Abstract
Inflammatory bowel diseases (IBD) are common inflammatory disorders of the gastrointestinal tract that include ulcerative colitis (UC) and Crohn’s disease (CD). The incidences of IBD are high in North America and Europe, affecting as many as one in 500 people. These diseases are associated with high morbidity and mortality. Colorectal cancer risk is also increased in IBD, correlating with inflammation severity and duration. IBD are now recognized as complex multigenetic disorders involving at least 32 different risk loci. In 2007, two different autophagy-related genes, ATG16L1 (autophagy-related gene 16-like 1) and IRGM (immunity-related GTPase M) were shown to be specifically involved in CD susceptibility by three independent genome-wide association studies. Soon afterwards, more than forty studies confirmed the involvement of ATG16L1 and IRGM variants in CD susceptibility and gave new information on the importance of macroautophagy (hereafter referred to as autophagy) in the control of infection, inflammation, immunity and cancer. In this review, we discuss how such findings have undoubtedly changed our understanding of CD pathogenesis. A unifying autophagy model then emerges that may help in understanding the development of CD from bacterial infection, to inflammation and finally cancer. The Pandora's box is now open, releasing a wave of hope for new therapeutic strategies in treating Crohn's disease.
Keywords: Autophagy, Crohn's disease, infection, inflammation, immunity, cancer
I. INTRODUCTION
Crohn’s disease [MIM266600] and ulcerative colitis [MIM 191390], two major forms of inflammatory bowel diseases, are common inflammatory disorders of the gastrointestinal tract. Their incidences are high in North America and Europe, affecting as many as one in 500 people [1, 2]. Diagnosis of IBD occurs between the age of 15 and 30 years, with ~30% of patients presenting before the age of 20. Patients with UC and CD have an increased risk of developing small-bowel, colon, and colorectal cancers [3, 4]. These cancers are distinct from those observed in the general population as they are diagnosed earlier, are more aggressive and are associated with a poor prognosis.
Considerable advances have been made recently in the treatment of IBD, based on the neutralization of the immune system by corticosteroids or the suppression of the action of pro-inflammatory cytokinessuch as tumor necrosis factor-α (TNF-α) [5]. However, none of these expensive treatments is able to cure IBD, resulting at best in symptomatic improvements with severe side toxicity and opportunistic infections. Therefore, there is an urgent need for improving our understanding of IBD pathogenesis, with the ultimate goal of therapeutic intervention.
I.A. Pathogenesis of Crohn’s Disease
Within the past ten years, epidemiological, clinical and basic studies have highlighted the importance of genetic, immunological and environmental factors in CD pathogenesis. Histologically, this disease is characterized by chronic inflammation with a massive infiltration of leukocytes into the intestinal mucosa. It is thus widely accepted that the development of CD results from an exaggerated inflammatory response to a hitherto undefined lumenal antigen, probably derived from the microbial flora. Given the therapeutic benefits of antibiotic treatment [5], intracellular bacteria appear to be involved in CD inflammation. However, the evidence for a causal pathogen remains elusive. The current consensus is that commensal gut bacteria rather than a particular pathogen may have a pivotal role in the maintenance of chronic inflammation, and that a genetic host defect may initiate the intestinal inflammation.
Indeed, there is a strong genetic basis for CD, as ~20% of people with CD have a family relative with CD, and 36% of monozygotic twins share the disease [4]. Such familial history of the disease, together with the observation that the inflammation can affect any part of the gastrointestinal tract, suggests germline mutations that predispose an individual to CD.
In 2001, the first gene was identified that correlates with susceptibility to CD, NOD2 (Nucleotide-binding Oligomerization Domain containing 2, also known as CARD15 for caspase recruitment domain-containing protein 15 [6–8]), which is an intracellular sensor of bacterial infection that drives via NF-κB the production of proinflammatory cytokines in macrophages and α-defensins in intestinal Paneth cells [9, 10]. However, NOD2 mutations per se are not sufficient to cause the expression of disease, as there are healthy individuals homozygous or compound heterozygous for NOD2 risk alleles. Moreover, the age of diagnosis and the disease aggressiveness differ significantly between family members having the same NOD2 mutation [11].
Since then, IBD have become recognized as complex multigenetic disorders. At least 32 different risk loci have been identified by eleven genome-wide associations (GWA) [12–22]. Some of these susceptibility genes appear to be specific to UC or CD, whereas others are non-specific IBD loci (Wellcome Trust Case Control Consortium-WTCCC [23]). IL23R was the first gene to be significantly associated with CD by GWA [12]. Among the pathways that emerged to be involved in CD is autophagy [24, 25]. In 2007, two major hits were observed in two different autophagy-related genes, ATG16L1 (autophagy-related 16-like 1) and IRGM (Immunity-Related GTPase M) by three independent GWA studies [14, 15, 18]. Soon afterward, more than forty studies confirmed these observations and provided new information on the importance of autophagy in the control of inflammation, immunity and cancer (Tables 1 and 2).
Table 1.
Summary of ATG16L1 GWA and Replication Susceptibility Studies
| Associations observed | Populations | Sample Size Cases- Controls |
SNPs analyzed | p values | Associated with CD |
Interaction with CD loci |
Associated with UC |
||
|---|---|---|---|---|---|---|---|---|---|
| Genes | chr | Snp | |||||||
| Atg16L1 | 2q37.1 | Rs2241880 T300A |
German [14] | 735 CD 788 UC 368 C |
GWA–19,779 nsSNP (SNPlex) |
4 10−8 | Yes | Weak | No |
| CARD15 (not replicated) | |||||||||
| Homozygous GG |
498 CD 380 CD3 1032 C |
Case-control family (TaqMan) |
1.6 10−5 | ||||||
| British [14] | 509 CD 656 C |
Case-control | 0.0004 | ||||||
| N. American (European) [12, 15] |
946 ileal CD 977 C |
GWA–NIDDK IBDGC–304K (Illumina HumanHap300) |
6.38 10−8 | Yes Ileal |
No | No | |||
| 353 ileal CD 530 ileal CD3 207 C |
Case-control family (iPlex, TaqMan) |
4.1 10−8 | |||||||
| N. American [133] |
555 CD 486 C |
TaqMan | 0.001 | Yes | |||||
| N. American [68] |
213 CD 117 UC 310 C |
TaqMan | 0.001 | Yes Specific |
Yes IBD5, IL23R |
No | |||
| N. American [134] |
142 CD children 281 C |
Illumina HumanHap500 BeadChip |
6.93×10−;4 | Yes childhood- onset |
|||||
| Rs10210302 | British [16, 18] | 1748 CD 2938 C |
GWA–369K (Affymetrix 500k) |
7.1 10−14 | Yes Ileal |
No | No | ||
| Rs2241880 | British [34] | 645 CD 676 UC 1190 C |
Case–control family (Mass Spectrometry) |
2.33 10−7 | Yes | No | No | ||
| British [135] | 727 CD 877 UC 579 C |
TaqMan | 2.4 10−6 | Yes Ileal D ≤ 16 years |
No | Weak (not replicated) |
|||
| British [136] | 652 CD 1156 C |
PCR-SSP iPLEX platform |
Yes | ||||||
| No association with isolated colonic CD (135) | |||||||||
| British [24] | 1841 UC 1465 C |
GWA– WTCCC 10886 SNP |
Yes Specific | No | |||||
| British Scottish [137] |
361 CD children 360 adult CD 345 C |
TaqMan | Yes | Yes | No | ||||
| GG – Pure ileal / No association with childhood- onset | |||||||||
| German [25] | 1850 CD 1103 UC 1817 C |
GWA (SNPex) | 5.82 10- 8 | Yes Specific | No | ||||
| German [138]. |
768 CD 507 UC 1,615 C |
PCR-FRET | 3.7 × 10−6 | Yes | No | No | |||
| Dutch [139] | 311 CD 207 UC 893 C |
Case–control family (Mass Spectrometry) |
0.0017 | Yes | No | No | |||
| Dutch [123, 139] | 1684 CD 1120 UC 1350 C |
TaqMan | 3.6 10−4 | Yes | No | No | |||
| Stricturing, perianal disease | |||||||||
| Associations observed | Populations | Sample Size Cases- Controls |
SNPs analyzed |
p values |
Associated with CD |
Interaction with CD loci |
Associated with UC |
||
|---|---|---|---|---|---|---|---|---|---|
| Genes | chr | Snp | |||||||
| Atg16L1 | 2q37.1 | Rs2241880 T300A |
Polish [140] | 60 CD 140 C |
Taqman | 0.022 | Yes – Weak |
||
| Belgian [17] | 547 CD 246 UC 928 C |
GWA HumanHap300 |
2 × 10−4 | Yes | No | ||||
| Hungarian [141] | 266 CD 149 UC 149 C |
TaqMan | 0.027 | Yes | No | ||||
| GG – Colonic disease | |||||||||
| Australian [121] | 823 CD 543 UC 1664 C |
Mass Array Sequenom, TaqMan |
<0.001 | Yes Ileal |
No | Yes Protective |
|||
| Stricturing, penetrating disease GG – Smoking : 7x CD Risk | |||||||||
| New Zealand (Caucasians) [142] |
496 CD 466 UC 591 C |
Taqman | 0.0001 | Yes | No | No | |||
| Brazilian [31] | 187 CD 255 C |
TaqMan | 0.2 | No | |||||
| Japanese [30] | 484 CD 439 C |
TaqMan | No | ||||||
| Chinese [26] | 40 CD 40 UC 50 C |
No | |||||||
| Italian [28] | 163 CD 160 C |
TaqMan | No | No | |||||
| Italian [29] | 763 CD 843 UC 749 C |
TaqMan | 0.003 | Yes | No | No | |||
A total of 28 studies (4 GWA) were reported for Atg16L1, including ~13,000 cases and 17,000 controls (C). NIDDK: North American National Institute of Diabetes and Digestive and Kidney Diseases, IBDGC: IBD Genetics Consortium. WTCCC: Wellcome Trust Case Control Consortium. D: Age at diagnosis. y: years. CD3: trios (parent–offspring). PCR-FRET: PCR and melting curve analysis using fluorescence resonance energy transfer (FRET) probes.
Table 2.
Summary of IRGM GWA and Replication Susceptibility Studies
| Associations observed | Populations | Cases- Controls |
SNPs analyzed | p values | Associated with CD |
Interaction with CD loci |
Associated with UC |
||
|---|---|---|---|---|---|---|---|---|---|
| Genes | chr | Snp | |||||||
| IRGM | 5q33.1 | rs13361189 | British [16, 18] | 1748 CD 2938 C |
GWA– WTCCC 369K (Affymetrix) |
2.1 10−10 | Yes | No | |
| rs4958847 | 1182 CD 2024 C |
Cases-Control Replication (Taqman) |
3.8 10−9 | Yes | No | ||||
| rs10065172 tag-SNP 313T→C |
[18] | 769 CD 705 C |
Cases-Control | 0.008 | Yes Ileal | No | No | ||
| 20-kb Deletion | North American [32] |
172 CD 171 UC 344 C |
WGA– NIDDK IBDGC Cases- Control |
Yes | No | ||||
| rs13361189 | N. American (European)[133] |
555 CD D: 11.7 y 486 C |
TaqMan | 0.036 | Yes | ||||
| rs13361189 | New Zealand (Caucasians) [33] |
507 CD 475 UC 576 C |
Taqman | 0.0017 | Yes ileal- specific |
No | |||
| rs13361189 | British [24] | 1841 UC 1465 C |
GWA– WTCCC 10886 SNP |
Yes Specific |
No | ||||
| rs13361189 rs4958847 |
German [25] | 1850 CD 1103 UC 1817 C |
53 SNP | 1.27 10−5 3.95 10−7 |
Yes Specific |
No | |||
7 studies (3 GWA) were reported for IRGM, including ~6,700 cases and 8,900 controls (C).
In this review, we discuss how such findings have undoubtedly opened up a new vista on CD pathogenesis. A unifying autophagy model then emerges that may help in understanding the development of CD from bacterial infection, to inflammation and then to cancer. The Pandora's box is now open, releasing a wave of hope for new therapeutic strategies to treat Crohn's disease.
I.B. ATG16L1 and IRGM as New CD-Specific Susceptibility Loci
The strong association of the rs2241880 single-nucleotide polymorphism (Thr300→Ala ATG16L1) first identified by Hampe et al. through GWA [14] was confirmed by twenty-five studies in several northern Caucasian CD cohorts (Table 1). However, no significant association is found in the Japanese, Chinese, and Brazilian populations, and conflicting data are reported in two Italian studies [26–31]. Further genetic studies are thus required to confirm the association of the ATG16L1 rs2241880 variant with ethnically divergent populations. Similarly, several IRGM risk polymorphisms are associated with CD and replicated in independent cohorts of North America, North Europe and New Zealand (Table 2) [12, 14–16, 18, 24, 25, 32–34]. Compelling evidence indicates that the ATG16L1 and IRGM loci do not interact with other susceptibility loci for CD (CARD15; IL23R), and no significant association with specific CD sub-phenotype (inflammatory, stricturing, penetrating disease, etc.) has been identified. Of particular importance, a meta-analysis of all WGA indicates that ATG16L1 and IRGMlike NOD2are involved specifically in ileal CD inflammation [23]. Therefore, at least some types of CD can now be viewed as an autophagy disease similar to infection, neurodegenerative diseases and cancers [35].
II. XENOPHAGY – A SPECIALIZED FORM OF AUTOPHAGY– IS SPECIFICALLY DISRUPTED IN CD
As indicated by its acronym, during autophagy, organelles and long-lived proteins are sequestered by a double-membrane vesicle, called an “autophagosome”. Subsequently, the autophagosome rapidly fuses with an endosome and/or a lysosome to ultimately form an "autolysosome" where its content is degraded and recycled [36]. Physiologically, the autophagy pathway controls cell remodelling throughout development and prevents premature cell aging. This latter housekeeping function of autophagy occurs continuously at basal levels and degrades damaged proteins and organelles that would otherwise accumulate during the life span of the cell. In response to environmental stresses such as nutrient starvation, hypoxia and infections, this catabolic process is dramatically upregulated to provide the supply of energy needed for cell survival and repair [37, 38]. Especially, cells coping with microbes use a dedicated form of autophagy termed “xenophagy” as a host defense mechanism to engulf and degrade intracellular pathogens [39] (Fig. 1).
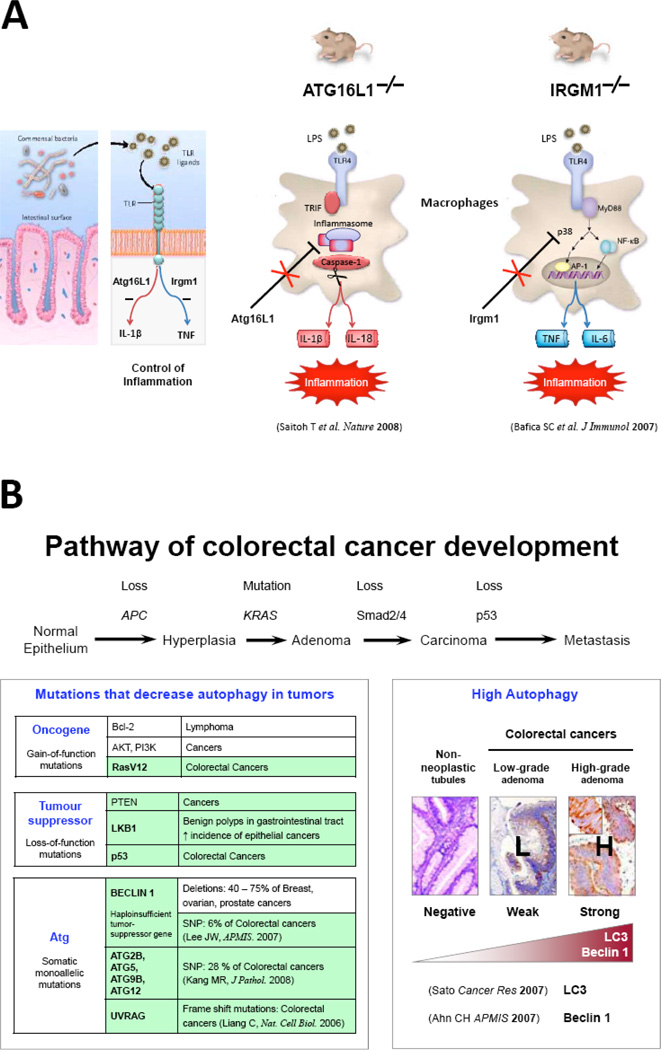
Fig. (1). Functions of Autophagy in Innate Immunity.
(A) Structures of the Atg16L1 and IRGM proteins. The two autophagy genes ATG16L1 and IRGM are broadly expressed in intestinal epithelium (colon, small bowel) and in hematopoietic cells (thymus, T cells, and macrophages) [14, 15]. Human Atg16L1 is a platform with an N-terminal domain that interacts with the autophagy proteins Atg5 and Atg12, a coiled-coil domain that mediates homodimeric interactions, and seven WD repeats that are believed to interact with an unknown ligand(s). In contrast, IRGM has a unique, truncated guanosine triphosphatase (GTPase) domain. Several splice isoforms of ATG16L1 and IRGM have been identified. At present, nothing is known about the functions of these different isoforms in autophagy and CD susceptibility. (B) Proposed functions of Atg16L1 and IRGM in xenophagy. Autophagy is a vesicular pathway, enabling cells to digest their own cytosol or invasive bacteria. During autophagy, a portion of cytosol is sequestered within a double-membrane vesicle, called an autophagosome. The autophagosome is formed by expansion of a phagophore, the origin of which remains unknown. During the maturation step, the autophagosome acquires an acidic pH and hydrolases by fusing with a lysosome to generate an autolysosome in which the content is degraded. At the molecular level, a family of at least 16 autophagy-related (Atg) proteins is required for autophagosome formation and maturation. Of these, it is proposed that IRGM may protect the host cells against bacteria by (a) driving vesiculation and disruption of the phagosome, releasing the pathogen out of its protective niche into the cytosol (inset), (b) directing the engulfment of bacteria by large double-membrane autophagosomes, and (c) targeting the pathogen for lysosomal degradation by promoting autolysomal maturation. During autophagosome formation, Atg12 is activated by Atg7 and conjugated by Atg10 to Atg5. The Atg5 (•) / Atg12(•) conjugate then associates with Atg16L1 (•) to form a ~800-kDa multimeric complex (referred to as the Atg16L complex). A fraction of the Atg16L complex localizes to the phagophore and mediates binding of the LC3/Atg8 (•)-phosphatidylethanolamine (ζ) conjugate to promote elongation of the phagophore. Upon completion of the autophagosome, the Atg16L complex and most of the LC3 are released from the membrane. Shown here are the steps controlled by IRGM and Atg16L1 (blue arrows), and the consequences of small-interfering RNA (siRNA)-mediated knockdown of ATG16L1 and IRGM on xenophagy. Images reproduced with permission from [61].
The identification of the two autophagy genes ATG16L1 and IRGM as new CD susceptibility loci has met at the beginning some skepticism. At first glance, it may seem puzzling that a general autophagy defect may be associated with CD. Indeed, the autophagy process is absolutely required during development and metabolism. Consistent with these roles, ATG16L1-deficient mice die within the first day of delivery, similar to ATG5- or ATG7-null mice [40–42]. Moreover, the high prevalence of the CD risk-variants ATG16L1 T300A (51%) and IRGM variant (10%) in unaffected populations argues against a general deleterious autophagy phenotype as being the cause of IBD. As a result, one might expect that disease-associated ATG16L1 and IRGM variants would confer subtle effects on autophagy or disturb one of its functions [43].
So far, very little information is currently available about the functions of the human Atg16L1 and IRGM, and even less about the consequences of their variations. We have learned from mouse models the critical roles of autophagy in the handling of invasive bacteria. While recent work in human epithelial cells supports the functions of the human Atg16L1 and IRGM in xenophagy [32, 43, 44], it should be emphasized that the lessons from the mouse models are only suggestive and might not be merely translated into the roles of Atg variants in human diseases. Here, we summarize recent insights into the roles of human Atg16L1 and IRGM as well as the phenotype of autophagy–deficient animal models that are particularly consistent with the CD pathogenesis.
Strikingly, mice that are null for IRGM1 develop normally, have no apparent phenotype and yet, they are extremely susceptible to bacterial infection, all dying at 11–16 weeks postinfection [45]. Similarly, expression of the Atg16L1 T300A variant in human epithelial cells has no effect on constitutive or starvation-induced autophagy, whereas it dramatically impairs the clearance of bacteria by autophagy [43]. This suggests that the losses of IRGM– and Atg16L1– dependent autophagy are dispensable under normal physiological conditions and rather impair xenophagy. Therefore, these defects of xenophagy might lead to persistent infection, a feature particularly relevant for CD.
III. ROLE OF XENOPHAGY IN INNATE IMMUNITY: ARSENAL AGAINST INVASIVE BACTERIA
The innate immune system is the first line of defense against bacteria, and it ensures host survival until the onset of adaptive immunity. Compelling evidence has helped to unravel the critical role of xenophagy in innate defense as it immediately recognizes, captures and kills invading pathogens [46–49]. Prompt compartmentalization of bacteria within autophagosomes is a powerful strategy a) to restrict nutrient availability and thereby bacterial growth; b) to trap the pathogen within an enclosed compartment where the cell can target a surprise oxidative burst; c) to alter the environment—within autolysosomes, the pathogen faces an acidic pH and hydrolases, such as cathepsin D that have antibacterial activity. Furthermore, d) before lysis, the autophagic pathway generates ubiquitin-derived peptides that kill bacteria efficiently; a further security to resolve infection [50]. In addition to these defensive functions against invasive bacteria, autophagy protects cells against bacterial toxins and bacterially-induced apoptosis [51]. Through such an arsenal, this ubiquitous pathway ensures the survival of professional phagocytes (macrophages and neutrophils) but also of intestinal epithelial cells.
III.A. IRGM: A Safeguard Against Bacterial Infection
The human IRGM is a poorly understood regulator of xenophagy. It belongs to the p47 Immunity-Related GTPase (IRG) family, one of the most powerful innate resistance mechanisms against intracellular pathogens (Gram-positive and Gram-negative bacteria, mycobacteria, and protozoans) [52]. It derives its name from the unique presence of a methionine within its GTP-binding motif [53, 54]. The mouse homologs, Irgm1 to Irgm3, are GTPases that upon GTP binding form regulatory homodimers and heterodimers with the other IRG proteins [55–57]. However, the human IRGM has a truncated GTP-binding domain and its GTP-binding properties have not been yet explored.
In uninfected cells, the IRG proteins reside in the endoplasmic reticulum (ER) and cis-Golgi complex [58–60]. Upon infection, the murine Irgm1 proteins translocate within minutes to the plasma membrane at the phagocytic cup as a pathogen (e.g., M. tuberculosis, L. monocytogenes) enters into the cell, and remain associated with the pathogen-containing phagosome as it matures [49, 59, 60]. Consistently, expression of human IRGM confers resistance against bacterial infection [32, 44]. It has been proposed that IRGM may protect the host against intracellular bacteria (a) by driving vesiculation and disruption of the phagosome, releasing the pathogen out of its protective niche into the cytosol (inset (Fig. 1B)) [57, 60, 61]; (b) then it may help drive the engulfment of bacteria into large autophagic vesicles through addition of IRGM-containing vesicles [61]; and (c) finally it may target the pathogen for lysosomal degradation by recruiting the autophagic machinery [44, 49, 60, 61]. At the time of writing, the exact roles of human IRGM, its regulation and its autophagic partners remain to be identified.
Consequences of IRGM Variation
Two polymorphisms of IRGM have been highly correlated with CD risk: a “silent” tag-SNP variation within the coding region (rs10065172, C313T)[18]; and a 20-kb deletion upstream of the IRGM gene [32] (Fig. 1A). Of particular interest, the deletion polymorphism is in perfect linkage disequilibrium (r2 = 1.0) with the tag-SNP; the CD risk haplotype carries therefore both the deletion and the T allele, whereas the protective (reference) haplotype carries the C allele. As a result, the consequence of these variations on IRGM expression is likely a combination of both: given its location (2.7 kb before the IRGM transcription start), the deletion polymorphism may lie within regulatory sequences that affect IRGM mRNA transcription, whereas the exonic (C313T) tag-SNP might have no effect, or might affect protein translation.
Remarkably, these variations were found to stimulate IRGM expression in some cell types while suppressing it in others. Indeed, HeLa cells heterozygous for IRGM haplotypes almost exclusively express the C allele arising from the protective haplotype. Similarly, lymphoblastoid cells from heterozygous individuals express the C allele more strongly than the T allele. By contrast, colon carcinoma cells, HCT116, express the CD risk haplotype more strongly than the C allele [32]. Therefore, the pattern of IRGM expression might be different between individuals carrying the risk or protective alleles, even though the IRGM protein is identical in both populations. The reason for such unique expression profiles will undoubtedly be the subject of future studies.
III.B. Atg16L1: An Essential Component of the Autophagic Machinery Involved in Host Defense and Paneth Cell Biology
Atg16L1 is a scaffold protein with an N-terminal protein interaction domain, a coiled coil domain and seven C-terminal WD repeats [62–64] (Fig. 1A). It interacts constitutively with Atg12–Atg5 protein conjugates via its N-terminal domain, and Atg16L1 self-assembles via its coiled-coil domain, thus forming a high molecular weight complex (Atg12–Atg5-Atg16) of ~800 kDa. Within this platform, Atg16L1 is absolutely required for the localization of the complex to the phagophore and the formation of autophagosomes via the conjugation of LC3 to phosphatidylethanolamine [65] (Fig. 1B). As a result, the loss of Atg16L1 severely impairs autophagosome formation, degradation of long-lived proteins and the clearance of bacteria within both immune and epithelial cells [15, 32, 40, 43, 44, 66].
Recently, Cadwell et al. (2008) provide the unexpected demonstration that Atg16L1, and likely the entire autophagy process, is important for the biology of the Paneth cell, a specialized epithelial cell that releases antimicrobial peptides (such as lysozyme and α-defensin) into the intestinal lumen [66]. Indeed, ATG16L1–deficient Paneth cells exhibit not only defective granule exocytosis but also unexpected overexpression of two adipocytokines, leptin and adiponectin, known to directly influence intestinal injury responses. Similarly, CD patients homozygous for the ATG16L1 CD risk allele display comparable Paneth cell granule defects and increased leptin levels [66].
Consequences of ATG16L1 Variation
The consequences of the T300A variant on Atg16L1 function are just beginning to be understood. The nature of the amino acid exchange (polar threonine to nonpolar alanine) and its location (close to a WD domain) suggest that this polymorphism would modify Atg16L1 conformation and interaction [14]. However, the region surrounding T300 is not required for Atg16L1 dimerization or binding to Atg12–Atg5, and consistent with this observation, T300A variants are fully competent in the formation of autophagosomes in both constitutive and starvation-induced conditions in atg16Δ yeast mutant and ATG16L1−/− fibroblasts [43, 67]. Strikingly, Kuballa et al. demonstrate that Atg16L1 T300A-expressing cells are specifically defective in the capture of internalized Salmonella within autophagosomes [43]. This raises the attractive possibility that WD-repeats of Atg16L1 may provide a platform for the Atg16L1 complex to interact with other proteins required for xenophagy, the nature of which remains to be identified. Of particular interest, such defects are only observed in the absence of the wild-type ATG16L1 gene and upon bacterial infection. Therefore, people homozygous for the ATG16L1 T300A allele, who have an increased CD risk, are likely to be competent for the homeostatic functions of autophagy, yet may exhibit altered responses to bacterial infection in the intestinal microenvironment, where the microbial burden is high.
Intriguingly, another recent study reports that the ATG16L1 T300A variant has little, if any, effect on xenophagy in fibroblasts [67]. This likely illustrates that the subtle consequence of the T300A variation might be critically dependent on the cell system examined; in this case, human gut epithelial Caco2 cells [43] versus murine fibroblast [67]. Alternatively, we cannot rule out the possibility that T300A may be a tag-SNP for another causal variant.
III.C. Dosage Effects of Atg16L1 and IRGM Variants in CD Disease
Compelling evidence indicates that a critical threshold of IRGM and Atg16L1 expression needs to be reached to trigger full xenophagy. Indeed, homozygotes for the ATG16L1 T300A variant have a greater risk than heterozygotes, suggesting a potential gene dosage effect [68]. Exposure to pathogens increases the expression of autophagy machinery such as LC3B, Beclin 1, and Irgm1 to upregulate xenophagy. Consistent with this finding, overexpression of human IRGM confers protection against bacterial infection and, conversely, its removal by siRNA renders macrophages and epithelial cells more permissive for microbial replication [32, 44]. Similarly, the bacterial load within autophagic vesicles is reduced by ATG16L1 disruption [15]. A lower rate of xenophagy is correlated with a decrease in both Atg16L1 T300A and Atg5 expression levels [43]. Therefore, both IRGM and Atg16L1 expression levels may serve to regulate the rate of xenophagy. However, the cooperative roles of IRGM and Atg16L1 in xenophagy remain unknown.
Altogether, this points to the attractive pathogenesis model that the germline ATG16L1 and IRGM variations observed in CD patients would impair in vivo innate resistance in all sentinel cells (immune cells, Paneth cells, and enterocytes) and thereby trigger excessive inflammation as a result of increased bacterial load.
IV. ROLE OF AUTOPHAGY IN ADAPTIVE IMMUNE RESPONSES
IV.A. Bacterial Antigen Presentation on MHC Class II
The role of autophagy in immunity is not limited to pathogen clearance. Autophagy also promotes the second wave of adaptive immune responses by delivering cytosolic antigens to autolysosomes, where they are degraded and loaded onto major histocompatibility complex class II (MHC II) molecules [69–72]. Peptide–MHC II complexes are then presented onto the cell surface and recognized by T cells with their specific T-cell receptor (TCR) [73]. One can therefore assume that the autophagic degradation of intracellular pathogens would provide an important source of bacterial antigens for MHC class II presentation. Such antigen delivery by autophagy might be relevant for professional antigen-presenting cells (dendritic cells, macrophages) and more particularly for epithelial cells with little or no phagocytic capacity.
IV.B. Autophagy as a Regulator of T Cell Life and Death
Inflammatory bowel diseases are associated with sustained TH1, TH2 or TH17 cell responses. As a result, most of the attention in treating IBD has focused on neutralizing the immune response. Normally, homeostasis of the immune response is tightly regulated by a balanced T cell growth and death in which T cell numbers drastically increase in response to their specific antigens and decrease upon antigen clearance.
Of particular importance, autophagy has recently emerged as a key player in the control of T-cell life span. The first indication for a pro-death function of autophagy comes from in vitro studies linking the induction of autophagy to conditions that model T cell homeostasis. Indeed, autophagy is induced upon TCR, IL-2 stimulation as well as growth-factor withdrawal in both TH1 and TH2 cells. Interestingly, the induction of autophagy by growth-factor depletion is much more robust and persistent in TH2 than in TH1 cells, leading to TH2 cell death [74]. In contrast, another study demonstrates that activation of autophagy ensures in vivo T cell survival. Autophagy-deficient (Atg5−/−) mice exhibit multiple defects, including a severe reduction of thymocytes (40% of control) and of peripheral T- and B-lymphocytes (only 10% of control), and an inability to undergo TCR-induced proliferation [75]. Therefore, autophagy may control the duration and strength of adaptive responses by executing both T cell life and death decisions.
A challenge for the future will be to define the consequences of the Atg16L1 and IGRM variants on T cell immune responses. Recently, analyses of IRGM1−/− mice reveal a critical role for Irgm1 in the survival of mature CD4+ T cells [76] and in the renewal of hematopoietic stem cells upon infection [77]. Strikingly, this GTPase was dispensable for haematopoiesis under steady-state conditions. As a result, the IRGM1 null mice have no apparent phenotype, but develop upon infection a profound lymphopenia [45, 60, 78]. Thus, the murine Irgm1 is absolutely required for the expansion of antigen-specific TH1 populations to combat effectively intracellular infection. From a therapeutic perspective, it will be imperative to find out to what extent a decreased autophagy, due to ATG16L1 and IGRM variants, might affect T cell survival upon infection.
IV.C. Autophagy in Tolerance and Autoimmunity
High levels of autophagy are observed in thymic epithelial cells [79], and consistent with this finding autophagy is involved in the presentation of self-antigens and the maintenance of CD4+ T-cell tolerance through thymic T-cell selection [80]. Similar to CD pathogenesis, a defect in autophagy in thymic epithelial cells is sufficient to lead to a breakdown in self-tolerance and the development of autoimmune diseases in the colon [80]. It is therefore tempting to speculate that decreased macroautophagy caused by ATG16L1 or IRGM variants might lead to insufficient tolerance against commensals or self-antigens in the gut; a possible explanation for some of the T cell– driven pathologies observed in Crohn's disease.
V. CROSSTALK BETWEEN AUTOPHAGY AND INFLAMMATION: STRUGGLE FOR LIFE
V.A. Activation of Autophagy by Inflammatory and Immune Signals
Crohn's disease is characterized by increased production of TH1 cytokines such as TNF-α and IFN-γ. Interestingly, the relationship between autophagy and immunity is reciprocal: autophagy does enhance adaptive immune responses, and in turn cytokines and receptors involved in innate and adaptive immunity upregulate autophagy. Immune and inflammatory signals that positively regulate autophagy include the bacterial LPS [81], the TH1 cytokines IFN-γ [44, 49, 61], and members of TNF family (TNF-α, TRAIL and CD40L) [82–87]. Interestingly, both IFN-γ and LPS induce autophagy in infected epithelial cells through upregulation of murine Irgm1 [44, 49, 61, 81, 82]. Such an amplification loop connecting autophagy and immunity might contribute to an effective resolution of infection [73].
Of note, autophagy is negatively regulated by the TH2-type cytokines, IL-4 and IL-13 both in epithelial [83, 88–90] and immune cells [91], a finding that may help explain the negative role of TH2-cell response in the control of intracellular pathogens.
V.B. Control of Inflammation by the Autophagy Pathway: Pandora's Box
Autophagy may help to control the intensity and duration of the inflammatory responses not only by eliminating pathogens but also by blocking cell necrosis, protecting cells from oxidative stress and limiting the production of inflammatory cytokines.
Engulfment of Apoptotic Cells
The rapid removal of apoptotic corpses is crucial for prevention of unwanted inflammation [92]. Recently, it was demonstrated that the activation of autophagy in dying cells is essential for the exposure of an “eat-me” signal, phosphatidylserine on the cell surface, and the secretion of the “come-get-me” signal, lysophos-phatidylcholine [93]. Indeed, autophagy-deficient Atg5−/− embryos that show impaired clearance of apoptotic cells, display increased inflammation in lung [93].
Protecting Cells From Oxidative Stress
The intestinal mucosa of CD-affected patients shows a massive oxidative damage. To minimize such an oxidative environment, the generation of reactive oxygen species (ROS) within immune and epithelial cells upregulates autophagy to efficiently degrade the pathogen, the damaged toxic proteins and the mitochondria [94]. Consistent with this observation, cells with defective autophagy accumulate dysfunctional mitochondria that generate higher levels of ROS, even under normal growth conditions [95].
Inactivation of TLR4-Induced Inflammation (Fig. 2A)
Fig. (2). Roles of Autophagy in Chronic Inflammation and Cancer Development.
(A) Shown here are the phenotypes of ATG16L1 and IRGM transgenic mice on the production of proinflammatory cytokines. (B) Complex Roles of Autophagy in Cancer Development. Shown in green are the oncogenes, tumor suppressors and ATG genes mutated in human colorectal cancers. In the right panel, enhanced expression of key Atg proteins, LC3 and Beclin 1, is observed in early and advanced stages of CRC. Images reproduced with permission from [110].
In the last couple of months, autophagy has emerged as a key mechanism in inflammation resolution, specifically in response to TLR4 activation. Toll-like receptor-4 (TLR4) is the master sensor of bacterial infection that promotes the expression of pro-inflammatory cytokines such as interleukin-1β (IL1-β) and TNF-α. Of particular interest, Saitoh et al. show that following stimulation with LPS, a ligand for TLR4, Atg16L1-deficient macrophages produce high amounts of the inflammatory cytokines IL1-β and IL18 [40]. Moreover, mice lacking ATG16L1 in hematopoietic cells are highly susceptible to dextran sulfate sodium-induced acute colitis, which is alleviated by injection of IL1-β and IL18 antibodies [40]. Similarly, a defect in IRGM1 contributes in mice not only to overproduction of IL6 and TNF by LPS-activated macrophages [96] but also to sustained recruitment of neutrophils at the site of infection, and granuloma [97, 98]. At the molecular level, Atg16L1 selectively suppresses the production of IL1-β and IL18 via the TRIF/inflammasome arm of TLR4 [40], whereas Irgm1 selectively downregulates the production of TNF-α and IL6 through the MyD88-dependent arm [96]. Such specific and complementary suppression of TLR4 signalling by Atg16L1 and Irgm1 may benefit the host by preventing the excessive production of inflammatory cytokines. In this context, one can assume that a defective autophagy by ATG16L1 and IRGM variants would initiate excessive inflammation in response to infection.
This crosstalk might have important physiological and pathological implications. Clearly, the resolution of bacterial infection requires the concerted action of inflammation and autophagy that are simultaneously activated. However, cells do not cannibalize themselves by an uncontrolled autophagic process. Likewise, both inflammation and autophagy are transient, reaching maximal level during bacterial infection and then gradually decaying upon bacterial clearance. We therefore propose that this sophisticated cross-inhibition between autophagy and inflammation may safeguard against uncontrolled autophagy and inflammation.
VI. AUTOPHAGY: A “TWO-FACED” PATHWAY IN COLORECTAL CANCER
Colon cancer is the second most common cause of tumor-related death worldwide with 900,000 new cases and over 500,000 deaths per year [99, 100]. Of particular interest, it is well recognized that colorectal cancers (CRC) often arise at sites of chronic inflammation, with IBD patients having a six-fold higher risk of developing this cancer. However, there is no specific treatment and no prognostic marker predictive of cancer in this risk population
Interestingly, autophagy seems to be at the heart of tumorigenesis, acting as a potent tumor suppressor pathway (Fig. 2B). During tumorigenesis, autophagy is downregulated by several oncogenes (i.e., PI3K/Akt, Ras) and tumor suppressors (i.e., PTEN and p53) that are frequently mutated in CRC (for review see [101]). Moreover, several ATG genes such as the ultraviolet radiation resistance-associated gene (UVRAG), Atg2, Atg4b and Atg12 are mutated or downregulated in CRC [102–105]. One consensus that therefore emerges is that autophagy suppresses tumor initiation. Conversely, an enhanced autophagy may also promote the progression of established CRC. Enhanced expression of key Atg proteins, LC3 and Beclin 1, is observed in early and advanced stages of CRC [106–108], enabling likely cancer cells to endure hypoxia, and nutrient limitation in the inner area of the tumor [109, 110] and later to survive anoikis during metastasis [111]. Not surprisingly, depending on the conditions, upregulation of autophagy may protect colorectal cancer cells against anticancer treatments such as 5-fluorouracil [112], or may alternatively result in cell death [113–115].
Despite the huge amount of work, there is no marker to predict the course of Crohn's disease, its relapse or recurrence, and the risk of cancer complications. It will be therefore important to determine whether ATG16L1 and IRGM variants may predispose to colorectal cancer. In this regard, evidence of a frequent overexpression of Atg16L1 protein during oral carcinogenesis is of particular interest [116].
VII. ROLE OF AN AUTOPHAGY DEFECT IN CROHN’S DISEASE: TOWARDS A UNIFYING MODEL FROM BACTERIAL INFECTION, AND INFLAMMATION TO CANCER
Altogether, these findings suggest that ATG16L1 and IRGM variants might contribute to Crohn's disease pathogenesis through a xenophagy defect. Especially, the ATG16L1 and IRGM1-deficient mice have abnormalities in innate immunity, T-cell immune responses, intestinal Paneth cells, and inflammation, phenotypic features that are particularly relevant to intestinal CD inflammation.
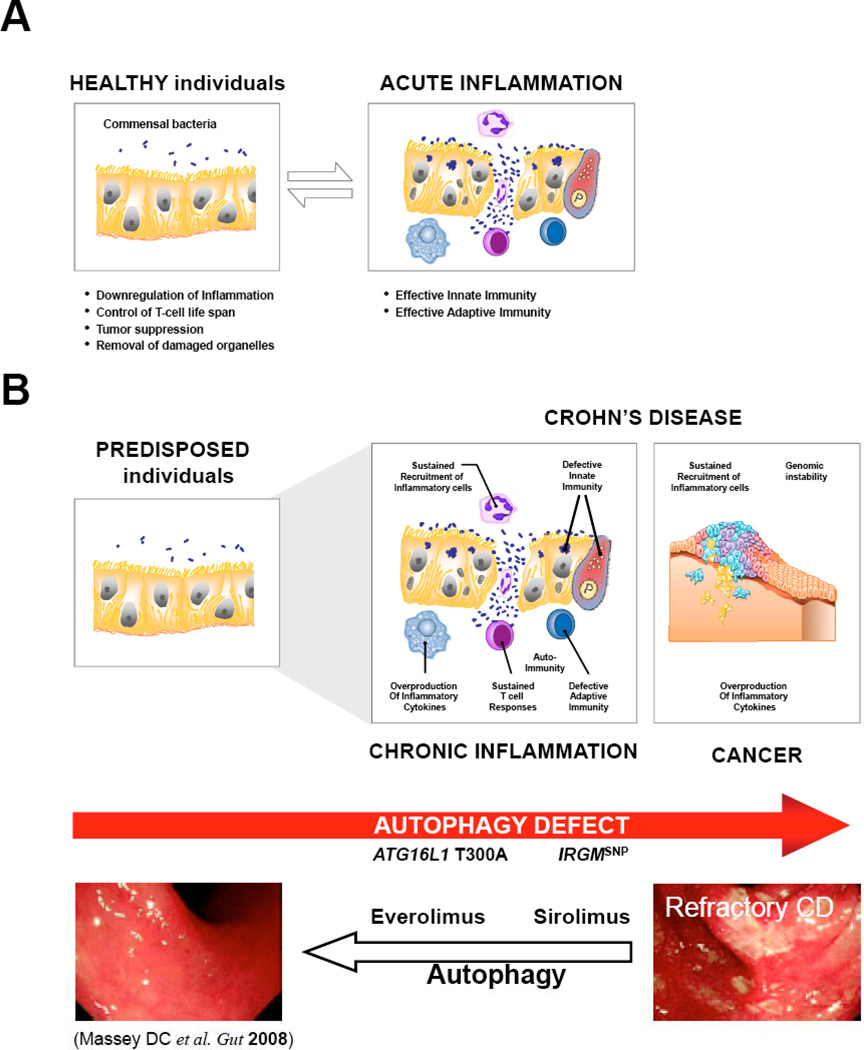
We therefore propose an autophagy model that may help in understanding the CD pathogenesis from impaired bacterial clearance, to exaggerated adaptive response and inflammation, to the development of colorectal cancers with a worsening prognosis (Fig. 3B). (a) It is likely that defective autophagy might underlie the persistence of commensal gut bacteria, particularly invasive E. coliwithin tissue macrophages and intestinal epithelial cells of individuals with Crohn’s disease [117]. (b) Such autophagic failure to clear intracellular bacteria would adversely affect processing and presentation of antigen peptide by MHC II. (c) Thereby, defects in autophagy would trigger excessive inflammation by the sustained production of inflammatory cytokines and the recruitment of immune cells. (d) In turn, the chronic inflammation with a massive infiltration of neutrophils would create a genotoxic microenvironment that damages intestinal epithelial cells. (e) The housekeeping function of autophagy would be particularly critical in situations of oxidative stress. Autophagy–deficient cells would fail not only to remove damaged proteins and mitochondria but also to limit genome damage, a feature that may contribute to cell transformation and tumor initiation.
Fig. (3). Proposed Model of How a Dysregulated Autophagy May Play an Essential Role in Crohn’s Disease Pathogenesis.
See text for a detailed explanation. Images reproduced with permission from [129].
VIII. FUTURE DIRECTIONS: TRANSLATIONAL CHALLENGES TOWARDS INDIVIDUALIZED HEALTH CARE…
Despite recent progress, much work remains to be done to understand the functions of human IRGM and Atg16L1 in autophagy and CD pathogenesis. Indeed, most studies have focused on mouse proteins, however, although informative, the IRGM1 and ATG16L1 transgenic mice could not be used as an experimental model of human CD. Indeed, there are important differences between human and mouse gene regulation and, hence, the functions of IRGM and Atg16L1 might be slightly different. While there are 23 Irg genes in the mouse genome [118], only two identifiable IRG genes are seen in the human genome. Moreover, murine Irg genes are inducible by IFN-α, whereas the human IRGM is constitutively expressed and seems not to be responsive to IFN-α [118]. In addition, there are five splice isoforms of human IRGM, with various sequences added C-terminally to the partially truncated G-protein [118]. Similarly, at least six Atg16L1 isoforms have been identified in human, with different sequences between the coiled-coil region and WD repeats [119]. All these isoforms carry the variation (Fig. 1A); however, nothing is known about the function of these different IRGM and Atg16L1 isoforms during xenophagy or the consequence of the CD variants. Furthermore, the possibility that Atg16L1 and IRGM isoforms fulfill an autophagic-independent role cannot be excluded yet. Along these lines, it would be critical to explore interactions between the environment and the expression of these two autophagic risk factors. Indeed, autophagy is an adaptive response to environmental stresses. Of note, cigarette smoking is a well-established autophagy inducer and a risk factor for CD [120] and the risk of CD is significantly increased for smokers who are homozygous at the T300A/A locus [121]. It is worth noting that the pathogens studied so far are all highly virulent bacteria that are not the causative agents of CD: Mycobacterium tuberculosisgroup A Streptococcusand Salmonella enterica serovar Typhimurium the causative agent of tuberculosis, strep throat, and gastroenteritis respectively. Whatever the pathogen, the delivery of bacteria to autolysosomal degradation is improved when autophagy is upregulated by physiological (starvation), immune (TLR4, IFN-γ), or pharmacological (rapamycin) signals [81, 122]. Therefore, a better understanding of the regulation of autophagy could expedite the development of new therapeutic strategies for CD.
Prognosis
CD is a chronic disorder with an unpredictable disease course. Most likely, such clinical heterogeneity results from differences in genetic susceptibility and exposure to environmental factors. Along these lines, we should keep in mind that CD is a complex disorder that involves at least 30 distinct susceptibility loci (meta-analysis of GWA [23]). Of particular importance, all identified polymorphisms are independently associated, and alone only moderately increase the risk for disease development [23]. Individuals carrying each of 7 different risk alleles (including NOD2, IBD5, ATG16L1and IL23R) have 25 x-increased risks for CD development with a more severe disease course (necessity of operations and younger age of onset, <40 years) [123]. In the near future, it might be possible to detect a genetic risk signature that predicts the course of CD and the drug response. Then, clinicians might be able to identify patients with a poor prognosis and to adjust their treatment accordingly.
Effective Therapeutic Strategies
Treatments for CD include corticosteroid in the acute phase. Patients who relapse frequently require immune suppression (usually with azathioprine, 6-mercaptopurine or methotrexate) or anti-TNF antibody treatments such as infliximab. Although the majority of patients can be maintained in remission with the use of such treatments, a significant portion are non-responders at the beginning or lose response after a period on therapy [124]. As such, resectional surgery remains the sole option in up to 80% of CD patients over the long term [125]. However, even surgical procedures carry appreciable morbidity, as the bowel function seldom returns to a pre-disease level.
The recent identification of risk variants in autophagy genes provides the rationale to prioritize this pathway as a new potential target for drug development. Clinically available drugs that upregulate autophagy such as sirolimus and everolimus (two rapamycin analogs) may be helpful for treating CD [126]. This possibility was supported by the efficacy of everolimus in the prevention/treatment of colitis in IL10−/− mice; a well-characterized mouse model of CD [127]. In 2008, Dumortier et al. and Massey et al. reported two cases of patients with severe refractory CD who were successfully treated with everolimus (4 mg/day – 18 months [128]) or sirolimus (4 mg/day – 6 months [129]). Both studies described a sustained improvement in CD symptoms with normalization of Harvey–Bradshaw index, of serum inflammation markers and of endoscopic appearance (Fig. 3). These encouraging first case reports open the avenue where pharmacological manipulation of autophagy might be used for prevention of CD inflammation. But there remain, undoubtedly, many burning issues to address. In particular, it will be important to evaluate the efficacy, safety and long-term outcomes of upregulating autophagy. While sirolimus and erolimus were effective and well tolerated during the first months [128, 129], long-term systemic autophagy induction might be associated with important side-effects [130, 131]. Of particular concern, such a strategy might exacerbate the progression of established colorectal cancers, as suggested by certain studies [110, 132]. In contrast to these initial reports, a double-blind randomized multicenter study of everolimus (6 mg/day) has failed to demonstrate benefit versus placebo in patients with active CD [131]. Despite these caveats, we guess that pharmacogenetics would rapidly optimize the clinical response of such a strategy.
In conclusion, in less than two years we have witnessed with Crohn's disease the most successful application of GWA from the disease gene association to a therapeutic strategy. The challenge will be now to identify those patients who are more likely to respond and the combination of autophagy inducers that would be most effective to improve/slow down CD development.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. R. Bocciardi and I. Ceccherini for their helpful comments on the manuscript. PB is funded by grants from INCA, EAC is funded by the Alfred Benzon Foundation, AC and AB are funded by Infectiopôle and Ademe (Convention n° 08 62 C 0044).
REFERENCES
- 1.Jess T, Riis L, Vind I, Winther KV, Borg S, Binder V, Langholz E, Thomsen OO, Munkholm P. Inflamm. Bowel Dis. 2007;13:481–489. doi: 10.1002/ibd.20036. [DOI] [PubMed] [Google Scholar]
- 2.Stone M, Mayberry J, Baker R. Eur. J. Gastroenterol. Hepatol. 2003;15:1275–1280. doi: 10.1097/00042737-200312000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein CN, Blanchard JF, Kliewer E, Wajda A. Cancer. 2001;91:854–862. doi: 10.1002/1097-0142(20010215)91:4<854::aid-cncr1073>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 4.Jess T, Riis L, Jespersgaard C, Hougs L, Andersen PS, Orholm MK, Binder V, Munkholm P. Am. J. Gastroenterol. 2005;100:2486–2492. doi: 10.1111/j.1572-0241.2005.00224.x. [DOI] [PubMed] [Google Scholar]
- 5.Baumgart DC, Sandborn WJ. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 6.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 8.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher SA, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, Bridger S, van Deventer S, Forbes A, Nikolaus S, Lennard-Jones JE, Foelsch UR, Krawczak M, Lewis C, Schreiber S, Mathew CG. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 9.Bonen DK, Ogura Y, Nicolae DL, Inohara N, Saab L, Tanabe T, Chen FF, Foster SJ, Duerr RH, Brant SR, Cho JH, Nunez G. Gastroenterology. 2003;124:140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 10.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 11.Halfvarson J, Bresso F, D'Amato M, Jarnerot G, Pettersson S, Tysk C. Dig. Liver Dis. 2005;37:768–772. doi: 10.1016/j.dld.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kugathasan S, Baldassano R, Bradfield J, Sleiman P, Imielinski M, Guthery S, Cucchiara S, Kim C, Frackelton E, Annaiah K, Glessner J, Santa E, Willson T, Eckert A, Bonkowski E, Shaner J, Smith R, Otieno F, Peterson N, Abrams D, Chiavacci R, Grundmeier R, Mamula P, Tomer G, Piccoli D, Monos D, Annese V, Denson L, Grant S, Hakonarson H. Nat.Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 15.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wellcome Trust Case Control Consortium. Nature. 2007;447:661–678. [Google Scholar]
- 17.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, Demarche B, Gut I, Heath S, Foglio M, Liang L, Laukens D, Mni M, Zelenika D, Van Gossum A, Rutgeerts P, Belaiche J, Lathrop M, Georges M. PLoS.Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raelson J, Little R, Ruether A, Fournier H, Paquin B, Van Eerdewegh P, Bradley W, Croteau P, Nguyen-Huu Q, Segal J, Debrus S, Allard R, Rosenstiel P, Franke A, Jacobs G, Nikolaus S, Vidal J, Szego P, Laplante N, Clark H, Paulussen R, Hooper J, Keith T, Belouchi A, Schreiber S. Proc. Natl. Acad. Sci. USA. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T, Saito S, Sekine A, Iida A, Takahashi A, Tsunoda T, Lathrop M, Nakamura Y. Hum. Mol. Genet. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Zhang H, Kugathasan S, Annese V, Bradfield J, Russell R, Sleiman P, Imielinski M, Glessner J, Hou C, Wilson D, Walters T, Kim C, Frackelton E, Lionetti P, Barabino A, Van Limbergen J, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant S, Satsangi J, Baldassano R, Hakonarson H. Am. J. Hum. Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhernakova A, Festen E, Franke L, Trynka G, van Diemen C, Monsuur A, Bevova M, Nijmeijer R, van 't Slot R, Heijmans R, Boezen H, van Heel D, van Bodegraven A, Stokkers P, Wijmenga C, Crusius J, Weersma R. Am. J. Hum. Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, RJ X, NIDDK IBD Genetics Consortium. Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, E L, Consortium. B-FI, Wellcome Trust Case Control Consortium. Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly M. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C, Barrett JC, Cummings FR, Drummond H, Lees CW. Nat. Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M, Schreiber S. Nat. Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 26.Zhi J, Zhi FC, Chen ZY, Yao GP, Guan J, Lin Y, Zhang YC. Nan. Fang. Yi. Ke. Da. Xue. Xue. Bao. 2008;28:649–651. [PubMed] [Google Scholar]
- 27.Zhang HF, Qiu LX, Chen Y, Zhu WL, Mao C, Zhu LG, Zheng MH, Wang Y, Lei L, Shi J. Hum. Genet. 2009;125:627–631. doi: 10.1007/s00439-009-0660-7. [DOI] [PubMed] [Google Scholar]
- 28.Perricone C, Borgiani P, Romano S, Ciccacci C, Fusco G, Novelli G, Biancone L, Calabrese E, Pallone F. Gastroenterology. 2008;134:368–370. doi: 10.1053/j.gastro.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Latiano A, Palmieri O, Valvano MR, D'Incà R, Cucchiara S, Riegler G, Staiano AM, Ardizzone S, Accomando S, de Angelis GL, Corritore G, Bossa F, Annese V. World J. Gastroenterol. 2008;14:4643–4651. doi: 10.3748/wjg.14.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. J. Hum.. Genet. 2007;52:575–583. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]
- 31.Baptista ML, Amarante H, Picheth G, Sdepanian VL, Peterson N, Babasukumar U, Lima HC, Kugathasan S. Inflamm. Bowel Dis. 2008;14:674–679. doi: 10.1002/ibd.20372. [DOI] [PubMed] [Google Scholar]
- 32.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Gene.s Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 34.Cummings JR, Cooney R, Pathan S, Anderson CA, Barrett JC, Beckly J, Geremia A, Hancock L, Guo C, Ahmad T, Cardon L, Jewell DP. Inflamm. Bowel Dis. 2007;13:941–946. doi: 10.1002/ibd.20162. [DOI] [PubMed] [Google Scholar]
- 35.Levine B, Kroemer G. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hait WN, Jin S, Yang JM. Clin. Cancer Res. 2006;12:1961–1965. doi: 10.1158/1078-0432.CCR-06-0011. [DOI] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Emr SD. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkegaard K, Taylor MP, Jackson WT. Nat. Rev. Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine B. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 41.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 42.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh SB, Davis AS, Taylor GA, Deretic V. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 45.Feng CG, Collazo-Custodio CM, Eckhaus M, Hieny S, Belkaid Y, Elkins K, Jankovic D, Taylor GA, Sher A. J. Immunol. 2004;172:1163–1168. doi: 10.4049/jimmunol.172.2.1163. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 47.Rich KA, Burkett C, Webster P. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 48.Cossart P, Sansonetti PJ. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 49.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 50.Alonso S, Pethe K, Russell DG, Purdy GE. Proc. Natl. Acad.. Sci. USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutierrez MG, Saka HA, Chinen I, Zoppino FC, Yoshimori T, Bocco JL, Colombo MI. Proc. Natl. Acad. Sci. USA. 2007;104:1829–1834. doi: 10.1073/pnas.0601437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor GA, Feng CG, Sher A. Nat. Rev. Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 53.Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, Fütterer A, Pfeffer K, Howard JC. J. Immunol. 1998;161:6715–6723. [PubMed] [Google Scholar]
- 54.Taylor GA, Jeffers M, Largaespada DA, Jenkins NA, Copeland NG, Woude GF. J. Biol. Chem. 1996;271:20399–20405. doi: 10.1074/jbc.271.34.20399. [DOI] [PubMed] [Google Scholar]
- 55.Uthaiah R, Praefcke GJM, Howard JC, Herrmann C. J. Biol. Chem. 2003;278:29336–29343. doi: 10.1074/jbc.M211973200. [DOI] [PubMed] [Google Scholar]
- 56.Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC. EMBO J. 2008;27:2495–2509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC. PLoS. Pathogens. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor GA, Stauber R, Rulong S, Hudson E, Pei V, Pavlakis GN, Resau JH, Vande Woude GF. J. Biol. Chem. 1997;272:10639–10645. doi: 10.1074/jbc.272.16.10639. [DOI] [PubMed] [Google Scholar]
- 59.Martens S, Sabel K, Lange R, Uthaiah R, Wolf E, Howard JC. J. Immunol. 2004;173:2594–2606. doi: 10.4049/jimmunol.173.4.2594. [DOI] [PubMed] [Google Scholar]
- 60.MacMicking J, Taylor GA, McKinney J. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 61.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. J. Exp. Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 63.Mizushima N, Noda T, Ohsumi Y. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 65.Fujita N, Itoh T, Fukuda M, Noda T, Yoshimori T. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW., 4th Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.037671. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okazaki T, Wang MH, Rawsthorne P, Sargent M, Datta LW, Shugart YY, Bernstein CN, Brant SR. Inflamm. Bowel Dis. 2008;14:1528–1541. doi: 10.1002/ibd.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crotzer VL, Blum JS. Proc. Natl. Acad. Sci. USA. 2005;102:7779–7780. doi: 10.1073/pnas.0503088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S. Proc. Natl. Acad. Sci. USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dorfel D, Appel S, Grunebach F, Weck MM, Muller MR, Heine A, Brossart P. Blood. 2005;105:3199–3205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 72.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 73.Schmid D, Pypaert M, Münz C. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. J. Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 75.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. J. Exp. Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng CG, Zheng L, Jankovic D, Báfica A, Cannons JL, Watford WT, Chaussabel D, Hieny S, Caspar P, Schwartzberg PL, Lenardo MJ, Sher A. Nat. Immunol. 2008;9:1279–1287. doi: 10.1038/ni.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng CG, Weksberg DC, Taylor GA, Sher A, Goodell MA. Cell Stem Cell. 2008;2:83–89. doi: 10.1016/j.stem.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santiago HC, Feng CG, Bafica A, Roffe E, Arantes RM, Cheever A, Taylor G, Vierira LQ, Aliberti J, Gazzinelli RT, Sher A. J. Immunol. 2005;175:8165–8172. doi: 10.4049/jimmunol.175.12.8165. [DOI] [PubMed] [Google Scholar]
- 79.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. Mol. Biol.Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 81.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. J. Clin. Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, Pierron G, Codogno P. J. Biol. Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 84.Jia G, Cheng G, Gangahar DM, Agrawal DK. Immunol. Cell Biol. 2006;84:448–454. doi: 10.1111/j.1440-1711.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 85.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Proc. Natl. Acad.. Sci. USA. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Subauste CS, Andrade RM, Wessendarp M. Autophagy. 2007;3:245–248. doi: 10.4161/auto.3717. [DOI] [PubMed] [Google Scholar]
- 87.Thorburn J, Moore F, Rao A, Barclay WW, Thomas LR, Grant KW, Cramer SD, Thorburn A. Mol. Biol. Cell. 2005;16:1189–1199. doi: 10.1091/mbc.E04-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. J. Biol. Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 89.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 90.Wright K, Ward SG, Kolios G, Westwick J. J. Biol. Chem. 1997;272:12626–12633. doi: 10.1074/jbc.272.19.12626. [DOI] [PubMed] [Google Scholar]
- 91.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 92.Maderna P, Godson C. Biochim.. Biophys. Acta. 2003;1639:141–151. doi: 10.1016/j.bbadis.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 93.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 94.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Proc. Natl. Acad. Sci. USA. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bafica A, Feng CG, Santiago HC, Aliberti J, Cheever A, Thomas KE, Taylor GA, Vogel SN, Sher A. J. Immunol. 2007;179:5514–5522. doi: 10.4049/jimmunol.179.8.5514. [DOI] [PubMed] [Google Scholar]
- 97.Miyairi I, Tatireddigari VR, Mahdi OS, Rose LA, Belland RJ, Lu L, Williams RW, Byrne GI. J. Immunol. 2007;179:1814–1824. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- 98.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, Oliver T, Taylor GA. J. Immunol. 2007;179:6963–6972. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 99.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. CA Cancer J. Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 100.Boyle P, Ferlay J. Nat. Clin. Pract. Oncol. 2005;2:424–425. doi: 10.1038/ncponc0288. [DOI] [PubMed] [Google Scholar]
- 101.Hippert MM, O'toole PS, Thorburn A. Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 102.Lee JW, Jeong EG, Lee SH, Yoo NJ, Lee SH. APMIS. 2007;115:750–756. doi: 10.1111/j.1600-0463.2007.apm_640.x. [DOI] [PubMed] [Google Scholar]
- 103.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH. J. Pathol. 2009;217:702–706. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- 104.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 105.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Hum. Pathol. 2008;39:1059–1063. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 106.Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, Takiguchi S, Fujiwara Y, Uchiyama Y, Monden M. Int. J. Oncol. 2008;33:461–468. [PubMed] [Google Scholar]
- 107.Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. APMIS. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- 108.Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, Zhu XF, Zhang XS. Autophagy. 2009;5:303–306. doi: 10.4161/auto.5.3.7491. [DOI] [PubMed] [Google Scholar]
- 109.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 111.Fung C, Lock R, Gao S, Salas E, Debnath J. Mol. Biol. Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li J, Hou N, Faried A, Tsutsumi S, Takeuchi T, Kuwano H. Ann. Surg. Oncol. 2009;16:761–771. doi: 10.1245/s10434-008-0260-0. [DOI] [PubMed] [Google Scholar]
- 113.Trincheri NF, Follo C, Nicotra G, Peracchio C, Castino R, Isidoro C. Carcinogenesis. 2008;29:381–389. doi: 10.1093/carcin/bgm271. [DOI] [PubMed] [Google Scholar]
- 114.Comes F, Matrone A, Lastella P, Nico B, Susca FC, Bagnulo R, Ingravallo G, Modica S, Lo Sasso G, Moschetta A, Guanti G, Simone C. Cell Death Differ. 2007;14:693–702. doi: 10.1038/sj.cdd.4402076. [DOI] [PubMed] [Google Scholar]
- 115.Ellington AA, Berhow MA, Singletary KW. Carcinogenesis. 2006;27:298–306. doi: 10.1093/carcin/bgi214. [DOI] [PubMed] [Google Scholar]
- 116.Nomura H, Uzawa K, Yamano Y, Fushimi K, Ishigami T, Kouzu Y, Koike H, Siiba M, Bukawa H, Yokoe H, Kubosawa H, Tanzawa H. Hum. Pathol. 2009;40:83–91. doi: 10.1016/j.humpath.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 117.Darfeuille-Michaud A. Int. J. Med. Microbiol. 2002;292:185–193. doi: 10.1078/1438-4221-00201. [DOI] [PubMed] [Google Scholar]
- 118.Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn M, Glowalla E, Leptin M, Howard JC. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng H, Ji C, Li J, Jiang H, Ren M, Lu Q, Gu S, Mao Y, Xie Y. DNA Seq. 2004;15:303–305. doi: 10.1080/10425170400004104. [DOI] [PubMed] [Google Scholar]
- 120.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, Nakahira K, Pilewski JM, Lee JS, Zhang Y, Ryter SW, Choi AM. PLoS ONE. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fowler EV, Doecke J, Simms LA, Zhao ZZ, Webb PM, Hayward NK, Whiteman DC, Florin TH, Montgomery GW, Cavanaugh JA, Radford-Smith GL. Am. J.Gastroenterol. 2008;103:2519–2526. doi: 10.1111/j.1572-0241.2008.02023.x. [DOI] [PubMed] [Google Scholar]
- 122.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. Cell Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 123.Weersma RK, Stokkers PC, van Bodegraven AA, van Hogezand RA, Verspaget HW, de Jong DJ, van der Woude CJ, Oldenburg B, Linskens RK, Festen E, van der Steege G, Hommes DW, Crusius JB, Wijmenga C, Nolte IM, Dijkstra G, Dutch Initiative on Crohn and Colitis ICC. Gut. 2009;58:388–395. doi: 10.1136/gut.2007.144865. [DOI] [PubMed] [Google Scholar]
- 124.Baert F, Noman M, Vermeire S, Van Assche G, D' Haens G, Carbonez A, Rutgeerts P. N. Engl. J. Med. 2003;348:601–608. doi: 10.1056/NEJMoa020888. [DOI] [PubMed] [Google Scholar]
- 125.Cosnes J, Nion-Larmurier I, Beaugerie L, Afchain P, Tiret E, Gendre JP. Gut. 2005;54:237–241. doi: 10.1136/gut.2004.045294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Webster AC, Lee VW, Chapman JR, Craig JC. Transplantation. 2006;81:1234–1248. doi: 10.1097/01.tp.0000219703.39149.85. [DOI] [PubMed] [Google Scholar]
- 127.Matsuda C, Ito T, Song J, Mizushima T, Tamagawa H, Kai Y, Hamanaka Y, Inoue M, Nishida T, Matsuda H, Sawa Y. Clin. Exp. Immunol. 2007;148:348–359. doi: 10.1111/j.1365-2249.2007.03345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dumortier J, Lapalus M, Guillaud O, Poncet G, Gagnieu M, Partensky C, Scoazec J. Inflamm. Bowel Dis. 2008;14:874–877. doi: 10.1002/ibd.20395. [DOI] [PubMed] [Google Scholar]
- 129.Massey DC, Bredin F, Parkes M. Gut. 2008;57:1294–1296. doi: 10.1136/gut.2008.157297. [DOI] [PubMed] [Google Scholar]
- 130.Zhu J, Wu J, Frizell E, Liu SL, Bashey R, Rubin R, Norton P, Zern MA. Gastroenterology. 1999;117:1198–1204. doi: 10.1016/s0016-5085(99)70406-3. [DOI] [PubMed] [Google Scholar]
- 131.Reinisch W, Panés J, Lémann M, Schreiber S, Feagan B, Schmidt S, Sturniolo G, Mikhailova T, Alexeeva O, Sanna L, Haas T, Korom S, Mayer H. Am. J. Gastroenterol. 2008;103:2284–2292. doi: 10.1111/j.1572-0241.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- 132.Kahan BD, Yakupoglu YK, Schoenberg L, Knight RJ, Katz SM, Lai D, Van Buren CT. Transplantation. 2005;80:749–758. doi: 10.1097/01.tp.0000173770.42403.f7. [DOI] [PubMed] [Google Scholar]
- 133.Peterson N, Guthery S, Denson L, Lee J, Saeed S, Prahalad S, Biank V, Ehlert R, Tomer G, Grand R, Rudolph C, Kugathasan S. Gut. 2008;57:1336–1337. doi: 10.1136/gut.2008.152207. [DOI] [PubMed] [Google Scholar]
- 134.Baldassano RN, Bradfield JP, Monos DS, Kim CE, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Kanterakis S, Shaner JL, Smith RM, Eckert AW, Robinson LJ, Onyiah CC, Abrams DJ, Chiavacci RM, Skraban R, Devoto M, Grant SF, Hakonarson H. Gut. 2007;56:1171–1173. doi: 10.1136/gut.2007.122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A, Mansfield JC, Lewis CM, Schreiber S, Mathew CG. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 136.Hancock L, Beckly J, Geremia A, Cooney R, Cummings FR, Pathan S, Guo C, Warren BF, Mortensen N, Ahmad T, Jewell D. Inflamm. Bowel Dis. 2008;14:1667–1677. doi: 10.1002/ibd.20517. [DOI] [PubMed] [Google Scholar]
- 137.Van Limbergen J, Russell RK, Nimmo ER, Drummond HE, Smith L, Anderson NH, Davies G, Gillett PM, McGrogan P, Weaver LT, Bisset WM, Mahdi G, Arnott ID, Wilson DC, Satsangi J. Inflamm. Bowel Dis. 2008;14:338–346. doi: 10.1002/ibd.20340. [DOI] [PubMed] [Google Scholar]
- 138.Glas J, Konrad A, Schmechel S, Dambacher J, Seiderer J, Schroff F, Wetzke M, Roeske D, Török HP, Tonenchi L, Pfennig S, Haller D, Griga T, Klein W, Epplen JT, Folwaczny C, Lohse P, Göke B, Ochsenkühn T, Mussack T, Folwaczny M, Müller-Myhsok B, Brand S. Am. J. Gastroenterol. 2008;103:682–691. doi: 10.1111/j.1572-0241.2007.01694.x. [DOI] [PubMed] [Google Scholar]
- 139.Weersma RK, Zhernakova A, Nolte IM, Lefebvre C, Rioux JD, Mulder F, van Dullemen HM, Kleibeuker JH, Wijmenga C, Dijkstra G. Am. J. Gastroenterol. 2008;103:621–627. doi: 10.1111/j.1572-0241.2007.01660.x. [DOI] [PubMed] [Google Scholar]
- 140.Gaj P, Habior A, Mikula M, Ostrowski J. BMC Med.Genet. 2008;9:81. doi: 10.1186/1471-2350-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lakatos PL, Szamosi T, Szilvasi A, Molnar E, Lakatos L, Kovacs A, Molnar T, Altorjay I, Papp M, Tulassay Z, Miheller P, Papp J, The Hungarian IBD Study Group. Tordai A, Andrikovics H. Dig. Liver Dis. 2008;40:867–873. doi: 10.1016/j.dld.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 142.Roberts RL, Gearry RB, Hollis-Moffatt JE, Miller AL, Reid J, Abkevich V, Timms KM, Gutin A, Lanchbury JS, Merriman TR, Barclay ML, Kennedy MA. Am. J. Gastroenterol. 2007;102:2754–2761. doi: 10.1111/j.1572-0241.2007.01525.x. [DOI] [PubMed] [Google Scholar]