163–175 [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S 2007. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 8: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 26: 207–210 [DOI] [PubMed] [Google Scholar]
- Dempwolff F, Wischhusen HM, Specht M, Graumann PL 2012. The deletion of bacterial dynamin and flotillin genes results in pleiotrophic effects on cell division, cell growth and in cell shape maintenance. BMC Microbiol 12: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J 2009. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol 186 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Allan VJ, Grierson AJ, Sheetz MP 2005. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr Biol 15: 678–683 [DOI] [PubMed] [Google Scholar]
- Dorn GW 2nd, Clark CF, Eschenbacher WH, Kang MY, Engelhard JT, Warner SJ, Matkovich SJ, Jowdy CC 2011. MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ Res 108: 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr M, Escobar-Henriques M, Merz S, Geimer S, Langer T, Westermann B 2006. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol Biol Cell 17: 3745–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E 2002. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J Mol Biol 323: 835–843 [DOI] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Oka T, Mihara K 2006. Identification of a novel protein that regulates mitochondrial fusion by modulating mitofusin (Mfn) protein function. J Cell Sci 119: 4913–4925 [DOI] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell 1: 515–525 [DOI] [PubMed] [Google Scholar]
- Frank M, Duvezin-Caubet S, Koob S, Occhipinti A, Jagasia R, Petcherski A, Ruonala MO, Priault M, Salin B, Reichert AS 2012. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta 1823: 2297–2310 [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, et al. 2006. OPA1 controls apoptotic Cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK 2011. ER tubules mark sites of mitochondrial division. Science 334: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S, Weinbach N, Westermann B 2003. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol Biol Cell 14: 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465: 502–506 [DOI] [PubMed] [Google Scholar]
- Gilson PR, Yu XC, Hereld D, Barth C, Savage A, Kiefel BR, Lay S, Fisher PR, Margolin W, Beech PL 2003. Two Dictyostelium orthologs of the prokaryotic cell division protein FtsZ localize to mitochondria and are required for the maintenance of normal mitochondrial morphology. Eukaryot Cell 2: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL 2006. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol 173: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC 2005. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol 170: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM 2004. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem 279: 18792–18798 [DOI] [PubMed] [Google Scholar]
- Griparic L, Kanazawa T, van der Bliek AM 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 178: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Farh L, Marshall TK, Deschenes RJ 1993. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr Genet 24: 141–148 [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90: 121–129 [DOI] [PubMed] [Google Scholar]
- Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, Matsushita M 2008. CaM kinase I α-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol 182: 573–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 187: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M, Vogel F, Bornhövd C, Neupert W, Reichert AS 2003. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem 278: 27781–27788 [DOI] [PubMed] [Google Scholar]
- Herlan M, Bornhovd C, Hell K, Neupert W, Reichert AS 2004. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol 165: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM 1998. Mitochondrial fusion in yeast requires the trans-membrane GTPase Fzo1p. J Cell Biol 143: 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J 2007. The machines that divide and fuse mitochondria. Annu Rev Biochem 76: 751–780 [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto M, Tachibana I, Urrutia R 1998. Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase vps1p. J Cell Sci 111: 1341–1349 [DOI] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J 2005. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol 170: 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Jofuku A, Eura Y, Mihara K 2003. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem Biophys Res Commun 301: 891–898 [DOI] [PubMed] [Google Scholar]
- Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, Grimm S 2011. Fis1 and Bap31 bridge the mitochondria–ER interface to establish a platform for apoptosis induction. EMBO J 30: 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Walsh ML, Bockus BJ, Chen LB 1981. Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J Cell Biol 88: 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BA, Fangman WL 1992. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev 6: 380–389 [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Nagai Y, Onogi H, Muro Y, Wakabayashi T, Hagiwara M 1998. Dymple, a novel dynamin-like high molecular weight GTPase lacking a proline-rich carboxyl-terminal domain in mammalian cells. J Biol Chem 273: 1044–1051 [DOI] [PubMed] [Google Scholar]
- Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith ED, Buttle KF, McDonald K, Mannella CA, van der Bliek AM 2008. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet 4: e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ 2006. Role of Bax and Bak in mitochondrial morphogenesis. Nature 443: 658–662 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ 2007. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol 178: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefel BR, Gilson PR, Beech PL 2004. Diverse eukaryotes have retained mitochondrial homologs of the bacterial division protein FtsZ. Protist 155: 105–115 [DOI] [PubMed] [Google Scholar]
- Koirala S, Bui HT, Schubert HL, Eckert DM, Hill CP, Kay MS, Shaw JM 2010. Molecular architecture of a dynamin adaptor: Implications for assembly of mitochondrial fission complexes. J Cell Biol 191: 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Osman C, Walter P 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci 108: 14151–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Ramabhadran V, Higgs HN 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339: 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC 2004. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305: 858–862 [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zapaterra M, Rube DA, van der Bliek AM 1999. C. elegans dynamin-related protein drp-1 controls severing of the mitochondrial outer membrane. Mol Cell 4: 815–826 [DOI] [PubMed] [Google Scholar]
- Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM 2012. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MR, Lewis WR 1914. Mitochondria (and other cytoplasmic structures) in tissue cultures. Am J Anat 17: 339–401 [Google Scholar]
- Liesa M, Palacin M, Zorzano A 2009. Mitochondrial dynamics in mammalian health and disease. Physiol Rev 89: 799–845 [DOI] [PubMed] [Google Scholar]
- Lokireddy S, Wijesoma IW, Teng S, Bonala S, Gluckman PD, McFarlane C, Sharma M, Kambadur R 2012. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab 16: 613–624 [DOI] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC 2013. Fis1, Mff, MiD49 and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 24: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Lowe J 2009. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139: 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10: 839–850 [DOI] [PubMed] [Google Scholar]
- McQuibban GA, Lee JR, Zheng L, Juusola M, Freeman M 2006. Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr Biol 16: 982–989 [DOI] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE 2011. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol 18: 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127: 383–395 [DOI] [PubMed] [Google Scholar]
- Meglei G, McQuibban GA 2009. The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion. Biochemistry 48: 1774–1784 [DOI] [PubMed] [Google Scholar]
- Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Lower B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, Langer T 2008. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev 22: 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill RA, Dagda RK, Dickey AS, Cribbs JT, Green SH, Usachev YM, Strack S 2011. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol 9: e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, et al. 2010. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol 151: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S 2006. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep 7: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A, Youle RJ 2005. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J Biol Chem 280: 18598–18603 [DOI] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U 2005. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: New implications for Charcot–Marie–Tooth disease. J Cell Biol 170: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A 2012. Mitochondria: In sickness and in health. Cell 148: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, Emorine LJ, Descoins E, Pelloquin L, Brichese L, Gas N, Guillou E, Delettre C, Valette A, Hamel CP, et al. 2002. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett 523: 171–176 [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 278: 7743–7746 [DOI] [PubMed] [Google Scholar]
- Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, Lenaers G 2007. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ 14: 682–692 [DOI] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Zhivotovsky B, Orrenius S 2007. Role of cardiolipin in cytochrome c release from mitochondria. Cell Death Differ 14: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT 2011. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT 2004. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Qi X, Disatnik MH, Shen N, Sobel RA, Mochly-Rosen D 2011. Aberrant mitochondrial fission in neurons induced by protein kinase Cδ under oxidative stress conditions in vivo. Mol Biol Cell 22: 256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, et al. 2009. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol 185: 1047–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci 108: 10190–10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem 273: 20150–20155 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, De Giorgi F, Rossi R, Heim R, Tsien RY, Pozzan T 1996. Double labelling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr Biol 6: 183–188 [DOI] [PubMed] [Google Scholar]
- Rowland AA, Voeltz GK 2012. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat Rev Mol Cell Biol 13: 607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Fuller MT 2001. Control of mitochondrial morphology by a human mitofusin. J Cell Sci 114: 867–874 [DOI] [PubMed] [Google Scholar]
- Schafer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J 2006. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem 281: 15370–15375 [DOI] [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: A control point for apoptosis. Science 300: 135–139 [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE 2004. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem 279: 28298–28303 [DOI] [PubMed] [Google Scholar]
- Sesaki H, Southard SM, Hobbs AE, Jensen RE 2003. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem Biophys Res Commun 308: 276–283 [DOI] [PubMed] [Google Scholar]
- Shin HW, Shinotsuka C, Torii S, Murakami K, Nakayama K 1997. Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J Biochem 122: 525–530 [DOI] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G 2005. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J 24: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM 1998. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol 143: 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12: 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K 2007. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282: 11521–11529 [DOI] [PubMed] [Google Scholar]
- Takahara M, Takahashi H, Matsunaga S, Sakai A, Kawano S, Kuroiwa T 1999. Two types of ftsZ genes isolated from the unicellular primitive red alga Galdieria sulphuraria. Plant Cell Physiol 40: 784–791 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Youle RJ 2008. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol Cell 29: 409–410 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Okreglak V, Naylor K, Nunnari J 2002. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol 158: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28: 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Graf SA, Wikstrom JD, Mohamed H, Haigh SE, Elorza A, Deutsch M, Zurgil N, Reynolds N, Shirihai OS 2006. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am J Physiol Cell Physiol 291: C176–C184 [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Payne GS 2010. Dynamin subunit interactions revealed. Dev Cell 18: 687–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang Y, Long J, Wang J, Haudek SB, Overbeek P, Chang BH, Schumacker PT, Danesh FR 2012. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab 15: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B 2010. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884 [DOI] [PubMed] [Google Scholar]
- White KE, Davies VJ, Hogan VE, Piechota MJ, Nichols PP, Turnbull DM, Votruba M 2009. OPA1 deficiency associated with increased autophagy in retinal ganglion cells in a murine model of dominant optic atrophy. Invest Ophthalmol Vis Sci 50: 2567–2571 [DOI] [PubMed] [Google Scholar]
- Wienke DC, Knetsch MLW, Neuhaus EM, Reedy MC, Manstein DJ 1999. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in dictyostelium discoideum. Mol Biol Cell 10: 225–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Gorsich SW, McCaffery JM, Shaw JM, Nunnari J 2000. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol 151: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W, Monserrate J, Tong JJ, Tse S, Le PK, Nguyen K, Brachmann CB, Wallace DC, Huang T 2008. The molecular mechanisms of OPA1-mediated optic atrophy in Drosophila model and prospects for antioxidant treatment. PLoS Genet 4: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, Dahan S, McNiven MA 1998. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol 140: 779–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP 2011. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM 2012. Mitochondrial fission, fusion, and stress. Science 337: 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M 2011. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J 30: 2762–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]