Abstract
Protein quality control (PQC) functions to minimize the level and toxicity of misfolded proteins in the cell. PQC is performed by intricate collaboration among chaperones and target protein degradation. The latter is carried out primarily by the ubiquitin-proteasome system and perhaps autophagy. Terminally misfolded proteins that are not timely removed tend to form aggregates. Their clearance requires macroautophagy. Macroautophagy serves in intracellular quality control also by selectively segregating defective organelles (e.g., mitochondria) and targeting them for degradation by the lysosome. Inadequate PQC is observed in a large subset of failing human hearts with a variety of etiologies and its pathogenic role has been experimentally demonstrated. Multiple post-translational modifications (PTMs) can occur to substrate proteins and/or PQC machineries, promoting or hindering the removal of the misfolded proteins. This article highlights recent advances in PTMs-mediated regulation of intracellular quality control mechanisms and its known involvement in cardiac pathology.
Keywords: chaperones, the ubiquitin proteasome system, autophagy, chaperone-mediated autophagy, neddylation, deubiquitination, phosphorylation
Genetic mutations and environmental factors, especially those associated with increased stress and pathological conditions, can compromise the integrity of proteins and organelles in the cell, which, if not properly controlled, can be catastrophic to cell function and survival. Hence, the cell has evolved intracellular quality control (QC) mechanisms at protein and organelle levels to minimize the level and toxicity of misfolded proteins and defective organelles in the cell. Inadequate QC is associated with many forms of heart disease in their progression to congestive heart failure (CHF) and implicated in cardiac pathogenesis.1, 2 Hence, improving intracellular QC may potentially become a new therapeutic strategy for cardiac protection. A better understanding of the molecular mechanisms of intracellular QC will facilitate the development of such a novel strategy. Intracellular QC is regulated by transcriptional, translational, and post-translational mechanisms. Post-translational modifications (PTMs), such as phosphorylation, ubiquitination, nitrosylation, oxidation, etc., expand the size of the proteome exponentially and are pivotal in the regulation of protein stability, distribution, and function. Emerging evidence supports a major role of PTMs in regulating multiple pathways of intracellular QC.3 After a brief introduction into the general processes of intracellular QC, this review highlights recent advances in defining the impact of various PTMs on major QC pathways in the cell, especially those with promising roles in cardiac (patho)physiology.
1. An Introduction into Protein Quality Control
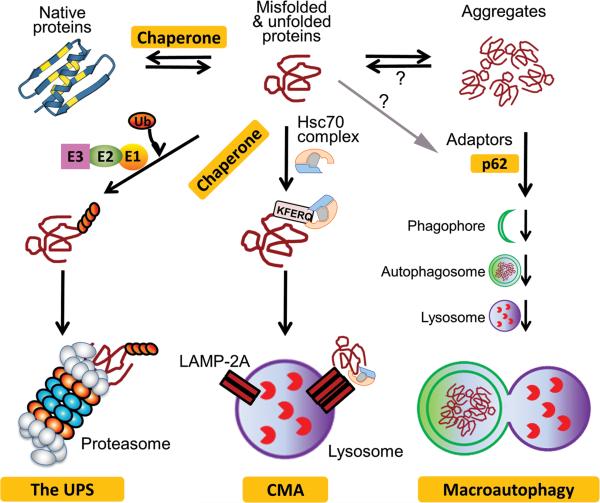
Protein quality control (PQC) is a set of molecular mechanisms ensuring that misfolded and damaged proteins are either repaired or removed in a timely fashion, thereby minimizing the toxic effects of misfolded proteins (Figure 1). The QC of proteins targeted for the secretary pathway (i.e., proteins passing through the endoplasmic reticulum [ER]) is performed by ER-associated PQC, this involves retro-translocation of misfolded proteins from ER lumen to the cytosol and degradation of them via ER-associated degradation (ERAD) pathways. ER-independent PQC is responsible for the QC of all non-ER proteins. In both cases, PQC is carried out by molecular chaperones and target protein degradation. Chaperones serve as the sensor of misfolded proteins and, in some cases, attempt to repair the misfolding by unfolding/refolding. If the repair fails, the misfolded protein is then termed as a terminally misfolded protein and will be escorted by chaperones for degradation by primarily the ubiquitin-proteasome system (UPS) and perhaps by chaperone-mediated autophagy (CMA). When misfolded proteins escape the surveillance of chaperones and target degradation, they tend to form aberrant aggregates. The intermediate, highly unstable, soluble species of aggregates are very toxic to the cell.4 The small aggregates assimilate into larger ones that are insoluble and perhaps less toxic to the cell. And finally, with the assistance from the microtubule transportation system, the small aggregates may be translocated to the microtubule organizing center and fuse with one another to form large inclusion bodies termed, by some, aggresomes.5 The insoluble aggregates and aggresomes are unlikely to be accessible to the proteasome and CMA, both of which can only degrade proteins individually. Hence, the removal of aggregated proteins requires a different mechanism that is capable of bulk degradation of substrates, a role filled by macroautophagy.
Figure 1. An illustration of protein quality control in the cell.
Chaperones help fold nascent polypeptides, unfold misfolded proteins and refold them, and channel terminally misfolded proteins for degradation by the ubiquitin-proteasome system (UPS) or chaperone-mediated autophagy (CMA). When escaped from targeted degradation, misfolded proteins form aggregates via hydrophobic interactions. Aggregated proteins can be selectively targeted by macroautophagy to, and degraded by, the lysosome.
1.1 The UPS
UPS-mediated protein degradation consists of two main steps: ubiquitination of a target protein molecule and degradation of the ubiquitinated protein by the proteasome. Ubiquitination is by itself an important and common form of post-translational modification (PTM). It is a cascade of enzymatic reactions that attach the carboxyl terminus of ubiquitin (Ub) to the ε-amino group on the side chain of a lysine residue on either the substrate molecule or a preceding Ub via an isopeptidyl bond. The reaction cascade is catalyzed by the Ub activating enzyme (E1), the Ub conjugating enzyme (E2), the Ub ligase (E3), and sometimes a Ub chain elongation factor (E4). The E3 is most critical because it confers substrate specificity. The proteasome, consisting of a proteolytic core (i.e., the 20S proteasome) and regulatory particles (the 19S and/or the 11S proteasome), mediates the degradation of polyubiquitinated proteins. Most cellular proteins are degraded by the UPS.6 In addition to degrading misfolded proteins for PQC, the UPS participates in the regulation of virtually all aspects of cell functions by timely degradation of normal proteins that are no longer needed. PTMs regulate UPS-mediated proteolysis at multiple levels (see Section 3).
1.2 Autophagy
Autophagy mechanisms function by sequestering cytoplasmic components that are degraded by lysosomes. Based on the route via which a substrate enters the lysosomal compartment, autophagy is classified into three types: microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy. In microautophagy, a small part of cytoplasm is internalized by invagination of lysosomal membrane and pinched off as a vesicle into the lysosomal lumen wherein it is degraded.
In CMA, a cytosolic protein molecule with a KFERQ motif or KFERQ-like motif is recognized and specifically bound by heat shock cognate 70 (Hsc70) and co-chaperones, targeted to lysosomal membrane protein (LAMP) 2A, and unfolded and translocated into lysosome lumen via LAMP-2A.7 CMA delivers substrate protein molecules to lysosomes one by one; hence, like the UPS, CMA is capable of target degradation of misfolded proteins. Approximately 30% cytosolic proteins contain the KFERQ motif in their primary sequences, not including those potentially generated by PTMs.7 The KFERQ motifs of many proteins in their native forms are likely buried in the interior, incapable of triggering CMA; however, misfolding, partial unfolding, or in some case conformational changes resulting from PTMs can all potentially expose the CMA-targeting motif and trigger CMA.8 Therefore, CMA may potentially play an important role in PQC by selectively targeting misfolded proteins with a KFERQ motif to the lysosome. CMA activities are reduced during aging due mainly to decreases of LAMP-2A. Restoration of CMA by overexpressing LAMP-2A in livers improves the ability of hepatocytes to handle protein damage and preserves liver function in aged mice.9 Inhibition of CMA leads to accumulation of oxidized proteins and protein aggregates in a cell and renders the cell more vulnerable to various stressors.10 CMA is constitutively activated in cells with impaired macroautophagy.11 Several neurodegenerative disease associated proteins, in their wild-type form, are degraded by CMA but their mutant forms often impair CMA.12, 13 Chronic exposure to a high-fat diet or acute exposure to a cholesterol-enriched diet both were shown to inhibit CMA via destabilizing LAMP-2A on lysosomal membrane,14 similar to aging-associated CMA decline. The role of CMA in cardiac (patho)physiology remains to be explored.
Macroautophagy delivers bulky cytoplasmic materials including organelles into lysosomes via formation of a double-membrane bound vesicle known as an autophagosome for fusion with, and degradation by, the lysosomes. Self-digestion of a portion of cytoplasm provides energy and essential amino acids for the synthesis of important proteins; hence, macroautophagy is essential to cell survival during nutrient deprivation. Macroautophagy is the most studied type of autophagy and in the past several years it has attracted extensive attention. Manipulating macroautophagy has been proposed to treat certain human diseases. Macroautophagy can selectively degrade defective organelles, such as depolarized mitochondria, as well as perhaps aberrant protein aggregates; hence, macroautophagy is considered a major player in both organelle QC and PQC in the cell (see Section 4).
1.3 Mitochondrial PQC
At the organelle level, the removal of defective mitochondria by macroautophagy (mitophagy) represents an important layer of QC mechanism for cell survival and cell functioning. At the suborganelle or molecular level, proteostasis within mitochondria is critical to the normal functioning of individual mitochondria. The cytosolic UPS might play a role in target degradation of misfolded proteins in the outer membrane of mitochondria but the interior compartments of mitochondria do not appear to contain the UPS.15 The refolding of unfolded polypeptides imported from the cytosol, the folding of nascent polypeptides synthesized within mitochondria, and the assembly of stoichiometric protein complexes are monitored by a mitochondrial PQC system localized in different mitochondrial compartments. Analogous to cytosolic PQC, the QC of mitochondrial proteins is performed by mitochondrial chaperones (e.g. mitochondrial HSP70 and HSP60) and proteases. In mitochondria, multiple proteases are responsible for the degradation of misfolded mitochondrial proteins. Most known mitochondrial proteases belong to the AAA (ATPase associated with diverse cellular activities) family, including Lon and Clp protease complexes of the matrix, as well as the two AAA proteases housed in the inner membrane.15
The research into cardiac mitochondrial PQC is just beginning to emerge. Lau et al reports that oxidative damage reduces the proteolytic capacity of cardiac mitochondria and decreases substrate availability for mitochondrial proteases. Results of this study further suggest that the 20S proteasome may contribute to cardiac mitochondrial proteostasis by degrading specific substrates of the mitochondria.16 Further investigation into the physiological and pathophysiological significance of mitochondrial PQC in the heart is well-justified, given that mitochondrial dysfunction has been linked to a variety of heart diseases.
1.4 Inadequate PQC in cardiac pathogenesis
Inadequate QC occurs in diseased hearts and may play an important role in cardiac pathogenesis. Improving QC protects the heart under stress conditions. The accumulation of ubiquitinated proteins and the presence of aberrant protein aggregation in the form of formation of pre-amyloid oligomers in a majority of failing human hearts represent compelling evidence for the existence of PQC inadequacy in the progression of common forms of heart disease to CHF.1 The pathophysiological significance of PQC inadequacy is best illustrated by cardiac proteinopathy which is exemplified by desmin-related cardiomyopathy (DRC). DRC, which can be caused by mutations in the desmin, αB-crystallin (CryAB), and several other genes, is featured by the presence of aberrant protein aggregates in myocytes.17 Inadequate PQC as represented by proteasome functional insufficiency (PFI) is associated with DRC. Recent experimental studies have demonstrated that PFI plays major pathogenic role in DRC.2 Similarly, PFI was recently unraveled in mouse hearts with acute regional I/R and demonstrated to be important mediator of myocardial I/R injury.2, 18 Cardiac expression of chaperones, such as heat shock proteins (HSPs), is often upregulated with stress, such as ischemia/reperfusion (I/R) or pressure overload, overexpression of HSPs is cardioprotective under the stress conditions.19-21 This suggests chaperone functional inadequacy during the stress condition. Impaired macroautophagy has been observed in I/R hearts and enhancing macroautophagy has been shown to protect I/R hearts in a large animal model.22, 23 Hence, improving QC in the cell by increasing chaperones, enhancing UPS-mediated degradation of misfolded proteins, or upregulation of selective autophagy, are potentially novel therapeutic strategies to protect the heart against proteotoxic stress.
2. PTMs Regulate Chaperone Functions in PQC
A complex network of molecular chaperones promotes efficient protein folding, neutralizes protein damage, and limits protein misfolding and aggregation. Molecular chaperones are defined as proteins that facilitate: the folding or unfolding of individual proteins, the assembly or disassembly of macromolecular complexes, but are not permanent constituents of those complexes. Chaperones play a major role in de novo protein folding, in stabilizing unfolded, newly synthesized proteins, and in preventing aberrant interactions, protein misfolding and aggregation. Chaperones also function in the unfolding of proteins for translocation across membranes of organelles. Chaperone function is important for restoring proteostasis. Chaperones help traffic terminally misfolded and aggregated proteins for degradation by the UPS or autophagy.6, 24 Many chaperones are HSPs, derived from the fact that cellular stresses, including heat shock cause protein denaturation, misfolding and aggregation.
2.1 HSP90
HSP 90 is a highly abundant chaperone that uses ATP hydrolysis to facilitate the efficient folding of proteins involved in signal transduction, receptor maturation and protein trafficking.25 The diverse functions that HSP90 influences rely on the >20 co-chaperones, >40 kinases, and receptors HSP90 interacts with. HSP90 function is modulated by several PTMs, including acetylation, nitrosylation and phosphorylation. Acetylation of HSP90 on Lys294 inhibits the binding of co-chaperones and client protein.26, 27 Nitrosylation of Cys597 of HSP90β inhibits eNOS activation.28, 29 Sites of HSP90 phosphorylation may inhibit (Ser225 and Ser254) or enhance client protein binding (Tyr300).30, 31 Phosphorylation of co-chaperones also influences HSP90 function.32 Thus, HSP90 chaperone function is regulated by its PTMs and interaction partners.
2.2 HSP70
The HSP70 family consists of 6 members that rely on ATP hydrolysis for activity. HSP70 is responsive to a host of cardiac stresses such as I/R, and hypertrophic stimuli including angiotensin II delivery, aortic banding, isoproterenol administration, and swimming exercise.24 HSP70 also interacts directly with histone deacetylase2 (HDAC2) and is necessary for HDAC2-dependent hypertrophy.33 However, few studies exist on the role of PTMs on HSP70 chaperone function.
2.3 Small heat shock proteins
HSP22, HSP27 and αB-crystallin (CryAB) are small HSPs that respond to stress. CryAB functions as a chaperone for desmin and perhaps myofibrillar proteins. Mutations in CryAB result in desmin-related myopathies, characterized by protein misfolding and aggregation, suggesting that CryAB chaperone-like activity plays an important role in maintaining cardiac structure and function. CryAB may also play a protective role through inhibition of cardiac hypertrophic signaling in response to pressure-overload.20 CryAB is phosphorylated at three residues (Ser19, Ser45 and Ser59) with stress. A phospho-mutant CryABS19A, S45A, S59E that mimics phosphorylation of CryAB at residue 59 protects against osmotic and ischemic stresses and stabilizes mitochondrial membrane potential, in vitro.34, 35 Thus phosphorylation may affect the chaperone function and cardioprotective actions of CryAB.
HSP27 expression protects against doxorubicin and LPS-induced cardiac dysfunction.36, 37 HSP27 is phosphorylated at three residues (Ser15, Ser78 and Ser82) in response to ischemic models. Phospho-mutant, HSP27 S15A, S78A, S82A transgenic mice is protective against I/R injury.38 Therefore phosphorylation may inhibit HSP27 chaperone-like activity. PQC is regulated by diverse signals acting on chaperones through a host of PTMs to regulate proteostasis. Better understanding of the mechanisms regulating chaperone function may lead to therapies to counter cardiac pathology.
3. PTMs Regulate the UPS
At multiple levels, PTMs regulate UPS-mediated proteolysis. First, ubiquitination is by itself a PTM crucial for targeting substrate proteins for degradation by the proteasome as well as for many nonproteolytic fates of the substrate; second, for most substrate proteins, other PTMs are required for their ubiquitination; third, PTMs regulate the process of ubiquitination by acting on Ub ligases; and finally the assembly and activity of the proteasome are heavily regulated by PTMs.
3.1 Ubiquitination regulates both stability and activity of substrate proteins
Ubiquitination regulates not only the stability, but also the localization and function of a substrate protein, depending on the length and topology of the Ub moieties. A chain of 4 or more Ubs is often required to target the substrate protein for proteasomal degradation. The 7 lysine residues (K6, K11, K27, K29, K33, K48, K63) of Ub can all be utilized to form polyubiquitin chains but K48 and K63 are the most commonly utilized two. K48-linked polyubiquitination usually targets the modified proteins for proteasomal degradation whereas K63-linked polyubiquitination often regulates the subcellular distribution, partner protein binding, and/or the activity of the modified protein.6 For example, TRAF6-mediated K63-linked polyubiquitination of Akt (protein kinase B) promotes Akt membrane translocation and activation.39 Unlike polyubiquitination, monoubiquitination usually does not trigger degradation but rather signals for non-proteolytic processes, such as signal transduction and transcription regulation. For instance, monoubiquitination of Smad4 at K519 inhibits Smad4 by impeding interaction with phospo-Smad2, playing an important role in TGFβ signaling.40
3.2 Regulation of ubiquitination by other PTMs
The initiation of ubiquitination obviously requires direct interaction of the substrate with an active specific Ub E3. The signal in a substrate protein to trigger ubiquitination is named a degron. A degron does not seem to be a single consensus segment of primary sequence but rather a conformational signal that mediates the binding of Ub E3 to the substrate. The degron of a native protein often is immature or inaccessible to its specific Ub E3. PTMs of substrates and/or E3 ligases are usually required to trigger ubiquitination. A misfolded or otherwise damaged protein may trigger ubiquitination by exposing hydrophobicity, exposing a buried degron, or generating a new degron.41
Ubiquitination can be regulated by other PTMs. This is best exemplified by cross-talk between phosphorylation and ubiquitination. Phosphorylation can regulate ubiquitination in at least three ways. First, phosphorylation of the substrate produces a phosphodegron which is a short motif that contains phosphorylated residues and mediates phosphorylation-dependent recognition by a Ub E3. Second, phosphorylation of the substrate brings it to the same subcellular location as its specific Ub E3. Third, phosphorylation of E3s modulates Ub ligase activity.
3.2.1 PTMs regulate E3 ligase activities
The specificity of ubiquitination is conferred by the Ub E3 which recognizes and specifically binds a mature degron on the substrate protein. An E3 enzyme harbors the HECT, the RING, or the U-box domain. A Ub E3 can be a single protein (e.g., HECT E3) but it is more often composed of multiple proteins. A number of E3s have recently been identified to target misfolded proteins for proteasomal degradation, functioning in ERAD, ribosome-associated PQC, or PQC in the mitochondrial, cytosolic, and nuclear compartments.42
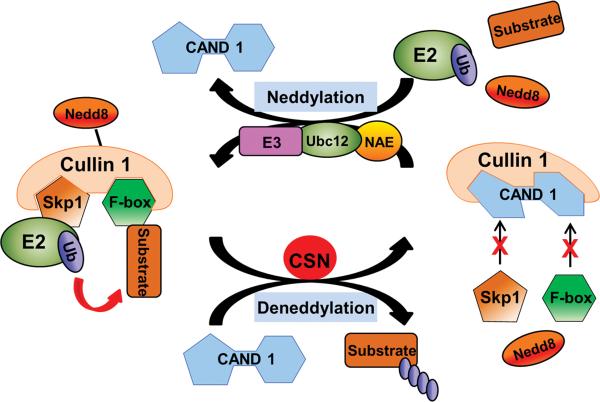
PTMs can regulate Ub E3s via many mechanisms, including altering their assembly, solubility, stability, subcellular localization, or binding affinity towards E2 enzymes and/or substrates. Cullin-RING ligases (CRLs) are the largest known class of Ub ligases. As exemplified by the SCF (Skp1-Cullin1-Fbox) E3 complex (Figure 2), a CRL usually consists of a scaffolding cullin, an E2-interacting RING protein, and a substrate-recognizing subunit (e.g., an F-box protein). Utilizing various substrate-specific adaptors and a specific cullin, CRLs control the ubiquitination and thus the stability of a wide spectrum of substrate proteins.43 The assembly of a functional CRL is triggered by the conjugation of NEDD8 to the cullin family proteins via neddylation, a ubiquitination-analogous process that uses distinct NEDD8 E1-E2-E3 enzymes.44 The cullin neddylation induces conformational changes in cullins, which displaces CAND1 (cullin-associated NEDD8-dissociated protein 1) and brings the Ub-charged E2 and substrates to a close proximity to facilitate the transfer of Ub from the E2 to the substrate.43 The conjugated NEDD8 can be removed by a deneddylating enzyme, such as the COP9 signalosome (CSN) and SENP8/NEDP1. The cycling of cullin neddylation and deneddylation is essential to the catalytic dynamics of CRLs (Figure 2). Disruption of the cycling, for instance by inactivating the CSN, resulted in auto-ubiquitination and subsequent destabilization of the CRL components.45
Figure 2. A prevalent model for the regulation of cullin RING ligases (CRLs) by neddylation-deneddylation.
Analogous to ubiquitination, neddylation covalently attaches NEDD8 to a lysine residue of a target protein (e.g., cullin). It is catalyzed by NEDD8 activating enzyme (NAE), conjugating enzyme (Ubc12), and presumably specific ligases (NEDD8 E3). As illustrated in the SCF (Skp1-Cullin1-Fbox) ubiquitin (Ub) ligase, cullin neddylation displaces CAND1 (cullin-associated NEDD8-dissociated protein 1), which triggers the assembly of an active CRL complex and brings the adaptor-bound substrate to a close proximity to Ub-charged E2 and allows efficient transfer of the Ub from E2 to the substrate. Deneddylation counters neddylation and is done by deneddylases. The COP9 signalosome (CSN) is the deneddylase responsible for cullin deneddylation. Cullin deneddylation triggers the disassembly of the CRL-substrate complex, releases ubiquitinated substrates, and recycles NEDD8.
An intact NEDD8 modification system is essential to embryonic development. Genetic deletion of a key component of this pathway all led to embryonic lethality in multiple model organisms including mice.44 The importance of neddylation in cardiac PQC is recently revealed by the studies on the CSN. The CSN comprises of 8 distinct subunits (CSN1 through CSN8). CSN-mediated deneddylation requires the CSN holocomplex composed of all 8 subunits. Cardiac-specific ablation of the CSN8 gene (CSN8KO) in mice accumulated the neddylated forms of cullins and non-cullin proteins and impaired UPS proteolytic function in the heart.46 CSN8KO resulted in cardiac hypertrophy, which quickly progressed to lethal dilated cardiomyopathy in mice.46 It remains to be elucidated how CSN8KO compromises the UPS function. Interestingly, the accumulation of ubiquitinated proteins and elevated proteasomal activities coexisted in CSN8KO hearts, implying a possible defect in the delivery of the ubiquitinated substrates to the proteasome.
The mammalian protein CHIP is a U-box domain-containing cytosolic PQC E3 ligase that facilitates the degradation of misfolded proteins with the help of chaperones HSP70 or HSP90.47 In the heart, CHIP has a strong cardioprotective effect, as demonstrated by inhibition of apoptosis following I/R injury,48 angiotensin II-induced cardiac remodeling,49 and myocardial infarction.50 It was recently found that monoubiquitination of CHIP by an E2 enzyme Ube2w promotes the interaction of CHIP with Ataxin-3, a deubiquitinating enzyme (DUB). CHIP-associated Ataxin-3 promotes the efficacy of Ub labeling on CHIP substrates, probably by preventing the lengthy Ub chain extension on CHIP substrates. Once the substrates have been polyubiquitinated, Ataxin-3 deubiquitinates Ub-CHIP, leading to the termination of CHIP-mediated substrate ubiquitination.51 Therefore, modulation of mono-ubiquitination of CHIP by the paring of Ube2w and Ataxin-3 regulates the transition of the E3 ligase activity between active and inactive state.
Parkin is a well-characterized RING Ub ligase. Mutations of the Parkin gene cause a common familial form of Parkinson's disease (PD), likely due to Parkin loss of function. Parkin plays a crucial role in mitochondrial quality control (see Section 5). Many PTMs modify Parkin Ub ligase activity. For example, S-nitrosylation of Parkin may contribute to pathogenesis of PD by inhibiting Parkin Ub ligase activity likely via increasing autoubiquitination of Parkin.52, 53 Neddylation of Parkin enhances its interaction with Ub E2 as well as its substrates, increasing Parkin Ub ligase activity.54, 55 Although loss of Parkin function has been associated with neuronal and mitochondrial dysfunction,56, 57 the role of Parkin in the heart has just begun to be unveiled.58
3.2.2 PTMs of substrates regulate recognition and binding of Ub E3 ligases
The discovery that phosphorylation of a substrate protein can create a phosphodegron represents a major insight into inducible UPS-mediated protein degradation. IκB (inhibitor of NFκB) is the best example. Phosphorylation of Ser32 and Ser36 of IκB by IκB kinase (IKK) generates a phosphodegron (DpSGLDpS) that is recognized by F-box protein β-TrCP in the SCF type of Ub E3 complex and targets IκB for ubiquitination and degradation, playing a critical role in NFκB activation.59 A second scenario is that phosphorylation of a residue outside of the degron induces a conformational change that exposes a degron. For instance, Ser345 phosphorylation exposes a phosphate-free degron in the regulatory region of Chk1faciliates its degradation by the UPS.60 Instead of promoting ubiquitination, phosphorylation can also prevent a substrate from being recognized by its E3.59
Besides phosphorylation, many other PTMs may also regulate ubiquitination at the substrate level. For example, acetylation or sumoylation may compete with ubiquitination for the same lysine residue in the substrate proteins. Cross-linking and conformational changes induced by oxidation may conceivably create degrons de novo and thereby activate UPS-mediated degradation of oxidized proteins.41
Spatial separation of Ub E3 ligases from their substrates can be a mechanism by which ubiquitination is regulated. Hence, PTMs may promote ubiquitination by bringing substrates and specific E3 ligases into the same subcellular location. A good example is p27Kip1. Phosphorylation of Ser10 in p27Kip1 triggers its translocation from the nucleus to cytoplasm where p27Kip1 is degraded by the UPS.61
3.3 Deubiquitination
Ubiquitination is countered by deubiquitination. The latter removes Ub from ubiquitinated proteins by DUBs. A search for DUB-specific domains in the human genome identified ~100 putative DUB-encoding genes. DUBs are Ub proteases, with the majority belonging to the cysteine family of proteases and the rest being metalloproteases. DUBs of the cysteine family are further classified into 4 sub-families, based on their Ub-protease domains: ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), Otubain protease (OTU), and Machado-Joseph disease protease (MJD). DUBs of the metalloprotease family all have a JAMM (JAB1/MPN/Mov34 metalloenzyme) domain. The function, targets, and regulation of most DUBs are not understood. No reported studies have examined the role of DUBs in cardiac PQC; however, studies from other cells/organs have demonstrated an important role in (patho)physiology.
DUBs appear to pair with E3 ligases with a relative specificity to regulate ubiquitination,62 analogous to the kinase-phosphatase pairing in modulating phosphorylation. From the point of view of PQC, the DUB-E3 pairing between Ataxin-3 and CHIP (see also Section 3.2) or Parkin is of particular interest, as polyglutamine (polyQ) track expansion of Ataxin-3 is linked to human Machado-Joseph disease (MJD) and Parkin mutations cause PD. MJD and PD are closely related neurodegenerative disease with overlapping clinical and pathological features. A major pathogenic role of impaired PQC has been suggested in most neurodegenerative diseases, including MJD, PD, and Huntington's disease. Ataxin-3 is a DUB of the MJD subfamily. Ataxin-3 interacts directly with Parkin and suppresses Parkin autoubiquitination via likely impeding the efficient charging of the E2 with Ub.63 Interestingly, PolyQ-expanded Ataxin-3 promotes degradation of Parkin via autophagy,64 which helps explain why Parkin protein levels are decreased in the brain of transgenic mice overexpressing polyQ-expanded but not wild type Ataxin-3.64
There are at least three DUBs associated with the mammalian 19S proteasome: Rpn11/POH1, Ubp6/USP14, and UCH37. POH1, a JAMM containing DUB, is a stoichiometric subunit of the 19S proteasome. POH1 is responsible for the removal of the Ub chain and Ub recycling during proteasome-mediated degradation of polyubiquitinated proteins. POH1 removes the Ub chain en bloc by cutting at the base of the Ub chain, frees the substrate for translocation into the 20S proteasome. Hence, inhibition of POH1 would non-specifically suppress proteasome-mediated degradation of polyubiquitinated proteins. In contrast, USP14 and UCH37 trim the Ub chain from its tip distal to the substrate, therefore shortening the chain rather than removing the chain en bloc. This Ub editing function of DUBs might provide an additional layer of regulation on proteasomal degradation of ubiquitinated proteins. A small chemical compound (IU1) capable of inhibiting USP14 deubiquitination was recently described for its enhancement of proteasome mediated degradation of some substrates in cultured cells.65 This represents the discovery of the first pharmacological proteasome activator. It will be important to test whether IU1 or alike can enhance proteasome function in animals, and whether IU1 can effectively treat disease with PFI.
3.4 PTMs regulate the proteasome
It is now clear that PTMs regulate both assembly and activities of the proteasome. The identification of PTMs on the proteasome has been greatly expanded by recent advances in functional proteomics.3 In addition to proteolytic cleavage, at least 11 different types of PTMs on proteasome subunits are identified, including phosphorylation, acetylation, sumoylation, ubiquitination, HNE modification, oxidation, glycosylation, poly-ADP ribosylation, O-GlcNAc modification, nitrosylation, and N-myristoylation.3 Phosphorylation at specific amino acid residues has been identified in many subunits of the 20S proteasome isolated from the heart using a combination of elegant proteomic approaches.66 The biological effect of these phosphorylation sites and the kinases and phosphatases that regulate these sites remain largely undetermined. Nonetheless, the significance of at least some PTMs has been suggested in cardiac proteasomes by recent studies.
3.4.1 N-terminal cleavage during proteasome assembly
All three peptidase subunits (β5, β2, and β1) of the 20S proteasome are synthesized as a precursor form in the cell (i.e., pre-β5, pre-β2, and pre-β1). The N-terminal proteolytic processing of these precursors is required to expose their catalytic N-terminal Threonine (e.g., T76 of pre-β5 in yeast, T60 of pre-β5 in mice) and to generate the catalytically competent form of these subunits during the final step of 20S proteasome assembly where two hemi-proteasomes merge to form a full 20S proteasome. The functional significance of this N-terminal processing of pre-β5 has recently been demonstrated in cardiomyocytes and mouse hearts. Transgenic expression of a T60A mutant pre-β5 in cardiomyocytes dose-dependently inhibited proteasome chymotrypsin-like activity (the activity normally arises from β5) and the overall proteasome proteolytic function.18
3.4.2 Phosphorylation regulates proteasome assembly and activities
Cyclic AMP-dependent protein kinase (PKA) mediates β-adrenergic stimulation on the heart, in which phosphorylation on calcium handling and myofilament proteins by PKA impacts profoundly on cardiac function and cardiomyocyte growth. Interestingly, PKA and protein phosphatase 2A (PP2A) can be co-purified with cardiac 20S proteasomes.67 Further in vitro testing showed that PKA is capable of phosphorylating multiple subunits of the 20S and increasing proteasome activities.67 Subsequently, a study using nuclear proteasomes from MDA-486 cells demonstrates that PKA phosphorylates Ser120 of Rpt6, a 19S proteasome subunit, and this phosphorylation fully accounts for the stimulating effect of PKA on proteasome function in these cells.68 The pathophysiological significance of PKA-mediated regulation on cardiac proteasomes is illustrated by two more recent reports. Asai et al. reported that PKA activation by either β-adrenergic stimulation of the heart or ischemic preconditioning (IPC) rapidly increases the assembly and activity of the 26S proteasome in canine hearts and the increased proteasome function may contribute to IPC-triggered reduction of total ubiquitinated proteins in I/R hearts.69 In a mouse cardiac hypertrophy model induced by chronic infusion of isoproterenol, Drews et al. showed that the activity and assembly of the 26S proteasome are increased but the caspase-like and trypsin-like activities of the 20S proteasome are depressed in the hypertrophic hearts. More importantly, they showed that the depressed 20S proteasome activity can be restored in vitro by activating endogenous PKA.70
Calcium/calmodulin dependent kinase II (CAMKII) regulates proteasome function in neuronal cells. In cultured hippocampal neurons, the positive regulation of proteasome function by neuronal activity depends on CAMKII activity. Overexpression of a constitutive active CAMKII stimulated the degradation of a reporter protein (GFPu) in HEK cells and stimulates 26S proteasome activity in hippocampal neurons. CAMKII can directly phosphorylate Rpt6 in vitro.71 More recently, it was shown that CAMKIIα phosphorylates Ser120 of Rpt6 and enhances proteasome activity. This study also reveals that Ser286 autophosphorylated CAMKIIs serves as a scaffold to recruit proteasomes to dendritic spines, promoting synaptic activity-induced redistribution of the proteasome in hippocampal neurons.72 Notably, CAMKII activation has been shown to simultaneously affect mechanical and electrical properties of cardiomyocytes and may play a role in the pathogenesis of arrhythmia and CHF.73 It will be interesting to determine whether CAMKII regulates proteasome function in cardiomyocytes.
It has been shown in animal cells (e.g., COS-7) that casein kinase II (CK-II) can phosphorylate α7 subunit of the 20S proteasome at Ser243 and Ser250. This phosphorylation appears to be constitutive and show no effect on the peptidase activity but it increases the stability of the 20S-19S proteasome complex. A decrease in α7 phosphorylation during interferon γ stimulation seems to destabilize the 26S proteasome, allowing more 20S proteasomes to form complexes with 11S proteasomes. 74 Notably, upregulation of 11S proteasomes by overexpression of PA28α, likely via increasing 11S association with the 20S, facilitates the removal of misfolded proteins in cardiomyocytes and protects cardiomyocytes from oxidative and proteotoxic stresses.2, 75
PKN, also referred to as protein kinase C–related kinase 1 (PRK1), is a stress-activated serine/threonine kinase. PKN is activated during myocardial I/R and plays a protective role against I/R injury. The protection appears to be mediated by improving PQC, specifically by increasing CryAB phosphorylation and cytoskeletal translocation as well as by increasing proteasome activity.76 It remains to be determined whether PKN directly phosphorylates proteasomes.
3.4.3 Ubiquitination of proteasome subunits
Rpn10/S5a is a 19S proteasome subunit but also exists free in the cytosol. Through its two Ub interacting motifs (UIMs), Rpn10/S5a binds ubiquitinated proteins. It is believed to be a main intraproteasomal receptor for polyubiquitinated proteins. It was found that S5a can be polyubiquitinated by all E3 ligases tested and rapidly degraded.77 More recently, monoubiquitination at K84 of yeast Rpn10 has been reported.78 Rpn10 monoubiquitination is regulated by Ub E3 Rsp5 and DUB Ubp2. K84-monubiquitinated Rpn10 shows reduced affinity to polyubiquitinated proteins and decreased proteasome proteolytic function. Interestingly, Rpn10 monoubiquitination decreases upon proteolytic stress.78 These lines of evidence support a biologically relevant role of Rpn10 monoubiquitination in regulating proteasome function. Ubiquitination on several other proteasome subunits has also been identified in various non-cardiac cell types but their biological function is unknown.3
3.4.5 Oxidative modifications of the proteasome
Many pathological conditions increase oxidative stress in the heart. The most common one is myocardial I/R which can occur in both the natural course and therapeutic intervention of ischemic heart disease (IHD), open heart surgery, and heart transplantation. It has been demonstrated that cardiac proteasome function is depressed by I/R and this depression plays an important role in I/R injury.2, 18, 79, 80 Hence, it is important to determine the mechanism underlying proteasome impairment during I/R. Reperfusion of ischemic myocardium results in an increase in production of highly reactive free radicals which can cause oxidative modifications to cellular components and damage them. Indeed, analyses of 20S proteasomes purified from I/R myocardium revealed that the lipid peroxidation product 4-hydroxy-2-nonenal (HNE) selectively modifies α1, α2 and α4 subunits of the 20S.79 More recently, oxidative modification of Rpt3 and Rpt5 is associated with impaired proteasome activity in I/R myocardium. IPC attenuates the modification and proteasome impairment as it diminishes I/R injury, suggesting that protection against oxidative damage to the 19S proteasome is a potential mechanism for IPC to reduce I/R injury.80 The 26S proteasome activities in crude myocardial protein extracts were found significantly decreased in the failing human hearts compared with non-failing controls. The reduction of proteasome activity was accompanied by oxidative modification of Rpt5 in the failing human hearts.81 The sufficiency of oxidative modifications to decrease proteasome function has been suggested by studies showing that exposure of purified proteasomes to oxidants, such as hydrogen peroxide and peroxynitrite, inactivates the proteasome.82-84
It has been shown that oxidized proteins can be degraded by the 20S proteasome in a Ub and ATP–independent manner.85 The 20S is more resistant to oxidative damages than the 26S.82 In addition to oxidative modification of proteasome subunits, oxidized proteins as the substrate can impair proteasome function.86 Conversely, proteasome inhibition can induce oxidative stress.87 Hence, oxidative stress and proteasome malfunction may form a vicious cycle.
4. PTMs Regulate Macroautophagy
Macroautophagy is upregulated with cellular stress, providing an alternative energy source, recycling macromolecules from digested substrates, and heightening intracellular QC. The structures involved in macroautophagic degradation are well defined. The initial step in macroautophagy is the formation and elongation of an isolation membrane, which encompasses cytoplasmic material for degradation. Once structures are completely engulfed by a double-membrane vesicle, they are called autophagosomes. Autophagosomes travel down microtubules and may fuse with vesicular structures to form amphisomes. Autophagosomes and amphisomes then fuse with primary lysosomes, dumping their contents into the lumen of the autolysosome full of hydrolytic enzymes, which degrades the delivered cargo.
Macroautophagy was shown to be an essential process in mouse development, when beclin 1-/- mice died early in embryogenesis.88 Subsequent loss of autophagy models demonstrated that macroautophagy was a critical process to survive starvation, atg5-/- and atg7-/- mice survived until birth, and then died during the neonatal starvation period, due to autophagic deficiency.89, 90 Atg7-null mice revealed that macroautophagy played a necessary role in PQC, when newborns had organs containing Ub-positive inclusions.90 Since then, numerous studies have tried to define the role of macroautophagy in the heart.
4.1 Beclin 1
Beclin 1 is a component of the Class III PI3K complex of autophagosome nucleation, along with Vps34 and p150, responsible for isolation membrane formation.91 To circumvent the embryonic lethality of beclin 1-/- mice, heterozygous (beclin 1+/-) mice were used to study the loss of beclin 1 effects in the heart. Hearts from beclin 1+/- mice showed reduced autophagosome levels with starvation and with pressure-overload conditions which showed diminished of systolic function after 3 weeks of banding.92 Cardiac-specific beclin 1 transgenic mice were created to study beclin 1 overexpression in the heart.92 Hearts overexpressing beclin 1 showed increased autophagosome levels with starvation and pressure-overload.92 However, aortic banding led to cardiac malfunction, ventricular hypertrophy and chamber dilation in beclin 1 transgenic hearts, relative to controls.92 These paradoxical results led to the hypothesis that beclin 1-dependent macroautophagy may function across a continuum where too little or too much may be detrimental.93
Beclin 1 protein levels were upregulated in hearts during reperfusion, following ischemia. To evaluate the role of beclin 1 with ischemic stress, beclin 1+/- mice were subjected to I/R injury.94 Hearts from beclin 1+/- mice developed smaller infarct sizes than wild-type mice, demonstrating the loss of beclin 1 function was cardioprotective against I/R injury.94
Many types of stress are associated with the accumulation of ubiquitinated, misfolded, and aggregated proteins, including pressure-overload stress.95 These accumulations may be due either to insufficient upregulation of proteasome function or inadequate autophagy. To define the role of beclin 1-dependent macroautophagy in PQC, beclin 1+/- mice were crossed mutant (R120G) human αB-crystallin (CryAB) mice. The resulting hCryABR120G/beclin 1+/- mice developed heart failure prematurely and accumulated more ubiquitinated proteins than hCryABR120G expressing hearts.96 Therefore the loss of beclin 1 exacerbates proteotoxic pathology and cardiac dysfunction in hCryABR120G cardiomyopathy.
Simple overexpression of beclin 1 may not induce macroautophagy. Beclin 1 interacts with a host co-factors (ATG14L, UVRAG, BIF-1, Rubicon, Ambra1, HMGB1, nPIST, VMP1, SLAM, IP3R, PINK, survivin, etc.), whose interactions can modulate macroautophagy.97 Beclin 1 can be phosphorylated by death associated protein kinase (DAPK), on Thr119, which causes the dissociation of BCL-2, permitting macroautophagy.98 Beclin 1 is also ubiquitinated (Lys117) by tumor necrosis factor receptor-associated factor 6 (TRAF6), which is negative regulated by the DUB A20.99 Future work will need to define the dominant post-translational modifications and interaction partners responsible for activating/inactivating beclin 1-dependent macroautophagy in the heart.
4.2 BCL-2
Beclin 1 was identified in cells through its interaction with BCL-2. Both BCL-2 and BCL-XL inhibit beclin 1-dependent macroautophagy by sequestering beclin 1 away from the autophagy nucleation complex.100, 101 Phosphorylation of BCL2 (Thr69, Ser70, Ser87) by JNK1 causes BCL-2 dissociation from beclin1, permitting association with the PI3K complex and macroautophagy.102 High mobility group box 1 (HMGB1) also promotes the phosphorylation of BCL-2 by extracellular signal-regulated kinase (ERK1/2), activating starvation-induced macroautophagy.103
Autophagosome levels in cardiac and skeletal muscle increase with exercise duration.104 Knock-in mice with bcl-2 phosphorylation sites mutated (Thr69Ala, Ser70Ala and Ser84Ala; bcl-2 AAA mice) were used to define the role of beclin 1-dependent macroautophagy with exercise.104 Skeletal and cardiac muscles from bcl-2 AAA mice were unable to induce autophagosome levels with starvation or treadmill exercise.104 The bcl-2 AAA mice were exercise intolerant, with impaired glucose metabolism.104 Thus BCL-2 phosphorylation is required for beclin 1-adaptation to exercise.
4.3 AMPK
AMP-dependent protein kinase (AMPK) is activated with low ATP or cell stress to activate macroautophagy. AMPK phosphorylates TSC2 (tuberous sclerosis complex-2) and raptor, triggering the inhibition of mTORC1.105, 106 Inhibition of mTORC1 stimulates autophagosome synthesis. AMPK also interacts with and phosphorylates ULK1 (Ser317, Ser467, Ser555, Ser574, Ser637, Ser722, Ser757, Ser777, Ser799).107-109 ULK1 (Atg1) forms a kinase complex with ATG13, ATG101 and FIP200 responsible for activating macroautophagy under nutrient-deficient conditions. The ULK1 complex is activated when phosphorylated by AMPK and inactivated by mTORC1-dependent phosphorylation. AMPK also stimulates the nuclear accumulation of FOXO3, increasing macroautophagic gene expression.110, 111
Ischemic conditions increase AMPK phosphorylation (Thr172) and autophagosome levels.94 Transgenic mice expressing dominant negative AMPK fail to increase autophagosome levels with ischemia and develop larger infarct sizes than wild-type mice.112 Thus AMPK stimulates autophagosomal content in response to ischemia and starvation.
4.4 ATG5 and ATG7
ATG5 is a necessary component for macroautophagy. When conjugated to ATG12 it participates in isolation membrane maturation. ATG5 is necessary for macroautophagy in the adult mouse heart. Conditional atg5-deficient mice, caused a loss of cardiac function, dilation, and accumulation of ubiquitinated proteins with pressure-overload, suggesting that macroautophagy may be cardioprotective.113 The lack of sufficiency of Atg5 to increase autophagic function, has limited its further study.
ATG7 is a key regulator of autophagosome formation, it serves two necessary enzymatic functions: in conjugating ATG12 to ATG5 and in adding a phosphatidylethanolamine to ATG8 (LC3).114 Similar to the loss of atg5 in vivo, knockdown of atg7 led to cardiomyocyte hypertrophy in vitro.113Atg7 overexpression in cardiomyocytes is sufficient to increase autophagic flux, in fed cells.115 ATG7-stimulated macroautophagy successfully reduces misfolded protein and aggregate accumulation in cardiomyocytes overexpressing CryABR120G,115 a misfolded protein linked to DRC. Atg7 overexpression has also been shown to rescue autophagic deficiency in other models, in vitro.116, 117 ATG7 is acetylated by p300 with starvation.118 The p300 acetyltransferase also acetylates ATG4, ATG8 (LC3), and ATG12, which is associated with decreased autophagosome content.118 Other work shows that NAD+ also facilitates the acetylation of ATG7, ATG5 and ATG8 (LC3) which can be deacetylated by SIRT1.119
4.5 SIRT1
SIRT1 is a NAD-dependent deacetylase that is upregulated with starvation.120 SIRT1 acts as a sensor to detect insufficient nutrition and triggers physiological changes linked to health and longevity.120 SIRT1 functions to deacetylate ATG5, ATG7 and ATG8 (LC3).119 SIRT1 also deacetylates FOXO proteins, altering FOXO-dependent expression of macroautophagy genes.121 SIRT1 deacetylation of FOXO1 increases autophagosome levels in glucose-deprived cardiomyocytes.122
To establish the role of Sirt1 in vivo, cardiac-specific sirt1-/- mice were exposed to I/R and sirt1-/-hearts developed larger infarctions than wild-types.123 Conversely, Sirt1-overexpression mice developed smaller infarct sizes than controls following I/R.123 Thus, SIRT1 is cardioprotective against I/R injury, though more work is needed to define whether FOXO1 acetylation and/or macroautophagic function is required.
4.6 FOXO1/FOXO3
FOXO transcription factors function in many biological processes, including regulation of the expression of ATG genes. FOXO1/FOXO3 are subject to multiple PTMs, including phosphorylation, ubiquitination and acetylation.121 FOXO1 acetylation is modulated by the balance of histone acetylases and deacetylases. Mutation of three FOXO1 lysine residues to arginine (Lys262Arg, Lys265Arg, and Lys274Arg) showed that acetylation of FOXO1 was required for macroautophagy. Acetylation of FOXO1 is required for interaction with ATG7.124 However, whether acetylated FOXO1 interaction with ATG7 is required for macroautophagy activation remains to be defined.
FOXO3 expression activates LC3 and Bnip3 expression and increases autophagosome levels in skeletal muscle cells.111 Overexpression of constitutively active FoxO3 (FoxO3-CA) in the heart causes a 25% reduction in heart mass.125 However, FoxO3-CA mice were unable to counter the pathological hypertrophy from pressure-overload.125 The mechanisms of FOXO PTMs to activate autophagy-related genes remain to be defined.121
4.7 The COP9 signalosome regulates autophagy in the heart
Neddylation appears to be involved in regulating autophagy. Disruption of deneddylation by CSN8KO in mouse hearts caused accumulation of autophagosomes and reduced autophagy flux, indicating compromised autophagic degradation.126 The autophagic dysfunction was at least partially due to a defect in the fusion of autophagosomes with lysosomes. Downregulation of Rab7 in CSN8KO hearts was likely attributed to the defect.126 Given the critical role of the CSN in regulating both the UPS and autophagy, an intact NEDD8 system may be essential to cardiac PQC. Interestingly, suppression of neddylation by MLN4924, a potent inhibitor of NEDD8 activating enzyme (NAE),127 activated autophagy in cancer cells, which appeared to be a survival response.128 MLN4924-induced autophagy was accompanied by accumulation of Deptor, an inhibitor of mTOR, and attenuated by down-regulation of Deptor, indicative of involvement of mTOR signaling. Hence, fine-tuned neddylation and deneddylation may regulate macroautophagy, warranting further investigations.
5. UPS-Autophagy Crosstalk
It becomes increasingly apparent that the UPS and autophagy cross-talk to each other and this cross-talk plays an important role in intracellular QC. The cross-talk is best exemplified by ubiquitination-triggered autophagic removal of protein aggregates (aggrephagy) or defective mitochondria (mitophagy).
Polyubiquitination linked by any lysine other than K63 can target a protein for proteasomal degradation in yeast.129 K63-linked ubiquitination was shown to promote the formation and autophagic removal of aggresomes from misfolded proteins.130 p62/SQSTM-1 may mediate the aggregation and autophagic targeting of polyubiquitinated proteins. Through its C-terminal Ub associating domain (UBA), p62 binds polyubiquitin chains, preferentially K63-linked Ub chain. p62 polymerizes via its N-terminal PB1 domain. p62 also harbors a LC3 interacting region (LIR), which may help recruit the LC3-positive phagophore and promote autophagosome formation.131 The function of p62 in selective autophagy is regulated by its phosphorylation status. Phosphorylation at Ser403 in p62 UBA domain mediated by CK-II increases p62 binding to polyubiquitin chains, enhancing inclusion body formation and efficient autophagic degradation of the polyubiquitinated proteins.132 Remarkable upregulation of p62 transcript and protein levels as well as macroautophagy were observed in mouse hearts overexpressing misfolded proteins.133 In cultured cardiomyocytes overexpressing misfolded proteins, p62 knockdown by small interference RNA decreases aggresome formation, autophagosome marker LC3-II, and cell survival.133 In macroautophagy impaired mice, p62 mediates the formation of Ub-positive inclusion body but its impact on cell survival appears to be cell type-dependent.134 Additionally, p62 seems to play a role in mediating autophagic activation by PFI and in chronic autophagic inhibition induced impairment of degradation of ubiquitinated proteins by the proteasome.135 The role of p62 in cardiac PQC remains to be investigated in vivo.
Given that defective mitochondria can endanger the cell by, for example, increasing production of reactive oxygen species and releasing pro-cell death factors, timely removal defective mitochondria is an integral part of cytoplasmic QC. The removal of damaged mitochondria is performed primarily by mitophagy. Mounting evidence suggests a critical role of the UPS in triggering mitophagy. In the best characterized pathway for mitophagy, mitophagy is mediated by PTEN-induced putative protein kinase 1 (PINK1) and E3 ligase Parkin. PINK1 accumulated on damaged mitochondria recruits cytosolic Parkin to the depolarized mitochondria.136 The mitochondrial Parkin then promotes ubiquitination of mitochondrial proteins. Parkin can catalyze the formation of both K48- and K63-linked Ub chains,137 both of which seem to be able to trigger mitophagy. According to one model, Parkin mediates K48-linked ubiquitination and proteasomal degradation of mitochondrial membrane proteins including VDAC1, Mfn1/2, and other proteins,138 which may subsequently facilitates mitochondrial fragmentation, triggering mitophagy.139 Supporting this model, removal of damaged mitochondria is blocked by proteasome inhibition.139, 140 A second model posits that Parkin mediated K63-linked polyubiquitination of mitochondrial substrate(s) recruits p62. p62 then mediates the clustering of damaged mitochondria and subsequent mitophagy in a manner analogous to the aggregation of polyubiquitinated proteins and aggrephagy. However, a recent study shows that p62 is required for Parkin-triggered mitochondrial clustering but is dispensable for mitophagy.141 There is also reported evidence that PINK1 and Parkin may directly recruit autophagic machinery to the mitochondria.137 A critical role for Parkin and p62 mediated mitophagy in cardiac protection by IPC is recently shown by Huang et al.58
6. Conclusion Remarks
Existing evidence strongly indicates that PQC becomes inadequate during the progression of many forms of heart disease to CHF. With virtually no exception, it remains unknown when PQC inadequacy occurs and what is primarily responsible for the inadequacy in a given disease. For instance, defect in the delivery of ubiquitinated proteins to proteolytic chambers is associated with PQC inadequacy in the heart of DRC mouse models.142 It will be important to determine whether the defect is causative to PQC inadequacy in the DRC hearts and if so, what its molecular basis is.
It is clear that PTMs to either misfolded proteins or the various components of QC machinery including chaperones, the UPS, and autophagy, impact profoundly on intracellular QC. However, in most cases, the molecular pathways that regulate these PTMs and/or the significance of the PTMs remain to be defined. Given that a major pathogenic role of inadequate QC in cardiac pathology has been demonstrated, there is a highly compelling rationale to improve our understanding of the regulation of intracellular QC, including the role of PTMs in PQC. Recent advancement in the field of proteomics has made it possible to globally and quantitatively characterize simultaneous changes in PTMs on multiple subunits of the same protein complex at a given condition.3 This is expected to help identify new PTMs and facilitate the quantification of PTM changes in both the substrates and the machinery of QC at baseline and during pathological conditions. Site-specific mutagenesis to abolish or mimic a PTM on the specific residue of a modified protein will continue to be an essential tool to help define the impact of the specific PTM on the function and/or fate of the modified protein. It will also be important to determine the functional interaction among different PTMs simultaneously occurred to the target protein. Dissection of the upstream pathways that regulate the PTM will be crucial to identify novel targets or strategies for developing pharmaceutics to improve QC in the cell.
It is anticipated that comprehensive investigations into intracellular QC in cardiac physiology and pathology will give rise to new therapeutics to better battle heart diseases, the leading cause of death of humans. On macroautophagy, there is a good body of evidence supporting that activation of macroautophagy improves PQC and thereby protects the heart;23 nonetheless, excessive macroautophagy upon certain conditions such as reperfusion may be detrimental.94
Some studies, but not others, have shown that pharmacologically induced ubiquitous proteasome inhibition protects against I/R injury and pressure overloaded cardiac hypertrophy.143, 144 However, genetically-induced moderate proteasome inhibition in cardiomyocytes was recently shown to exacerbate acute I/R injury in mice.18 Moreover, administration of bortezomib, a proteasome inhibitor, to multiple myeloma patients can cause reversible heart failure.145 Conversely, recent genetic studies reveal that proteasome function enhancement in the cardiomyocytes of diseased hearts can slow down the progression of a bona fide cardiac proteinopathy and minimizes acute I/R injury in mice.2 Hence, it is envisioned that enhancing proteasome proteolytic function may be a potential new strategy to treat heart diseases with increased proteolytic stress. Unfortunately, only one compound (IU-1) has been shown in cultured non-cardiac cells to enhance proteasome function.65 It is unclear whether IU-1 functions as a proteasome activator in vivo. Therefore, research aiming at facilitating the development of proteasome activators is urgently needed and its success will have great impact.
Table.
Examples of post-translational modifications in intracellular quality control
| Targets | PTMs | Regulating enzymes | Biological Function | References |
|---|---|---|---|---|
| Chaperones | ||||
| HSP90 | Acetylation at K294 | Suppress substrate binding. | 24, 25 | |
| Nitrosylation at C597 | eNOS activation. | 26, 27 | ||
| Phosphorylation | Alter substrate binding. | 30, 31 | ||
| CryAB | Phosphorylation at S59 | MAPKAP-K2 | Inhibit apoptosis | 32, 33 |
| HSP27 | Phosphorylation | Exacerbate I/R injury. | 38 | |
| Ubiquitin ligases | ||||
| CRLs | Neddylation | Facilitate the assembly of CRLs | 43 | |
| CHIP | Mono-ubiquitination | Ube2W/Ataxin-3 | Promote E3 ligase activity. | 51 |
| Parkin | Neddylation | Increase E3 ligase activity. | 54, 55 | |
| UPS substrates | ||||
| IκB | Phosphorylation at S32 and S36. | IKK | Form a phospho-degron. | 59 |
| Chkl | Phosphorylation at S345 | ATR | Expose the degron for ubiquitination. | 60 |
| Oxidized proteins | Oxidative modification | Generate new degrons. | 41 | |
| p27Kip1 | Phosphorylation at S10 | Translocate to cytoplasm for degradation. | 61 | |
| Proteasome subunits | ||||
| 20S subunits | Phosphorylation | PKA/PP2A | Increase proteasome activities. | 67 |
| Rpt6 | Phosphorylation at S120 | PKA/CAMKII | Increase proteasome activities. | 68, 69, 71 |
| α7 | Phosphorylation at S243 and S250. | CK-II | Increase the interaction of 19S-20S. | 74 |
| Rpnl0 | polyubiquitination | Degradation of Rpn10 | ||
| Rpnl0 | Mono-ubiquitination at K84 | Rsp5/Ubp2 | Reduce proteasome proteolytic function. | 78 |
| α1, α2, α4 | Oxidative modification | Impair proteasome activities. | 79 | |
| Rpt3, Rpt5 | Oxidative modification | Impair proteasome activities. | 80 | |
| Autophagy | ||||
| Beclin-1 | Phosphorylation at T119. | DAPK | Dislodge BCL-2 and activate autophagy. | 98 |
| Ubiquitination at K117 | TARF6/A20 | Unknown | 99 | |
| BCL-2 | Phosphorylation at T69, S70, S87 | JNK1/ERK1/2 | Disrupt BCL-2/Beclin-1 interaction; activate autophagy. | 102, 103 |
| ULK1 | Phosphorylation at multiple sites | AMPK | Activate autophagy. | 107-109 |
| ATG7 | Acetylation | p300/SIRT1 | Suppress autophagy | 118, 119 |
MAPKAP-K2, mitogen-activated protein kinase-activated protein kinase 2; ATR, ataxia-telangiectasia- and rad3-related; DAPK, death associated protein kinase.
Acknowledgments
Source of Funding:
This work is in part supported by NIH grants R01HL072166, R01HL085629, and R01HL068936, and American Heart Association grants 0740025N (to X. W.), and 11SDG6960011 (H.S.).
Non-standard abbreviations and non-standard acronyms
- AAA
ATPase associated with diverse cellular activities
- AMPK
AMP-dependent protein kinase
- CAMKII
calcium/calmodulin dependent kinase II
- CHF
congestive heart failure
- CHIP
c-terminus of heat shock protein 70 interacting protein
- CK-II
casein kinase II
- CMA
chaperone-mediated autophagy
- CRLs
Cullin-RING ligases
- CryAB
αB-crystallin
- CSN
the COP9 signalosome
- DRC
desmin-related cardiomyopathy
- DUB
deubiquitinating enzyme
- ER
the endoplasmic reticulum
- HSP
heat shock protein
- I/R
ischemia/reperfusion
- IPC
ischemic preconditioning
- LAMP-2A
lysosome-associated membrane protein 2A
- PD
Parkinson's disease
- PFI
proteasome functional insufficiency
- PKA
cyclic AMP-dependent protein kinase
- PQC
protein quality control
- PTM
posttranslational modification
- QC
quality control
- SCF
Skip1-Cullin1-F-box
- Ub
ubiquitin
- UPS
ubiquitin-proteasome system
Footnotes
Disclosures: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301:H2207–2219. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scruggs SB, Zong NC, Wang D, Stefani E, Ping P. Post-translational modification of cardiac proteasomes: Functional delineation enabled by proteomics. Am J Physiol Heart Circ Physiol. 2012;303:H9–H18. doi: 10.1152/ajpheart.00189.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, Robbins J. Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation. 2008;117:2743–2751. doi: 10.1161/CIRCULATIONAHA.107.750232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 6.Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: The ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res. 2010;106:463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, Hodara R, Fredenburg R, Wu DC, Follenzi A, Dauer W, Przedborski S, Ischiropoulos H, Lansbury PT, Sulzer D, Cuervo AM. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Navarro JA, Kaushik S, Koga H, Dall'Armi C, Shui G, Wenk MR, Di Paolo G, Cuervo AM. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci. 2011;36:254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Lau E, Wang D, Zhang J, Yu H, Lam MP, Liang X, Zong N, Kim TY, Ping P. Substrate- and isoform-specific proteome stability in normal and stressed cardiac mitochondria. Circ Res. 2012;110:1174–1178. doi: 10.1161/CIRCRESAHA.112.268359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLendon PM, Robbins J. Desmin-related cardiomyopathy: An unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Z, Zheng H, Li J, Li YF, Su H, Wang X. Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circ Res. 2012;111:532–542. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumarapeli AR, Wang X. Genetic modification of the heart: Chaperones and the cytoskeleton. J Mol Cell Cardiol. 2004;37:1097–1109. doi: 10.1016/j.yjmcc.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Kumarapeli AR, Su H, Huang W, Tang M, Zheng H, Horak KM, Li M, Wang X. Alpha b-crystallin suppresses pressure overload cardiac hypertrophy. Circ Res. 2008;103:1473–1482. doi: 10.1161/CIRCRESAHA.108.180117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian J, Vafiadaki E, Florea SM, Singh VP, Song W, Lam CK, Wang Y, Yuan Q, Pritchard TJ, Cai W, Haghighi K, Rodriguez P, Wang HS, Sanoudou D, Fan GC, Kranias EG. Small heat shock protein 20 interacts with protein phosphatase-1 and enhances sarcoplasmic reticulum calcium cycling. Circ Res. 2011;108:1429–1438. doi: 10.1161/CIRCRESAHA.110.237644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Przyklenk K, Undyala VV, Wider J, Sala-Mercado JA, Gottlieb RA, Mentzer RM., Jr Acute induction of autophagy as a novel strategy for cardioprotection: Getting to the heart of the matter. Autophagy. 2011;7:432–433. doi: 10.4161/auto.7.4.14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis MS, Patterson C. Hold me tight: Role of the heat shock protein family of chaperones in cardiac disease. Circulation. 2010;122:1740–1751. doi: 10.1161/CIRCULATIONAHA.110.942250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taipale M, Jarosz DF, Lindquist S. Hsp90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 26.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. Hdac6 regulates hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An acetylation site in the middle domain of hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Retzlaff M, Stahl M, Eberl HC, Lagleder S, Beck J, Kessler H, Buchner J. Hsp90 is regulated by a switch point in the c-terminal domain. EMBO Rep. 2009;10:1147–1153. doi: 10.1038/embor.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S. S-nitrosylation of hsp90 promotes the inhibition of its atpase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duval M, Le Boeuf F, Huot J, Gratton JP. Src-mediated phosphorylation of hsp90 in response to vascular endothelial growth factor (vegf) is required for vegf receptor-2 signaling to endothelial no synthase. Mol Biol Cell. 2007;18:4659–4668. doi: 10.1091/mbc.E07-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogiso H, Kagi N, Matsumoto E, Nishimoto M, Arai R, Shirouzu M, Mimura J, Fujii-Kuriyama Y, Yokoyama S. Phosphorylation analysis of 90 kda heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry. 2004;43:15510–15519. doi: 10.1021/bi048736m. [DOI] [PubMed] [Google Scholar]
- 32.Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein unc-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- 33.Kee HJ, Eom GH, Joung H, Shin S, Kim JR, Cho YK, Choe N, Sim BW, Jo D, Jeong MH, Kim KK, Seo JS, Kook H. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ Res. 2008;103:1259–1269. doi: 10.1161/01.RES.0000338570.27156.84. [DOI] [PubMed] [Google Scholar]
- 34.Morrison LE, Hoover HE, Thuerauf DJ, Glembotski CC. Mimicking phosphorylation of alphab-crystallin on serine-59 is necessary and sufficient to provide maximal protection of cardiac myocytes from apoptosis. Circ Res. 2003;92:203–211. doi: 10.1161/01.res.0000052989.83995.a5. [DOI] [PubMed] [Google Scholar]
- 35.Jin JK, Whittaker R, Glassy MS, Barlow SB, Gottlieb RA, Glembotski CC. Localization of phosphorylated alphab-crystallin to heart mitochondria during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2008;294:H337–344. doi: 10.1152/ajpheart.00881.2007. [DOI] [PubMed] [Google Scholar]
- 36.You W, Min X, Zhang X, Qian B, Pang S, Ding Z, Li C, Gao X, Di R, Cheng Y, Liu L. Cardiac-specific expression of heat shock protein 27 attenuated endotoxin-induced cardiac dysfunction and mortality in mice through a pi3k/akt-dependent mechanism. Shock. 2009;32:108–117. doi: 10.1097/SHK.0b013e318199165d. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Zhang X, Qian B, Min X, Gao X, Li C, Cheng Y, Huang J. Over-expression of heat shock protein 27 attenuates doxorubicin-induced cardiac dysfunction in mice. Eur J Heart Fail. 2007;9:762–769. doi: 10.1016/j.ejheart.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Hollander JM, Martin JL, Belke DD, Scott BT, Swanson E, Krishnamoorthy V, Dillmann WH. Overexpression of wild-type heat shock protein 27 and a nonphosphorylatable heat shock protein 27 mutant protects against ischemia/reperfusion injury in a transgenic mouse model. Circulation. 2004;110:3544–3552. doi: 10.1161/01.CIR.0000148825.99184.50. [DOI] [PubMed] [Google Scholar]
- 39.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK. The e3 ligase traf6 regulates akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. Fam/usp9x, a deubiquitinating enzyme essential for tgfbeta signaling, controls smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 41.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fredrickson EK, Gardner RG. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. Semin Cell Dev Biol. 2012;23:530–537. doi: 10.1016/j.semcdb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome biology. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson IR, Irwin MS, Ohh M. Nedd8 pathways in cancer, sine quibus non. Cancer Cell. 2011;19:168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Kato JY, Yoneda-Kato N. Mammalian cop9 signalosome. Genes Cells. 2009;14:1209–1225. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- 46.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, Wang X. Perturbation of cullin deneddylation via conditional csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. Chip-mediated stress recovery by sequential ubiquitination of substrates and hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naito AT, Okada S, Minamino T, Iwanaga K, Liu ML, Sumida T, Nomura S, Sahara N, Mizoroki T, Takashima A, Akazawa H, Nagai T, Shiojima I, Komuro I. Promotion of chip-mediated p53 degradation protects the heart from ischemic injury. Circ Res. 2010;106:1692–1702. doi: 10.1161/CIRCRESAHA.109.214346. [DOI] [PubMed] [Google Scholar]
- 49.Yang K, Zhang TP, Tian C, Jia LX, Du J, Li HH. Carboxyl terminus of heat shock protein 70-interacting protein inhibits angiotensin ii-induced cardiac remodeling. Am J Hypertens. 2012;25:994–1001. doi: 10.1038/ajh.2012.74. [DOI] [PubMed] [Google Scholar]
- 50.Zhang C, Xu Z, He XR, Michael LH, Patterson C. Chip, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. 2005;288:H2836–2842. doi: 10.1152/ajpheart.01122.2004. [DOI] [PubMed] [Google Scholar]
- 51.Scaglione KM, Zavodszky E, Todi SV, Patury S, Xu P, Rodriguez-Lebron E, Fischer S, Konen J, Djarmati A, Peng J, Gestwicki JE, Paulson HL. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase chip. Mol Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic parkinson's disease: S-nitrosylation of parkin regulates its e3 ubiquitin ligase activity. Proc Natl Acad Sci U S A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 54.Choo YS, Vogler G, Wang D, Kalvakuri S, Iliuk A, Tao WA, Bodmer R, Zhang Z. Regulation of parkin and pink1 by neddylation. Hum Mol Genet. 2012;21:2514–2523. doi: 10.1093/hmg/dds070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. Neddylation positively regulates the ubiquitin e3 ligase activity of parkin. J Neurosci Res. 2012;90:1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- 56.Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, Kruger R, Winklhofer KF, Vogel F, Reichert AS, Auburger G, Kahle PJ, Schmid B, Haass C. Loss-of-function of human pink1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitada T, Tong Y, Gautier CA, Shen J. Absence of nigral degeneration in aged parkin/dj-1/pink1 triple knockout mice. J Neurochem. 2009;111:696–702. doi: 10.1111/j.1471-4159.2009.06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by parkin and p62/sqstm1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunter T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. Genotoxic stress targets human chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 61.Besson A, Gurian-West M, Chen X, Kelly-Spratt KS, Kemp CJ, Roberts JM. A pathway in quiescent cells that controls p27kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 2006;20:47–64. doi: 10.1101/gad.1384406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durcan TM, Kontogiannea M, Bedard N, Wing SS, Fon EA. Ataxin-3 deubiquitination is coupled to parkin ubiquitination via e2 ubiquitin-conjugating enzyme. J Biol Chem. 2012;287:531–541. doi: 10.1074/jbc.M111.288449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durcan TM, Kontogiannea M, Thorarinsdottir T, Fallon L, Williams AJ, Djarmati A, Fantaneanu T, Paulson HL, Fon EA. The machado-joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum Mol Genet. 2011;20:141–154. doi: 10.1093/hmg/ddq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of usp14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, Li X, Whitelegge J, Drews O, Yang PY, Ping P. Revealing the dynamics of 20s proteasome phosphoproteome: A combined cid and etd approach. Mol Cell Proteomics. 2008;7:2073–2089. doi: 10.1074/mcp.M800064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zong C, Gomes AV, Drews O, Li X, Young GW, Berhane B, Qiao X, French SW, Bardag-Gorce F, Ping P. Regulation of murine cardiac 20s proteasomes: Role of associating partners. Circ Res. 2006;99:372–380. doi: 10.1161/01.RES.0000237389.40000.02. [DOI] [PubMed] [Google Scholar]
- 68.Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic amp-dependent protein kinase through phosphorylation of rpt6. J Biol Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 69.Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, Takahama H, Sasaki H, Higo S, Asakura M, Takashima S, Hori M, Kitakaze M. Pka rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol. 2009;46:452–462. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Drews O, Tsukamoto O, Liem D, Streicher J, Wang Y, Ping P. Differential regulation of proteasome function in isoproterenol-induced cardiac hypertrophy. Circ Res. 2010;107:1094–1101. doi: 10.1161/CIRCRESAHA.110.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase ii. J Biol Chem. 2009;284:26655–26665. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated camkiialpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 73.Swaminathan PD, Purohit A, Hund TJ, Anderson ME. Calmodulin-dependent protein kinase ii: Linking heart failure and arrhythmias. Circ Res. 2012;110:1661–1677. doi: 10.1161/CIRCRESAHA.111.243956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20s proteasome alpha subunit c8 (alpha7) stabilizes the 26s proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J. 2004;378:177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Powell SR, Wang X. Enhancement of proteasome function by pa28α overexpression protects against oxidative stress. FASEB J. 2011;25:883–893. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takagi H, Hsu CP, Kajimoto K, Shao D, Yang Y, Maejima Y, Zhai P, Yehia G, Yamada C, Zablocki D, Sadoshima J. Activation of pkn mediates survival of cardiac myocytes in the heart during ischemia/reperfusion. Circ Res. 2010;107:642–649. doi: 10.1161/CIRCRESAHA.110.217554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uchiki T, Kim HT, Zhai B, Gygi SP, Johnston JA, O'Bryan JP, Goldberg AL. The ubiquitin-interacting motif protein, s5a, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. J Biol Chem. 2009;284:12622–12632. doi: 10.1074/jbc.M900556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B. Monoubiquitination of rpn10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 80.Divald A, Kivity S, Wang P, Hochhauser E, Roberts B, Teichberg S, Gomes AV, Powell SR. Myocardial ischemic preconditioning preserves postischemic function of the 26s proteasome through diminished oxidative damage to 19s regulatory particle subunits. Circ Res. 2010;106:1829–1838. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 81.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, Pagani F, Powell SR, Day SM. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20s and 26s proteasome to oxidative stress. Biochem J. 1998;335(Pt 3):637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amici M, Lupidi G, Angeletti M, Fioretti E, Eleuteri AM. Peroxynitrite-induced oxidation and its effects on isolated proteasomal systems. Free Radic Biol Med. 2003;34:987–996. doi: 10.1016/s0891-5849(02)01369-2. [DOI] [PubMed] [Google Scholar]
- 84.Osna NA, Haorah J, Krutik VM, Donohue TM., Jr Peroxynitrite alters the catalytic activity of rodent liver proteasome in vitro and in vivo. Hepatology. 2004;40:574–582. doi: 10.1002/hep.20352. [DOI] [PubMed] [Google Scholar]
- 85.Davies KJ. Degradation of oxidized proteins by the 20s proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 86.Friguet B, Stadtman ER, Szweda LI. Modification of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal. Formation of cross-linked protein that inhibits the multicatalytic protease. J Biol Chem. 1994;269:21639–21643. [PubMed] [Google Scholar]
- 87.Papa L, Gomes E, Rockwell P. Reactive oxygen species induced by proteasome inhibition in neuronal cells mediate mitochondrial dysfunction and a caspase-independent cell death. Apoptosis. 2007;12:1389–1405. doi: 10.1007/s10495-007-0069-5. [DOI] [PubMed] [Google Scholar]
- 88.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 90.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pyo JO, Nah J, Jung YK. Molecules and their functions in autophagy. Experimental & molecular medicine. 2012;44:73–80. doi: 10.3858/emm.2012.44.2.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 95.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr., Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kang R, Zeh HJ, Lotze MT, Tang D. The beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. Dap-kinase-mediated phosphorylation on the bh3 domain of beclin 1 promotes dissociation of beclin 1 from bcl-xl and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi CS, Kehrl JH. Traf6 and a20 regulate lysine 63-linked ubiquitination of beclin-1 to control tlr4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 101.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Functional and physical interaction between bcl-x(l) and a bh3-like domain in beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. Jnk1-mediated phosphorylation of bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, Lotze MT. Endogenous hmgb1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced bcl2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. Ampk phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Inoki K, Zhu T, Guan KL. Tsc2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 107.Lee JW, Park S, Takahashi Y, Wang HG. The association of ampk with ulk1 regulates autophagy. PLoS One. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ulk1 (hatg1) by amp-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim J, Kundu M, Viollet B, Guan KL. Ampk and mtor regulate autophagy through direct phosphorylation of ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chiacchiera F, Matrone A, Ferrari E, Ingravallo G, Lo Sasso G, Murzilli S, Petruzzelli M, Salvatore L, Moschetta A, Simone C. P38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from hif1alpha- to foxo-dependent transcription. Cell Death Differ. 2009;16:1203–1214. doi: 10.1038/cdd.2009.36. [DOI] [PubMed] [Google Scholar]
- 111.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. Foxo3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 112.Takagi H, Matsui Y, Hirotani S, Sakoda H, Asano T, Sadoshima J. Ampk mediates autophagy during myocardial ischemia in vivo. Autophagy. 2007;3:405–407. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 113.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 114.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian atg8 modifiers, lc3, gabarap, and gate-16. J Biol Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 115.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in cryabr120g cardiomyocytes. Circ Res. 2011;109:151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS, Robbins J, Martinez J, Tabas I. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15:545–553. doi: 10.1016/j.cmet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, Condorelli G, Sadoshima J. Rheb is a critical regulator of autophagy during myocardial ischemia: Pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the nad-dependent deacetylase sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guarente L, Picard F. Calorie restriction--the sir2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 121.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of foxo transcription factors by the sirt1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 122.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of foxo by sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic foxo1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 125.Schips TG, Wietelmann A, Hohn K, Schimanski S, Walther P, Braun T, Wirth T, Maier HJ. Foxo3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res. 2011;91:587–597. doi: 10.1093/cvr/cvr144. [DOI] [PubMed] [Google Scholar]
- 126.Su H, Li F, Ranek MJ, Wei N, Wang X. Cop9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of nedd8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 128.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The nedd8-activating enzyme inhibitor mln4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 129.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, Gygi SP, Lee MK, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 131.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/sqstm1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 133.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 135.Su H, Wang X. P62 stages an interplay between the ubiquitin-proteasome system and autophagy in the heart of defense against proteotoxic stress. Trends Cardiovasc Med. 2011;21:224–228. doi: 10.1016/j.tcm.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. Pink1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2012 Jun 29; doi: 10.1038/cdd.2012.81. doi: 10.1038/cdd.2012.81. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walden H, Martinez-Torres RJ. Regulation of parkin E3 ubiquitin ligase activity. Cell Mol Life Sci. 2012;69:3053–3067. doi: 10.1007/s00018-012-0978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. P62/sqstm1 is required for parkin-induced mitochondrial clustering but not mitophagy; vdac1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 143.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, Hong C, Hittinger L, Ghaleh B, Sadoshima J, Vatner DE, Vatner SF, Madura K. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 144.Tang M, Li J, Huang W, Su H, Liang Q, Tian Z, Horak KM, Molkentin JD, Wang X. Proteasome functional insufficiency activates the calcineurin-nfat pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88:424–433. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yeh ET, Bickford CL. Cardiovascular complications of cancer therapy: Incidence, pathogenesis, diagnosis, and management. J Am Coll Cardiol. 2009;53:2231–2247. doi: 10.1016/j.jacc.2009.02.050. [DOI] [PubMed] [Google Scholar]