Abstract
The human pathogens enterohemorrhagic and enteropathogenic Escherichia coli (EHEC and EPEC), as well as the mouse pathogen Citrobacter rodentium encode type III secretion system (T3SS) effector proteins to promote their survival in the infected host. The mechanisms of action and the host targets of T3SS effectors are under active investigation because of their importance to bacterial virulence. The non-locus of enterocyte effacement (LEE)-encoded protein F, NleF, contributes to E. coli and C. rodentium colonization of piglets and mice, respectively. Here we sought to characterize the host binding partners of NleF. Using a yeast two-hybrid screen, we identified Tmp21, a type-I integral membrane protein and COPI-vesicle receptor involved in trans-Golgi network function, as an NleF-binding partner. We confirmed this interaction using immunoprecipitation and bimolecular fluorescence complementation (BiFC). We expressed a temperature-sensitive vesicular stomatitis virus glycoprotein (tsVSVG) to monitor protein trafficking and determined that NleF slows the intracellular trafficking of tsVSVG from the endoplasmic reticulum to the Golgi.
Keywords: Effector, Enterohemorrhagic E. coli, NleF, Tmp21, Type III secretion
1. Introduction
Enterohemorrhagic Escherichia coli (EHEC) and other Shiga-like toxin-producing E. coli (STEC) are endemic health threats and major causes of food borne disease (Clarke, 2001). These human pathogens cause hemorrhagic colitis and are a leading cause of pediatric renal failure. In addition to the Shiga-like toxin, STEC encode a large number of virulence proteins, which they translocate into intestinal epithelial cells using a type III secretion system [T3SS (Cornelis, 2010)], a mechanism conserved among the other attaching/effacing (A/E) pathogens, enteropathogenic E. coli (EPEC) and Citrobacter rodentium. While it is known that these translocated proteins (effectors) subvert host cell function to promote diarrheal disease and bacterial transmission, the biochemical mechanisms and host targets of many of these effectors are unknown.
The mammalian protein secretory pathway relies on vesicular trafficking to transport cargo between different organelles. Transport vesicles composed of protein coat complexes known as COPI and COPII mediate protein trafficking between the endoplasmic reticulum (ER) and Golgi (Wessels et al., 2006). Proteins are transported from the ER to the Golgi (anterograde transport) by COPII carrier vesicles that bud from the ER (Kaiser and Ferro-Novick, 1998). Proteins arriving at the Golgi are modified and sorted into transport vesicles destined for the plasma membrane (PM), the endo/lysosomal system, or to secretory granules. A subset of proteins is recycled to the ER from the Golgi (retrograde transport) in COPI vesicles (Orci et al., 2000).
Bacterial exploitation of eukaryotic secretory pathways is of interest. For example, Salmonella recruits exocytic transport vesicles to the Salmonella-containing vacuole, possibly to interfere with antigen presentation (Kuhle et al., 2006). Brucella abortus utilizes the endoplasmic reticulum (ER) GTPase Sar1 for intracellular replication (Celli et al., 2005). The E. coli effector NleA inhibits COPII-dependent protein export from the ER by binding to Sec24 (Kim et al., 2007).
We previously identified the NleF protein in a proteomic screen of EHEC and C. rodentium secreted proteins (Deng et al., 2004). NleF is found in the E. coli lineages associated with human disease (Zhang et al., 2007) and with epidemic potential (Coombes et al., 2008). NleF is present with 100% amino acid identity in EPEC E2348/69 and an ortholog is also found in C. rodentium. Our previous experiments demonstrated that NleF is a T3SS-translocated effector (Echtenkamp et al., 2008). Infection experiments using C. rodentium in mice and EHEC in gnotobiotic piglets indicated that NleF contributes to bacterial colonization of the host (Echtenkamp et al., 2008). The goal of this study was to determine the mammalian binding partner of NleF. We demonstrate here that NleF binds to the human Tmp21 protein and subsequently disrupts intracellular protein trafficking.
2. Materials and methods
2.1. Chemicals and antibodies
Chemicals and antibodies were used according to manufacturer’s recommendations. GFP and Tmp21 antibodies were obtained from Cell Signaling. Golgin-97 antibody was obtained from Invitrogen. Calnexin, HA, and His antibodies were obtained from Sigma. NleF antiserum was described previously (Echtenkamp et al., 2008). pEGFP-VSVG was provided by J. Lippincott-Schwartz [Addgene #11912 (Presley et al., 1997)].
2.2. Bacterial strains, cell culture, and infection experiments
The bacterial strains and plasmids used in this study are described in Table 1. HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. Cells were transfected using TransPass (New England Biolabs). Bacteria were cultured in Luria-Bertani broth at 37 °C for 18 h without shaking. Overnight LB cultures were diluted 1:10 into DMEM, followed by a further incubation for 3 h at 37 °C, 5% CO2. Cell culture medium was replaced with DMEM prior to infection and bacteria were added at a multiplicity of infection of 25–50.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference |
|---|---|---|
| Yeast strains | ||
| S. cerevisiae AH109 |
MATa,trp1-901,leu2-3,112,ura3-52,his3-200,gal4D, gal80D,LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2,URA3::MEL1UAS-MEL1TATA-lacZ |
Clontech |
| S. cerevisiae Y187 |
MATα,ura3-52,his3-200,ade2-101,trp1-901,leu2-3,112,
gal4D,met−,gal80D,URA3::GAL1UAS-GAL1TATA-lacZ |
Clontech |
| yPRH-5 | HeLa cDNA library in Y187 | Clontech |
| yPRH-11 | AH109/nleF-Gal4DBD-Myc | This study |
| Bacterial strains | ||
| C. rodentium DBS100/NleF-FLAG | C. rodentium ATCC 51459 expressing FLAG-tagged NleF | This study |
| E. coli BL21(DE3) | E. coli F− ompT hsdSB (rB−mB−) gal dcm (DE3) | Novagen |
| E. coli BL21(DE3)/NleF-FLAG | NleF-FLAG | This study |
| E. coli BL21(DE3)/Tmp21-His | Tmp21-His | This study |
| E. coli BL21(DE3)/NleF-FLAG/Tmp21-His | NleF-FLAG + Tmp21-His coexpression | This study |
| Plasmids | ||
| pGBKT7 | Two-hybrid bait plasmid | Clontech |
| pGADT7 | Two-hybrid library plasmid | Clontech |
| nleF-Gal4DBD-Myc | NleF-Gal4-Myc bait plasmid | This study |
| pFLAG-CTC | Bacterial FLAG fusion protein expression | Sigma |
| NleF-pFLAG-CTC | NleF-FLAG | Echtenkamp et al. (2008) |
| pET28a | Bacterial hexahistidine fusion protein expression | Novagen |
| Tmp21-pET28a | Tmp21-His | This study |
| VN | Venus fluorescence protein (AAs 1–173) | Gao et al. (2009) |
| VC | Venus fluorescence protein (AAs 155–238) | Gao et al. (2009) |
| VN-actin | Venus 1–173 fused to human actin | Gao et al. (2009) |
| VC-actin | Venus 155–238 fused to human actin | (Gao et al. (2009) |
| VN-NleF | Venus 1–173 fused to NleF | This study |
| VC-NleF | Venus 155–238 fused to NleF | This study |
| VC-NleF (1–162) | Venus 155–238 fused to NleF (AAs 1–162) | This study |
| VC-NleF (1–117) | Venus 155–238 fused to NleF (AAs 1–117) | This study |
| VC-NleF (1–84) | Venus 155–238 fused to NleF (AAs 1–84) | This study |
| VC-NleF (1–65) | Venus 155–238 fused to NleF (AAs 1–65) | This study |
| VN-Tmp21 | Venus 1–173 fused to Tmp21 | This study |
| VC-Tmp21 | Venus 155–238 fused to Tmp21 | This study |
| VC-Tmp21 (1–180) | Venus 155–238 fused to Tmp21 (AAs 1–180) | This study |
| VC-Tmp21 (1–150) | Venus 155–238 fused to Tmp21 (AAs 1–150) | This study |
| VC-Tmp21 (1–124) | Venus 155–238 fused to Tmp21 (AAs 1–124) | This study |
| VC-Tmp21 (1–63) | Venus 155–238 fused to Tmp21 (AAs 1–63) | This study |
| pEGFP-VSVG | VSVG in pEGFP-N1 | Presley et al. (1997) |
2.3. Protein purification
NleF and Tmp21 were cloned into pFLAG-CTC and pET28a, respectively, and expressed in E. coli BL21(DE3). Bacterial cultures were grown to an OD600 of 0.3 and then induced with 1 mM IPTG for 2 h. Cells were centrifuged, lysed by sonication, applied to α-FLAG M2 beads and Ni-NTA agarose, respectively, and incubated at 4 °C overnight. After washing, proteins were eluted with either 0.1 M glycine HCl, pH 3.5, or with imidazole and then analyzed on SDS-12% PAGE.
2.4. Yeast two-hybrid assay
NleF was cloned into the yeast two-hybrid GAL4 DNA binding domain vector pGBKT7 to generate the ‘bait’ plasmid. The NleF bait was used to screen a pre-transformed human HeLa cell cDNA library for proteins interacting with NleF according to protocols in the BD Matchmaker Pre-transformed Libraries User Manual (Clontech). Yeast clones containing library plasmids encoding human proteins interacting with NleF were purified by restreaking on selective media and retested for growth phenotypes. β-Galactosidase activity was calculated using Eq. (1) where t refers to the incubation time (min) and v refers to the concentration factor.
| (1) |
2.5. Pull down assay
Purified proteins (~20 μg) were applied to α-FLAG M2 beads and incubated with rotation for 5 h at 4 °C. Beads were washed three times with PBS and resuspended in SDS-PAGE buffer. The samples were interrogated for the presence of His- and FLAG-tagged proteins by immunoblotting.
2.6. Immunoblotting
Cells were lysed in RIPA buffer [150 mM NaCl, 50 mM Tris pH 8.0, 0.5% sodium deoxycholate, 0.1% SDS, 1% Nonident P-40], incubated on ice for 30 min, and centrifuged. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, blocked in Odyssey blocking buffer (Li-Cor) and then probed with appropriate primary and secondary antibodies. After rinsing in PBS, blots were imaged using an Odyssey infrared imaging system.
2.7. Bimolecular fluorescence complementation
HeLa cells were co-transfected with two BiFC plasmids (250 ng each) representing NleF and Tmp21 sequences cloned as fusions to the N- or C-terminus of Venus eYFP (designated VN and VC). The fluorescence derived from BiFC was visualized using Eclipse 80i fluorescence microscope (Nikon) after 24 h incubation and was quantified using a fluorescence plate reader with appropriate filters (excitation: 500/20 nm; emission: 535/30 nm).
2.8. VSVG trafficking and immunofluorescence microscopy
HeLa cells were grown on glass coverslips in 24-well tissue culture plates. For VSVG experiments, cells were transfected in 4 replicates with pEGFP-VSVG in the presence or absence of NleF-HA. After 24 h, one replicate was left at 37 °C while the other replicates were transferred to 19 °C, 32 °C, or 40 °C. Three hours after temperature shift, cells were rinsed 3 times with PBS, fixed in 3.7% formaldehyde and permeabilized in 0.2% saponin in PBS, blocked with 10% goat serum, and incubated with primary antibodies for 1 h at room temperature. Cells were washed with PBS and probed with Alexa Fluor-conjugated secondary antibodies for 1 h. Coverslips were mounted in Mowiol and samples were visualized using Eclipse 80i fluorescence or Eclipse C1Si confocal microscopes (Nikon).
2.9. Statistical analyses
β-Galactosidase data were analyzed using t-tests. BiFC data were analyzed using one-way ANOVA. VSVG trafficking data were evaluated with Fisher’s exact test. p-Values < 0.05 were considered significant.
3. Results and discussion
3.1. NleF binds to Tmp21
We used a yeast two-hybrid (Y2H) screening assay to determine the mammalian binding partners of NleF. EHEC NleF was cloned as a fusion to the Gal4 DNA-binding domain (DBD) and co-expressed in the yeast reporter strain AH109 with a HeLa cell cDNA library. Co-transformants were plated on synthetic quadruple-dropout (QDO) medium to select for interactions between NleF and a library component that activated the transcriptional reporter. We isolated 153 colonies after 21 days growth on QDO plates, 26 of which had a high level of reporter gene activation as measured in β-galactosidase assays (data not shown).
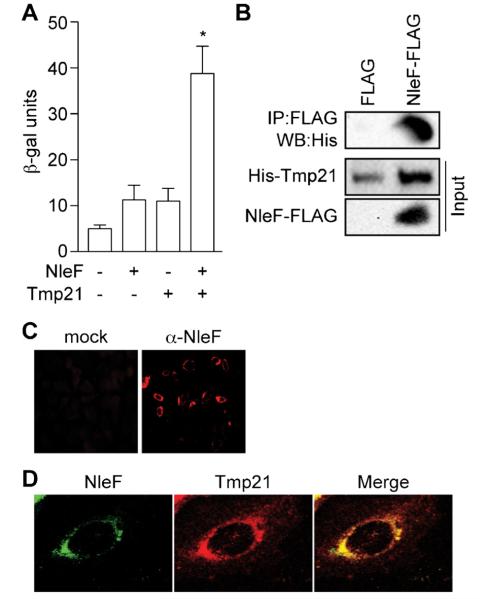
We identified 3 different human cDNA sequences (Tmp21, CD151, PAIP2) that may encode proteins that interact with NleF. Among these, we further studied Tmp21 (also named p23/p24d) as a putative NleF binding partner. We confirmed the Tmp21–NleF interaction using direct Y2H assays and by quantifying β-galactosidase activity resulting from NleF–Tmp21 co-expression (Fig. 1A).
Fig. 1.
NleF binds Tmp21. (A) β-Galactosidase assays. NleF was expressed in yeast in the presence or absence of human Tmp21 and β-galactosidase activity was quantified. Asterisks indicate significantly different β-galactosidase activity as compared with untransformed yeast (p < 0.05, t-test). (B) Coimmunoprecipitation. His-Tmp21 was co-expressed with NleF-FLAG in E. coli BL21(DE3). NleF was immunoprecipitated and its binding to His-Tmp21 binding was assessed using immunoblotting. (C) Immunofluorescence microscopy. HeLa cells were infected with C. rodentium DBS100/pnleF-FLAG and stained with an α-NleF antibody (red). (D) NleF and Tmp21 co-localize. HeLa cells were infected with C. rodentium/pnleF-FLAG, transfected with Tmp21-HA and stained with α-NleF (green) and α-HA (red) antibodies.
Tmp21 is a 219 amino acid integral type I transmembrane protein that functions as an integral receptor for the COPI-vesicle coat (Blum et al., 1999). Tmp21 is a member of the p24 (p24/gp25L/emp24/Erp) protein family. These proteins provide cargo receptors to proteins (Anantharaman and Aravind, 2002) and regulate protein packaging into COPI vesicles in concert with a small GTPase, the ADP-ribosylation factor 1 (Arf1). p24 proteins are assembled into heteromeric complexes that cycle between the ER and the Golgi and recruit Arf1 in early stages of vesicle formation (Gommel et al., 2001). p24 proteins thus play active roles in retrograde protein transport from Golgi to ER by facilitating the formation of COPI-coated vesicles (Aguilera-Romero et al., 2008).
By expressing and purifying recombinant forms of NleF and Tmp21 in E. coli BL21(DE3) and then using these proteins in pulldown assays, we confirmed that NleF and Tmp21 bind directly in vitro (Fig. 1B). We also used immunofluorescence microscopy to determine the extent of NleF–Tmp21 colocalization. To do this, we used polyclonal NleF antisera (Echtenkamp et al., 2008) to detect NleF after its translocation into HeLa cells during C. rodentium infection (Fig. 1C). Co-staining for Tmp21 revealed that both proteins colocalized in a perinuclear location (Fig. 1D).
3.2. BiFC
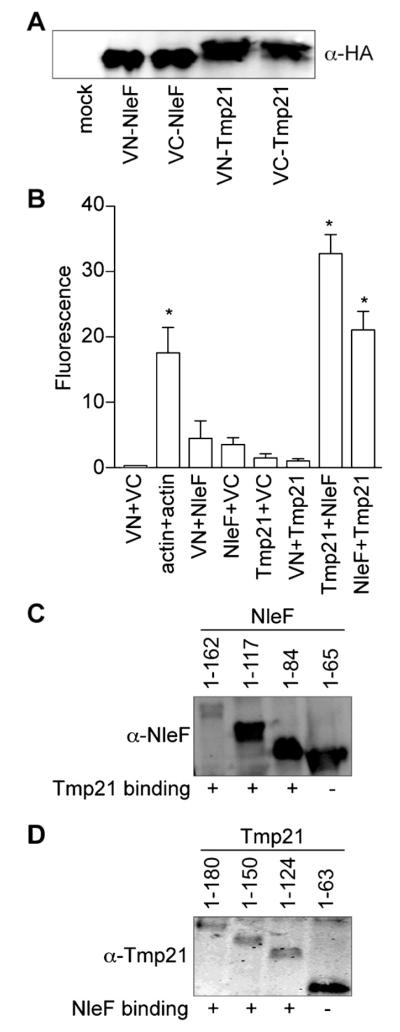
To determine whether NleF and Tmp21 interact when they are co-expressed in mammalian cells, we used bimolecular fluorescence complementation (BiFC) assays. This technology utilizes the reconstitution of two fragments of the enhanced yellow fluorescent protein (eYFP) to generate a fluorescent signal at the site of the protein–protein interaction under investigation (Hu et al., 2002). We generated protein chimeras with split N- and C-terminal fragments (VN and VC, respectively) of eYFP fused to either NleF or Tmp21 (Fig. 2A). Co-expressing eYFP chimeras of Tmp21 and NleF reconstituted YFP fluorescence, to a similar magnitude as the reconstitution of the actin positive control (Fig. 2B). By contrast, transfecting individual plasmids did not reconstitute YFP fluorescence (Fig. 2B), suggesting that NleF binds to Tmp21 in mammalian cells.
Fig. 2.
BiFC. (A) NleF and Tmp-21 expression. NleF and Tmp21 were cloned into eYFP-VN and eYFP-VC vectors and protein expression was evaluated by immunoblotting. (B) BiFC quantification. Relative fluorescence intensity after co-transfecting indicated NleF- and Tmp21-eYFP plasmid combinations (n = 3). Asterisks indicate significantly different fluorescence intensity as compared with untransfected samples (p < 0.05, ANOVA). (C) NleF truncations binding to Tmp21. NleF truncations were cloned into eYFP-VC vectors and protein expression was evaluated by immunoblotting. Binding to Tmp21-eYFP-VN was measured using BiFC and is scored as positive (+) or negative (−) in the respective columns. (D) Tmp21 truncations binding to NleF. Tmp21 truncations were cloned into eYFP-VC vectors and protein expression was evaluated by immunoblotting. Binding to NleF-eYFP-VN was measured using BiFC and is scored as positive (+) or negative in the respective columns.
The N-terminal luminal domain of Tmp21 mediates cargo uptake into transport vesicles, whereas the KKLIE cytoplasmic tail at the Tmp21 carboxy-terminus mediates COPI-dependent transport vesicle formation (Blum and Lepier, 2008). To map the binding domain of NleF on Tmp21, we carried out a BiFC study with C-terminal deletions of NleF. These experiments revealed that deleting the NleF C-terminus beyond amino acid 84 eliminated NleF binding to full length Tmp21, as indicated by loss of fluorescence with the sequentially truncated constructs (Fig. 2C). A similar BiFC analysis revealed that the C-terminal region of Tmp21, amino acids 63–180, was required for binding to NleF (Fig. 2D). Overall, these data suggest that NleF and Tmp21 associate through their respective C-termini.
3.3. NleF alters VSVG trafficking
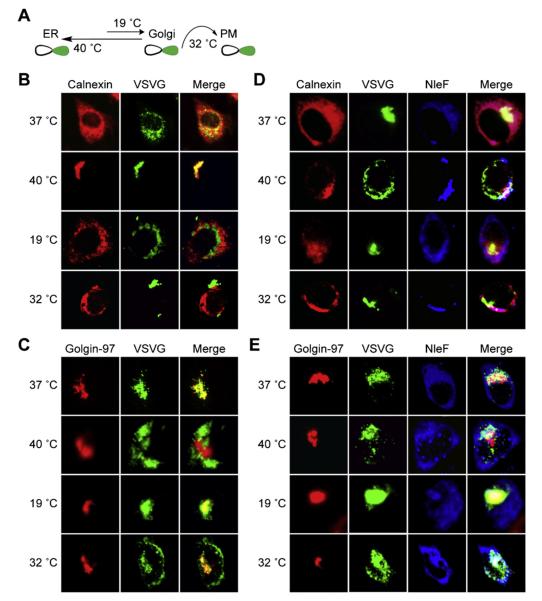
We tested the hypothesis that NleF binding to Tmp21 would cause defects in protein trafficking by characterizing the localization of a vesicular stomatitis virus glycoprotein (VSVG)-GFP fusion as a function of NleF expression and of temperature. VSVG localization is commonly used to study mammalian protein trafficking (Wessels et al., 2005). This glycoprotein is essential for viral envelope fusion with the host PM and traffics intracellularly via the ER and Golgi (Lippincott-Schwartz et al., 2000).
VSVG-GFP transport can be manipulated by incubating cell cultures at different temperatures (Toomre et al., 1999). If incubated at 40 °C, VSVG-GFP becomes reversibly misfolded and retained in the ER. If shifted to 19 °C, VSVG-GFP refolds and can be transported to the TGN. A further temperature shift to 32 °C allows subsequent trafficking to the PM (Fig. 3A).
Fig. 3.
NleF alters VSVG-GFP trafficking. (A) VSVG-GFP localization schematic. At 40 °C, VSVG-GFP becomes reversibly misfolded and retained in the ER. Incubation at 19 °C allows VSVG-GFP refolding and transport to the TGN. Further incubation at 32 °C allows VSVG-GFP trafficking to the PM. (B) VSVG-GFP colocalization with calnexin. HeLa cells were transfected with VSVG-GFP and then incubated at 37 °C or shifted to 19 °C, 32 °C, or 40 °C. Cells were stained with an α-calnexin antibody. (C) VSVG-GFP colocalization with golgin-97. Experiment performed as in (B). Cells were stained with an α-golgin-97 antibody. (D) Impact of NleF on VSVG-GFP colocalization with calnexin. HeLa cells were cotransfected with both VSVG-GFP and NleF-HA and then incubated at 37 °C or shifted to 19 °C, 32 °C, or 40 °C. Cells were stained with α-calnexin (red) and α-HA (blue) antibodies. (E) Impact of NleF on VSVG-GFP colocalization with golgin-97. Experiment performed as in (D). Cells were stained with α-golgin-97 (red) and α-HA (blue) antibodies.
We first transfected HeLa cells with VSVG-GFP and then evaluated VSVG-GFP localization using immunofluorescence microscopy. As expected, when cells were shifted from 37 °C to 40 °C, VSVG-GFP fluorescence became localized primarily in the ER, as shown by its colocalization with the ER protein calnexin (Fig. 3B). When cells were subsequently shifted to 19 °C, VSVG-GFP localization with calnexin was reduced and VSVG-GFP was primarily redistributed to the Golgi, as shown by its colocalization with the Golgi protein golgin-97 (Fig. 3C). Incubating cells at 32 °C also resulted in the expected subsequent shift of VSVG-GFP from the Golgi to the PM.
We predicted that if NleF disrupts Tmp21 function, VSVG-GFP redistribution to the Golgi would either be delayed or blocked by NleF. After transfecting NleF and performing temperature shift experiments, we observed that VSVG-GFP was retained in the ER at 19 °C in 74% of cells examined, rather than redistributing to the Golgi (p < 0.05, Fisher’s exact test; Fig. 3D). Similarly, in the presence of NleF, VSVG-GFP was mislocalized to the Golgi at 32 °C in 58% of cells examined, rather than trafficking to the PM (p < 0.05, Fisher’s exact test; Fig. 3E).
4. Conclusions
Overall, the yeast two-hybrid, immunoprecipitation, and BiFC data suggest that NleF binds to Tmp21. Expressing NleF disrupted VSVG-GFP localization, suggesting that the NleF–Tmp21 interaction disrupts intracellular protein trafficking. The functional significance of these data in subverting host cells in the context of E. coli and C. rodentium infection awaits further experimentation.
Acknowledgement
The project described was supported by Grant Number AI087923 from the National Institute of Allergy and Infectious Diseases (NIAID). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID.
Abbreviations
- A/E
attaching and effacing
- BiFC
bimolecular fluorescence complementation
- EHEC
enterohemorrhagic E. coli
- EPEC
enteropathogenic E. coli
- ER
endoplasmic reticulum
- LEE
locus of enterocyte effacement
- NleF
non-LEE encoded protein F
- STEC
Shiga-like toxin-producing E. coli
- T3SS
type III secretion system
References
- Aguilera-Romero A, Kaminska J, Spang A, Riezman H, Muniz M. The yeast p24 complex is required for the formation of COPI retrograde transport vesicles from the Golgi apparatus. J. Cell Biol. 2008;180:713–720. doi: 10.1083/jcb.200710025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-research0023. research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Lepier A. The luminal domain of p23 (Tmp21) plays a critical role in p23 cell surface trafficking. Traffic. 2008;9:1530–1550. doi: 10.1111/j.1600-0854.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- Blum R, Pfeiffer F, Feick P, Nastainczyk W, Kohler B, Schafer KH, Schulz I. Intracellular localization and in vivo trafficking of p24A and p23. J. Cell Sci. 1999;112(Pt 4):537–548. doi: 10.1242/jcs.112.4.537. [DOI] [PubMed] [Google Scholar]
- Celli J, Salcedo SP, Gorvel JP. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1673–1678. doi: 10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SC. Diarrhoeagenic Escherichia coli – an emerging problem? Diagn. Microbiol. Infect. Dis. 2001;41:93–98. doi: 10.1016/s0732-8893(01)00303-0. [DOI] [PubMed] [Google Scholar]
- Coombes BK, Wickham ME, Mascarenhas M, Gruenheid S, Finlay BB, Karmali MA. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 2008;74:2153–2160. doi: 10.1128/AEM.02566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome, a complex nano-machine for intracellular ‘toxin’ delivery. Biol. Chem. 2010;391:745–751. doi: 10.1515/BC.2010.079. [DOI] [PubMed] [Google Scholar]
- Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O’Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtenkamp F, Deng W, Wickham ME, Vazquez A, Puente JL, Thanabalasuriar A, Gruenheid S, Finlay BB, Hardwidge PR. Characterization of the NleF effector protein from attaching and effacing bacterial pathogens. FEMS Microbiol. Lett. 2008;281:98–107. doi: 10.1111/j.1574-6968.2008.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, Lenardo MJ, Hardwidge PR. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 2009;5:e1000708. doi: 10.1371/journal.ppat.1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommel DU, Memon AR, Heiss A, Lottspeich F, Pfannstiel J, Lechner J, Reinhard C, Helms JB, Nickel W, Wieland FT. Recruitment to Golgi membranes of ADP-ribosylation factor 1 is mediated by the cytoplasmic domain of p23. EMBO J. 2001;20:6751–6760. doi: 10.1093/emboj/20.23.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Ferro-Novick S. Transport from the endoplasmic reticulum to the Golgi. Curr. Opin. Cell Biol. 1998;10:477–482. doi: 10.1016/s0955-0674(98)80062-8. [DOI] [PubMed] [Google Scholar]
- Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, Metalnikov P, Rizg K, Thomas NA, Lee SF, Hartland EL, Hardwidge PR, Pawson T, Strynadka NC, Finlay BB, Schekman R, Gruenheid S. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe. 2007;2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Kuhle V, Abrahams GL, Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic. 2006;7:716–730. doi: 10.1111/j.1600-0854.2006.00422.x. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Volchuk A, Engel T, Gmachl M, Amherdt M, Perrelet A, Sollner TH, Rothman JE. Anterograde flow of cargo across the golgi stack potentially mediated via bidirectional “percolating” COPI vesicles. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10400–10405. doi: 10.1073/pnas.190292497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Toomre D, Keller P, White J, Olivo JC, Simons K. Dual-color visualization of trans-Golgi network to plasma membrane traffic along microtubules in living cells. J. Cell Sci. 1999;112(Pt 1):21–33. doi: 10.1242/jcs.112.1.21. [DOI] [PubMed] [Google Scholar]
- Wessels E, Duijsings D, Niu TK, Neumann S, Oorschot VM, de Lange F, Lanke KH, Klumperman J, Henke A, Jackson CL, Melchers WJ, van Kuppeveld FJ. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Wessels E, Duijsings D, Notebaart RA, Melchers WJ, van Kuppeveld FJ. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-golgi transport. J. Virol. 2005;79:5163–5173. doi: 10.1128/JVI.79.8.5163-5173.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Laing C, Steele M, Ziebell K, Johnson R, Benson AK, Taboada E, Gannon VP. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics. 2007;8:121. doi: 10.1186/1471-2164-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]