Abstract
Candida albicans and C. glabrata are the two most common pathogenic yeasts of humans, yet they are phylogenetically, genetically and phenotypically very different. In this review, we compare and contrast the strategies of C. albicans and C. glabrata to attach to and invade into the host, obtain nutrients and evade the host immune response. Although their strategies share some basic concepts, they differ greatly in their outcome. While C. albicans follows an aggressive strategy to subvert the host response and to obtain nutrients for its survival, C. glabrata seems to have evolved a strategy which is based on stealth, evasion and persistence, without causing severe damage in murine models. However, both fungi are successful as commensals and as pathogens of humans. Understanding these strategies will help in finding novel ways to fight Candida, and fungal infections in general.
Introduction
Fungi infect billions of people every year, but still remain largely under-appreciated as pathogens of humans (Brown et al., 2012). In fact, some fungal diseases have an extremely high mortality rate and fungi kill at least as many people as tuberculosis or malaria (Brown et al., 2012). Over the last decades, they have become a major problem especially in the clinical setting (Perlroth et al., 2007). Candida albicans and C. glabrata, for example, are ubiquitous commensals of humans, and can be found especially in the oral cavity and the gastrointestinal tract of most healthy humans (Cole et al., 1996; Fidel et al., 1999). On the other hand, they are also the most important pathogenic yeasts. The majority of the population is asymptomatically colonized by either of the two species, or even both (Li et al., 2007b). However, under certain predisposing factors, such as treatment with antibiotics, diabetes, cancer, extreme age, immunosuppression, intravenous catheters or long-term hospitalization, these fungi can lead to superficial or even life-threatening systemic infections, with high morbidity and mortality (Perlroth et al., 2007). Among the Candida species, C. albicans and C. glabrata rank first and second in isolation frequency, respectively, and, together, are responsible for approximately 65%–75% of all systemic candidiasis, followed by C. parapsilosis and C. tropicalis (Perlroth et al., 2007).
While C. albicans, C. parapsilosis and C. tropicalis are relatively closely related members of the so-called CUG clade (sharing a unique codon exchange from leucine to serine), C. glabrata is, as a Candida species, in fact a ‘misnomer’: this yeast is actually much more closely related to the baker's yeast Saccharomyces cerevisiae than to C. albicans (Dujon et al., 2004), and this is reflected by a number of differences. For example, unlike C. albicans, the progenitor of C. glabrata and S. cerevisiae experienced a whole-genome duplication event (Dujon et al., 2004). Additionally, C. albicans is a diploid, polymorphic fungus, switching readily from yeast to hyphal (and pseudohyphal) growth and back. In contrast, C. glabrata is strictly haploid and normally grows only in the yeast form (Kaur et al., 2005). The morphological flexibility of C. albicans seems to play fundamental roles in several aspects of infection (Sudbery, 2011). In contrast, the pathogenicity of C. glabrata seems to be independent of morphology. Yet, both fungi are closely associated with humans, and similarly successful as commensals and as pathogens. But how similar are their commensal and pathogenic strategies?
The phylogenetic tree, in which C. albicans (and the CUG clade) and C. glabrata are separated by several non-pathogenic yeasts, strongly implies that the ability to infect humans has evolved independently in the two species. Therefore, a closer look at the similarities and differences in the persistence and infection strategies of C. albicans and C. glabrata may help to understand general principles of fungal infections.
Adhesion
For any successful commensal and pathogen, adhesion to host cells is essential. Many bacteria possess elaborate systems to detect the presence of the host by environmental cues, in order to express specific adhesins (Kline et al., 2009). These factors in turn allow the microorganisms to attach firmly to a host cell, and help to prevent them from being washed away. Adhesins are also critical for the formation of biofilms: Both C. albicans and C. glabrata can form biofilms on abiotic substrates, especially medical devices such as catheters (Iraqui et al., 2005; Nobile and Mitchell, 2006).
Candida albicans and C. glabrata have large protein families of adhesins at their disposal: for C. albicans, the Als proteins with their Agglutinin-Like Sequences, are crucial adhesins (Hoyer et al., 2008). Especially the Als3 protein seems to play a vital role in adhesion, since its deletion strongly reduces adhesion to epithelial cells (Hoyer et al., 2008; Wächtler et al., 2012). In addition, this protein is also important for iron acquisition from ferritin (Almeida et al., 2008), formation of mixed-species biofilms (Silverman et al., 2010) and induction of endocytosis by host cells (Phan et al., 2007; Wächtler et al., 2012 and see also next section). As Als3 is hypha-associated (Argimon et al., 2007), it is expressed as the fungus forms filaments, for example upon physical contact with host cells, at body temperature and at ambient neutral pH. Expression of another important adhesin, Hwp1, is likewise hypha-associated (Staab et al., 1999), strengthening the concept of induced, hypha-associated adhesion, as opposed to the ‘ad hoc’ non-induced adhesion of attaching yeast cells (Wilson and Hube, 2010; Wächtler et al., 2011).
The main adhesins of C. glabrata, the Epa proteins, are related to the Flo proteins of S. cerevisiae, which are responsible for flocculation during the brewing process. In C. glabrata, this family of approximately 17–23 genes (depending on the isolate) allows attachment to epithelial cells (Cormack et al., 1999; Castano et al., 2005) and macrophages (Kuhn and Vyas, 2012). The predominant adhesin during in vitro interaction with epithelial cells is Epa1 (Cormack et al., 1999), whereas other Epa and non-Epa adhesins can mediate attachment to other cell types (de Groot et al., 2008; Desai et al., 2011; Kraneveld et al., 2011). Additionally, it has been suggested that the high expression heterogeneity of Epa1, and likely other adhesins, leads to distinct subsets of Epa-expressing cells in any population, each with different adhesion properties (Halliwell et al., 2012).
A unique mechanism for sensing the host environment has been described for the genes encoding C. glabrata's Epa proteins: their expression is regulated by Sir-complex-mediated transcriptional silencing (De Las Penas et al., 2003; Castano et al., 2005). This system requires NAD+ as a cofactor, and since C. glabrata is auxotrophic for the NAD+ precursor nicotinic acid (NA), the silencing activity is indirectly influenced by external NA concentrations. Because urine is low in nicotinic acid, EPA genes are derepressed in the urinary tract, promoting adhesion of C. glabrata in this niche (Domergue et al., 2005). Interestingly, no systematic studies have so far investigated whether the physiological body temperature has similar effects on the transcriptional programme, as is clearly the case in C. albicans.
In summary, both fungi have independently evolved specific adhesins which rely on different cues to detect the presence of the host. While the details differ, the basic principle is the same – upon detection of a ‘host’ environment, adhesins are expressed to attach to host cells.
Invasion
After attachment, the next step in Candida pathogenesis is invasion, normally into epithelial cell layers. For C. albicans, invasion can occur via two mechanisms: induced endocytosis by host cells, and active penetration by C. albicans hyphae (Wächtler et al., 2011b2012). Als3 is one of the invasins which trigger endocytosis by inducing host cytoskeletal rearrangements (Phan et al., 2007). This process does not require viable fungi and does not cause damage per se, but seems to be important at the early stages of invasion (Wächtler et al., 2011; 2012; Zhu et al., 2012). Yet, the dominant route of invasion is active penetration of host cells. Hyphae penetrate tissue by a combination of physical forces exerted by the extending filaments, the secretion of hydrolytic enzymes and as yet unknown damaging factors, which finally leads to disruption of host cell membranes (Wächtler et al., 2012). However, hyphae formation comes at a price: epithelial cells react to the hyphal surface, and to the inflicted damage, by secreting pro-inflammatory cytokines, demarcating the transition to a pathogenic lifestyle (Schaller et al., 2002; Moyes et al., 2010b2012). These events in turn attract macrophages and neutrophils (Schaller et al., 2004), which can fight and kill invading C. albicans.
Hypha-mediated penetration is widely assumed to play an important role in gaining access to deeper tissue and the bloodstream. How can C. glabrata enter the circulatory system without hyphae? A possible route to reach the bloodstream is the accidental or iatrogenic breach of natural barriers via trauma, catheters, surgery or parenteral nutrition (Perlroth et al., 2007). However, even in the absence of such breaches, C. glabrata yeasts invade into deeper tissues and readily disseminate in a chicken embryo model of fungal infections (Jacobsen et al., 2011). The mode of this invasion is unclear, although S. cerevisiae cells are known to form agar-invasive pseudohyphal under in vitro starvation conditions (Gimeno et al., 1992) and one report even described pseudohyphae of C. glabrata in vitro (Csank and Haynes, 2000). However, in vivo, it may rely on endocytosis, in lieu of active penetration, with close to no host cell damage (Li et al., 2007a). Most likely because of low host damage, the cytokine profile of epithelia infected with C. glabrata differs dramatically from that of C. albicans-infected cells: C. glabrata induces more GM-CSF than C. albicans, but nearly none of the other inflammatory cytokines like IL-1α or Il-8 (Schaller et al., 2002; Li et al., 2007a). This is in good agreement with the cytokine pattern observed during in vivo murine infections (Jacobsen et al., 2010). Consequently, strong neutrophil infiltration is characteristic for C. albicans infections, while C. glabrata either does not stimulate or is able to suppress neutrophil attraction and is rather associated with mononuclear cells (Westwater et al., 2007; Jacobsen et al., 2010). Overall, the presence of hyphae and host cell damage in C. albicans infections leads to a stronger pro-inflammatory cytokine response than in C. glabrata infections.
Interaction with immune cells
Macrophages are part of the first line of defence by the innate immune system. These phagocytes recognize invading fungi in the tissue via a subset of their PRRs (pattern recognition receptors) which specifically bind to fungal PAMPs (pathogen associated molecular pattern), such as β-glucan (Taylor et al., 2007), mannan or chitin (van der Meer et al., 2010; Mora-Montes et al., 2011). Interestingly, C. glabrata is recognized and ingested by macrophages at a much higher rate than C. albicans, and C. albicans yeasts more than hyphae (Keppler-Ross et al., 2010). These preferences seem to be mannan dependent, but independent of glucan or chitin (Keppler-Ross et al., 2010). Without intervention by the phagocytosed microbe, the phagosome normally matures via a series of fusion and fission events, and the pathogens are killed in the matured phagolysosome. Furthermore, by releasing cytokines upon recognition of pathogens, macrophages help to orchestrate the immune responses.
However, when C. glabrata is internalized by macrophages, it modifies the normal phagosome maturation process. It remains in a non-acidified organelle which lacks typical lysosomal markers such as cathepsin D (Seider et al., 2011). Similar to Histoplasma capsulatum (Woods, 2003; Seider et al., 2010) and bacteria like Mycobacterium spp. (de Chastellier, 2009), C. glabrata not only survives, but replicates inside the phagosome until the phagocyte finally bursts (Kaur et al., 2007; Roetzer et al., 2010; Seider et al., 2011). Other survival strategies include expression of a highly active catalase (Cuellar-Cruz et al., 2008) and pigment production, which may counteract the oxidative burst of phagocytes (Brunke et al., 2010). In addition, the cytokine response of macrophages to C. glabrata in vitro is much lower than during interaction with C. albicans (Seider et al., 2011). Consistently, murine and chicken embryo infections with C. glabrata lead only to a transient pro-inflammatory cytokine response, and only a minor influx of immune effector cells (Jacobsen et al., 2010; 2011). During in vitro experiments, C. albicans also appears to delay phagosome maturation and to induce recycling of late maturation markers like LAMP-1 (Fernández-Arenas et al., 2009). However, soon after these initial events, C. albicans starts to form hyphae which disrupt the macrophage membranes, effectively killing the phagocyte and allowing the fungus to rapidly escape (Lo et al., 1997; McKenzie et al., 2010). The macrophage phagosome is a nutrient-poor and harmful environment. Escape from phagocytes may therefore be a strategy of C. albicans to quickly escape this detrimental environment (Hummert et al., 2010), while C. glabrata depends on fungal autophagy to survive inside macrophages (Roetzer et al., 2010). Interestingly, even autophagy-deficient mutants of C. albicans can still escape the phagocytes and subsequently gain access to the external nutrients (Palmer et al., 2007). The in vitro observations of a quick escape from macrophages by C. albicans have been challenged recently by experiments in live zebrafish. In this model, C. albicans remained viable after phagocytosis, but hyphae formation was not observed (Brothers et al., 2011). How far this reflects the situation in the mammalian host, however, is largely unknown, especially since mammals differ from the ectothermic zebrafish model by their high body temperature – an important hypha-inducing factor. Additionally, a recent study even reported a rare non-lytic escape of C. albicans yeasts cells from macrophages (Bain et al., 2012), in a manner reminiscent of the process described for Cryptococcus neoformans (Alvarez and Casadevall, 2006). Thus, the precise in vivo escape strategies of C. albicans from phagocytes require further research.
Nutrient acquisition within the host
During infection, a molecular tug-of-war takes place between the host, which tries to restrict access to essential nutrients, and the pathogen, which needs these nutrients to survive and multiply. Acquisition of nutrients by the fungus is therefore central to establishing and maintaining infection. Interestingly, due to its frequent gene losses (Dujon et al., 2004), C. glabrata lacks many of the metabolic pathways known in other yeasts, including S. cerevisiae and C. albicans. It cannot catabolize galactose (due to its loss of genes GAL1, 7, 10) and allantoin (DAL1–4, 7), and is auxotrophic for pyridoxine (SNO1, 2, 3), nicotinic acid (BNA1–6) and thiamine (Dujon et al., 2004; Wong and Wolfe, 2005). These restrictions must be overcome in the specific host niches conquered by C. glabrata. In contrast, C. albicans does not have any known auxotrophies, can metabolize a broad range of sugars and can use all amino acids as sole nitrogen sources (Odds, 1988; Kaur et al., 2005; and own observations). In addition, C. albicans possesses several families of secreted hydrolases and transmembrane transporters with central roles in virulence (Butler et al., 2009). Secreted aspartic proteases (Saps), for example, have the potential to destroy host tissue and liberate oligopeptides and amino acids, which are taken up by the fungus via oligopeptide and amino acid transportes (Naglik et al., 2004). Contrarily, C. glabrata exhibits low extracellular protease activity in vitro (Kaur et al., 2005).
Micronutrients are another prerequisite for a successful infection. Especially metals like iron and zinc are subject to a process called ‘nutritional immunity’, where the host actively sequesters these elements from invading microorganisms (Hood and Skaar, 2012). Iron levels in human serum are held as low as 10−24 M, severely restricting its availability to pathogens. To counteract this limitation, C. albicans has a plethora of iron acquisition systems at its disposal (Almeida et al., 2009): It can utilize siderophores from other microorganisms without producing its own (via Sit1/Arn1, Heymann et al., 2002), and bind host transferrin (via an unknown receptor, Knight et al., 2005) and ferritin (via Als3, Almeida et al., 2008). In addition, C. albicans can express haemolysins that disrupt red blood cells (Watanabe et al., 1999) and then bind and utilize haemoglobin via the Rbt5/Hmx1 system (Pendrak et al., 2004; Weissman and Kornitzer, 2004). Free iron, if available, is taken up via the reductive pathway with its large gene families of reductases, oxidases and iron permeases (Almeida et al., 2009). Together, these systems enable C. albicans to effectively use nearly all natural iron sources both of the host during infection and of surrounding microbes during commensal growth.
Candida glabrata, on the other hand, has no known receptors for haem (Nevitt and Thiele, 2011); however, haemolysin expression has been reported (Luo et al., 2004). In addition, although the reductive pathway is present, C. glabrata is not known to use host ferritin or transferrin as iron sources. Interestingly, like C. albicans, it can bind hydroxamate-type xenosiderophores of fungal origin, like ferrichrome, ferrirubin or coprogen, but unlike C. albicans not bacterial siderophores (Nevitt and Thiele, 2011). This binding significantly increases fitness and survival inside macrophages after subsequent phagocytosis (Nevitt and Thiele, 2011). Therefore, C. glabrata's in vivo choice of iron sources would appear to be limited in comparison with C. albicans.
Zinc, a central cofactor for many proteins, is also actively limited by the host during infections (Corbin et al., 2008), but it can be scavenged by C. albicans via a recently discovered ‘zincophore’ system using Pra1 (Citiulo et al., 2012). This protein is secreted, binds zinc and delivers it back to the pathogen in a manner reminiscent of the iron-carrying siderophores. C. glabrata is missing both Pra1 and its proposed binding partner, the high-affinity zinc transporter Zrt1 (Citiulo et al., 2012). Possibly, C. glabrata acquires Zn via its two homologues of the low-affinity S. cerevisiae zinc transporter Zrt2, but this still requires experimental confirmation. In addition, other micronutrients like manganese (Kehl-Fie and Skaar, 2010) or copper (Hodgkinson and Petris, 2012) may play important roles in host–pathogen interactions, and their uptake systems in Candida species are still largely unknown.
It seems, therefore, that the high metabolic flexibility of C. albicans may be part of its infection strategy, enabling this fungus to survive and grow in the many different and changing host niches it encounters. C. glabrata, on the other hand, appears more specialized in its metabolic requirement, possibly requiring a more stable environment, where these needs are met.
Conclusion
The problems and obstacles faced by both C. albicans and C. glabrata during infections are essentially the same, but the solutions employed differ in many ways (Fig. 1): overall, C. glabrata seems to follow a strategy of stealth and concealment in infection. It does not cause extensive epithelial damage, probably due to its lack of an invasive growth form. It does not elicit a strong immune response in murine models or in in vitro reconstituted human epithelia, and it can reside within macrophages without immediately destroying them. When it comes to nutrient supply in the host, C. glabrata relies on autophagy and some so far uncharacterized nutrient uptake mechanisms, but it would not appear to elicit rapid tissue damage to release nutrients from host cells. Genomically, C. glabrata displays some of the hallmarks of a specialized commensal or pathogen of humans. The loss of many metabolic pathways is indicative of a more stable environment than the one faced by C. albicans. Although it is unclear how far these (mostly) in vitro observations translate into infections of the human host, C. glabrata can persist within the internal organs of mice for a surprisingly long time without any clinical symptoms. A possible scenario is that C. glabrata attracts macrophages, which it subverts to use as a ‘trojan horse’, to hide from immune surveillance and to spread to the organs, analogous maybe to the strategy employed by C. neoformans (Charlier et al., 2009).
Fig. 1.
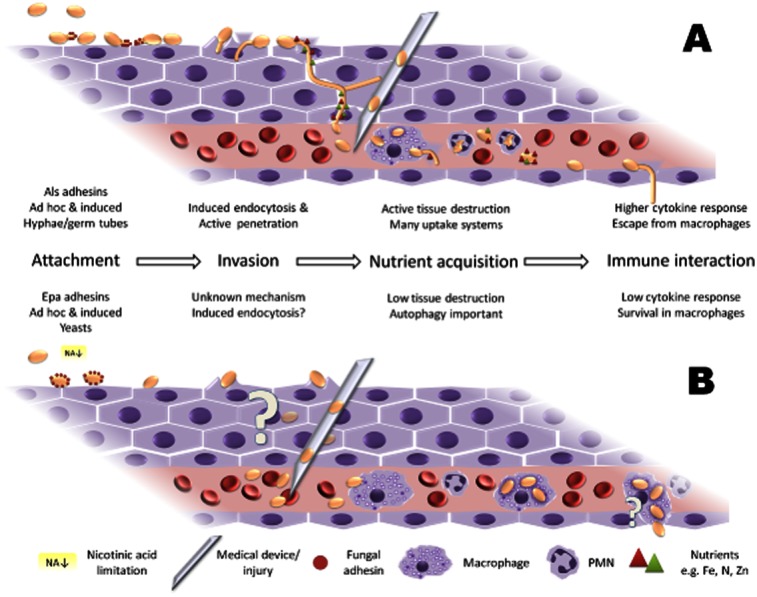
Schematic overview of the two different infection strategies of C. albicans and C. glabrata.
A. C. albicans forms hyphae and aggressively destroys tissue, eliciting a strong immune response.
B. Many aspects of C. glabrata pathogenicity are still unknown, like the precise mechanism of invasion. Active host tissue damage is low, as is the immune response. For a detailed description of the individual steps, see the related sections in the text.
Candida albicans, on the other hand, follows a strategy which can be described as ‘shock and awe’ when it has changed from commensal to pathogen. It actively invades epithelia when the circumstances permit and elicits stronger immune responses, which it seems to counteract in many cases. Macro- and micronutrients from damaged host tissue are taken up by a broad range of acquisition systems, and defending macrophages may be killed by formation of hyphae. Tissue damage by the fungus and the activated immune system can lead to severe disease, and death. In some cases, C. glabrata may benefit from this strategy of C. albicans. In oral candidiosis, for example, co-infections by both fungi are common (Redding et al., 2002; Coco et al., 2008), and C. glabrata may exploit the tissue destruction caused by C. albicans to gain nutrients, and possibly even to access the bloodstream. Moreover, this concept of C. albicans paving the way for C. glabrata may have serious clinical consequences: once C. glabrata reaches internal organs, its inherently high resistance to many commonly used antifungals makes treatment more problematic.
These strategies may be over-simplified for the purpose of this review, and many nuances were necessarily left out. However, much of the evidence shows that these two fungi use very different pathways to obtain the same goal – to survive and proliferate during infection of the human host. Understanding these strategies will hopefully help in finding novel ways to fight Candida, and fungal infections in general.
Acknowledgments
Our own research is supported by the German Federal Ministry of Education and Research (BMBF: ERA Net PathoGenoMics CandiCol, 0315901B; Center for Sepsis Control and Care – CSCC, FKZ 01EO1002), the Deutsche Forschungsgemeinschaft (DFG Hu 528/14, 15, 16 and 17; including the Priority Program SPP1580 ‘Intracellular compartments as places of pathogen–host-interactions’), the European Union (FinSysB MCRTN FP7-214004), Bayer Vital and the Jena graduate schools ‘International Leibniz Research School for Microbial and Biomolecular Interactions’ (ILRS) and the ‘Jena School for Microbial Communication’ (JSMC). We thank Katja Seider, Ilse Jacobsen and Duncan Wilson for critical reading of the manuscript.
References
- Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, et al. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RS, Wilson D, Hube B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009;9:1000–1012. doi: 10.1111/j.1567-1364.2009.00570.x. [DOI] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Argimon S, Wishart JA, Leng R, Macaskill S, Mavor A, Alexandris T, et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot Cell. 2007;6:682–692. doi: 10.1128/EC.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM, Lewis LE, Okai B, Quinn J, Gow NA, Erwig LP. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet Biol. 2012;49:677–678. doi: 10.1016/j.fgb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers KM, Newman ZR, Wheeler RT. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot Cell. 2011;10:932–944. doi: 10.1128/EC.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;336:647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- Brunke S, Seider K, Almeida RS, Heyken A, Fleck CB, Brock M, et al. Candida glabrata tryptophan-based pigment production via the Ehrlich pathway. Mol Microbiol. 2010;76:25–47. doi: 10.1111/j.1365-2958.2010.07052.x. [DOI] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun. 2009;77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chastellier C. The many niches and strategies used by pathogenic mycobacteria for survival within host macrophages. Immunobiology. 2009;214:526–542. doi: 10.1016/j.imbio.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Citiulo F, Jacobsen ID, Miramón P, Schild L, Brunke S, Zipfel P, et al. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog. 2012;8:e1002777. doi: 10.1371/journal.ppat.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco BJ, Bagg J, Cross LJ, Jose A, Cross J, Ramage G. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol Immunol. 2008;23:377–383. doi: 10.1111/j.1399-302X.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- Cole GT, Halawa AA, Anaissie EJ. The role of the gastrointestinal tract in hematogenous candidiasis: from the laboratory to the bedside. Clin Infect Dis. 1996;22(Suppl. 2):S73–S88. doi: 10.1093/clinids/22.supplement_2.s73. [DOI] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Ghori N, Falkow S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science. 1999;285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- Csank C, Haynes K. Candida glabrata displays pseudohyphal growth. FEMS Microbiol Lett. 2000;189:115–120. doi: 10.1111/j.1574-6968.2000.tb09216.x. [DOI] [PubMed] [Google Scholar]
- Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castano I, De Las Penas A. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell. 2008;7:814–825. doi: 10.1128/EC.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Penas A, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Mavrianos J, Chauhan N. Candida glabrata Pwp7p and Aed1p are required for adherence to human endothelial cells. FEMS Yeast Res. 2011;11:595–601. doi: 10.1111/j.1567-1364.2011.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue R, Castano I, De Las Penas A, Zupancic M, Lockatell V, Hebel JR, et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Fernández-Arenas E, Bleck CK, Nombela C, Gil C, Griffiths G, Diez-Orejas R. Candida albicans actively modulates intracellular membrane trafficking in mouse macrophage phagosomes. Cell Microbiol. 2009;11:560–589. doi: 10.1111/j.1462-5822.2008.01274.x. [DOI] [PubMed] [Google Scholar]
- Fidel PL, Jr, Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- de Groot PW, Kraneveld EA, Yin QY, Dekker HL, Gross U, Crielaard W, et al. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot Cell. 2008;7:1951–1964. doi: 10.1128/EC.00284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell SC, Smith MC, Muston P, Holland SL, Avery SV. Heterogeneous expression of the virulence-related adhesin Epa1 between individual cells and strains of the pathogen Candida glabrata. Eukaryot Cell. 2012;11:141–150. doi: 10.1128/EC.05232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun. 2002;70:5246–5255. doi: 10.1128/IAI.70.9.5246-5255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson V, Petris MJ. Copper homeostasis at the host–pathogen interface. J Biol Chem. 2012;287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family – a sticky pursuit. Med Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummert S, Hummert C, Schroter A, Hube B, Schuster S. Game theoretical modelling of survival strategies of Candida albicans inside macrophages. J Theor Biol. 2010;264:312–318. doi: 10.1016/j.jtbi.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Garcia-Sanchez S, Aubert S, Dromer F, Ghigo JM, d'Enfert C, Janbon G. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol Microbiol. 2005;55:1259–1271. doi: 10.1111/j.1365-2958.2004.04475.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen ID, Brunke S, Seider K, Schwarzmuller T, Firon A, d'Enfert C, et al. Candida glabrata persistence in mice does not depend on host immunosuppression and is unaffected by fungal amino acid auxotrophy. Infect Immun. 2010;78:1066–1077. doi: 10.1128/IAI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen ID, Grosse K, Berndt A, Hube B. Pathogenesis of Candida albicans infections in the alternative chorio-allantoic membrane chicken embryo model resembles systemic murine infections. PLoS ONE. 2011;6:e19741. doi: 10.1371/journal.pone.0019741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci USA. 2007;104:7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Ross S, Douglas L, Konopka JB, Dean N. Recognition of yeast by murine macrophages requires mannan but not glucan. Eukaryot Cell. 2010;9:1776–1787. doi: 10.1128/EC.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host–microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Knight SA, Vilaire G, Lesuisse E, Dancis A. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect Immun. 2005;73:5482–5492. doi: 10.1128/IAI.73.9.5482-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraneveld EA, de Soet JJ, Deng DM, Dekker HL, de Koster CG, Klis FM, et al. Identification and differential gene expression of adhesin-like wall proteins in Candida glabrata biofilms. Mycopathologia. 2011;172:415–427. doi: 10.1007/s11046-011-9446-2. [DOI] [PubMed] [Google Scholar]
- Kuhn DM, Vyas VK. The Candida glabrata adhesin Epa1p causes adhesion, phagocytosis, and cytokine secretion by innate immune cells. FEMS Yeast Res. 2012;12:398–414. doi: 10.1111/j.1567-1364.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- Li L, Kashleva H, Dongari-Bagtzoglou A. Cytotoxic and cytokine-inducing properties of Candida glabrata in single and mixed oral infection models. Microb Pathog. 2007a;42:138–147. doi: 10.1016/j.micpath.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Redding S, Dongari-Bagtzoglou A. Candida glabrata: an emerging oral opportunistic pathogen. J Dent Res. 2007b;86:204–215. doi: 10.1177/154405910708600304. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Luo G, Samaranayake LP, Cheung BP, Tang G. Reverse transcriptase polymerase chain reaction (RT-PCR) detection of HLP gene expression in Candida glabrata and its possible role in in vitro haemolysin production. APMIS. 2004;112:283–290. doi: 10.1111/j.1600-0463.2004.apm11204-0509.x. [DOI] [PubMed] [Google Scholar]
- McKenzie CG, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, et al. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect Immun. 2010;78:1650–1658. doi: 10.1128/IAI.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer JW, van de Veerdonk FL, Joosten LA, Kullberg BJ, Netea MG. Severe Candida spp. infections: new insights into natural immunity. Int J Antimicrob Agents. 2010;36(Suppl. 2):S58–S62. doi: 10.1016/j.ijantimicag.2010.11.013. [DOI] [PubMed] [Google Scholar]
- Mora-Montes HM, Netea MG, Ferwerda G, Lenardon MD, Brown GD, Mistry AR, et al. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect Immun. 2011;79:1961–1970. doi: 10.1128/IAI.01282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes DL, Runglall M, Murciano C, Shen C, Nayar D, Thavaraj S, et al. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe. 2010;8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes DL, Murciano C, Runglall M, Kohli A, Islam A, Naglik JR. Activation of MAPK/c-Fos induced responses in oral epithelial cells is specific to Candida albicans and Candida dubliniensis hyphae. Med Microbiol Immunol (Berl) 2012;201:93–101. doi: 10.1007/s00430-011-0209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik J, Albrecht A, Bader O, Hube B. Candida albicans proteinases and host/pathogen interactions. Cell Microbiol. 2004;6:915–926. doi: 10.1111/j.1462-5822.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- Nevitt T, Thiele DJ. Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog. 2011;7:e1001322. doi: 10.1371/journal.ppat.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006;8:1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. London: Baillière Tindall; 1988. [DOI] [PubMed] [Google Scholar]
- Palmer GE, Kelly MN, Sturtevant JE. Autophagy in the pathogen Candida albicans. Microbiology. 2007;153:51–58. doi: 10.1099/mic.0.2006/001610-0. [DOI] [PubMed] [Google Scholar]
- Pendrak ML, Chao MP, Yan SS, Roberts DD. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. J Biol Chem. 2004;279:3426–3433. doi: 10.1074/jbc.M311550200. [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding SW, Kirkpatrick WR, Coco BJ, Sadkowski L, Fothergill AW, Rinaldi MG, et al. Candida glabrata oropharyngeal candidiasis in patients receiving radiation treatment for head and neck cancer. J Clin Microbiol. 2002;40:1879–1881. doi: 10.1128/JCM.40.5.1879-1881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A, Gratz N, Kovarik P, Schuller C. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol. 2010;12:199–216. doi: 10.1111/j.1462-5822.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M, Mailhammer R, Grassl G, Sander CA, Hube B, Korting HC. Infection of human oral epithelia with Candida species induces cytokine expression correlated to the degree of virulence. J Invest Dermatol. 2002;118:652–657. doi: 10.1046/j.1523-1747.2002.01699.x. [DOI] [PubMed] [Google Scholar]
- Schaller M, Boeld U, Oberbauer S, Hamm G, Hube B, Korting HC. Polymorphonuclear leukocytes (PMNs) induce protective Th1-type cytokine epithelial responses in an in vitro model of oral candidosis. Microbiology. 2004;150:2807–2813. doi: 10.1099/mic.0.27169-0. [DOI] [PubMed] [Google Scholar]
- Seider K, Heyken A, Lüttich A, Miramón P, Hube B. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol. 2010;13:392–400. doi: 10.1016/j.mib.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Seider K, Brunke S, Schild L, Jablonowski N, Wilson D, Majer O, et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol. 2011;187:3072–3086. doi: 10.4049/jimmunol.1003730. [DOI] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtler B, Wilson D, Haedicke K, Dalle F, Hube B. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS ONE. 2011;6:e17046. doi: 10.1371/journal.pone.0017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtler B, Citiulo F, Jablonowski N, Förster S, Dalle F, Schaller M, et al. Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS ONE. 2012;7:e36952. doi: 10.1371/journal.pone.0036952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takano M, Murakami M, Tanaka H, Matsuhisa A, Nakao N, et al. Characterization of a haemolytic factor from Candida albicans. Microbiology. 1999;145(Part 3):689–694. doi: 10.1099/13500872-145-3-689. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53:1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- Westwater C, Schofield DA, Nicholas PJ, Paulling EE, Balish E. Candida glabrata and Candida albicans; dissimilar tissue tropism and infectivity in a gnotobiotic model of mucosal candidiasis. FEMS Immunol Med Microbiol. 2007;51:134–139. doi: 10.1111/j.1574-695X.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- Wilson D, Hube B. Hgc1 mediates dynamic Candida albicans-endothelium adhesion events during circulation. Eukaryot Cell. 2010;9:278–287. doi: 10.1128/EC.00307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- Woods JP. Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr Opin Microbiol. 2003;6:327–331. doi: 10.1016/s1369-5274(03)00080-8. [DOI] [PubMed] [Google Scholar]
- Zhu W, Phan QT, Boontheung P, Solis NV, Loo JA, Filler SG. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc Natl Acad Sci USA. 2012;109:14194–14199. doi: 10.1073/pnas.1117676109. [DOI] [PMC free article] [PubMed] [Google Scholar]


