SUMMARY
A canonical quantitative view of transcriptional regulation holds that the only role of operator sequence is to set the probability of transcription factor binding, with operator occupancy determining the level of gene expression. In this work, we test this idea by characterizing repression in vivo and the binding of RNA polymerase in vitro in experiments where operators of various sequences were placed either upstream or downstream from the promoter in Escherichia coli. Surprisingly, we find that operators with a weaker binding affinity can yield higher repression levels than stronger operators. Repressor bound to upstream operators modulates promoter escape, and the magnitude of this modulation is not correlated with the repressor-operator binding affinity. This suggests that operator sequences may modulate transcription by altering the nature of the interaction of the bound transcription factor with the transcriptional machinery, implying a new layer of sequence dependence that must be confronted in the quantitative understanding of gene expression.
INTRODUCTION
Cells control how much, when and where to express a gene in response to changes in their intracellular and extracellular environments. A variety of mechanisms are employed to exert this control at each of the steps along the path from DNA to active protein (Alberts, 2008). An important mechanism of gene regulation in bacteria acts through transcription factors that bind to specific sites in the promoter region, the sequence of DNA immediately upstream of genes, where RNA polymerase binds. As a result of interactions or steric interference between transcription factors bound to these sites and RNA polymerase, activation or repression of transcription ensues (Bintu et al., 2005b; Ptashne and Gann, 2002). Indeed, an important activity of modern genome science is finding transcription factor binding sites and determining the rules by which promoter architecture, i.e., the position and sequence of these binding sites, dictates the level of the gene expression (Buchler et al., 2003; Segal and Widom, 2009).
It is often assumed that that the role of operators is simply to act as docking sites for transcription factors, recruiting them to the promoter region (Meijsing et al., 2009). In this view, which we here term the “occupancy hypothesis” the sequence of the operator simply determines its binding affinity for its target transcription factor. This binding affinity, together with the concentration of active transcription factors and the interactions with other DNA-binding proteins, determines the occupancy of the operator which, in turn, is thought to influence the level of transcriptional regulation exerted by the transcription factor (Alberts, 2008; Bintu et al., 2005b; Buchler et al., 2003; Davidson, 2006). For example, in Figure 1A we consider promoters presenting binding sites of different affinities for a repressor. Given these binding affinities the intracellular number of active repressors will determine the probability of finding the repressor bound to each one of the operators. As a result, the shape of the input-output function, that is, the level of output gene expression as a function of the input concentration of repressors, will reach the same level of repression at different repressor concentrations, which are determined by the binding affinities of the operators. A promoter that contains a strong operator, a site on the DNA that binds the transcription factor tightly, is expected to require a lower intracellular concentration of the transcription factor to reach the same level of repression (or activation) as a promoter that has a weaker operator (Bintu et al., 2005a, 2005b; Buchler et al., 2003; Vilar and Leibler, 2003). Quantitatively, this can be expressed by saying that the level of transcription is determined by the binding probabilities of these transcription factors to the DNA, as shown in Figure 1A. As a result, a key prediction of the occupancy hypothesis is that when plotting the level of gene expression as a function of the operator occupancy the curves corresponding to different operators, regardless of their affinity, will all collapse onto a master curve as shown in Figure 1A.
Figure 1. The Occupancy Hypothesis for the Action of Transcription Factors and Its Predictions for Gene Regulation.
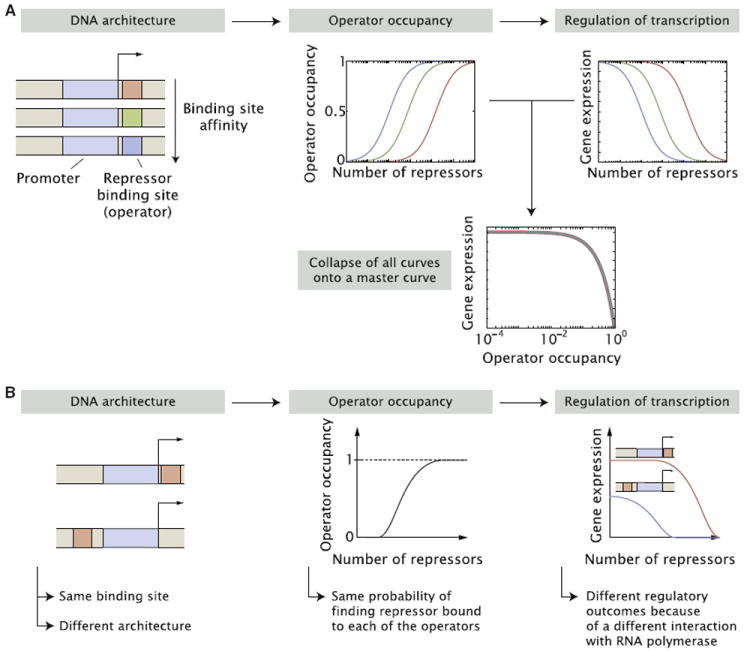
(A) Binding sites of different affinity lead to different occupancies as a function of the number of repressors with weaker operators requiring higher repressor number to achieve the same occupancy. The shapes of both the occupancy and gene expression curves as a function of the number of repressors depend on the operator binding affinity. However, all these curves collapse onto a single universal curve when the level of gene expression is plotted as a function of operator occupancy. Note that the use of repression to make this point is inconsequential and that the same point can be made using activation of gene expression.
(B) An alternative modulation of regulatory architecture can stem from moving the binding site with respect to the promoter by keeping the operator binding affinity constant. In this case, the probability of finding repressor bound to the operator (operator occupancy) should presumably be the same for both constructs. The positioning of the operator, however, is expected to modulate the nature of the interaction between the repressor and the transcriptional machinery leading to different input-output functions for each regulatory architecture.
The effect of the modulation of the affinity of an operator as the surrounding sequence context is kept constant is not to be confused with the effect of moving a given operator with respect to the promoter while leaving its affinity constant, shown diagrammatically in Figure 1B. Because the affinity is kept constant one would, in principle, expect the probability of finding the transcription factor bound to the operator in each of the constructs exemplified in Figure 1B to be the same as a function of the transcription factor concentration. However, we expect the relative positioning of the operator and promoter to modulate the nature of the interaction between the transcription factor and the transcriptional machinery. As a result, the shape of the input-output function of each regulatory architecture will, in principle, differ.
From the point of view of transcriptional regulation described above it follows that once the occupancy of binding sites by transcription factors within a regulatory region is determined and the effect of the bound transcription factor on the transcriptional machinery is known, the resulting level of gene expression can be calculated (Bintu et al., 2005b; Buchler et al., 2003; Segal et al., 2008; Vilar and Leibler, 2003). This view of transcriptional regulation has been challenged by recent results in eukaryotic cells that demonstrated that the affinity of transcription factors for different cofactors can be modulated by the sequence of the transcription factor binding site (Lefstin and Yamamoto, 1998; Meijsing et al., 2009). This suggests that in order to fully understand the function of a regulatory region, the effect of operators on the nature of the interaction between transcription factors and the transcriptional machinery may have to be considered in addition to finding their position in the genome and their affinity for transcription factors.
Despite this recent evidence, many quantitative studies both in the bacterial and eukaryotic context, make explicit or implicit use of the occupancy hypothesis in order to describe the action of transcription factors on the level of gene expression (Ackers et al., 1982; Amit et al., 2011; Davidson, 2006; Garcia and Phillips, 2011; Gertz et al., 2009; Ptashne and Gann, 2002; Segal et al., 2008; Zinzen et al., 2009). Indeed, every time that the rate of protein production is written in terms of Hill functions, for example, this occupancy hypothesis has been made implicitly (Cağatay et al., 2009; Elowitz and Leibler, 2000; Fowlkes et al., 2008; Gardner et al., 2000; Klumpp et al., 2009; Kuhlman et al., 2007; Novák and Tyson, 2008; Tsai et al., 2008). As a result, such quantitative descriptions of transcriptional regulation are at least potentially incomplete and not on par with our current qualitative knowledge of the nuanced role of operator sequence beyond that of determining binding affinity.
In the remainder of the study, we demonstrate a form of modulation in transcriptional regulation that is at odds with the traditional operator occupancy viewpoint, suggesting that the canonical picture is incomplete. We do this by adopting a synthetic biology approach, in which we deliberately tune operator position, operator strength and transcription factor copy number in order to systematically traverse the parameter space of the simple repression architecture (i.e., the case in which a repressor regulates a promoter through the presence of a single binding site in its vicinity) by Lac repressor. This repressor is one of the best understood transcription factors (Müller-Hill, 1996). Through systematic in vivo gene expression measurements, in vitro single molecule experiments, and theoretical modeling we show that when the repressor binds upstream from the promoter the choice of the sequence of its operator binding site influences the rate of synthesis of mRNA, but that the extent of repression does not respect the rank ordering of the strength of the different operators in the way predicted by the occupancy-based model of regulation (Figure 1A). As a result, we expand the quantitative view for the role of operator sequence in the context of the paradigmatic Lac repressor-operator interaction. In this context, operator sequence acts not only to determine the occupancy of DNA binding proteins, but also affects the nature of the interactions between the transcription factors and the transcriptional machinery.
RESULTS
Operator Position Leads to Different Repression Behaviors
In Escherichia coli, genome-wide studies have resulted in an atlas of binding sites for both repressors and activators that give a picture of the diversity of binding site arrangements even in the case of simple repression. For example, in Figure 2A we show a histogram of the positions of repressor binding sites that regulate promoters through simple repression in E. coli (Gama-Castro et al., 2008; Madan Babu and Teichmann, 2003). As can be seen from this histogram the simple repression motif may be able to act over a wide range of positions relative to the polymerase start site.
Figure 2. Operator Position Leads to Different Repression Behavior Upstream and Downstream from the Promoter.
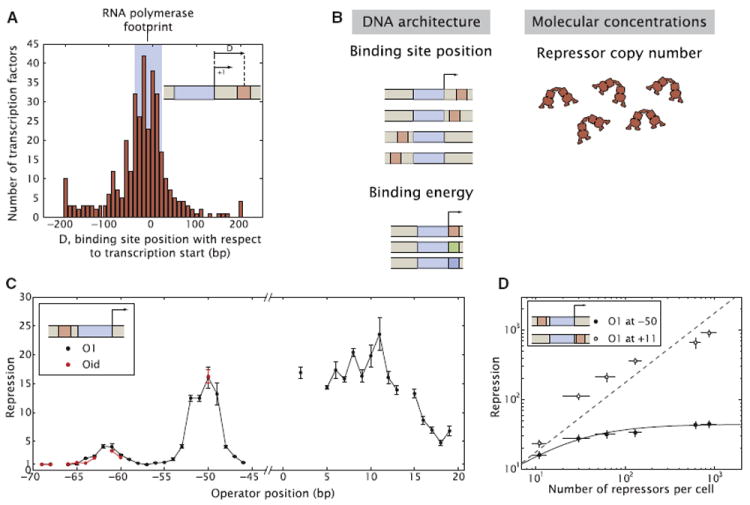
(A) Histogram of the positions of transcription factor binding with respect to the transcription start site of the regulated promoter (Gama-Castro et al., 2008). For reference, the DNase footprint of RNA polymerase in the open complex is overlaid in light blue (Straney and Crothers, 1987).
(B) Schematic of the parameters that may be tuned in vivo in order to elicit different levels of gene expression within the simple repression architecture.
(C) Repression as a function of operator position for an O1 and an Oid operator sequence.
(D) Unlike the case of repression for an operator located at +11, the input-output function of repression at −50 shows saturation. The lines are fits of the +11 data to Equation S6 and of the −50 data to Equation S7. The positions are defined as the distance between the center of the operator and the lacUV5 transcription start site. Error bars are SD over multiple repeats.
See also Figures S1 and S2.
In order to investigate the effects of lac operator position relative to the polymerase binding site, we carried out systematic gene expression measurements for different operators as a function of their position relative to the transcription start site with single base pair resolution. Examples of the parameters varied in the construct library used to assay the effect of operator positioning on repression are shown schematically in Figure 2B, whereas a more detailed version including the sequences is shown in Figures S1A–S1C. We used the lacUV5 promoter, which is a mutant of the lac promoter that does not require activation by CRP (Müller-Hill, 1996). This promoter controls the expression of the YFP or lacZ gene, which we use to quantify the level of gene expression. We measure the regulatory effect of Lac repressor as repression, which is defined as
| (Equation 1) |
where R is the intracellular number of repressors. When the operator is moved downstream from the transcriptional initiation site, the level of repression is relatively independent of position until the center of the operator reaches +16 as shown in Figure 2C. At these downstream positions, Lac repressor might be acting by the same mechanism as it does at +11, where it blocks open complex formation or an earlier step in initiation (Sanchez et al., 2011; Schlax et al., 1995). However, it is also possible that as the repressor is moved from its wild-type position repression might be realized through different mechanisms, as has already been shown for a variety of transcription factors (Hochschild and Dove, 1998; Pavco and Steege, 1990, 1991; Rojo, 1999), possibly affecting any of the various steps in transcription initiation or elongation as shown schematically in Figures S2A and S2B (Elledge and Davis, 1989; Lopez et al., 1998).
By way of contrast, when the operator is moved to positions upstream from the initiation site, the level of repression strongly depends on the location of the operator and the variation in repression is substantial, with at least a 15-fold effect between the peaks and valleys and with the valleys corresponding to no repression (Figure 2C). Interestingly, the repression profile shows two peaks, with a separation between them of ~10–11 bp, intriguingly close to the helical period of the double stranded DNA helix (Amit et al., 2011; Becker et al., 2005; Lee and Schleif, 1989; Müller et al., 1996). We find a maximum repression level when the operator is centered at −50 base pairs upstream from the initiation site, with another smaller peak at −61. Our results are qualitatively consistent with previous studies on the effect of lac operator position on repression (Besse et al., 1986; Bond et al., 2010; Elledge and Davis, 1989). For a detailed comparison of previous results to our work please refer to the Extended Experimental Procedures and Figures S1D and S1E.
In order to better understand how operator location affects the input-output function, we measured repression as a function of the intracellular number of repressors over almost two orders of magnitude in the number of repressors (Garcia and Phillips, 2011) for two selected locations, one downstream at +11, the wild-type position of O1 in the lac promoter, and one at −50, where the peak of maximum upstream repression lies. As shown in Figure 2D, the two operator locations differ both qualitatively and quantitatively in the nature of their input-output function. For the +11 location, the repression factor grows linearly with the intracellular number of repressors. This behavior is expected for the repression mechanism based on blocking of open complex formation or of an earlier step in initiation, as discussed by Garcia and Phillips (2011); Sanchez et al. (2011); Vilar and Leibler (2003) and shown in Equation S6. A detailed description of this model in the context of simple repression for the constructs described above can be found in the Extended Experimental Procedures, Figure S3A, and Garcia and Phillips (2011).
In contrast, for the operator at −50 we found that the repression factor grows with repressor copy number only until it saturates. This result cannot be explained by a competition between Lac repressor and RNA polymerase, and suggests that RNA polymerase can bind to the promoter and initiate transcription even when Lac repressor is bound, although the overall transcription rate (i.e., the number of mRNA transcripts produced per unit of time) is reduced by the presence of the repressor to a low, basal level, about 40-fold less than the unregulated level. Thus, a direct prediction of this hypothesis is that Lac repressor does not completely inhibit the formation of stable RNA polymerase-promoter complexes when bound at −50.
It is important to note, however, that the data obtained with the operator at +11 could also accommodate a saturating behavior. In the Extended Experimental Procedures and Figure S2C we discuss this scenario in detail and conclude that, if that was the case, this would signal a violation of the occupancy hypothesis in repression at this well-studied operator location as well.
Single Molecule Imaging In Vitro Suggests that Lac Repressor Located at −50 and RNA Polymerase Are Bound Simultaneously
In order to test the hypothesis outlined above, and gain insight into the mechanism of repression when the lac operator is at −50, we performed single-molecule experiments where the occupancy of RNA polymerase on individual DNA molecules can be observed directly. Fluorescently labeled RNA polymerase and fluorescently labeled DNA were incubated together prior to adding heparin, which sequesters RNA polymerase molecules that have not formed an open complex. Finally, the reaction was introduced into a flow chamber yielding the arrangement shown in Figure 3A (Sanchez et al., 2011). We used multi-wavelength single molecule total internal reflection fluorescence (TIRF) microscopy to determine the fraction of DNA molecules tethered to the surface of the chamber that were occupied by RNA polymerase. A similar experiment was performed in which Lac repressor was preincubated with the DNA prior to the addition of RNA polymerase.
Figure 3. Upstream Repression through Lac Repressor-RNA Polymerase Ternary Complex Formation.
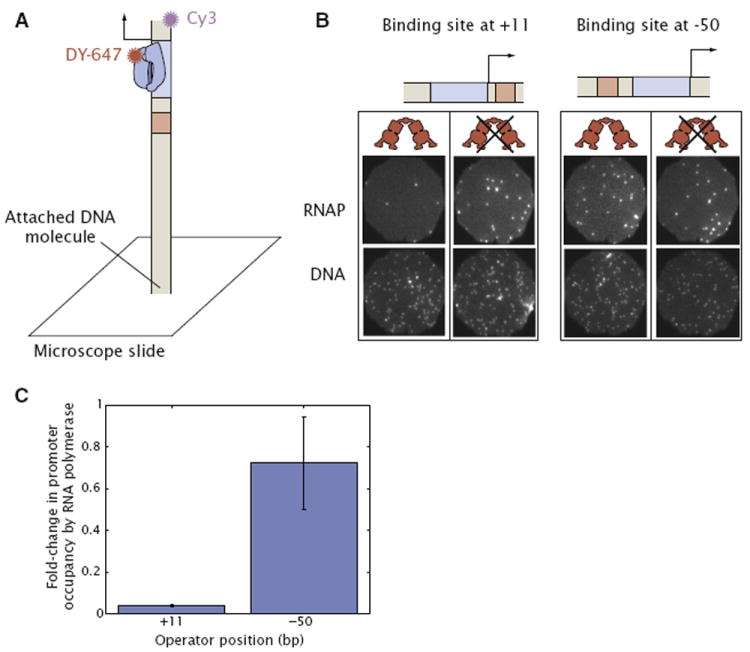
(A) In vitro single molecule experiments in which the occupancy of RNA polymerase on the promoter DNA is quantified by fluorescently labeling each molecular species (Sanchez et al., 2011).
(B) Representative fields of view are shown where the occupancy of RNA polymerase on DNA is measured for the constructs with O1 located at +11 and −50 preincubated with and without Lac repressor.
(C) The analysis of several images such as those shown in (B) confirms the presence of a promoter bound RNA polymerase for the −50 construct because of the small change in RNA polymerase occupancy caused by the presence of repressor. In contrast, there is a significant fold-change in the promoter occupancy by RNA polymerase for the +11 construct, consistent with a model of inhibition of the open complex and previous work (Sanchez et al., 2011). More than 500 molecules were counted for each condition. Error bars are the SD over two replicates for each condition.
See also Figure S2.
Representative fields of view for the experiment performed on both +11 and −50 constructs are shown in Figure 3B. We see that Lac repressor causes a significant change in DNA occupancy by RNA polymerase when the operator is located at +11, indicating that Lac repressor excludes the formation of stable RNA polymerase-DNA open complexes. In contrast, when the operator is located at −50 there is little change, which suggests that Lac repressor is not able to prevent formation of stable RNA polymerase-DNA open complexes when the repressor is bound at this location. At +11 the presence of stably bound polymerase on the DNA is not completely abolished by repressor due to the existence of nonpromoter polymerase binding site and presumably a similar effect occurs with our −50 constructs as described below (for details please see the Extended Experimental Procedures and Sanchez et al., 2011).
Many such fields of view were imaged for each construct. By counting the number of RNA polymerase-DNA complexes that form in the absence and the presence of Lac repressor (and correcting for the fraction of those events that correspond to RNA polymerase bound to a nonpromoter location) we can calculate the fold-change in promoter occupancy by polymerase induced by repressor. The results are shown in Figure 3C, and summarize the average occupancies obtained in different replicates of the experiment with different preparations of all the reagents. From Figure 3C we see again that Lac repressor bound at +11 largely inhibits RNA polymerase occupancy on the promoter DNA. In this construct repressor reduces the formation of RNA polymerase-promoter open complexes down to (4.0 ± 0.4)% of the number of complexes that form in the absence of Lac repressor. This reduction is consistent with recent measurements with the same promoter (Sanchez et al., 2011), which revealed that Lac repressor works by inhibiting open complex formation at the lacUV5 promoter, and indicates that under the conditions of our in vitro experiments, and for the concentrations of repressor (200 nM) and RNA polymerase (80 nM) we use, the O1 operator is almost saturated with repressor (~96%). By way of contrast, Lac repressor bound at −50 reduces open complex formation only modestly, down to (72 ± 22)% of that in the absence of repressor.
These quantitative results indicate that RNA polymerase occupancy on the promoter is affected only slightly by repressor bound at −50. If Lac repressor at −50 reduces open complex formation by <2-fold in vitro, how can we observe a 40-fold reduction of gene expression in vivo? Because our results suggest that Lac repressor bound at −50 allows stable formation of open complexes by RNA polymerase at the promoter, they imply that the regulation of the level of gene expression comes from a substantial effect of repressor on steps occurring after open complex formation in the transcription initiation pathway.
In light of these results, we hypothesize that at −50 the repressor is directly affecting the overall rate of promoter escape, rather than just the occupancy of RNA polymerase on the promoter as an open complex. As a result we propose a thermodynamic model for in vivo upstream simple repression by Lac repressor that is schematized in Figure 4A and tested systematically in the following section.
Figure 4. General Thermodynamic Model of Simple Repression for an Upstream Repressor Binding Site.
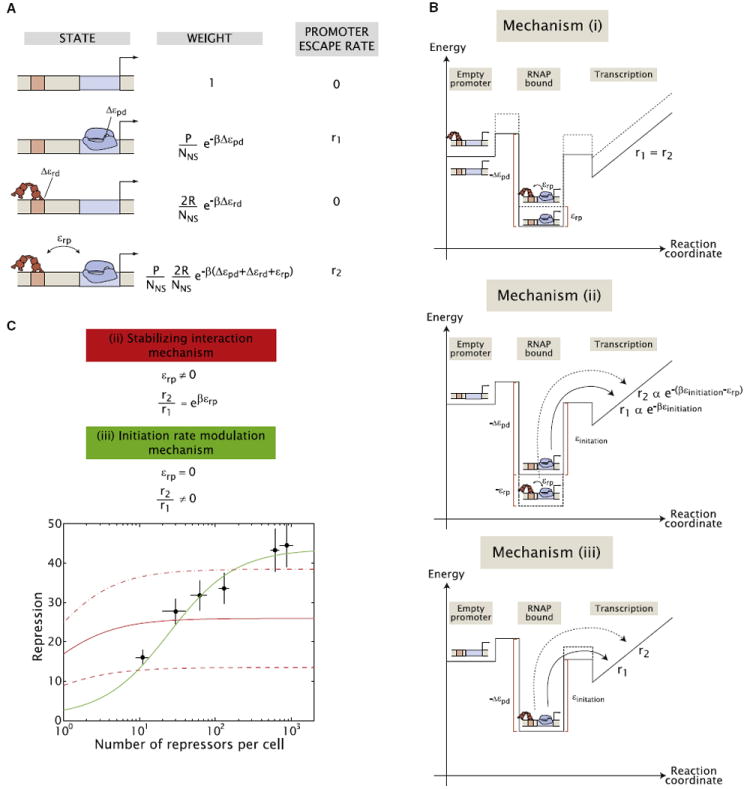
(A) A thermodynamic model inspired by our in vitro results allows for an interaction between Lac repressor (shown in red) and RNA polymerase (shown in blue) with an energy εrp. Additionally, it allows for different rates of transcriptional initiation in the absence or presence of repressor. P and R are the number of RNApolymerase and Lac repressor molecules inside the cell, respectively, and Δεpd and Δεrd are their corresponding interaction energies with their specific sites measured with respect to their nonspecific binding energies. NNS is the size of the bacterial genome in base pairs. β = (kB T)−1, with kB the Boltzmann constant and T the absolute temperature. The rate of promoter escape is different in the absence (r1) and presence (r2) of Lac repressor. For a detailed description of this model, refer to the Extended Experimental Procedures.
(B) Simple energy diagrams for the possible mechanisms of transcriptional regulation: (i) destabilization of the RNA polymerase-promoter complex, (ii) stabilization of the RNA polymerase-promoter complex impeding promoter escape, and (iii) modulation of the rate of promoter escape.
(C) Repression as a function of intracellular number of repressors for O1 located at −50. A mechanism that exclusively considers stabilizing interactions [mechanism (ii)] cannot fit the data regardless of the value adopted by its only free parameter, ξ (see Equation 2). The solid red line is the best fit of such a mechanism to the data whereas the dashed and dashed-dotted lines correspond to varying that parameter by a factor of two. On the other hand, a mechanism that just considers a modulation in the promoter escape rate [mechanism (iii)], also having only one free parameter, can fit the data. Details of these two mechanisms are given in the main text and the Extended Experimental Procedures. Error bars are SD over multiple repeats.
See also Figure S3.
The Input-Output Function of Simple Repression from Upstream Positions Involves Modulation of the Promoter Escape Rate by the Repressor
The general model for upstream repression proposed based on our experimental results and shown in Figure 4A covers three different mechanisms of regulation: (i) a direct, destabilizing interaction between RNA polymerase and Lac repressor that decreases occupancy of polymerase at the promoter when repressor is present, (ii) a direct, attractive interaction between RNA polymerase and Lac repressor in the closed and/or open complex that, by lowering the energy of the complex, effectively increases the amount of energy required for RNA polymerase to move forward on the pathway to transcription, and (iii) an increase in the activation energy for promoter escape, without any stabilization of RNA polymerase when Lac repressor is bound to the DNA. In the last case, Lac repressor does not affect the occupancy of the states, but only the kinetics of RNA polymerase escaping the promoter. These mechanisms are not mutually exclusive, but can act together to exert regulation depending on the values of the different parameters of the model. The different reaction diagrams corresponding to each one of these mechanisms are shown schematically in Figure 4B. In the following we explore these three cases through a quantitative comparison between theoretical predictions and expression data.
We start by considering mechanism (i). Qualitatively, this mechanism predicts a mutual destabilization between Lac repressor and RNA polymerase such that the occupancy of RNA polymerase on the promoter would be affected in the presence of repressor. However, our in vitro results shown in Figure 3C suggest that promoter occupancy is not affected significantly. We conclude that this effect, if present, will be of a small magnitude. As a result we do not consider this mechanism any further in this work. Further discussion of this point can be found in the Extended Experimental Procedures.
Next, we consider mechanism (ii), which leads to the following expression for the repression as a function of repressor copy number
| (Equation 2) |
where ξ is a function of the interaction parameter εrp, of the binding energy of polymerase to the promoter, and of the copy number of polymerases. Notice that the parameter ξ can only determine the maximum level of expression. However, it does not have an effect on the half-point, the repressor copy number at which the repression has reached half of its maximum value (this half-point is analogous to a dissociation constant, see Extended Experimental Procedures and Figures S3B and S3C for further details).
Finally, if we take mechanism (iii), where there is no stabilizing interaction between repressor and polymerase, but there is a change in the rate of promoter escape, we get the expression
| (Equation 3) |
This mechanism gives us a new parameter to consider: the ratio of the RNA polymerase escape rate in the presence of repressor to the rate in its absence, r2/r1, as shown in Figure 4A. However, unlike ξ in mechanism (ii), this parameter sets the value of both the half-point of the repression curve (notice the presence of r2/r1 in the denominator) as well as the maximum level of repression (see Extended Experimental Procedures and Figures S3B and S3C).
Continuing with the strategy employed in Figure 2D, we dissected simple repression upstream from the promoter in order to test the predictions of the different regulatory mechanisms posited by the model shown in Figure 4A. We created DNA constructs bearing all four lac operators (Oid, O1, O2, and O3, in order of high to low affinity) at −50 and we placed them in strains containing different intracellular numbers of Lac repressor that spanned nearly two orders of magnitude (Garcia and Phillips, 2011).
Figure 4C shows repression as a function of repressor number for O1 located at −50. As shown previously in Figure 2D, one of the surprising outcomes when comparing repression at −50 to repression at +11 is that repression at +11 grows with the number of repressors as called for by Equation S6 (see Figure S3A and Garcia and Phillips [2011]) whereas there is a saturation of repression at −50. This saturation is not consistent with the model embodied in Equation S6.
Given our previous knowledge of the in vivo binding energies of Lac repressor to the various operators (see Extended Experimental Procedures and Garcia and Phillips [2011]) the repression formulas for mechanisms (ii) and (iii) discussed above only have one free parameter each: ξ for mechanism (ii) and the ratio r2/r1 for mechanism (iii). In Figure 4C we show a fit of both mechanisms to our experimental data with O1 located at −50 shown in Figure 2D (for considerations on data fitting, please refer to the Extended Experimental Procedures). As indicated by the various red lines in that figure, mechanism (ii) shown in Equation 2 produces curves of the wrong shape and thus cannot fit the data regardless of the choice of parameter ξ. On the other hand, mechanism (iii), which leads to Equation 3, can fit the data as shown by the green line in Figure 4C.
Based on the analysis above we propose that the main mode of regulation by repressor is the modulation of promoter escape rate by RNA polymerase. This does not rule out a contribution from a stabilizing interaction between repressor and polymerase. In fact, a combination of both regulatory strategies can also fit the data as shown in Figure S3B. However, regulation of the escape rate constitutes a minimal mechanism that is sufficient to explain the data. We will assume this mechanism to further explore repression when the operator is located at −50.
Operator Strength Is Not Sufficient to Explain the Level of Repression
Given our knowledge of the modulation of the escape rate by Lac repressor obtained from the O1 data, we predict the shapes of the input-output functions for the remaining lac operators in Figure 5A. Under the occupancy hypothesis the model corresponding to this mechanism (mechanism (iii), shown in Equation 3) predicts that repression saturates at the same level regardless of the choice of operator because different operator sequences only change the affinity of Lac repressor to operator DNA. However, the operator choice determines the half-point of repression in a way that follows a clear rank-ordering based on the repressor binding affinity of the various operators considered.
Figure 5. Repression at −50 by Lac Repressor as a Function of Repressor Copy Number and Operator Affinity Is Inconsistent with the Occupancy Hypothesis.
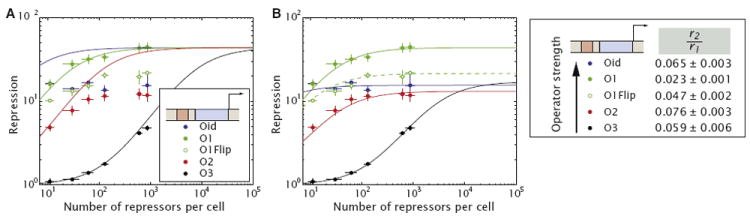
(A) Given the modulation in transcription rate obtained from the fit to the O1 data shown in Figure 4C and our previous knowledge of the in vivo binding energies we predict the shape of the input-output function for constructs where the different operators are used (lines). The measured repression (circles) is systematically inconsistent with the model predictions.
(B) By letting the modulation in transcription rate (r2/r1) be different for each operator we can successfully fit the data. Notice that the modulation does not bear a correlation with operator strength.
O1Flip is the reverse complement of O1 and the dashed green line corresponds to a fit to its resulting experimental data. Error bars are SD over multiple repeats.
See also Figures S4, S5, and S6.
In Figure 5A we also show the corresponding experimental data. It is clear from this plot that the model cannot describe the data. In particular, it is both intriguing and surprising that the data for different operators saturates at different levels and that this saturation does not follow the rank ordering of the in vivo and in vitro binding affinity of the operators. For example, Lac repressor binds to Oid ~20 times more strongly than O2, with the Kd for Oid at ~170 pM and the Kd for O2 at ~4 nM. Yet, these two operators have a comparable level of repression at a high number of repressors of ~900. On the other hand, Oid is also bound ~5 times stronger than O1, with O1 having a Kd of ~1 nM. Still, O1 presents a higher level of repression than Oid at the same intracellular number of repressors of 900. Perhaps even more interesting, if we replace the O1 binding site by its reverse complement, which should leave its binding affinity unaltered, we see a qualitatively different behavior from wild-type O1 suggesting that binding affinity alone is not sufficient to determine the different saturation levels.
If we abandon the view that the only role of the operator sequence is to set the binding affinity of Lac repressor to DNA, and adopt a view where it can modulate the transcription initiation rate in a sequence-dependent way (with a different choice of the parameter r2/r1) our model can account for all of the experimental data. For example, the choice of operator might modulate the nature of the interaction between repressor and RNA polymerase. Figure 5B shows that when we allow the parameter r2/r1 to change with operator sequence the model now accounts for the experimental data. Thus, the observed difference in modulation of initiation rate for the different constructs is at odds with the interpretation that the role of binding sites is exclusively to determine the probability of finding the repressor bound to the DNA, but is consistent with models where operator sequence can alter the nature of the repressor-polymerase interaction in a way that modulates the polymerase escape rate.
An alternative hypothesis is that the modification of the operator sequence leads to a change in the unregulated level of gene expression. In this case the differences in the observed r2/r1 ratios could be purely due to a change in r1 for each operator. In Figure S4 we show that there is no significant correlation between the unregulated levels of expression and the fitted r2/r1 values. We conclude that the observed effect of operator sequences cannot be explained by the change in the unregulated levels of expression.
An alternative way to examine the effect of operator sequence on the level of repression is to replot the data for repression as a function of operator occupancy. As described in the introduction, the occupancy hypothesis implies that all data should fall on the same curve, as shown in Figure 1A. In Figure S5 and the Extended Experimental Procedures we show that although the data for the +11 constructs collapses as expected from Figure 1A, the data corresponding to the −50 constructs does not, suggesting again that repressor occupancy is not sufficient to determine the level of repression.
The model used so far represents a simplified view of transcription initiation that combines both closed and open complexes into one effective complex. However, the exact same conclusions, without any loss of generality, can be reached when both complexes are considered independently (see Extended Experimental Procedures and Figure S6). Furthermore, the thermodynamic model used assumes quasi-equilibrium between states leading up to promoter escape. If we consider a full kinetic model in which no assumptions about equilibrium are made we nevertheless reach the same conclusions (see Extended Experimental Procedures): the different levels of repression observed for different operator sequences placed at the −50 location cannot be explained in the context of the occupancy hypothesis.
DISCUSSION
The occupancy hypothesis states that the role of operator sequence is to determine its occupancy by its target transcription factor. The nature of the interaction between the bound transcription factor and the transcriptional machinery is then determined by the spatial arrangements of binding sites and the DNA sequence context, i.e., the presence of DNA binding sites for other proteins in the vicinity, the particular mechanical properties of the surrounding DNA, etc. (Davidson, 2006; Ptashne and Gann, 2002). For example, the relative positioning between binding sites and the mechanical properties of the intervening DNA can have drastic effects on gene regulatory input-output functions (Aki et al., 1996; Amit et al., 2011; Belyaeva et al., 1998; Browning and Busby, 2004; Busby et al., 1994; Choy et al., 1995, 1997; Gaston et al., 1990; Hogan and Austin, 1987; Joung et al., 1994; Joung et al., 1993; Lilja et al., 2004; Mao et al., 1994; Ryu et al., 1998). Additionally, the nature of the promoter can modulate how a transcription factor will interact with its bound RNA polymerase (Monsalve et al., 1996, 1997). The majority of the current models of action of the diverse known interactions between transcription factors and the transcriptional machinery are based on assuming the applicability of the occupancy hypothesis (Bintu et al., 2005b; Buchler et al., 2003; Segal and Widom, 2009; Vilar and Leibler, 2003).
Several recent works have suggested that this canonical picture of transcriptional regulation is incomplete (see Haugen et al., 2008 and Voss et al., 2011 for two specific examples). More directly related to this work, the occupancy hypothesis has been suggested to be insufficient to describe regulation by MarA in bacteria (Martin et al., 2008; Wall et al., 2009) and examples where the occupancy hypothesis falls short have been found in the context of the regulation of cofactors by transcription factors in eukaryotes, as we describe below (Meijsing et al., 2009; Scully et al., 2000).
In this study, we quantitatively expanded our understanding of the paradigmatic Lac repressor and showed that the sequence of an operator located upstream from the promoter can dictate different gene regulatory input-output functions leading to different maximum repression values that cannot be explained by the occupancy of repressor on DNA. We used theoretical models of transcriptional regulation in order to qualitatively and quantitatively frame these conclusions. Whether thermodynamic models are used or a kinetic one in which no equilibrium assumption is invoked (see Figure S6), the conclusions are independent of the particular theoretical framework used to analyze the experimental results. As a result, in clear violation of the occupancy hypothesis, we conclude that the lac operator sequences encode more than just repressor binding affinity: they can also determine the nature of the “effective” interaction between repressor and RNA polymerase. We emphasize the word “effective” to make clear that our model cannot determine if the effect is due to a direct contact between repressor and RNA polymerase, due to information being transferred through the DNA in some “allosteric” way or due to some other, unknown mechanism.
What is the mechanistic nature of the effective interaction between Lac repressor and RNA polymerase? The −50 position, where we carried out our most detailed characterization of upstream repression, is within the footprint of the RNA polymerase alpha C-terminal domain (αCTD) subunit (Newlands et al., 1991). This suggests that αCTD might be involved in the repression mechanism through a direct contact with the repressor in a fashion analogous to class I activators (Busby and Ebright, 1999). A prediction of this direct contact hypothesis is that if we introduce mutations or deletions in αCTD the repression should be abolished. By way of contrast, in the allosteric hypothesis, such mutations should have little effect because the repression is mediated by binding to the DNA, not by protein-protein contacts. Previous experiments by Adhya and co-workers shed light on this issue (Choy et al., 1995, 1997; Roy et al., 2004). They found that Lac repressor (and also Gal repressor) bound at an operator at −60 (the position of the secondary peak of repression in Figure 2C) represses transcription of the galP1 promoter, and that deletion of the αCTD completely alleviates repression at −60 (Choy et al., 1995). In addition, mutations in the αCTD also abolished repression (Choy et al., 1997). Both of these experiments support the direct contact hypothesis. Furthermore, a mutant with a single point mutation in GalR was found to be able to bind to the operator at −60, but not to repress transcription (Roy et al., 2004). It is worth noting that all of these experiments were done for a different promoter than the one we have characterized here. However, their results support a mechanism based on direct contact between repressor and RNA polymerase.
None of the different mechanistic hypotheses discussed above can explain why different operator sequences determine the level of repression in a way that does not correlate with operator occupancy by repressor, which results in a violation of the occupancy hypothesis. One possible explanation is that these different regulatory outcomes result from subtle differences in the three-dimensional structures of the protein-DNA complexes or in the dynamics of these molecules. These differences could lead to altered interactions with RNA polymerase or the promoter region and result in the modulation of gene expression. In fact, differences in structure have been observed for the Lac repressor binding domain bound to its different operators as well as for the structural parameters of the intervening DNA such as twist, roll and base pair stacking, but their correlation with any phenotypic effects is unclear (Kalodimos et al., 2002, 2004; Romanuka et al., 2009). It is then also possible to speculate that the information about which operator is present is transferred through the DNA itself. However, because these studies resolved only the DNA binding-domain it remains unclear whether the conformation of the remaining protein was altered in any relevant way. Despite uncertainties about the detailed sequence-dependent molecular mechanism, the work reported here is a further step toward a more detailed understanding of the molecular interactions exerted by transcription factors.
A few studies in eukaryotic cells had previously found that DNA may act as more than simply a docking site for transcription factors; in addition, it may act as an allosteric ligand that conveys information about the mode of gene regulation (Geserick et al., 2005; Ma et al., 2010). These studies found that the specific sequence of a transcription factor binding site determined the affinity of the bound transcription factor for a different set of corepressors or coactivators. These changes in affinity may have profound physiological effects as has been suggested for the Pit-1 factor, the glucocorticoid receptor, and NF-κB (Lefstin and Yamamoto, 1998; Leung et al., 2004; Meijsing et al., 2009; Scully et al., 2000).
Our study demonstrates that modulation of transcription factor activity by the DNA sequence of its binding site may well be a much more general phenomenon, occurring as shown here in bacteria as well as in eukaryotes, despite the differences between transcriptional mechanisms in these two domains of life. Our study was performed in E. coli, where transcriptional regulation is thought to be much simpler than in eukaryotes, and we used a promoter that does not involve any cofactors. This simplicity has allowed us to find a direct mechanistic link between the DNA sequence of an operator and the transcriptional output. These results suggest that a similar effect of operator sequence on the modulation of promoter escape could arise in other bacterial transcription factors that either halt or enhance transcription at the same step as has recently been suggested for activation by MarA (Martin et al., 2008; Wall et al., 2009). Thus, MarA regulated promoters may be good candidate systems to further investigate the generality of our findings in bacterial gene regulation.
Much work that has focused on the dissection of gene regulatory regions has been based on assuming the applicability of the occupancy hypothesis (Ackers et al., 1982; Amit et al., 2011; Bintu et al., 2005a; Davidson, 2006; Gertz et al., 2009; Ptashne and Gann, 2002; Raveh-Sadka et al., 2012; Segal et al., 2008; Segal and Widom, 2009; Zinzen et al., 2009). This study gives further evidence for the existence of an additional layer of complexity to consider in transcriptional regulation given by the nature of the interaction between transcription factors and the transcriptional machinery imposed by transcription factor binding site sequence. Given the fact that a large number of repressors act on promoters by binding to a single site located upstream from the promoter region in E. coli (see Figure 2A and Gama-Castro et al., 2008) it is possible that this mechanism of repression might be widespread. Thus, knowing the list of operators and their strengths is not sufficient to predict the input-output function of a promoter. A detailed analysis of specific repressors will be necessary to determine how widespread the effects observed here may be.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and In Vivo Measurements
The construction of all plasmids and strains is described in the Extended Experimental Procedures. In short, we placed a YFP or lacZ reporter gene under the control of a lacUV5 promoter and the regulation of one of four lac operators at different positions with respect to the transcription start site. The different constructs used throughout the paper are shown schematically in Figures S1A–S1C. These constructs were integrated in the chromosome of E. coli strains bearing different intracellular numbers of Lac repressor (Garcia and Phillips, 2011).
Gene expression measurements were performed using a plate reader as described in the Extended Experimental Procedures and (Garcia et al., 2011).
Single Molecule Experiments
Single molecule experiments were performed as described in the Extended Experimental Procedures and (Sanchez et al., 2011). In short, fluorescently-and biotin-labeled DNA containing a promoter and a repressor binding site was incubated in the presence of RNA polymerase labeled with a second, spectrally distinct fluorophore. The DNA molecules were bound to a streptavidin coated glass slide and the fraction of RNA polymerase-bound DNA molecules was quantified. In order to assay the effect of repressor on the formation of RNA polymerase-bound complexes the DNA was pre-incubated with repressor before the addition of RNA polymerase and the resulting reaction was again imaged. As reported previously (Sanchez et al., 2011), not all RNA polymerase-bound DNA molecules correspond to stable, open complexes. This was taken into account in our analysis. Details pertaining to this point can be found in the Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to Jon Widom, Tom Kuhlman, Justin Kinney, Stephanie Johnson, Daniel Jones and Rob Brewster for helpful discussions. We dedicate this work to Prof. Widom who recently passed away unexpectedly. We would like to thank Larry Friedman for technical assistance with the single molecule experiments and for useful discussions. We thank Robert Landick, Rachel Mooney and Abbey Vangeloff for the generous gifts of purified SNAP-tagged core RNA polymerase and σ70. This work was supported by National Institutes of Health Pioneer award DP1 OD000217 (H.G.G., R.P.) and grants R01 GM085286 and R01 GM085286-01S (H.G.G., J.Q.B., R.P.), GM81648 and GM43369 (J.G., A.S., M.L.O.), La Fondation Pierre Gilles de Gennes (R.P.), and National Science Foundation award DMR-0706458 (J.K.) and MRSEC-0820492 (J.K., J.G.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.06.004
Publisher's Disclaimer: LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
References
- Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci USA. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aki T, Choy HE, Adhya S. Histone-like protein HU as a specific transcriptional regulator: co-factor role in repression of gal transcription by GAL repressor. Genes Cells. 1996;1:179–188. doi: 10.1046/j.1365-2443.1996.d01-236.x. [DOI] [PubMed] [Google Scholar]
- Alberts B. Molecular Biology of the Cell. Fifth Edition. New York: Garland Science; 2008. [Google Scholar]
- Amit R, Garcia HG, Phillips R, Fraser SE. Building enhancers from the ground up: a synthetic biology approach. Cell. 2011;146:105–118. doi: 10.1016/j.cell.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker NA, Kahn JD, Maher LJ., 3rd Bacterial repression loops require enhanced DNA flexibility. J Mol Biol. 2005;349:716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Belyaeva TA, Rhodius VA, Webster CL, Busby SJ. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase alpha subunits. J Mol Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- Besse M, von Wilcken-Bergmann B, Müller-Hill B. Synthetic lac operator mediates repression through lac repressor when introduced upstream and downstream from lac promoter. EMBO J. 1986;5:1377–1381. doi: 10.1002/j.1460-2075.1986.tb04370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R. Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev. 2005a;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R. Transcriptional regulation by the numbers: models. Curr Opin Genet Dev. 2005b;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond LM, Peters JP, Becker NA, Kahn JD, Maher LJ., 3rd Gene repression by minimal lac loops in vivo. Nucleic Acids Res. 2010;38:8072–8082. doi: 10.1093/nar/gkq755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Busby S, West D, Lawes M, Webster C, Ishihama A, Kolb A. Transcription activation by the Escherichia coli cyclic AMP receptor protein. Receptors bound in tandem at promoters can interact synergistically. J Mol Biol. 1994;241:341–352. doi: 10.1006/jmbi.1994.1511. [DOI] [PubMed] [Google Scholar]
- Cağatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- Choy HE, Park SW, Aki T, Parrack P, Fujita N, Ishihama A, Adhya S. Repression and activation of transcription by Gal and Lac repressors: involvement of alpha subunit of RNA polymerase. EMBO J. 1995;14:4523–4529. doi: 10.1002/j.1460-2075.1995.tb00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy HE, Hanger RR, Aki T, Mahoney M, Murakami K, Ishihama A, Adhya S. Repression and activation of promoter-bound RNA polymerase activity by Gal repressor. J Mol Biol. 1997;272:293–300. doi: 10.1006/jmbi.1997.1221. [DOI] [PubMed] [Google Scholar]
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Burlington, MA; San Diego, CA: Academic; 2006. [Google Scholar]
- Elledge SJ, Davis RW. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 1989;3:185–197. doi: 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Fowlkes CC, Hendriks CL, Keränen SV, Weber GH, Rübel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Jiménez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muñiz-Rascado L, Martínez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36(Database issue):D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG, Phillips R. Quantitative dissection of the simple repression input-output function. Proc Natl Acad Sci USA. 2011;108:12173–12178. doi: 10.1073/pnas.1015616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG, Lee HJ, Boedicker JQ, Phillips R. Comparison and calibration of different reporters for quantitative analysis of gene expression. Biophys J. 2011;101:535–544. doi: 10.1016/j.bpj.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gaston K, Bell A, Kolb A, Buc H, Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geserick C, Meyer HA, Haendler B. The role of DNA response elements as allosteric modulators of steroid receptor function. Mol Cell Endocrinol. 2005;236:1–7. doi: 10.1016/j.mce.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A, Dove SL. Protein-protein contacts that activate and repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- Hogan ME, Austin RH. Importance of DNA stiffness in protein-DNA binding specificity. Nature. 1987;329:263–266. doi: 10.1038/329263a0. [DOI] [PubMed] [Google Scholar]
- Joung JK, Le LU, Hochschild A. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc Natl Acad Sci USA. 1993;90:3083–3087. doi: 10.1073/pnas.90.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Koepp DM, Hochschild A. Synergistic activation of transcription by bacteriophage lambda cI protein and E. coli cAMP receptor protein. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- Kalodimos CG, Bonvin AM, Salinas RK, Wechselberger R, Boelens R, Kaptein R. Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain. EMBO J. 2002;21:2866–2876. doi: 10.1093/emboj/cdf318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman T, Zhang Z, Saier MH, Jr, Hwa T. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6043–6048. doi: 10.1073/pnas.0606717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Schleif RF. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci USA. 1989;86:476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Lilja AE, Jenssen JR, Kahn JD. Geometric and dynamic requirements for DNA looping, wrapping and unwrapping in the activation of E. coli glnAp2 transcription by NtrC. J Mol Biol. 2004;342:467–478. doi: 10.1016/j.jmb.2004.07.057. [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Guillerez J, Sousa R, Dreyfus M. On the mechanism of inhibition of phage T7 RNA polymerase by lac repressor. J Mol Biol. 1998;276:861–875. doi: 10.1006/jmbi.1997.1576. [DOI] [PubMed] [Google Scholar]
- Ma B, Tsai CJ, Pan Y, Nussinov R. Why does binding of proteins to DNA or proteins to proteins not necessarily spell function? ACS Chem Biol. 2010;5:265–272. doi: 10.1021/cb900293a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003;19:75–79. doi: 10.1016/S0168-9525(02)00039-2. [DOI] [PubMed] [Google Scholar]
- Mao C, Carlson NG, Little JW. Cooperative DNA-protein interactions. Effects of changing the spacing between adjacent binding sites. J Mol Biol. 1994;235:532–544. doi: 10.1006/jmbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- Martin RG, Bartlett ES, Rosner JL, Wall ME. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J Mol Biol. 2008;380:278–284. doi: 10.1016/j.jmb.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve M, Calles B, Mencía M, Salas M, Rojo F. Transcription activation or repression by phage psi 29 protein p4 depends on the strength of the RNA polymerase-promoter interactions. Mol Cell. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Mencía M, Salas M, Rojo F. Protein p4 represses phage phi 29 A2c promoter by interacting with the alpha subunit of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci USA. 1996;93:8913–8918. doi: 10.1073/pnas.93.17.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Hill B. The lac Operon: A Short History of a Genetic Paradigm. Berlin, New York: Walter de Gruyter; 1996. [Google Scholar]
- Müller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- Newlands JT, Ross W, Gosink KK, Gourse RL. Factor-independent activation of Escherichia coli rRNA transcription. II. characterization of complexes of rrnB P1 promoters containing or lacking the upstream activator region with Escherichia coli RNA polymerase. J Mol Biol. 1991;220:569–583. doi: 10.1016/0022-2836(91)90101-b. [DOI] [PubMed] [Google Scholar]
- Novák B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- Pavco PA, Steege DA. Characterization of elongating T7 and SP6 RNA polymerases and their response to a roadblock generated by a site-specific DNA binding protein. Nucleic Acids Res. 1991;19:4639–4646. doi: 10.1093/nar/19.17.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes and Signals. New York: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Tuning expression in a predictable manner by manipulating nucleosomes disfavoring sequences. Nat Rev Genet. 2012 doi: 10.1038/ng.2305. http://dx.doi.org/10.1038/ng.2305. [DOI] [PubMed]
- Rojo F. Repression of transcription initiation in bacteria. J Bacteriol. 1999;181:2987–2991. doi: 10.1128/jb.181.10.2987-2991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanuka J, Folkers GE, Biris N, Tishchenko E, Wienk H, Bonvin AM, Kaptein R, Boelens R. Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes. J Mol Biol. 2009;390:478–489. doi: 10.1016/j.jmb.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Roy S, Semsey S, Liu M, Gussin GN, Adhya S. GalR represses galP1 by inhibiting the rate-determining open complex formation through RNA polymerase contact: a GalR negative control mutant. J Mol Biol. 2004;344:609–618. doi: 10.1016/j.jmb.2004.09.070. [DOI] [PubMed] [Google Scholar]
- Ryu S, Fujita N, Ishihama A, Adhya S. GalR-mediated repression and activation of hybrid lacUV5 promoter: differential contacts with RNA polymerase. Gene. 1998;223:235–245. doi: 10.1016/s0378-1119(98)00237-6. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Osborne ML, Friedman LJ, Kondev J, Gelles J. Mechanism of transcriptional repression at a bacterial promoter by analysis of single molecules. EMBO J. 2011;30:3940–3946. doi: 10.1038/emboj.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlax PJ, Capp MW, Record MT., Jr Inhibition of transcription initiation by lac repressor. J Mol Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carrière C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- Straney SB, Crothers DM. Lac repressor is a transient gene-activating protein. Cell. 1987;51:699–707. doi: 10.1016/0092-8674(87)90093-6. [DOI] [PubMed] [Google Scholar]
- Tsai TY, Choi YS, Ma W, Pomerening JR, Tang C, Ferrell JE., Jr Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 2008;321:126–129. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar JM, Leibler S. DNA looping and physical constraints on transcription regulation. J Mol Biol. 2003;331:981–989. doi: 10.1016/s0022-2836(03)00764-2. [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall ME, Markowitz DA, Rosner JL, Martin RG. Model of transcriptional activation by MarA in Escherichia coli. PLoS Comput Biol. 2009;5:e1000614. doi: 10.1371/journal.pcbi.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


