Abstract
Breast cancer is the most common cancer in women in developed countries. To identify common breast cancer susceptibility alleles, we conducted a genome-wide association study in which 582,886 SNPs were genotyped in 3,659 cases with a family history of the disease and 4,897 controls. Promising associations were evaluated in a second stage, comprising 12,576 cases and 12,223 controls. We identified five new susceptibility loci, on chromosomes 9, 10 and 11 (P = 4.6 × 10−7 to P = 3.2 × 10−15). We also identified SNPs in the 6q25.1 (rs3757318, P = 2.9 × 10−6), 8q24 (rs1562430, P = 5.8 × 10−7) and LSP1 (rs909116, P = 7.3 × 10−7) regions that showed more significant association with risk than those reported previously. Previously identified breast cancer susceptibility loci were also found to show larger effect sizes in this study of familial breast cancer cases than in previous population-based studies, consistent with polygenic susceptibility to the disease.
Genome-wide association studies (GWAS) provide a powerful approach to identify common disease alleles. Recent GWAS have identified common variants at 12 loci that are associated with an increased risk of breast cancer, and an additional locus, CASP8 (specifically, a polymorphism resulting in a D302H substitution), has been identified through a candidate-gene association study1-8. However, because the risks associated with these variants are modest (per-allele odds ratios (OR) <1.3), they explain only a small fraction of the estimated twofold familial relative risk of breast cancer in first-degree relatives of affected women. Moreover, the GWAS conducted to date have been relatively small, and it is likely that many susceptibility variants have been missed due to lack of power in these studies. In an attempt to identify additional breast cancer loci, we conducted a GWAS that was substantially larger than those conducted to date.
We studied 3,960 cases of breast cancer from the UK, selected for a positive family history of breast cancer. We selected cases with a positive family history because, under a polygenic model of susceptibility, this is expected to increase the effect size and hence improve study power9. DNA samples from these women were genotyped using an Illumina Infinium 660k array. Case genotypes were compared with those from 5,069 controls, drawn from two UK population-based studies. After quality control exclusions, we utilized data on 582,886 SNPs in 3,659 cases and 4,897 controls (Online Methods).
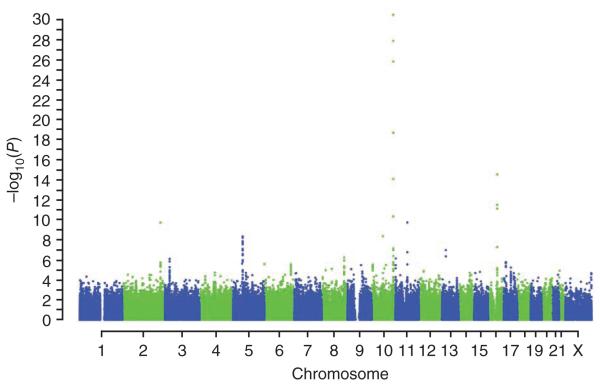
Genotype frequencies in cases and controls were compared using a 1-degree-of-freedom (d.f.) Cochran-Armitage trend test (Fig. 1; for the quantile-quantile plot see Supplementary Fig. 1). There was modest evidence for inflation in the test statistic (λ = 1.12, which is equivalent to λ1,000 = 1.03 for a study of 1,000 cases and 1,000 controls). Adjustment for differential population structure using the first two components based on a principal-components analysis of uncorrelated SNPs reduced the inflation to λ = 1.06 (Online Methods).
Figure 1.
Manhattan plot of 1-d.f. Cochran-Armitage P values for association by genomic position.
We observed evidence of association for all 12 of the susceptibility loci identified through previous GWAS, using the same SNP as that previously identified or a strongly correlated SNP (P = 0.02 to P = 3.6 × 10−31; Table 1). Seven of these loci reached P < 10−4, among which five have previously been evaluated in large collaborative analyses of case-control studies by the Breast Cancer Association Consortium (BCAC). The BCAC analyses involved more than 20,000 cases and 20,000 controls, providing a reliable estimate of the per-allele OR1,5,10. For each of these five SNPs, the per-allele OR in the current study was higher than that estimated from the population-based studies by BCAC by a factor of 1.46-fold to 1.75-fold (P < 0.05 for difference in OR for all SNPs except rs13281615; Supplementary Table 1). This enrichment is broadly consistent with the selection of cases with a family history, assuming a multiplicative polygenic model (which predicts a 1.5-fold higher excess relative risk for the associated SNP for women with one affected first-degree relative and a twofold higher excess relative risk for women with two affected first-degree relatives)9. The loci on 5p12 (rs7716600, a surrogate for rs10941679) and 1p11.2 do not conform to this pattern, having smaller ORs than those published previously (a 1.5-fold higher excess OR can be excluded here in each case, P = 0.018 and P = 0.015, respectively). These results suggest either that the initial effect sizes were overestimated (perhaps due to ‘winner’s curse’) or that these loci have weaker than expected effects in women with a family history due to a different model of susceptibility than is applicable for the other loci. We also found limited evidence in support of the association with the CASP8 D302H polymorphism (P = 0.14; Table 1)8. Consistent with previous results, the two loci showing the largest effect sizes and most significant associations in this GWAS were on chromosome 10, in intron 2 of FGFR2 (rs2981579, P = 3.6 × 10−31) and at the TOX3 locus on 16q (rs3803662, P = 3.2 × 10−15).
Table 1.
Associations in the current study at previously known breast cancer loci
| Strongest association in current study |
Published association |
Association for published SNP in current study |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Most signifcant SNP |
Allelesa | Per-allele OR (95% CI)b |
P | Published SNP |
Allelesa | (r2)c | Published OR | Best tag in GWAS (r 2)d | Allelesa | Per-allele OR (95% CI)b |
P |
| FGFR2 | rs2981579 | G/A (0.42) |
1.43 (1.35–1.53) |
3.6 × 10−31 | rs2981582e | G/A (0.38) |
1.0 | 1.26 (1.22–1.29)1 |
rs2981579 (r2 = 1.0) |
G/A (0.42) |
1.43 (1.35–1.53) |
3.6 × 10−31 |
| TOX3 | rs3803662 | G/A (0.26) | 1.30 (1.22–1.39) |
3.2 × 10−15 | rs3803662 | G/A (0.25) |
1.0 | 1.19 (1.15–1.23)1 |
rs3803662 | G/A (0.26) |
1.30 (1.22–1.39) |
3.2 × 10−15 |
| MAP3K1 | rs889312 | A/C (0.28) |
1.22 (1.14–1.30) |
4.6 × 10−9 | rs889312 | A/C (0.38) |
1.0 | 1.12 (1.08–1.16)1 |
rs889312 | A/C (0.28) |
1.22 (1.14–1.30) |
4.6 × 10−9 |
| 8q24 | rs1562430 | C/T (0.58) |
1.17 (1.10–1.25) |
5.8 × 10−7 | rs13281615 | A/G (0.40) |
0.42 | 1.08 (1.05–1.12)1 |
rs13281615 | A/G (0.41) |
1.14 (1.07–1.21) |
2.2 × 10−5 |
| 2q35 | rs13387042 | G/A (0.49) |
1.21 (1.14–1.29) |
2.0 × 10−10 | rs13387042 | G/A (0.49) |
1.0 | 1.12 (1.09–1.15)10 |
rs13387042 | G/A (0.49) |
1.21 (1.14–1.29) |
2.0 × 10−10 |
| LSP1 | rs909116 | C/T (0.53) |
1.17 (1.10–1.24) |
7.3 × 10−7 | rs3817198 | T/C (0.30) |
0.23 | 1.07 (1.04–1.11)1 |
rs3817198 | T/C (0.33) |
1.12 (1.05–1.19) |
0.0006 |
| 5pl2 | rs9790879 | T/C (0.40) |
1.10 (1.03–1.17) |
0.0032 | rs10941679 | (A/G) (0.25) |
0.48 | 1.19 (1.11–1.28)4 |
rs7716600 (r2 = 0.75) |
C/A (0.22) |
1.11 (1.04–1.19) |
0.0034 |
| 6q25.1 | rs3757318 | G/A (0.07) |
1.30 (1.17–1.46) |
2.9 × 10−6 | rs2046210 | G/A (0.34) |
0.088 | 1.15f (1.03–1.28)7 |
rs6900157 (r2 = 0.96) |
T/C (0.35) |
1.15 (1.08–1.22) |
1.8 × 10−5 |
| SLC4A7 | rs4973768 | C/T (0.47) | 1.16 (1.10–1.24) |
5.8 × 10−7 | rs4973768 | C/T (0.46) |
1.0 | 1.11 (1.08–1.13)5 |
Rs4973768 | C/T (0.47) |
1.16 (1.10–1.24) |
5.8 × 10−7 |
| COX11 | rs1156287 | A/G (0.29) |
0.91 (0.85–0.97) |
0.0058 | rs6504950 | G/A (0.27) |
0.91 | 0.95 (0.92–0.97)5 |
rs7222197 (r2 = 1.0) |
G/A (0.28) |
0.92 (0.86–0.99) |
0.021 |
| RAD51L1 | rs8009944 | C/A (0.75) |
0.88 (0.82–0.95) |
0.0004 | rs999737 | C/T (0.24) |
0.13 | 0.94 (0.88–0.99)6 |
rs999737 | C/T (0.25) |
0.89 (0.83–0.95) |
0.0009 |
| lpll.2 | rs11249433 | A/G (0.42) |
1.08 (1.02–1.15) |
0.010 | rs11249433 | A/G (0.39) |
1.0 | 1.16 (1.09–1.24)6 |
rs11249433 | A/G (0.42) |
1.08 (1.02–1.15) |
0.010 |
| CASP8 | rs10931936 | T/C (0.74) |
0.88 (0.82–0.94) |
0.00015 | rs1045485 | G/C (0.13) |
0.083 | 0.88 (0.84–0.92)8 |
rs17468277 (r2 = 1.0) |
C/T (0.13) |
0.93 (0.85–1.02) |
0.14 |
Allele (frequency of the second listed allele).
Per-allele OR for the second listed allele, relative to the first. In each case the second listed allele was that which correlated with the second-listed published allele.
r2 between the published SNP and most significant SNP in this study based on HapMap CEU.
r2 between the published SNP and the best tagSNP in this study based on HapMap CEU.
Note that fine-mapping and functional analyses suggest that the strongest association for breast cancer is with rs298157825. It is correlated with rs2981579 and rs2981582 at r2 = 0.85. No more strongly correlated tag for rs2981578 was typed in the GWAS.
Estimated OR in Europeans. Estimated OR in Chinese was 1.36.
For three loci (6q25.1, LSP1 and 8q24) we identified a SNP that showed a more significant association than the SNP originally reported associated to breast cancer susceptibility. The SNP with the lowest P value at 6q25.1 (rs3757318, P = 2.9 × 10−6) lies ~200 kb upstream of ESR1 in an intron of C6orf97. In Europeans, rs3757318 is only weakly correlated with rs2046210, which has previously been identified as a susceptibility SNP7 in a study from Shanghai (r2 = 0.088), though these two SNPs are more strongly correlated in an East Asian population (r2 = 0.48 in HapMap CHB). Both rs3757318 and rs6900157 (a surrogate for rs2046210 with r2 = 0.96) remained significantly associated with breast cancer after multiple logistic regression analysis (P = 0.0003 and P = 0.002, respectively). These results suggest either the presence of a single causal variant that is more strongly correlated with rs3757318 than rs2046210 in Europeans or the presence of two causal variants. The more strongly associated SNPs that we identified in the 8q24 and LSP1 regions lie within the same linkage disequilibrium (LD) blocks as the originally identified SNP, and in each case, the original SNP was not significantly associated with risk after adjusting for the new SNP. Thus, these results may reflect the same underlying association and should assist in narrowing the search for the true causal variants. A more strongly associated variant, rs10931936, was also identified at the CASP8 locus (P = 0.0014, r2 = 0.13).
After eliminating SNPs in previously identified susceptibility regions, we identified 28 SNPs in 13 regions of LD that were significant at P < 0.00001. After eliminating SNPs that were strongly correlated, we attempted to replicate these associations by genotyping 15 SNPs in a second stage involving 11,431 cases and 11,081 controls from four studies in the UK and The Netherlands (Online Methods). We also incorporated available data from 1,145 cases and 1,142 controls from the Cancer Genetic Markers of Susceptibility (CGEMS) study. Six SNPs from five regions on chromosomes 9,10 and 11 showed clear evidence of replication in stage 2 (P = 0.0017 or better and in the same direction as stage 1) and reached significance levels over both stages combined of P = 4.6 × 10−7 to P = 3.2 × 10−15 (Table 2 and Supplementary Tables 2 and 3). rs614367 and rs624797, which both showed strong evidence of association, were correlated, and rs624797 showed no independent association after adjustment for rs614367. The per-allele OR was higher in stage 1 than stage 2 for each SNP (P < 0.05 in each case; Supplementary Table 2). This may reflect either winner’s curse or the enrichment of stage 1 for cases with a positive family history. There was no evidence for heterogeneity in the per-allele ORs among the stage 2 samples, with the exception of the weak evidence shown for rs10995190 (P = 0.08; Supplementary Table 2). There was no evidence for departure from a log-additive model for any SNP (that is, the OR for rare homozygotes did not differ significantly from the square of the OR for heterozygotes). There was weak evidence of a decrease in the per-allele OR with age for rs1011970 and of an increase in the per-allele OR with age for rs614367 (P = 0.071 and P = 0.068; Supplementary Table 4). rs614367 and rs624797 (but no other SNPs) showed a consistently stronger association with a positive family history in both stages (for rs614367, P = 0.006 and P = 0.00016, respectively; for rs624797, P = 0.012 and P = 0.001, respectively; Supplementary Table 4). For four of the SNPs (rs10995190, rs1011970, rs614367 and rs624797), the estimated per-allele ORs were higher for estrogen receptor–positive disease and showed little association in estrogen receptor–negative breast cancer, consistent with the pattern seen for the majority of breast cancer loci identified to date. For rs2380205 and rs704010, the per-allele ORs for estrogen receptor–positive and estrogen receptor–negative disease were similar, but the number of estrogen receptor–negative cases used was too small to draw firm conclusions on the effect sizes for this subset (Supplementary Table 4).
Table 2.
Associations between genotype and breast cancer risk for six SNPs
| Marker | Chromosome position | Stagea | Cases/controls | MAFb | Per-allele OR (95% CI) | Heterozygous OR (95% CI) |
Homozygous OR (95%CI) |
Pvaluec |
|
|---|---|---|---|---|---|---|---|---|---|
| Stage | Combined | ||||||||
| rs1011970 G/T | 9 22,052,134 |
Stage 1 | 3,730/4,894 | 0.16 | 1.20 (1.11–1.30) |
1.19 (1.08–1.31) |
1.45 (1.13–1.86) |
2.6 × 10−5 | |
| Stage 2 | 12,253/12,000 | 0.17 | 1.09 (1.04–1.14) |
1.07 (1.01–1.13) |
1.29 (1.12–1.50) |
0.00026 | 2.5 × 10−8 | ||
| rs2380205 C/T | 10 5,926,740 |
Stage 1 | 3,730/4,895 | 0.44 | 0.86 (0.81–0.92) |
0.86 (0.78–0.95) |
0.75 (0.66–0.85) |
7.9 × 10−5 | |
| Stage 2 | 12,235/11,961 | 0.43 | 0.94 (0.91–0.98) |
0.95 (0.90–1.01) |
0.89 (0.82–0.95) |
0.0017 | 4.6 × 10−7 | ||
| rs10995190 G/A | 10 63,948,688 |
Stage 1 | 3,731/4,891 | 0.14 | 0.76 (0.70–0.84) |
0.77 (0.69–0.86) |
0.55 (0.40–0.77) |
6.1 × 10−8 | |
| Stage 2 | 12,261/12,000 | 0.15 | 0.86 (0.82–0.91) |
0.84 (0.79–0.89) |
0.83 (0.69–1.00) |
1.4 × 10−8 | 5.1 × 10−15 | ||
| rs704010 G/A | 10 80,511,154 |
Stage 1 | 3,726/4,893 | 0.39 | 1.15 (1.09–1.23) |
1.05 (0.95–1.15) |
1.38 (1.22–1.57) |
3.5 × 10−6 | |
| Stage 2 | 12,222/11,992 | 0.39 | 1.07 (1.03–1.11) |
1.11 (1.05–1.17) |
1.13 (1.04–1.21) |
0.00026 | 3.7 × 10−9 | ||
| rs614367 C/T |
11 69,037,945 |
Stage 1 | 3,723/4,882 | 0.15 | 1.30 (1.20–1.41) |
1.24 (1.13–1.37) |
2.02 (1.56–2.64) |
3.9 × 10−8 | |
| Stage 2 | 12,114/11,967 | 0.15 | 1.15 (1.10–1.20) |
1.16 (1.10–1.23) |
1.27 (1.10–1.47) |
1.3 × 10−8 | 3.2 × 10−15 | ||
Stage 2 includes genotype data in SEARCH, RBCS and FBCS together with publicly available data from CGEMS.
MAF, frequency of the minor (second listed) allele.
Adjusted 1-d.f. P trend (see Online Methods).
To examine whether there was evidence for a more strongly associated variant in any of the above regions, we used imputation to estimate the genotype probabilities in the stage 1 data at known SNPs in region using the HapMap CEU data as a framework. On chromosome 11, we identified four SNPs that showed a more significant association than rs614367 (most significantly associated SNP rs6610204; P = 4.6 × 10−14; Supplementary Table 5). In the other regions, no SNPs showed associations that were more significant than the original SNP. We also estimated the ORs associated with haplotypes of SNPs in each of the five regions (Supplementary Table 6). In each case, the association was present on more than one haplotype carrying the risk allele for the initially associated SNP, suggesting that the associations are unlikely to be driven by a single rare, high penetrance variant. For the chromosome 11 region, there was evidence of association with risk for two related haplotypes carrying the T allele of rs614367 with a combined frequency of 4%, suggesting that the causal variant may be somewhat rarer than the 15% minor allele frequency of rs614367.
SNP rs1011970 lies in a 180-kb block on 9p21 that includes CDKN2A and CDKN2B. These two genes encode cyclin-dependent kinase inhibitors and are frequently mutated or deleted in a wide variety of human tumors11. Germline mutations in CDKN2A predispose to malignant melanoma and pancreatic cancer12, and recent GWAS also identified rs1011970 to be associated with melanoma risk13; SNPs within this same region are associated with nevus density and melanoma14, basal cell carcinoma15, glioma16,17, diabetes18 and coronary heart disease19. This is the first example of the same common variant predisposing to breast cancer and another cancer type rs10757278, which is correlated with rs1011970 (r2 = 0.7), is associated with levels of expression in lymphocytes of CDKN2A, CDKN2B and a noncoding RNA in the same block, CDKN2BAS (also known as ANR1L)20.
rs614367 on 11q13 lies in an LD block of ~166 kb that contains no annotated genes. This region is frequently amplified in human tumors, including breast cancers21. Plausible genes flanking this block include: proximally, MYEOV, a gene overexpressed in myeloma; distally, CCND1, encoding cyclin D1, a protein critical for cell-cycle control that is somatically altered in many tumor types; ORAOV1, a gene overexpressed in oral cancer; and three genes encoding fibroblast growth factors, FGF19, FGF4 and FGF3. FGF3 and FGF4 are oncogenic growth factors that bind distinct FGFR2 isoforms, providing a possible link with the FGFR2 susceptibility locus22.
rs10995190 on chromosome 10 lies within intron 4 of ZNF365, which encodes zinc finger protein 365. An amino acid substitution in this gene has been associated with uric acid nephrolithiasis23. Recent GWAS have identified another variant within this gene, rs10995271, located 159 kb downstream of rs10995190, to be associated with Crohn’s disease24. rs2380205 lies in a 105-kb block on chromosome 10 containing the genes ANKRD16 (encoding ankyrin repeat domain 16) and FBXO18 (encoding the F-box protein, helicase 18). rs704010 on chromosome 10 lies in a 20-kb block 90 kb upstream of ZMIZ1 (encoding zinc finger MIZ-type containing 1).
Based on the estimated per-allele ORs from stage 2 of our study, the newly identified loci explain approximately 1.2% of the familial risk of breast cancer, though the overall contribution may be larger because the true causal variants may be more strongly associated with disease than the SNPs tagging them in this study. Taken together with estimates from previous studies, the 18 confirmed breast cancer susceptibility loci explain approximately 8% of the familial risk of breast cancer, whereas rarer mutations in the known high risk loci (principally BRCA1 and BRCA2) and moderate risk loci explain a further ~20%. This is by far the largest breast cancer GWAS to date and confirms that the FGFR2 and TOX3 loci (conferring per-allele ORs between 1.2 and 1.3) have the largest effect sizes from among the common susceptibility loci that are detectable with the current high-coverage genome-wide SNP sets. The residual familial risk is therefore likely to be due to a combination of a large number of common variants with smaller effects together with rarer variants not testable with current arrays. It is likely that many additional loci will be identifiable through more extensive follow-up of data from this and other GWAS.
URLs. CGEMS, http://cgems.cancer.gov/; Welcome Trust Case Control Consortium (WTCCC), http://www.wtccc.org.uk/; Nurses Health Study, http://www.channing.harvard.edu/nhs/; Mach, http://www.sph.umich.edu/csg/abecasis/MaCH/index.html; data access from this GWAS, http://www.srl.cam.ac.uk/genepi/.
ONLINE METHODS
Samples
Three thousand nine hundred and sixty breast cancer cases were used in stage 1, of which 3,652 were from cancer genetics clinics in the UK recruited through the Familial Breast Cancer Study (FBCS) and 308 were from oncology clinics in the UK recruited through the Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH) study. Cases were preferentially selected to have at least two affected first- or second-degree relatives. The majority of cases were screened and found to be negative for germline mutations, including large rearrangements, in BRCA1 and BRCA2. A minority of samples were not tested for BRCA1 or BRCA2 mutations. All carriers of disease-associated mutations in BRCA1 and BRCA2 were excluded. We also excluded all cases with self-reported non-European ancestry.
Controls for stage 1 were drawn from two sources: 2,930 controls were drawn from the 1958 Birth Cohort (1958BC), a population-based study in the United Kingdom of individuals born in 1 week in 1958 (ref. 26). The remaining 2,737 controls were identified through the UK National Blood Service (NBS)19. These samples were genotyped as part of the Wellcome Trust Case Control Consortium (WTCCC2; see URLs)27. The analyses presented here are based on 2,482 1958BC and 2,587 NBS controls for which genotype data were available at the time of analysis.
Samples for stage 2 were drawn from six sources: (i) the SEARCH study, a population-based study of cases from East Anglia (n = 6,640); controls (n = 6,832) were drawn from the European Prospective Investigation into Cancer and Nutrition (EPIC) study, a population-based cohort study of diet and cancer from general practices contributing to SEARCH; (ii) the Rotterdam breast cancer study (RBCS) (799 cases, 800 controls); (iii) the Familial Breast Cancer Study (FBCS), consisting of additional cases ascertained through UK cancer genetics clinics (n = 2,009); (iv) the RMH breast cancer series (n = 1,732); and (v) the Prospective study of Outcomes in Sporadic vs. Hereditary breast cancer (POSH) study (n = 251). The combined samples from these latter three series (n = 3,992) were analyzed in a single replication experiment together with additional controls selected through the 1958BC (n = 3,450), none of which were included in stage 1. For stage 2, we also incorporated data on the relevant SNPs from the CGEMS study (see URLs). CGEMS is based on 1,145 cases and 1,142 controls drawn from the Nurses Health Study (see URLs) which were genotyped using the Illumina 550k array.
All studies were approved by the appropriate ethics committees.
Genotyping
Genotypes for stage 1 cases were generated using a custom Illumina Infinium 670k array and controls were genotyped using an Illumina Infinium 1.2M array at the Wellcome Trust Sanger Institute. For this analysis, we analyzed data on 594,375 SNPs that were successfully genotyped on both arrays. Genotypes for both arrays were called using the Illuminus algorithm28. We used genotypes for which Illuminus generated a posterior probability of >0.95. Cluster plots were inspected manually for all SNPs considered for inclusion in stage 2.
Genotyping for stage 2 was performed by 5′ exonuclease assay (Taqman) using the ABI Prism 7900HT Sequence Detection System according to the manufacturer’s instructions. Primers and probes were supplied directly by Applied Biosystems as Assays-By-Design. Assays included at least two negative controls and 2% to 5% duplicates per plate. Genotyping for one marker, rs1866823, failed for the SEARCH and RBCS studies, and the marker was replaced by rs2246873 (r2 = 0.94 in HapMap CEU).
Analyses
We restricted analyses to individuals who were called on >97% of successfully genotyped SNPs. To identify close relatives, we computed identity-by-state (IBS) probabilities for all pairs. We confirmed 2 case monozygotic twin (MZ) pairs, 22 duplicate case pairs and 24 first-degree relative pairs (IBS > 0.86). We also identified 4 probable case-control and 44 probable control-control sibling pairs. We excluded the control from the case-control pairs and the sample with the lower call rate from the remaining pairs. By computing IBS scores between participants and individuals in HapMap and by using multidimensional scaling, we identified 89 individuals who appeared to have substantial Asian or African ancestry (defined as approximately >15% non-European ancestry, comprising 69 cases, 4 individuals from 1958BC and 16 NBS). After these exclusions, 3,659 cases and 4,897 controls were used in the final analysis.
We filtered out all SNPs with, in either cases or controls, a MAF < 1%, a call rate of < 99% and a MAF < 5%, or a call rate < 95% and MAF ≥ 5%. We also excluded SNPs whose frequencies departed from HWE at P < 0.00001 in controls or P < 10−12 in cases. After these exclusions, we used data on 582,886 SNPs. Duplicate concordance was 99.99%.
Statistical methods
We first assessed associations between each SNP and breast cancer at stage 1 using a 1-d.f. Cochran-Armitage trend test and a general 2-d.f. χ2 test. Inflation in the χ2 statistic was assessed using the genomic control approach; we derived an inflation factor λ by dividing the median of the lowest 90% of the 1-d.f. statistics by the 45% percentile of a 1-d.f. χ2 distribution (0.357). We have also presented the equivalent inflation factor for a study of 1,000 cases and 1,000 controls (λ1,000) calculated by λ1,000 = 1 + 500 (1 / Ncases+ 1 / Ncontrols) / (λ − 1), where Ncases and Ncontrols are the number of cases and controls, respectively.
To correct for potential inflation due to population structure, we performed a principal-components analysis based on the genotypes of a subset of 35,797 uncorrelated SNPs (r2 < 0.1)29. We then computed 1-d.f. score tests for each SNP, adjusting for progressively larger numbers of principal components as covariates. Adjustment for the first two components reduced the inflation slightly (to 1.06); however, adjustment for further components did not reduce the inflation further. Adjusted significance tests were therefore calculated from the score tests adjusted for two principal components. To allow for the residual inflation, we adjusted the resulting test statistics using the genomic control approach by dividing the test statistic by the inflation factor.
SNPs were selected for evaluation in stage 2 on the basis of a significance level of P < 10−5 based on the unadjusted 1-d.f. trend test. Where two or more SNPs were selected from the same region, we used multiple logistic regression to determine a minimal set of SNPs that showed evidence of association after adjustment for other SNPs. In practice, one SNP was selected in each region except in the case of one region, in which two SNPs were genotyped.
After stage 2, overall 1-d.f. and 2-d.f. tests of association were derived, stratified by stage and study. Adjusted tests of association were derived by adjusting in stage 1 for principal components and genomic control as described above. In the combined analysis, the effect size in stage 1 was weighted by a factor of 2 relative to that in stage 2, consistent with the effect size expected under a polygenic model. A criterion of P < 5 × 10−7 was used for genome-wide significance19, and ORs and 95% confidence limits were estimated using unconditional logistic regression, stratified by study. In the text, we have reported the combined tests of association over both stages, but we have emphasized the OR estimates from stage 2 to minimize the effect of winner’s curse. Tests of homogeneity of the ORs across strata were assessed using likelihood ratio tests. The associations between genotype and family history in stage 2, and between genotype and estrogen receptor status, were assessed using a case-only analysis (that is, treating family history or estrogen receptor status as the outcome variable and estimating a per-allele OR for each SNP using logistic regression). For stage 1, the effect of family history was analyzed using a family history score, derived as the total number of affected relatives weighted by their degree of relationship to the case. The effect of family history score on per-allele OR was assessed using constrained polytomous regression. Modification of the ORs by age at diagnosis was assessed using a case-only analysis, assessing the association between age and SNP genotype in the cases using polytomous regression. The contribution of the loci to the familial risk of breast cancer was estimated by first computing the familial risk to a daughter of an affected individual that was attributable to each locus (λ1) from the allele frequency and the estimated per-allele OR in the SEARCH study, which was largest study contributing to stage 2 and which is population based. The proportion of the familial risk due to each locus was then calculated as ln(λ1) / ln(2), assuming an overall familial relative risk of 2. The combined effect of all loci was then derived by summing the locus-specific contributions (that is, assuming a log-additive model). Imputed genotypes for non-typed SNPs were estimated using Mach (see URLs), using the HapMap CEU data as a framework. Haplotype analyses were conducted in haplo.stats30. Haplotypes were based on SNPs in each region that were significantly associated with breast cancer at P < 0.001, after eliminating perfectly correlated SNPs. Per-haplotype ORs were estimated using the haplo.cc routine. Other analyses were performed in R, principally using GenABEL31, and Stata (R, http://www.r-project.org/; Stata, http://www.stata.com/).
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust and by Cancer Research UK. D.F.E. is a Principal Research Fellow of Cancer Research UK. C.T. is funded by a Medical Research Council Clinical Research Fellowship. The samples were collected and screened for BRCA mutations through funding from Cancer Research UK; US Military Acquisition (ACQ) Activity, Era of Hope Award (W81XWH-05-1-0204) and the Institute of Cancer Research (UK). This study makes use of data generated by the Wellcome Trust Case Control Consortium (WTCCC) 2. A full list of the investigators who contributed to the generation of the data is available from the WTCCC website. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. Funding for this project was provided by the Wellcome Trust under award 085475. We thank the SEARCH team and Eastern Cancer Registry and Information Centre (ECRIC) for recruitment of the SEARCH cases. We acknowledge the clinicians from the Rotterdam Family Cancer Clinic who were involved in collecting the RBCS samples: C. Seynaeve, J. Klijn, J. Collee and R. Oldenburg.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS: D.F.E., N.R., M.R.S., P.D.P.P. and A.M.D. obtained funding for the study. D.F.E. designed the study and drafted the manuscript. D.F.E. and C.T. conducted the statistical analyses. J.M. provided data management and bioinformatics support. C.T. and N.R. coordinated the Familial Breast Cancer Study (FBCS). D.P., A.R., S.S., S.H., D.H., M.W.-P., C.T. and N.R. coordinated the FBCS genotyping. P.D.P.P. and D.F.E. coordinated Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH). S.A., M.M., M.G., C.S.H. and A.M.D. coordinated the stage 2 genotyping of the SEARCH and RBCS samples. M.H., M.S. and A.v.d.O. coordinated and provided samples and data from RBCS. C.L. coordinated the stage 1 genotyping. G.R. and R.H. provided data and samples from the Royal Marsden Hospital (RMH) study. W.T. and D.E. provided data and samples from the Prospective study of Outcomes in Sporadic vs. Hereditary breast cancer (POSH) study. D.E. and D.G.E. provided data and samples for FBCS. All authors provided critical review of the manuscript.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
References
- 1.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 4.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 9.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: implications for design of association studies. Genet. Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- 10.Milne RL, et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J. Natl. Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamb A, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 12.Kamb A, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat. Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 13.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falchi M, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stacey SN, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shete S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrensch M, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlseder J, et al. Patterns of DNA amplification at band q13 of chromosome 11 in human breast cancer. Genes Chromosom. Cancer. 1994;9:42–48. doi: 10.1002/gcc.2870090108. [DOI] [PubMed] [Google Scholar]
- 22.Ornitz DM, et al. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 23.Gianfrancesco F, et al. Identification of a novel gene and a common variant associated with uric acid nephrolithiasis in a Sardinian genetic isolate. Am. J. Hum. Genet. 2003;72:1479–1491. doi: 10.1086/375628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udler MS, et al. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum. Mol. Genet. 2009;18:1692–1703. doi: 10.1093/hmg/ddp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int. J. Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 27.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, et al. Principal components analysis corrects for stratification in genome- wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 30.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]