Abstract
Clearance of apoptotic cells is the final stage of programmed cell death. Uncleared corpses can become secondarily necrotic, promoting inflammation and autoimmunity. Remarkably, even in tissues with high cellular turnover, apoptotic cells are rarely seen because of efficient clearance mechanisms in healthy individuals. Recently, significant progress has been made in understanding the steps involved in prompt cell clearance in vivo. These include the sensing of corpses via “find me” signals, the recognition of corpses via “eat me” signals and their cognate receptors, the signaling pathways that regulate cytoskeletal rearrangement necessary for engulfment, and the responses of the phagocyte that keep cell clearance events “immunologically silent.” This study focuses on our understanding of these steps.
Phagocytes recognize dead cells through “find me” (e.g., fractalkine) and “eat me” (e.g., phosphatidylserine) signals. Signaling events within phagocytes then control the internalization and digestion of the dead cells.
Multicellular organisms execute the majority of unwanted cell populations in a regulated fashion via the process of apoptosis (Henson and Hume 2006; Nagata et al. 2010). Examples of unwanted cells include excess cells generated during development, cells infected with intracellular bacteria or viruses, transformed or malignant cells capable of tumorigenesis, and cells irreparably damaged by cytotoxic agents. Swift removal of these cells is necessary for maintenance of overall health and homeostasis and prevention of autoimmunity, pathogen burden, or cancer. Quick removal of dying cells is a key final step, if not the ultimate goal of the apoptotic program.
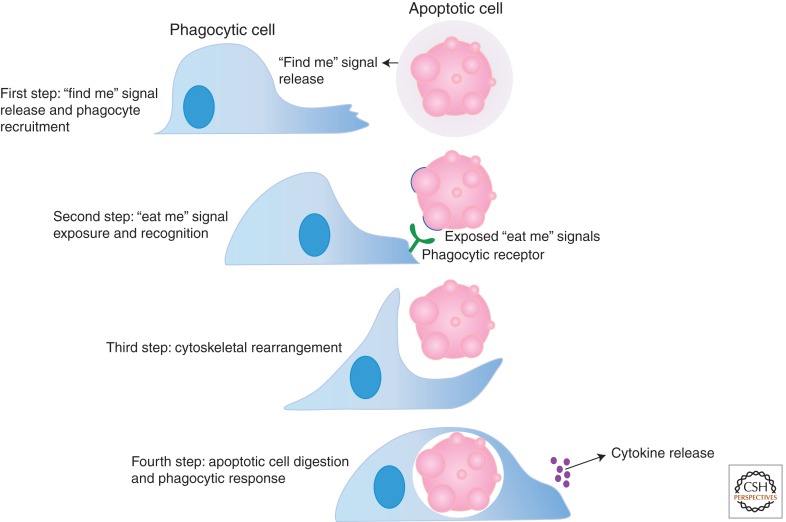
The term “phagocytosis” refers to an internalization process by which larger particles, such as bacteria and dead/dying cells, are engulfed and processed within a membrane-bound vesicle called the phagosome (Ravichandran and Lorenz 2007). A phagocyte is any cell that is capable of engulfment, including “professional” phagocytes such as macrophages, immature dendritic cells, and neutrophils. Metazoa have multiple mechanisms for clearing apoptotic cells, often depending on the tissue and apoptotic cell type (Gregory 2009). Macrophages and immature dendritic cells readily engulf dead or dying cells in tissues such as bone marrow (where a large number of new hematopoietic cells are generated), spleen (during or after an immune response), and the thymus (in young animals during T-lymphocyte development). In other tissues, neighboring “nonprofessional” phagocytes can also mediate the clearance of apoptotic targets. For example, in the mammary epithelium, viable mammary epithelial cells engulf apoptotic mammary epithelial cells after cessation of lactation (Monks et al. 2005, 2008). What distinguishes the phagocytosis of apoptotic cells from the phagocytosis of most bacteria or necrotic cells is the lack of a pro-inflammatory immune response (Henson 2005). This article discusses apoptotic cell engulfment, specifically the recruitment of phagocytes, through “find me” signals, the recognition of apoptotic cells by phagocytes via “eat me” signals, the internalization process and signaling pathways used for cytoskeletal rearrangement, and finally the digestion of apoptotic cells and phagocytic response to this process (Fig. 1).
Figure 1.
The steps of efficient apoptotic cell clearance. First, “find me” signals released by apoptotic cells are recognized via their cognate receptors on the surface of phagocytes. This is the sensing stage and stimulates phagocyte migration to the location of apoptotic cells. Second, phagocytes recognize exposed “eat me” signals on the surface of apoptotic cells via their phagocytic receptors, which leads to downstream signaling events culminating in Rac activation. Finally, further signaling events within the phagocyte regulate the digestion and processing of the apoptotic cell meal and the secretion of anti-inflammatory cytokines.
RECRUITMENT OF PHAGOCYTES TO THEIR APOPTOTIC MEAL
Remarkably, even in tissues with high cellular turnover, apoptotic cells are rarely seen in situ, which is thought to be due to efficient clearance mechanisms. Early studies in the nematode Caenorhabditis elegans suggested that apoptotic cells are recognized and cleared before they are “fully dead” (Hoeppner et al. 2001; Reddien et al. 2001). This work led to the idea that apoptotic cells advertise their status to local and distant phagocytes at their earliest stages of death, perhaps via the release of “find me” signals (Ravichandran 2003).
“Find Me” Signals: Establishing a Chemotactic Gradient to Direct Phagocyte Migration
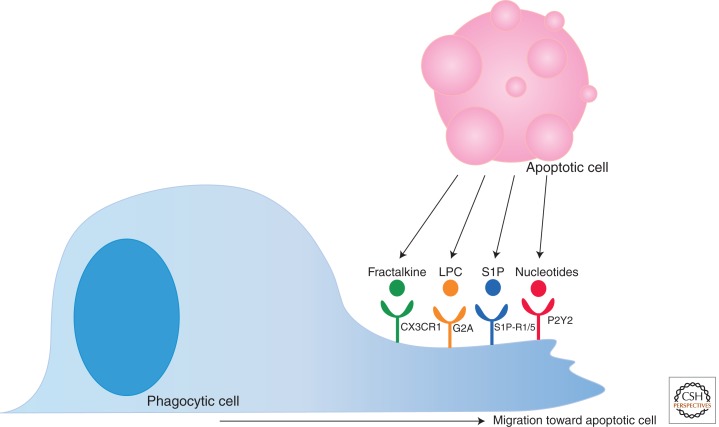
The role of “find me” signals is to establish a chemotactic gradient stimulating the migration of phagocytes to the apoptotic cell. To date, several proposed “find me” signals released by dying cells have been reported (Fig. 2). These include fractalkine, lysophosphatidylcholine (LPC), sphingosine-1-phosphate (S1P), and the nucleotides ATP and UTP (Lauber et al. 2003; Gude et al. 2008; Truman et al. 2008; Elliott et al. 2009).
Figure 2.
“Find me” signals and their receptors. Apoptotic cells release “find me” signals including fractalkine, LPC, S1P, and nucleotides. These molecules bind their cognate receptors (CX3CR1, G2A, S1P-R1/5, and P2Y2, respectively) present on the phagocyte surface. “Find me” signal recognition by the phagocyte stimulates migration toward the dying target.
Fractalkine (i.e., CXC3CL1) is currently the only classical chemokine “find me” signal identified (Peter et al. 2010). It is a membrane-associated protein that is released from apoptotic B cells and neurons by a yet unknown protease; the released fractalkine is sensed via CX3CR1, which, in turn, directs macrophages to the dying targets (Truman et al. 2008). During affinity maturation of an antibody response, B cells in germinal centers undergo a high rate of apoptosis, and in an experiment measuring the clearance of these apoptotic B cells, fractalkine/CX3CR1 attraction was shown to influence macrophage recruitment to germinal centers in vivo (Truman et al. 2008). Interestingly, no increase in the presence of apoptotic or secondarily necrotic cells was observed in the germinal centers of CX3CR1 knockout mice, suggesting that fractalkine-mediated macrophage attraction is not required for clearance. Expression of fractalkine is limited to only a few cell types, so other mechanisms of phagocyte attraction must also exist.
LPC was the first discovered lipid “find me” signal, and it is released from apoptotic cells by the caspase-3-dependent activation of phospholipase A2, leading to the conversion of phosphatidylcholine to LPC (Lauber et al. 2003). The recognition of LPC is thought to occur via the G-protein-coupled receptor G2A and thereby stimulates macrophage chemotaxis toward apoptotic cells (Peter et al. 2008). However, the concentration of LPC reported to be required for macrophage chemotaxis appears to be quite high. In addition, the amount of LPC present in circulation is higher than LPC levels released by apoptotic cells, making LPC an unlikely candidate for a chemotactic mediator (Nagata et al. 2010). S1P, another lipid reported to function as a “find me” signal for apoptotic cells, is produced by sphingosine kinase 1 (SphK1) and secreted by apoptotic cells (Gude et al. 2008). S1P stimulates chemotaxis by binding to the G-protein-coupled receptors S1P-R1 through S1P-R5. At this time, no one specific S1P-R has been primarily associated with phagocyte chemotaxis, and macrophages are known to express all five (Rosen and Goetzl 2005).
Recently, nucleotides ATP and UTP have been implicated as a new class of “find me” signals. Small amounts of intracellular ATP and UTP are released in a regulated manner early during apoptosis to establish a gradient for monocyte attraction in vitro and in vivo (Elliott et al. 2009); this small amount of nucleotide release (<0.1% of the cellular content) should be distinguished from the release of significantly higher levels of nucleotides after other types of cell death such as necrosis. Subsequent studies showed that the release of ATP and UTP is mediated via the pannexin channels, which are opened during apoptosis by caspase-dependent cleavage of their carboxy-terminal tail (Chekeni et al. 2010). How are the nucleotides sensed? Current evidence suggests that the P2Y2 receptor on monocytes binds the released nucleotides and stimulates phagocyte chemotaxis; consistent with a role for P2Y2 in vitro, disruption of the nucleotide/P2Y2 “find me” network resulted in uncleared apoptotic thymocytes after glucocorticoid injection in vivo (Elliott et al. 2009). Because nucleotides are readily degraded by extracellular nucleotidases, they are unlikely to serve as long-range “find me” signals to phagocytes in circulation; rather, they attract tissue resident macrophages (Ravichandran 2010).
The nature of these identified “find me” signals is varied both in composition and recognition. It is currently unclear whether they function together in an additive or synergistic manner during the phagocyte attraction to apoptotic cells.
Additional Roles for “Find Me” Signals: Whetting the Appetite and Keeping the Meal Quiet
Nucleotides have obvious cellular functions independent of their role as “find me” signals (i.e., DNA building blocks and as energy currency). It is not surprising that the other “find me” signals also serve additional roles, even in the context of apoptotic cell clearance. For example, fractalkine was shown to induce milk fat globule EGF factor 8 (MFG-E8) expression in peritoneal macrophages (Miksa et al. 2007). MFG-E8 (described below) is a bridging molecule that facilitates recognition of apoptotic cells by phagocytes. This priming of fractalkine-dependent MFG-E8 expression enhanced the clearance of apoptotic thymocytes in vitro. LPC was shown to induce the expression of MCP-1 and RANTES in human vascular endothelial cells, thereby potentiating the recruitment of monocytes toward apoptotic cells (Murugesan et al. 2003; Peter et al. 2010). In addition, LPC has been described as a surface-bound target for IgM and may serve an additional role during the removal of apoptotic cells as an “eat me” signal (Kim et al. 2002). “Find me” signals may also influence a non-immunogenic response to apoptotic cells as seen with S1P-enhanced secretion of IL-10 and PGE2 by tumor macrophages (Weigert et al. 2007; Johann et al. 2008; Ravichandran 2010). Why are there additional functions? Perhaps not every phagocyte requires “find me” signal-mediated chemotaxis to their target; there are many instances in which targets and phagocytes are in close proximity, and there is no need for the phagocyte to actually find the dying cell. In these cases, “find me” signals may have alternative functions such as enhancing the phagocytic machinery within the phagocyte or the ability to digest the ingested cargo. These possible functions remain to be addressed more carefully using different targets and different phagocytes.
“Stay Away” Signals
Phagocytosis of apoptotic cells is generally recognized as a non-immunogenic, non-inflammatory process with minimal, if any, neutrophil recruitment (Savill 1997; Savill et al. 2002). Of the known “find me” signals, however, LPC, S1P, and nucleotides are all known to stimulate neutrophil chemotaxis as well (Chen et al. 2006; Florey and Haskard 2009). Then how might the system avoid recruitment of neutrophils under these circumstances? One possibility is via the iron-binding glycoprotein lactoferrin (LTF), which is also released by apoptotic cells and appears to specifically inhibit neutrophil migration (Bournazou et al. 2009). This “stay away” signal is the first of its kind discovered, and currently mechanisms of LTF release and the identity of LTF receptor(s) on neutrophils remain unknown.
RECOGNITION OF CORPSES BY PHAGOCYTIC CELLS
Once in the proximity of their targets, phagocytes must be able to distinguish live from dead cells. Apoptotic cells display “eat me” signals on their surface, which phagocytes detect via receptors on their cell surface. Detection is, however, not always direct. Although certain phagocytic receptors can directly engage ligands on the dying cells, other receptors use soluble bridging molecules as intermediaries to recognize the “eat me” signals on apoptotic cells. Additionally, living cells have mechanisms in place to prevent their unwanted removal. The section below details the modes of specific recognition of apoptotic cells and the mechanisms used by live cells to avoid engulfment by phagocytes.
“Eat Me” Signals: Distinguishing the Live from the Dead
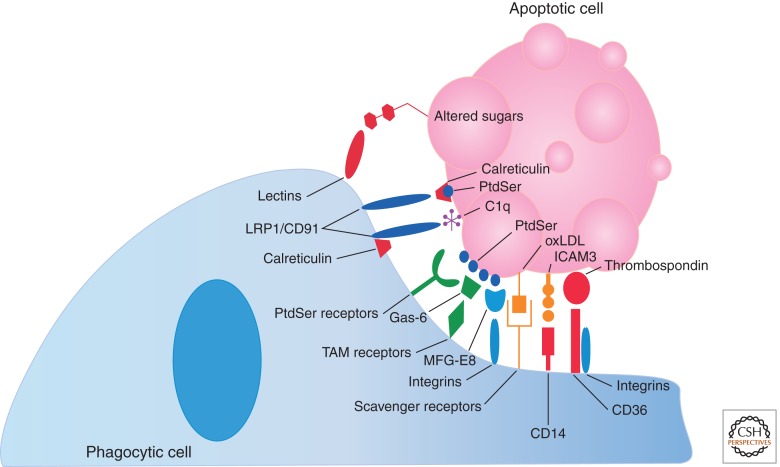
Numerous “eat me” signals have been identified to date including changes in glycosylation of surface proteins or changes in surface charge (Kinchen and Ravichandran 2007), binding of serum proteins such as thrombospondin and complement C1q to the apoptotic cell (Ravichandran and Lorenz 2007), expression of intercellular adhesion molecule 3 (ICAM3) and oxidized low-density lipoprotein (LDL)-like moiety (Devitt et al. 1998; Fadok et al. 1998b; Aderem and Underhill 1999; Schlegel et al. 1999), and the exposure of certain intracellular proteins such as calreticulin and annexin I (Arur et al. 2003; Gardai et al. 2005; Obeid et al. 2007a,b). However, the most widely studied and universally detected “eat me” signal is the exposure of the phospholipid phosphatidylserine (PtdSer) (Fig. 3) (Fadok et al. 1992, 1998a, 2001).
Figure 3.
Apoptotic cell “eat me” signals and phagocytic receptors. As apoptotic cells undergo programmed cell death, they begin to expose “eat me” signals on their surfaces. Phosphatidylserine (PtdSer) is the best studied “eat me” signal; however, several others are also pictured here. “Eat me” signals are recognized by phagocytic engulfment receptors either directly (as with PtdSer receptors including TIM-4, BAI1, and Stabilin-2) or indirectly via bridging molecules or accessory receptors (as with Gas-6/TAM receptors, MFG-E8/αvβ3/5, and αvβ3/5 in conjunction with CD36 in the recognition of thrombospondin).
In viable cells, PtdSer is kept exclusively on the inner leaflet of the lipid bilayer via ATP-dependent translocases (Balasubramanian and Schroit 2003). In apoptotic cells, the concentration of PtdSer on the outer leaflet of the lipid bilayer is estimated to increase by more than 280-fold within only a few hours after induction of apoptosis (Borisenko et al. 2003; Ravichandran 2010). The exact mechanism by which PtdSer exposure occurs is just beginning to be better defined, but it appears to be caspase dependent (Nagata et al. 2010). Recently, a member of the TMEM16 family of Ca2+-activated Cl− channels, TMEM16F, was shown to mediate Ca2+-dependent phospholipid scrambling (Suzuki et al. 2010). Overexpression and knockdown experiments determined that TMEM16F regulates the exposure of PtdSer and phosphatidylethanolamine (PtdEtn) to the outer leaflet of the plasma membrane in response to apoptotic stimuli, and a mutation of this channel was linked to a patient with Scott syndrome, a rare bleeding disorder, which results from defective phospholipid scrambling (Suzuki et al. 2010). Currently it is also unknown whether PtdSer recognition alone is sufficient to trigger phagocytosis; several studies have suggested that PtdSer recognition is both necessary and sufficient for clearance of apoptotic cells (Fadok et al. 1998a); however, some evidence suggests otherwise. For example, PtdSer exposure can occur on viable cells, yet these cells are not engulfed (van den Eijnde et al. 2001; Huppertz et al. 2006; Helming and Gordon 2009). Furthermore, viable cells forced to expose PtdSer (in equivalent amount to that detected on apoptotic cells) via expression of a constitutively active form of TMEM16F were not engulfed (Segawa et al. 2011). In addition, some phagocytes have a preference for certain targets over others, although both targets expose PtdSer similarly, as measured by annexin V staining (Ravichandran 2010). Perhaps apoptotic cells expose multiple “eat me” signals like those described above, and the combination of these with PtdSer may enhance engulfment. Calreticulin and annexin I have both been shown to colocalize with PtdSer on the surface of apoptotic cells and enhance the uptake of targets by phagocytes (Arur et al. 2003; Gardai et al. 2005). Furthermore, some reports have detected oxidized PtdSer on the surface of apoptotic cells (Kagan et al. 2002), so perhaps PtdSer modification is another method by which dead cells identify themselves.
Apoptotic Cell Engulfment Receptors
A plethora of receptors are expressed on the surface of phagocytic cells that recognize “eat me” signals displayed on dying cells (Fig. 3). These include the lectins that bind altered sugars on apoptotic cells (Ezekowitz et al. 1990), CD36 (in conjunction with integrins αvβ3 and αvβ5) that binds thrombospondin (Savill et al. 1990), LRP1/CD91 (in conjunction with calreticulin) that binds complement C1q (Ogden et al. 2001), CD14 that binds ICAM3 (Gregory et al. 1998), and the scavenger receptors that bind oxidized LDL (Gordon 1999). Furthermore, PtdSer is recognized by an assortment of bridging proteins and direct recognition receptors, adding complexity to the process of dying cell recognition. For example, LRP1/CD91 has also been shown to bind calreticulin colocalized with PtdSer (Gardai et al. 2005). In addition, secreted proteins such as MFG-E8, growth-arrest-specific 6 (Gas6), and protein S have also been shown to bind PtdSer on the surface of apoptotic cells and promote engulfment via their cognate receptors on phagocytes. MFG-E8 (originally discovered in milk fat globules in the mammary gland) is expressed and secreted by professional phagocytes, associates with the αvβ3/5 integrins on phagocytes via its RGD motif, and binds PtdSer on apoptotic cells via its C1 and C2 domains (Hanayama et al. 2002; Nagata et al. 2010). Gas6 and protein S bridge PtdSer on apoptotic cells with the Tyro-3-Axl-Mer family of receptors (TAM receptors) on phagocytes (Nakano et al. 1997; Nagata et al. 2010).
Recently, receptors that directly recognize PtdSer on apoptotic cells and promote phagocytosis have been discovered, each with a unique expression pattern. Brain-specific angiogenesis inhibitor 1 (BAI1) is a transmembrane protein belonging to the adhesion-type G-protein-coupled receptor family that binds PtdSer via thrombospondin type 1 repeats (Park et al. 2007). BAI1 is expressed in macrophage cell lines and immunological tissues such as the bone marrow and spleen; however, BAI1 is expressed at a much higher degree in the brain glia and neuronal cells (Mori et al. 2002). Another set of recently described PtdSer receptors, T-cell immunoglobulin and mucin-domain-containing molecule 4 (TIM-4) and TIM-1, are small transmembrane proteins that bind PtdSer via their IgV domain (Kobayashi et al. 2007; Miyanishi et al. 2007; Santiago et al. 2007). TIM-4 is highly expressed by professional phagocytes; however, TIM-1 is primarily expressed in kidney cells. In contrast to the other identified PtdSer receptors, TIM-4 (and perhaps TIM-1) does not mediate direct signaling for engulfment and therefore likely functions as a tethering receptor (Park et al. 2009). Stabilin-2 (also known as hyaluronic acid receptor for endocytosis, or HARE) is a large type I membrane protein that stereospecifically recognizes PtdSer on the surface of apoptotic cells via its epidermal growth factor-like domains (Park et al. 2008a,c). It is primarily expressed on sinusoidal endothelial cells; however, Stabilin-2 is also expressed on human monocyte-derived macrophages (Park et al. 2008a). The most recent PtdSer receptor described to date is the receptor for advanced glycation end products (RAGE), which binds PtdSer in both its membrane-bound and soluble forms (He et al. 2011). The soluble form of RAGE appears to act as a decoy receptor, blocking PtdSer recognition and engulfment mediated by the membrane-bound form and other PtdSer receptors. RAGE is primarily expressed in lung tissue, including alveolar macrophages; however, its expression is induced by inflammation, perhaps suggesting a role for RAGE in the resolution of inflammation (Armstrong and Ravichandran 2011). Finally, the protein referred to as simply the “phosphatidylserine receptor” (PSR), was originally described as capable of binding PtdSer and mediating engulfment; however, the identity of PSR has turned out to be that of a Jumonji domain-containing nuclear protein, which does not directly mediate phagocytosis of apoptotic cells (Bose et al. 2004; Bratton and Henson 2008). The signaling mechanisms activated by the aforementioned receptors are discussed in the following section.
“Don’t Eat Me” Signals
In addition to detecting the “eat me” signals on the surface of apoptotic cells, phagocytes can further distinguish between live and dead targets by the presence of “don’t eat me” signals on the surface of living cells. CD47 (also known as integrin-associated protein) is a membrane protein expressed on the surface of healthy cells, recognized by its cognate receptor, SIRPα, which inhibits engulfment by phagocytes even in the presence of PtdSer (Gardai et al. 2005; Tsai and Discher 2008). CD47 expression is suppressed or down-modulated during apoptosis, thereby permitting clearance. Recently, mouse hematopoietic stem cells were shown to transiently increase CD47 expression during inflammation-mediated migration in order to avoid clearance; however, constitutive CD47 expression was detected in both mouse and human myeloid leukemias (Jaiswal et al. 2009). This poses a clever mechanism by which cancer cells may evade immune recognition and clearance, and new clinical therapies for the treatment of acute lymphoblastic leukemia are currently being developed that use blocking antibodies for CD47 (Chao et al. 2011). Research has shown that CD47 blocking antibodies increases engulfment in vitro and inhibits tumor engraftment in vivo; however, one large caveat to such treatment is that CD47 expression is important for the survival of many cell types including erythrocytes (Oldenborg et al. 2000), suggesting that blocking its recognition, even temporarily, may cause severe side effects, including anemia. CD31 is another example of a “don’t eat me” signal (Brown et al. 2002); however, currently the CD47 literature is more advanced. Finally, the inhibitory receptor CD300a, broadly expressed on many hematopoietic cell types, has been recently shown to bind both PtdSer and PtdEtn on the surface of apoptotic cells and block their engulfment by macrophages (Simhadri et al. 2012). Interestingly, this form of regulation is intrinsic to the macrophage and independent of the apoptotic target.
ENGULFMENT SIGNALING PATHWAYS FOR CYTOSKELETON REARRANGEMENT
Once the apoptotic target is captured, the phagocyte undergoes cytoskeletal rearrangements necessary for corpse internalization. Genetic studies in Caenorhabditis elegans provided the initial insight into our understanding of the engulfment signaling pathways leading to the rearrangement of the phagocyte cytoskeleton necessary for corpse internalization (Kinchen 2010). The ongoing identification of the fly and mammalian homologs, as well as identification of new proteins exclusive to the mammalian contexts, are further defining signaling pathways in higher organisms (Table 1).
Table 1.
Evolutionarily conserved apoptotic cell engulfment signaling proteins
| C. elegans | Drosophila | Mammal | Function |
|---|---|---|---|
| CED-1 | DRPR | LRP1 (CD91) /MEGF10 | Surface receptors on phagocytes that recognize ligands on the surface of apoptotic cells |
| CED-2 | DCrk | CrkII | Adaptor protein that is proposed to localize the CED-5/CED-12 complex to the membrane; the precise role in apoptotic cell engulfment signaling is currently unclear |
| CED-5 | Myoblast City | Dock180 | RAC GEF containing a Docker (DHR2) domain that functions as a DH domain |
| CED-6 | Dced-6 | GULP | Adaptor proteins that bind CED-1; the precise role in apoptotic cell engulfment signaling is currently unclear |
| CED-7 | Unknown | ABCA1/ABCA7 | ABC transporter involved in cholesterol efflux downstream from apoptotic cell engulfment; the precise role is unknown, but its function is required in both the phagocyte and the apoptotic cell |
| CED-10 | DRac | Rac | Rho family GTPase; regulates Arp2/3 activation, actin polymerization, and cytoskeletal rearrangement via the Scar/WAVE complex |
| CED-12 | Dced-12 | ELMO | Adaptor protein containing a PH domain; binds to CED-5 and enhances its RAC GEF activity |
Rho-Family GTPases
GTPases are the molecular “switches” that turn signaling pathways on or off depending on the state of their bound guanine nucleotide. Active GTPase is the GTP-bound form, which is formed by the action of specific guanine-nucleotide-exchange factors (GEFs) that catalyze the GDP-to-GTP exchange. Conversely, GTPase-activating proteins (GAPs) stimulate the irreversible hydrolysis of GTP to GDP, effectively turning off the GTPase switch. The Rho family GTPases are members of the Ras superfamily of small signaling proteins with known functions in regulation of cellular movement (Ridley 2001). Members of this family include RhoA, Cdc42, and Rac. RhoA loss or suppression is associated with increased engulfment of apoptotic cells, and, conversely, forced activation is associated with inhibited engulfment (Tosello-Trampont et al. 2003; Nakaya et al. 2006). Regulation of clearance by RhoA is executed via the Rho-associated coiled-coil-containing protein kinase (ROCK). Active, or GTP-bound, Rho increases the kinase activity of ROCK, which, in turn, mediates phosphorylation of the myosin light chain (MLC) and promotes cell contraction (Riento and Ridley 2003). Contractility likely inhibits the extension of pseudopods and phagocytic cup formation, necessary for the early stages of engulfment. RhoA activation at subsequent stages of engulfment, however, is thought to promote apoptotic cell digestion by regulating the acidification of phagosomes (Erwig and Henson 2008). In contrast to RhoA, the current function of Cdc42 in the clearance of apoptotic cells is unclear; however, some reports suggest that it may serve a beneficial role in this process (Leverrier and Ridley 2001). Finally, GTP-bound Rac has an evolutionarily conserved positive effect on engulfment, and Rac activation at sites of apoptotic cell recognition subsequently leads to Arp2/3 activation/actin polymerization/cytoskeletal rearrangement via the Scar/WAVE complex (Miki et al. 1998; Castellano et al. 2000). Presently, at least three distinct signaling pathways governing apoptotic cell clearance have been described, which ultimately lead to downstream activation of Rac.
Rac Activation via the CrkII–Dock180–ELMO Pathway
A combination of studies in C. elegans, Drosophila melanogaster, and mammalian models led to the identification of a signaling pathway for Rac activation composed of the proteins Dock180 (homologs include CED-5 in C. elegans and Myoblast City in the fly), ELMO (C. elegans CED-12 and fly DCed-12), and CrkII (C. elegans CED-2 and fly DCrk) (Wu and Horvitz 1998b; Reddien and Horvitz 2000; Gumienny et al. 2001; Zhou et al. 2001a). This group of proteins is involved in several cytoskeletal rearrangement processes including cell migration, neurite growth, muscle fusion, and phagocytosis of apoptotic cells (Nagata et al. 2010). Dock180 was identified as a Rac GEF that lacks the traditional Dbl-homology and pleckstrin-homology (DH-PH) domains required among GEFs to mediate GTP exchange (Brugnera et al. 2002). Although Dock180 contains a Docker (DHR2) domain that functions as a DH domain, monomeric Dock180 exists in a “closed” confirmation, thereby preventing the interaction of the DHR2 domain with Rac (Lu et al. 2005). ELMO is an adaptor protein that binds directly to Dock180, creating an unconventional bipartite Rac GEF (Brugnera et al. 2002). There are at least three possible mechanisms by which ELMO enhances Dock180 activity. First, binding of ELMO to the carboxy-terminal region of Dock180 relieves a steric inhibition that blocks the Dock180 DHR2 domain interaction with Rac (Lu et al. 2005). Second, the PH domain of ELMO then stabilizes the bond between Rac and Dock180 (Lu et al. 2004). Finally, ELMO targets Dock180 to the phagocytic membrane via direct binding to the carboxyl terminus of the transmembrane receptor BAI1 (Park et al. 2007).
Our understanding of Dock180–ELMO regulation is currently evolving. Original reports suggested that CrkII, an adaptor protein, recruits Dock180 and ELMO to the cell membrane after cell surface receptor/ligand engagement (Gumienny et al. 2001). Further experiments showed that CrkII can be coimmunoprecipitated with the Dock180–ELMO complex and is indeed required for efficient engulfment; however, direct interaction between CrkII and Dock180 is not essential for the removal of dead cells (Tosello-Trampont et al. 2007). Moreover, it was shown that ELMO also possesses a membrane-targeting signal and that activation of RhoG by the GEF Trio leads to membrane recruitment of the Dock180–ELMO complex via binding of ELMO Armadillo (ARM) repeats to active RHOG (deBakker et al. 2004). Finally, other studies have determined that Dock180, too, has a membrane-targeting signal, raising further doubt regarding the requirement of CrkII for this purpose (Cote et al. 2005). Therefore, the exact role of CrkII in this pathway is presently unknown.
Several phagocytic receptors introduced in the previous section have been shown to use the CrkII–Dock180–ELMO pathway for clearance of apoptotic cargo. These include integrins αvβ3 and αvβ5, which recognize PtdSer via bridging molecules (Albert et al. 2000; Akakura et al. 2004; Hanayama et al. 2004), and the TAM receptor Mer, which recognizes PtdSer via Gas6 (Wu et al. 2005). Likewise, the PtdSer receptor BAI1 also binds ELMO (via its cytoplasmic tail) and activates this signaling pathway during cell clearance (Park et al. 2007).
Rac Activation via the LRP1/MEGF10–GULP–ABCA1/ABCA7 Pathway
The second evolutionarily conserved signaling pathway for the removal of apoptotic cells includes LRP1 (CD91)/MEGF10 (C. elegans homolog CED-1 and fly Draper), GULP (C. elegans CED-6 and fly dCed-6), and ABCA1/ABCA7 (C. elegans CED-7) (Liu and Hengartner 1998; Wu and Horvitz 1998a; Zhou et al. 2001b; Su et al. 2002; Manaka et al. 2004). Although this pathway leads to downstream Rac activation (Kinchen et al. 2005), the intermediate steps are unclear. LRP1/CD91 is a surface receptor expressed on many cells, including phagocytes that recognize “eat me” signals on the surface of the apoptotic cells. Ligands for LRP1 (including calreticulin and thrombospondin) and the fly homolog, Draper (including Pretaporter), are known (Kuraishi et al. 2009); however, the ligand(s) for the worm homolog, CED-1, remains unidentified. After apoptotic cell recognition, LRP1 directly interacts with the adaptor protein, GULP, and may lead to cytoskeletal rearrangements (Su et al. 2002). Interestingly, the PtdSer receptor Stabilin-2 was also shown to interact with GULP via its NPXY motif, and knocking down endogenous GULP significantly reduced Stabilin-2-mediated engulfment of apoptotic cells, suggesting that it uses this pathway for clearance (Park et al. 2008b). Presently, the role of ABCA1/ABCA7 in this pathway is less well characterized. ABCA1/ABCA7 are members of the ATP-binding cassette transporter (ABC-transporter) protein family. How these proteins function in this engulfment signaling pathway is currently unclear, although their function is required in both the target and phagocyte (Wu and Horvitz 1998a). Interestingly, these proteins function in the transport of cholesterol and maintenance of lipid subdomains on the plasma membrane and may thus play a role in cholesterol homeostasis in the phagocyte during apoptotic cell engulfment (Kiss et al. 2006a,b; Landry et al. 2006).
A Third Parallel Engulfment Signaling Pathway?
In addition to the two described parallel engulfment signaling pathways, the existence of a third pathway has long been proposed because of the remaining (albeit inefficient) engulfment that occurs in the C. elegans CED-1, CED-5, or CED-10 mutants (Kinchen 2010). In fact, a recently described pathway was proposed to participate in engulfment signaling, consisting of the tyrosine kinase Abl (C. elegans ABL-1) and the Abl-interacting protein Abi (C. elegans ABI-1) (Hurwitz et al. 2009). Using C. elegans, this group reported that ABI-1 is required for engulfment of apoptotic cells and proper distal tip cell migration. ABI-1 was proposed to act in parallel to the two known engulfment signaling pathways, and ABL-1 likely opposes apoptotic cell engulfment by inhibiting ABI-1 function. The molecular pathways functioning upstream and downstream of these proteins are not yet defined, and it is still unclear whether ABI-1 signals through Rac (CED-10) similar to the other signaling pathways described above.
APOPTOTIC CELL DIGESTION AND PHAGOCYTE RESPONSE
After recognition and engulfment, the process of apoptotic cell clearance is not complete. Recent studies suggest that events downstream from internalization (collectively termed phagosome maturation) influence the phagocytic capacity of a cell to internalize additional targets (Wu et al. 2000; Krieser et al. 2002; Schrijvers et al. 2005; Park et al. 2011). Proper recognition, clearance, and degradation of apoptotic cell material are vital to maintaining an environment that protects the host against unchecked inflammation and eventual autoimmunity. Unengulfed apoptotic cells have the propensity to leak their cellular contents over time (secondary necrosis), resulting in inflammation, exposure of self-antigens, and a break in tolerance (Franz et al. 2006). On the other hand, recognition of apoptotic cells can trigger the secretion of anti-inflammatory cytokines (such as TGF-β and IL-10) from the phagocyte, thereby dampening or resolving inflammation (Henson 2005; Elliott and Ravichandran 2010). Furthermore, the proper degradation and processing of apoptotic cell material by professional phagocytes, particularly dendritic cells, are imperative for self-antigen presentation necessary to establish and maintain tolerance (Delamarre et al. 2005; Erwig and Henson 2008). The steps involved in apoptotic cell digestion and the effects this event has on the phagocyte are discussed further below.
Phagosome Maturation: Preparing the Meal for Digestion
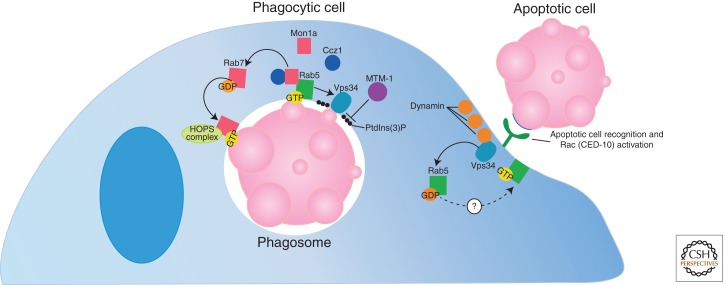
Presently, the process of phagosome maturation is actively studied, and our knowledge is evolving. Following internalization, phagosomes (membrane-bound compartments containing the phagocytosed target) become increasingly acidic, ultimately fusing with lysosomes, which contain the digestive enzymes required for degradation (Kinchen and Ravichandran 2008). This process begins with recruitment of the large GTPase, Dynamin (C. elegans DYN-1), to the apoptotic cell/phagocyte interface (Fig. 4) (Yu et al. 2006). Dynamin interaction with Vps34 [a phosphatidylinositol(3)-kinase] on the forming phagosome leads to recruitment of the GDP-bound small GTPase, Rab5, to the phagosome surface (Kinchen et al. 2008). Rab5 is subsequently activated by an unidentified protein (the currently known Rab5 GEFs are not required in this process). In mammalian cells, however, depletion of the GEF, Gapex-5, was shown to inhibit Rab5 activation at sites of engulfment (Kitano et al. 2008). GTP-bound, active Rab5 promotes Vps34 activation, leading to generation of PtdIns(3)P on the phagosome surface, which is subsequently removed by the lipid phosphatase MTM-1 (Zou et al. 2009; Kinchen 2010). Recent studies describe that Mon1a (C. elegans SAND-1) and its binding partner, Ccz1, tie Rab5 activation to Rab7 recruitment to the phagosome (Kinchen and Ravichandran 2010) and its subsequent activation (Nordmann et al. 2010). At the Rab7-positive stage, the HOPS complex is recruited to the phagosome, leading to Rab7 activation and, ultimately, fusion of the phagosome with the lysosomal network (Kinchen et al. 2008). It is at this stage that acidic proteases and nucleases get activated and the apoptotic cell targets are degraded (Lennon-Dumenil et al. 2002). Rab5 must also be inactivated, and PtdIns(3)P must be removed from the phagosome surface for maturation to proceed (Li et al. 2009; Zou et al. 2009).
Figure 4.
Phagosome maturation: The post-engulfment processing of apoptotic cells. In the first step of phagosome maturation, Dynamin is recruited to the apoptotic cell/phagocyte interface. Dynamin interacts with Vps34, leading to Rab5 recruitment and activation. Activated Rab5 further promotes Vps34 activation, leading to generation of PtdIns(3)P on the phagosome surface (subsequently removed by MTM-1). Mon1a and its binding partner, Ccz1, tie Rab5 activation and Rab7 recruitment to the phagosome. At this stage, the HOPS complex is recruited, leading to Rab7 activation and eventual fusion of the phagosome with the lysosome.
Degradation and Processing of Apoptotic Cells
After engulfment, apoptotic cells are digested into their basic cellular building blocks including nucleotides, fats, sterols, and peptides/amino acids. Phagocytes often consume multiple corpses at one time; therefore, they must efficiently process the apoptotic cell constituents to maintain homeostasis. Several mechanisms have been identified that help a phagocyte maintain homeostasis. The mitochondrial membrane protein UCP2, which uncouples oxidative phosphorylation from ATP synthesis and reduces mitochondrial membrane potential in cells, has been shown to positively regulate the engulfment capacity of phagocytes (Park et al. 2011). Overexpression of UCP2 effectively enhanced phagocytosis of apoptotic cells in vitro, and loss of UCP2 expression reduced phagocytosis both in vitro and in vivo, suggesting that mitochondrial membrane potential modulates engulfment. UCP2 may thus provide a mechanism by which the energy balance within phagocytes is maintained during the process of engulfment. DNase II is a lysosomal enzyme responsible for degrading DNA, and lack of this enzyme leads to accumulation of DNA fragments within phagocytes (Kawane et al. 2001, 2003). Furthermore, conditional deletion of DNase II in the mouse led to the development of polyarthritis and an increase of inflammatory cytokines in the joint tissues (Kawane et al. 2006). This inflammation is thought to result from the buildup of target cell DNA in macrophages, which, in turn, were shown to produce the inflammatory cytokine TNFα (Kawane et al. 2006). Cholesterol also serves several important functions within a cell (regulates plasma membrane fluidity, plays a role in cell signaling and transport, etc.) and is therefore strictly regulated. Intriguingly, studies show that apoptotic cell-derived cholesterol becomes part of the cholesterol pool within the phagocyte, and phagocytes handle the additional sterol load by increasing their basal efflux mechanism via ABCA1 (Gerbod-Giannone et al. 2006; Kiss et al. 2006a). Conversely, increased cholesterol efflux is not seen when macrophages are fed necrotic cells or when macrophages are forced to internalize apoptotic cells via the FcR, suggesting an apoptotic-cell-specific trigger. PtdSer on the surface of apoptotic cells appears to be one such trigger; blocking PtdSer recognition was shown to inhibit enhanced cholesterol efflux, whereas providing purified PtdSer liposomes to macrophages augments it (Kiss et al. 2006a). Finally, processing of apoptotic cell proteins is linked to establishment and maintenance of self-tolerance via cross-presentation of engulfed cell peptides by the major histocompatibility complex (MHC) class I molecules (Bellone et al. 1997; Albert et al. 1998; Huang et al. 2000). Presentation of apoptotic-cell-derived peptides appears to be excluded from MHC class II presentation, perhaps reflecting the endogenous source of antigen (Blander and Medzhitov 2006).
Consequences of Apoptotic Cell Recognition and Internalization
The hallmark feature of apoptotic cell clearance is the non-inflammatory nature of this process. Many human diseases are caused or worsened by inflammation; however, the inflammatory process can also be of benefit to host survival as in the case of infection or cancer (Henson 2005). It is currently understood that a balance occurs between immunogenic and tolerogenic cell death/clearance depending on considerations such as the phagocytic cell type that clears the debris and the factors produced by the engulfing cells in response to apoptotic cell recognition.
Apoptotic cell death occurs in healthy organisms as part of normal tissue turnover (Henson and Hume 2006). The location of cell death and the means by which the cells are cleared will affect the response (or lack thereof) of the immune system. For example, organs such as the spleen and liver are considered tolerogenic, and apoptotic cells localized to these tissues will not generally elicit an immune response (Green et al. 2009). In addition, phagocytes are “programmed” to elicit specific responses after recognition of targets, dependent on their cell type. For instance, engulfment of uninfected apoptotic cells by tissue resident macrophages is usually immunologically silent; however, engulfment of apoptotic cells via infiltrating macrophages and/or dendritic cells in an inflamed environment has the potential to drive an antigen-specific immune response and break tolerance (Geissmann et al. 2010; Hochreiter-Hufford and Ravichandran 2012). Signals on the surface of dying cells can direct specific phagocytic cell types (i.e., dendritic cells or macrophages) toward them for clearance; however, in a recent study, lipid oxidation by one type of phagocyte was shown to inhibit phagocytosis of apoptotic cells via another phagocytic cell type. Briefly, expression of 12/15-lipoxygenase by resident peritoneal macrophages led to exposure of oxidized lipid on their cell surface; this oxidized lipid sequestered the MFG-E8 necessary for inflammatory monocyte recognition and engulfment of apoptotic cells (Uderhardt et al. 2012). This is the first example of a phagocyte-intrinsic mechanism for regulating apoptotic cell sorting to specific phagocytes. Annexin V is a naturally occurring PtdSer binding protein that blocks phagocytosis of apoptotic cells by macrophages but not dendritic cells (Krahling et al. 1999; Munoz et al. 2007; Frey et al. 2009). Necrotic cell immunization of Annexin V-deficient mice did not lead to elicitation of an immune response, unlike wild-type mice; macrophages isolated from Annexin V-deficient mice were shown to secrete significantly higher levels of IL-10 in response to incubation with LPS and necrotic cells than macrophages from wild-type mice (Frey et al. 2009). The investigators suggest that Annexin V blocks PtdSer-mediated anti-inflammatory cytokine release as well as phagocytosis of necrotic cells by macrophages, thereby allowing engulfment by dendritic cells and the initiation of an immune response. Further studies will elucidate whether this mechanism functions in an in vivo setting.
As mentioned above, one method by which apoptotic cells induce tolerance is by stimulating phagocyte secretion of anti-inflammatory and immunosuppressive cytokines such as TGF-β and IL-10. These cytokines, in turn, induce differentiation of regulatory T-cells and T helper 2 cells important in prevention of an inflammatory response (Green et al. 2009). Likewise, apoptotic cells have been shown to repress inflammatory cytokine secretion by monocytes in vitro (Voll et al. 1997; Kim et al. 2004). Interestingly, internalization of apoptotic cells is not required for stimulated cytokine secretion. In fact, in in vitro studies, apoptotic cell membranes were sufficient to inhibit IL-12 secretion by LPS-induced macrophages, and PtdSer vesicles, alone, stimulated TGF-β secretion by macrophages (Huynh et al. 2002; Kim et al. 2004). Work is currently underway to piece together the receptors and signaling molecules that connect PtdSer recognition with cytokine secretion.
Although apoptotic cell clearance is typically considered immunologically silent, there are exceptions in which apoptotic cells can elicit an immune response. The clearance of apoptotic tumor cells is an example in which immunogenic cell death is beneficial to the host so that residual tumor cells and metastases are maintained under control. Engulfment of oncogenic cells can give rise to the presentation of altered self-peptides, which may be presented to the immune system (Segal et al. 2008); however, the tolerogenic nature of cell clearance can dampen a subsequent immune response. Certain forms of chemotherapy and radiotherapy have been linked to the exposure or release of factors by dying cells that can initiate a robust immune response. For example, treatment of tumor cells with antracyclins led to cell-surface exposure of calreticulin early during the death program, which was essential for phagocytosis of dying tumor cells by dendritic cells and the initiation of an immune response (Obeid et al. 2007b). Additionally, calreticulin exposure was linked to immunogenic cell death caused by irradiation and UVC-light treatment (Obeid et al. 2007a). Uncleared, late apoptotic or necrotic cells with “leaky” plasma membranes can release damage-associated molecular-pattern molecules (DAMPs), such as heat shock proteins, high mobility group box 1 (HMGB-1) protein, and nucleic acids. These released intracellular molecules activate Toll-like receptors (TLRs) on phagocytes, facilitating antigen presentation and the initiation of an immune response (Peter et al. 2010; Kepp et al. 2011).
CONCLUDING REMARKS
Apoptotic cell death is an integral part of cell turnover in many tissues, and proper corpse clearance is vital to maintaining tissue homeostasis in all multicellular organisms. All living tissues have some mechanism(s) in place to handle corpse clearance, and most cell types (not just professional phagocytes) possess the ability to phagocytose apoptotic cells, underlining the relevance of this process in metazoan health.
Defects in clearance are believed to play a role in a wide variety of human pathologies including autoimmune diseases (systemic lupus erythematosus [SLE] and rheumatoid arthritis); pulmonary diseases (chronic obstructive pulmonary disease [COPD] and asthma); cardiovascular diseases (atherosclerosis); neurological diseases (Alzheimer’s disease); infectious diseases (bacterial invasion by certain Shigella, Yersinia, and Salmonella species); and cancer (Elliott and Ravichandran 2010). Furthermore, the tolerogenic nature of apoptotic cell clearance has led to the emergence of potential new therapies using apoptotic cells for the prevention of transplant rejection and modulating autoimmune conditions such as type 1 diabetes (Castro et al. 2010; Morelli and Larregina 2010).
The field of apoptotic cell clearance is relatively young, and we are just beginning to better define the individual steps of this process and the consequences to the phagocyte, the tissue where it resides, and the organism. A better understanding of the sensing, recognition, engulfment, and processing of apoptotic cells will likely present new targets for therapy in the treatment of multiple immunological and metabolic diseases.
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
- Aderem A, Underhill DM 1999. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 17: 593–623 [DOI] [PubMed] [Google Scholar]
- Akakura S, Singh S, Spataro M, Akakura R, Kim JI, Albert ML, Birge RB 2004. The opsonin MFG-E8 is a ligand for the αvβ5 integrin and triggers DOCK180-dependent Rac1 activation for the phagocytosis of apoptotic cells. Exp Cell Res 292: 403–416 [DOI] [PubMed] [Google Scholar]
- Albert ML, Pearce SF, Francisco LM, Sauter B, Roy P, Silverstein RL, Bhardwaj N 1998. Immature dendritic cells phagocytose apoptotic cells via αvβ5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J Exp Med 188: 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert ML, Kim JI, Birge RB 2000. αvβ5 integrin recruits the CrkII–Dock180–rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol 2: 899–905 [DOI] [PubMed] [Google Scholar]
- Armstrong A, Ravichandran KS 2011. Phosphatidylserine receptors: What is the new RAGE? EMBO Rep 12: 287–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, Mohler W, Han DK 2003. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev Cell 4: 587–598 [DOI] [PubMed] [Google Scholar]
- Balasubramanian K, Schroit AJ 2003. Aminophospholipid asymmetry: A matter of life and death. Annu Rev Physiol 65: 701–734 [DOI] [PubMed] [Google Scholar]
- Bellone M, Iezzi G, Rovere P, Galati G, Ronchetti A, Protti MP, Davoust J, Rugarli C, Manfredi AA 1997. Processing of engulfed apoptotic bodies yields T cell epitopes. J Immunol 159: 5391–5399 [PubMed] [Google Scholar]
- Blander JM, Medzhitov R 2006. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature 440: 808–812 [DOI] [PubMed] [Google Scholar]
- Borisenko GG, Matsura T, Liu S-X, Tyurin VA, Jianfei J, Serinkan FB, Kagan VE 2003. Macrophage recognition of externalized phosphatidylserine and phagocytosis of apoptotic Jurkat cells—existence of a threshold. Arch Biochem Biophys 413: 41–52 [DOI] [PubMed] [Google Scholar]
- Bose J, Gruber AD, Helming L, Schiebe S, Wegener I, Hafner M, Beales M, Kontgen F, Lengeling A 2004. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J Biol 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazou I, Pound JD, Duffin R, Bournazos S, Melville LA, Brown SB, Rossi AG, Gregory CD 2009. Apoptotic human cells inhibit migration of granulocytes via release of lactoferrin. J Clin Invest 119: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton DL, Henson PM 2008. Apoptotic cell recognition: Will the real phosphatidylserine receptor(s) please stand up? Curr Biol 18: R76–R79 [DOI] [PubMed] [Google Scholar]
- Brown S, Heinisch I, Ross E, Shaw K, Buckley CD, Savill J 2002. Apoptosis disables CD31-mediated cell detachment from phagocytes promoting binding and engulfment. Nature 418: 200–203 [DOI] [PubMed] [Google Scholar]
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS 2002. Unconventional Rac-GEF activity is mediated through the Dock180–ELMO complex. Nat Cell Biol 4: 574–582 [DOI] [PubMed] [Google Scholar]
- Castellano F, Montcourrier P, Chavrier P 2000. Membrane recruitment of Rac1 triggers phagocytosis. J Cell Sci 113: 2955–2961 [DOI] [PubMed] [Google Scholar]
- Castro CN, Barcala Tabarrozi AE, Noguerol MA, Liberman AC, Dewey RA, Arzt E, Morelli AE, Perone MJ 2010. Disease-modifying immunotherapy for the management of autoimmune diabetes. Neuroimmunomodulation 17: 173–176 [DOI] [PubMed] [Google Scholar]
- Chao MP, Alizadeh AA, Tang C, Jan M, Weissman-Tsukamoto R, Zhao F, Park CY, Weissman IL, Majeti R 2011. Therapeutic antibody targeting of CD47 eliminates human acute lymphoblastic leukemia. Cancer Res 71: 1374–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, Armstrong AJ, Penuela S, Laird DW, Salvesen GS, et al. 2010. Pannexin 1 channels mediate “find-me” signal release and membrane permeability during apoptosis. Nature 467: 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Corriden R, Inoue Y, Yip L, Hashiguchi N, Zinkernagel A, Nizet V, Insel PA, Junger WG 2006. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science 314: 1792–1795 [DOI] [PubMed] [Google Scholar]
- Cote JF, Motoyama AB, Bush JA, Vuori K 2005. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat Cell Biol 7: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, et al. 2004. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and Armadillo repeats of CED-12/ELMO. Curr Biol 14: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307: 1630–1634 [DOI] [PubMed] [Google Scholar]
- Devitt A, Moffatt OD, Raykundalia C, Capra JD, Simmons DL, Gregory CD 1998. Human CD14 mediates recognition and phagocytosis of apoptotic cells. Nature 392: 505–509 [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS 2010. Clearance of apoptotic cells: Implications in health and disease. J Cell Biol 189: 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, et al. 2009. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461: 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig LP, Henson PM 2008. Clearance of apoptotic cells by phagocytes. Cell Death Differ 15: 243–250 [DOI] [PubMed] [Google Scholar]
- Ezekowitz RA, Sastry K, Bailly P, Warner A 1990. Molecular characterization of the human macrophage mannose receptor: Demonstration of multiple carbohydrate recognition-like domains and phagocytosis of yeasts in Cos-1 cells. J Exp Med 172: 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148: 2207–2216 [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Frasch SC, Warner ML, Henson PM 1998a. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ 5: 551–562 [DOI] [PubMed] [Google Scholar]
- Fadok VA, Warner ML, Bratton DL, Henson PM 1998b. CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (αvβ3). J Immunol 161: 6250–6257 [PubMed] [Google Scholar]
- Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL 2001. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J Biol Chem 276: 1071–1077 [DOI] [PubMed] [Google Scholar]
- Florey O, Haskard DO 2009. Sphingosine 1-phosphate enhances Fc γ receptor-mediated neutrophil activation and recruitment under flow conditions. J Immunol 183: 2330–2336 [DOI] [PubMed] [Google Scholar]
- Franz S, Gaipl US, Munoz LE, Sheriff A, Beer A, Kalden JR, Herrmann M 2006. Apoptosis and autoimmunity: When apoptotic cells break their silence. Curr Rheumatol Rep 8: 245–247 [DOI] [PubMed] [Google Scholar]
- Frey B, Munoz LE, Pausch F, Sieber R, Franz S, Brachvogel B, Poschl E, Schneider H, Rodel F, Sauer R, et al. 2009. The immune reaction against allogeneic necrotic cells is reduced in Annexin A5 knock out mice whose macrophages display an anti-inflammatory phenotype. J Cell Mol Med 13: 1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM 2005. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123: 321–334 [DOI] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K 2010. Development of monocytes, macrophages, and dendritic cells. Science 327: 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbod-Giannone MC, Li Y, Holleboom A, Han S, Hsu LC, Tabas I, Tall AR 2006. TNFα induces ABCA1 through NF-κB in macrophages and in phagocytes ingesting apoptotic cells. Proc Natl Acad Sci 103: 3112–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S 1999. Macrophage-restricted molecules: Role in differentiation and activation. Immunol Lett 65: 5–8 [DOI] [PubMed] [Google Scholar]
- Green DR, Ferguson T, Zitvogel L, Kroemer G 2009. Immunogenic and tolerogenic cell death. Nat Rev Immunol 9: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory C 2009. Cell biology: Sent by the scent of death. Nature 461: 181–182 [DOI] [PubMed] [Google Scholar]
- Gregory CD, Devitt A, Moffatt O 1998. Roles of ICAM-3 and CD14 in the recognition and phagocytosis of apoptotic cells by macrophages. Biochem Soc Trans 26: 644–649 [DOI] [PubMed] [Google Scholar]
- Gude DR, Alvarez SE, Paugh SW, Mitra P, Yu J, Griffiths R, Barbour SE, Milstien S, Spiegel S 2008. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a “come-and-get-me” signal. FASEB J 22: 2629–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, et al. 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107: 27–41 [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417: 182–187 [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y, Nagata S 2004. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304: 1147–1150 [DOI] [PubMed] [Google Scholar]
- He M, Kubo H, Morimoto K, Fujino N, Suzuki T, Takahasi T, Yamada M, Yamaya M, Maekawa T, Yamamoto Y, et al. 2011. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep 12: 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming L, Gordon S 2009. Molecular mediators of macrophage fusion. Trends Cell Biol 19: 514–522 [DOI] [PubMed] [Google Scholar]
- Henson PM 2005. Dampening inflammation. Nat Immunol 6: 1179–1181 [DOI] [PubMed] [Google Scholar]
- Henson PM, Hume DA 2006. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol 27: 244–250 [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford AE, Ravichandran KS 2012. Oxygenated lipids: A mode to WiPE out inflammation? Immunity 36: 699–701 [DOI] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, Schnabel R 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412: 202–206 [DOI] [PubMed] [Google Scholar]
- Huang FP, Platt N, Wykes M, Major JR, Powell TJ, Jenkins CD, MacPherson GG 2000. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J Exp Med 191: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz B, Bartz C, Kokozidou M 2006. Trophoblast fusion: Fusogenic proteins, syncytins and ADAMs, and other prerequisites for syncytial fusion. Micron 37: 509–517 [DOI] [PubMed] [Google Scholar]
- Hurwitz ME, Vanderzalm PJ, Bloom L, Goldman J, Garriga G, Horvitz HR 2009. Abl kinase inhibits the engulfment of apoptotic [corrected] cells in Caenorhabditis elegans. PLoS Biol 7: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh ML, Fadok VA, Henson PM 2002. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-β1 secretion and the resolution of inflammation. J Clin Invest 109: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138: 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johann AM, Weigert A, Eberhardt W, Kuhn AM, Barra V, von Knethen A, Pfeilschifter JM, Brune B 2008. Apoptotic cell-derived sphingosine-1-phosphate promotes HuR-dependent cyclooxygenase-2 mRNA stabilization and protein expression. J Immunol 180: 1239–1248 [DOI] [PubMed] [Google Scholar]
- Kagan VE, Gleiss B, Tyurina YY, Tyurin VA, Elenstrom-Magnusson C, Liu SX, Serinkan FB, Arroyo A, Chandra J, Orrenius S, et al. 2002. A role for oxidative stress in apoptosis: Oxidation and externalization of phosphatidylserine is required for macrophage clearance of cells undergoing Fas-mediated apoptosis. J Immunol 169: 487–499 [DOI] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S 2001. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292: 1546–1549 [DOI] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Yoshida H, Nagase H, Ohsawa Y, Uchiyama Y, Okada K, Iida T, Nagata S 2003. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat Immunol 4: 138–144 [DOI] [PubMed] [Google Scholar]
- Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S 2006. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443: 998–1002 [DOI] [PubMed] [Google Scholar]
- Kepp O, Galluzzi L, Martins I, Schlemmer F, Adjemian S, Michaud M, Sukkurwala AQ, Menger L, Zitvogel L, Kroemer G 2011. Molecular determinants of immunogenic cell death elicited by anticancer chemotherapy. Cancer Metastasis Rev 30: 61–69 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Gershov D, Ma X, Brot N, Elkon KB 2002. I-PLA(2) activation during apoptosis promotes the exposure of membrane lysophosphatidylcholine leading to binding by natural immunoglobulin M antibodies and complement activation. J Exp Med 196: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Elkon KB, Ma X 2004. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity 21: 643–653 [DOI] [PubMed] [Google Scholar]
- Kinchen JM 2010. A model to die for: Signaling to apoptotic cell removal in worm, fly and mouse. Apoptosis 15: 998–1006 [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS 2007. Journey to the grave: Signaling events regulating removal of apoptotic cells. J Cell Sci 120: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS 2008. Phagosome maturation: Going through the acid test. Nat Rev Mol Cell Biol 9: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS 2010. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464: 778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, Schnabel R, Hengartner MO 2005. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 434: 93–99 [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS 2008. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol 10: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss RS, Elliott MR, Ma Z, Marcel YL, Ravichandran KS 2006a. Apoptotic cells induce a phosphatidylserine-dependent homeostatic response from phagocytes. Curr Biol 16: 2252–2258 [DOI] [PubMed] [Google Scholar]
- Kiss RS, Ma Z, Nakada-Tsukui K, Brugnera E, Vassiliou G, McBride HM, Ravichandran KS, Marcel YL 2006b. The lipoprotein receptor-related protein-1 (LRP) adapter protein GULP mediates trafficking of the LRP ligand prosaposin, leading to sphingolipid and free cholesterol accumulation in late endosomes and impaired efflux. J Biol Chem 281: 12081–12092 [DOI] [PubMed] [Google Scholar]
- Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M 2008. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453: 241–245 [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, et al. 2007. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27: 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahling S, Callahan MK, Williamson P, Schlegel RA 1999. Exposure of phosphatidylserine is a general feature in the phagocytosis of apoptotic lymphocytes by macrophages. Cell Death Differ 6: 183–189 [DOI] [PubMed] [Google Scholar]
- Krieser RJ, MacLea KS, Longnecker DS, Fields JL, Fiering S, Eastman A 2002. Deoxyribonuclease IIα is required during the phagocytic phase of apoptosis and its loss causes perinatal lethality. Cell Death Differ 9: 956–962 [DOI] [PubMed] [Google Scholar]
- Kuraishi T, Nakagawa Y, Nagaosa K, Hashimoto Y, Ishimoto T, Moki T, Fujita Y, Nakayama H, Dohmae N, Shiratsuchi A, et al. 2009. Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. EMBO J 28: 3868–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry YD, Denis M, Nandi S, Bell S, Vaughan AM, Zha X 2006. ATP-binding cassette transporter A1 expression disrupts raft membrane microdomains through its ATPase-related functions. J Biol Chem 281: 36091–36101 [DOI] [PubMed] [Google Scholar]
- Lauber K, Bohn E, Krober SM, Xiao YJ, Blumenthal SG, Lindemann RK, Marini P, Wiedig C, Zobywalski A, Baksh S, et al. 2003. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 113: 717–730 [DOI] [PubMed] [Google Scholar]
- Lennon-Dumenil AM, Bakker AH, Maehr R, Fiebiger E, Overkleeft HS, Rosemblatt M, Ploegh HL, Lagaudriere-Gesbert C 2002. Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J Exp Med 196: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverrier Y, Ridley AJ 2001. Requirement for Rho GTPases and PI 3-kinases during apoptotic cell phagocytosis by macrophages. Curr Biol 11: 195–199 [DOI] [PubMed] [Google Scholar]
- Li W, Zou W, Zhao D, Yan J, Zhu Z, Lu J, Wang X 2009. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development 136: 2445–2455 [DOI] [PubMed] [Google Scholar]
- Liu QA, Hengartner MO 1998. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93: 961–972 [DOI] [PubMed] [Google Scholar]
- Lu M, Kinchen JM, Rossman KL, Grimsley C, deBakker C, Brugnera E, Tosello-Trampont AC, Haney LB, Klingele D, Sondek J, et al. 2004. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat Struct Mol Biol 11: 756–762 [DOI] [PubMed] [Google Scholar]
- Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J, Hengartner MO, Yajnik V, Ravichandran KS 2005. A steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr Biol 15: 371–377 [DOI] [PubMed] [Google Scholar]
- Manaka J, Kuraishi T, Shiratsuchi A, Nakai Y, Higashida H, Henson P, Nakanishi Y 2004. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem 279: 48466–48476 [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T 1998. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J 17: 6932–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksa M, Amin D, Wu R, Ravikumar TS, Wang P 2007. Fractalkine-induced MFG-E8 leads to enhanced apoptotic cell clearance by macrophages. Mol Med 13: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature 450: 435–439 [DOI] [PubMed] [Google Scholar]
- Monks J, Rosner D, Geske FJ, Lehman L, Hanson L, Neville MC, Fadok VA 2005. Epithelial cells as phagocytes: Apoptotic epithelial cells are engulfed by mammary alveolar epithelial cells and repress inflammatory mediator release. Cell Death Differ 12: 107–114 [DOI] [PubMed] [Google Scholar]
- Monks J, Smith-Steinhart C, Kruk ER, Fadok VA, Henson PM 2008. Epithelial cells remove apoptotic epithelial cells during post-lactation involution of the mouse mammary gland. Biol Reprod 78: 586–594 [DOI] [PubMed] [Google Scholar]
- Morelli AE, Larregina AT 2010. Apoptotic cell-based therapies against transplant rejection: Role of recipient’s dendritic cells. Apoptosis 15: 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Kanemura Y, Fujikawa H, Nakano A, Ikemoto H, Ozaki I, Matsumoto T, Tamura K, Yokota M, Arita N 2002. Brain-specific angiogenesis inhibitor 1 (BAI1) is expressed in human cerebral neuronal cells. Neurosci Res 43: 69–74 [DOI] [PubMed] [Google Scholar]
- Munoz LE, Franz S, Pausch F, Furnrohr B, Sheriff A, Vogt B, Kern PM, Baum W, Stach C, von Laer D, et al. 2007. The influence on the immunomodulatory effects of dying and dead cells of Annexin V. J Leukoc Biol 81: 6–14 [DOI] [PubMed] [Google Scholar]
- Murugesan G, Sandhya Rani MR, Gerber CE, Mukhopadhyay C, Ransohoff RM, Chisolm GM, Kottke-Marchant K 2003. Lysophosphatidylcholine regulates human microvascular endothelial cell expression of chemokines. J Mol Cell Cardiol 35: 1375–1384 [DOI] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K 2010. Autoimmunity and the clearance of dead cells. Cell 140: 619–630 [DOI] [PubMed] [Google Scholar]
- Nakano T, Ishimoto Y, Kishino J, Umeda M, Inoue K, Nagata K, Ohashi K, Mizuno K, Arita H 1997. Cell adhesion to phosphatidylserine mediated by a product of growth arrest-specific gene 6. J Biol Chem 272: 29411–29414 [DOI] [PubMed] [Google Scholar]
- Nakaya M, Tanaka M, Okabe Y, Hanayama R, Nagata S 2006. Opposite effects of Rho family GTPases on engulfment of apoptotic cells by macrophages. J Biol Chem 281: 8836–8842 [DOI] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Brocker C, Ostrowicz C, Engelbrecht-Vandre S, Ungermann C 2010. The Mon1–Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 20: 1654–1659 [DOI] [PubMed] [Google Scholar]
- Obeid M, Panaretakis T, Joza N, Tufi R, Tesniere A, van Endert P, Zitvogel L, Kroemer G 2007a. Calreticulin exposure is required for the immunogenicity of γ-irradiation and UVC light-induced apoptosis. Cell Death Differ 14: 1848–1850 [DOI] [PubMed] [Google Scholar]
- Obeid M, Tesniere A, Ghiringhelli F, Fimia GM, Apetoh L, Perfettini JL, Castedo M, Mignot G, Panaretakis T, Casares N, et al. 2007b. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med 13: 54–61 [DOI] [PubMed] [Google Scholar]
- Ogden CA, deCathelineau A, Hoffmann PR, Bratton D, Ghebrehiwet B, Fadok VA, Henson PM 2001. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med 194: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP 2000. Role of CD47 as a marker of self on red blood cells. Science 288: 2051–2054 [DOI] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS 2007. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450: 430–434 [DOI] [PubMed] [Google Scholar]
- Park SY, Jung MY, Kim HJ, Lee SJ, Kim SY, Lee BH, Kwon TH, Park RW, Kim IS 2008a. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ 15: 192–201 [DOI] [PubMed] [Google Scholar]
- Park SY, Kang KB, Thapa N, Kim SY, Lee SJ, Kim IS 2008b. Requirement of adaptor protein GULP during Stabilin-2-mediated cell corpse engulfment. J Biol Chem 283: 10593–10600 [DOI] [PubMed] [Google Scholar]
- Park SY, Kim SY, Jung MY, Bae DJ, Kim IS 2008c. Epidermal growth factor-like domain repeat of Stabilin-2 recognizes phosphatidylserine during cell corpse clearance. Mol Cell Biol 28: 5288–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Hochreiter-Hufford A, Ravichandran KS 2009. The phosphatidylserine receptor TIM-4 does not mediate direct signaling. Curr Biol 19: 346–351 [DOI] [PubMed] [Google Scholar]
- Park D, Han CZ, Elliott MR, Kinchen JM, Trampont PC, Das S, Collins S, Lysiak JJ, Hoehn KL, Ravichandran KS 2011. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature 477: 220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C, Waibel M, Radu CG, Yang LV, Witte ON, Schulze-Osthoff K, Wesselborg S, Lauber K 2008. Migration to apoptotic “find-me” signals is mediated via the phagocyte receptor G2A. J Biol Chem 283: 5296–5305 [DOI] [PubMed] [Google Scholar]
- Peter C, Wesselborg S, Herrmann M, Lauber K 2010. Dangerous attraction: Phagocyte recruitment and danger signals of apoptotic and necrotic cells. Apoptosis 15: 1007–1028 [DOI] [PubMed] [Google Scholar]
- Ravichandran KS 2003. “Recruitment signals” from apoptotic cells: Invitation to a quiet meal. Cell 113: 817–820 [DOI] [PubMed] [Google Scholar]
- Ravichandran KS 2010. Find-me and eat-me signals in apoptotic cell clearance: Progress and conundrums. J Exp Med 207: 1807–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran KS, Lorenz U 2007. Engulfment of apoptotic cells: Signals for a good meal. Nat Rev Immunol 7: 964–974 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol 2: 131–136 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Cameron S, Horvitz HR 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412: 198–202 [DOI] [PubMed] [Google Scholar]
- Ridley AJ 2001. Rho family proteins: Coordinating cell responses. Trends Cell Biol 11: 471–477 [DOI] [PubMed] [Google Scholar]
- Riento K, Ridley AJ 2003. Rocks: Multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol 4: 446–456 [DOI] [PubMed] [Google Scholar]
- Rosen H, Goetzl EJ 2005. Sphingosine 1-phosphate and its receptors: An autocrine and paracrine network. Nat Rev Immunol 5: 560–570 [DOI] [PubMed] [Google Scholar]
- Santiago C, Ballesteros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casasnovas JM 2007. Structures of T cell immunoglobulin mucin protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity 27: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savill J 1997. Apoptosis in resolution of inflammation. J Leukoc Biol 61: 375–380 [DOI] [PubMed] [Google Scholar]
- Savill J, Dransfield I, Hogg N, Haslett C 1990. Vitronectin receptor-mediated phagocytosis of cells undergoing apoptosis. Nature 343: 170–173 [DOI] [PubMed] [Google Scholar]
- Savill J, Dransfield I, Gregory C, Haslett C 2002. A blast from the past: Clearance of apoptotic cells regulates immune responses. Nat Rev Immunol 2: 965–975 [DOI] [PubMed] [Google Scholar]
- Schlegel RA, Krahling S, Callahan MK, Williamson P 1999. CD14 is a component of multiple recognition systems used by macrophages to phagocytose apoptotic lymphocytes. Cell Death Differ 6: 583–592 [DOI] [PubMed] [Google Scholar]
- Schrijvers DM, De Meyer GR, Kockx MM, Herman AG, Martinet W 2005. Phagocytosis of apoptotic cells by macrophages is impaired in atherosclerosis. Arterioscler Thromb Vasc Biol 25: 1256–1261 [DOI] [PubMed] [Google Scholar]
- Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, Allison JP 2008. Epitope landscape in breast and colorectal cancer. Cancer Res 68: 889–892 [DOI] [PubMed] [Google Scholar]
- Segawa K, Suzuki J, Nagata S 2011. Constitutive exposure of phosphatidylserine on viable cells. Proc Natl Acad Sci 108: 19246–19251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simhadri VR, Andersen JF, Calvo E, Choi SC, Coligan JE, Borrego F 2012. Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood 119: 2799–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su HP, Nakada-Tsukui K, Tosello-Trampont AC, Li Y, Bu G, Henson PM, Ravichandran KS 2002. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J Biol Chem 277: 11772–11779 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Umeda M, Sims PJ, Nagata S 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468: 834–838 [DOI] [PubMed] [Google Scholar]
- Tosello-Trampont AC, Nakada-Tsukui K, Ravichandran KS 2003. Engulfment of apoptotic cells is negatively regulated by Rho-mediated signaling. J Biol Chem 278: 49911–49919 [DOI] [PubMed] [Google Scholar]
- Tosello-Trampont AC, Kinchen JM, Brugnera E, Haney LB, Hengartner MO, Ravichandran KS 2007. Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ 14: 963–972 [DOI] [PubMed] [Google Scholar]
- Truman LA, Ford CA, Pasikowska M, Pound JD, Wilkinson SJ, Dumitriu IE, Melville L, Melrose LA, Ogden CA, Nibbs R, et al. 2008. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood 112: 5026–5036 [DOI] [PubMed] [Google Scholar]
- Tsai RK, Discher DE 2008. Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J Cell Biol 180: 989–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uderhardt S, Herrmann M, Oskolkova OV, Aschermann S, Bicker W, Ipseiz N, Sarter K, Frey B, Rothe T, Voll R, et al. 2012. 12/15-lipoxygenase orchestrates the clearance of apoptotic cells and maintains immunologic tolerance. Immunity 36: 834–846 [DOI] [PubMed] [Google Scholar]
- van den Eijnde SM, van den Hoff MJ, Reutelingsperger CP, van Heerde WL, Henfling ME, Vermeij-Keers C, Schutte B, Borgers M, Ramaekers FC 2001. Transient expression of phosphatidylserine at cell–cell contact areas is required for myotube formation. J Cell Sci 114: 3631–3642 [DOI] [PubMed] [Google Scholar]
- Voll RE, Herrmann M, Roth EA, Stach C, Kalden JR, Girkontaite I 1997. Immunosuppressive effects of apoptotic cells. Nature 390: 350–351 [DOI] [PubMed] [Google Scholar]
- Weigert A, Tzieply N, von Knethen A, Johann AM, Schmidt H, Geisslinger G, Brune B 2007. Tumor cell apoptosis polarizes macrophages role of sphingosine-1-phosphate. Mol Biol Cell 18: 3810–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR 1998a. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93: 951–960 [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR 1998b. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392: 501–504 [DOI] [PubMed] [Google Scholar]
- Wu YC, Stanfield GM, Horvitz HR 2000. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev 14: 536–548 [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Singh S, Georgescu MM, Birge RB 2005. A role for Mer tyrosine kinase in αvβ5 integrin-mediated phagocytosis of apoptotic cells. J Cell Sci 118: 539–553 [DOI] [PubMed] [Google Scholar]
- Yu X, Odera S, Chuang CH, Lu N, Zhou Z 2006. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell 10: 743–757 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR 2001a. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell 1: 477–489 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR 2001b. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56 [DOI] [PubMed] [Google Scholar]
- Zou W, Lu Q, Zhao D, Li W, Mapes J, Xie Y, Wang X 2009. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet 5: e1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]