Abstract
The intracellular nucleotide-binding oligomerization domain-2 (NOD2) receptor detects bacteria-derived muramyl dipeptide (MDP) and activates the transcription factor NF-κB. Here we describe the regulatome of NOD2 signaling using a systematic RNAi screen. Using three consecutive screens, we identified a set of 20 positive NF-κB regulators including the known pathway members RIPK2, RELA, and BIRC4 (XIAP) as well as FRMPD2 (FERM and PDZ domain-containing 2). FRMPD2 interacts with NOD2 via leucine-rich repeats and forms a complex with the membrane-associated protein ERBB2IP. We demonstrate that FRMPD2 spatially assembles the NOD2-signaling complex, hereby restricting NOD2-mediated immune responses to the basolateral compartment of polarized intestinal epithelial cells. We show that genetic truncation of the NOD2 leucine-rich repeat domain, which is associated with Crohn disease, impairs the interaction with FRMPD2, and that intestinal inflammation leads to down-regulation of FRMPD2. These results suggest a structural mechanism for how polarity of epithelial cells acts on intestinal NOD-like receptor signaling to mediate spatial specificity of bacterial recognition and control of immune responses.
Keywords: intestinal epithelium, nuclear factor kappaB, innate immune responses
Innate host defense relies on effective sensing of danger signals arising in different cellular compartments. Nucleotide-binding and oligomerization domain (NOD)–like receptor (NLR) proteins exert pivotal roles in innate immunity as sensors for exogenous, as well as endogenous, danger signals (1). Their characteristic tripartite domain structure with a central NOD, C-terminal leucine-rich repeats (LRRs), and an N-terminal effector-binding domain (EBD) has been preserved throughout evolution (2). The prototypical NLR-family member NOD2 recognizes the intracellular presence of the bacterial cell-wall component muramyl dipeptide (MDP) and subsequently triggers a signal cascade leading to nuclear factor kappaB (NF-κB) activation (3, 4). In recent years, a substantial number of integral components of the NOD2-dependent NF-κB activation pathway have been identified: In response to binding of MDP to the NOD2 LRR domain (5), the adaptor protein RIPK2 is recruited to NOD2 through homotypic caspase activation and recruitment domain (CARD)–CARD interactions (3, 6). The subsequent activation of the IκB–kinase complex through NEMO (7, 8) leads to IκB degradation. NF-κB translocates to the nucleus and induces transcription of proinflammatory target genes (e.g., IL-8 and IL-1β) (9) and antimicrobial effectors (10, 11). Genetic variants in the NOD2 gene are associated with Crohn disease (CD), a subentity of inflammatory bowel disease and other chronic inflammatory diseases of barrier organs (12). A paradigmatic NOD2 variant that contributes to manifestation of CD causes a frameshift through a single-nucleotide insertion in the coding sequence (L1007fsinsC), which leads to a truncated LRR (13–15). Cells expressing this variant fail to activate NF-κB upon stimulation with MDP (16, 17). Several reports have described NOD2 localizing at the membrane (18, 19). It was shown that membrane targeting is required for MDP-induced NF-κB activation (18), yet no direct mechanism has been proposed so far. To identify modulators of NOD2 signaling, we performed a high-throughput siRNA screen targeting 7,783 genes. Among the genes identified was FRMPD2 (FERM and PDZ domain protein-containing 2), a protein involved in basolateral membrane targeting in epithelial cells (20). We show here that FRMPD2 supports basolateral specificity of MDP recognition by NOD2 in polarized intestinal epithelial cells (IECs) through recruitment of the LRR of NOD2 to the basolateral membrane compartment. We propose that FRMPD2 acts as a membrane-scaffolding complex that provides a spatial control mechanism for NOD2-mediated immune responses.
Results
Identification and Functional Network Analysis of NOD2-Signaling Modulators.
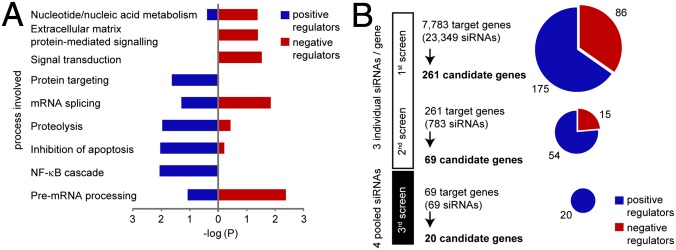
To identify modulators of the NOD2-signaling pathway, we targeted 7,783 genes using a commercially available “druggable” genome siRNA library. The screen was performed in HEK293 cells, which were transfected with siRNA, plasmids encoding NOD2, and dual–NF-κB luciferase reporter constructs. Luciferase activity was measured 12 h after MDP stimulation (Fig. S1A). Results showed a reproducible six- to eightfold induction of NF-κB activation in cells transfected with control siRNA. Independent knockdown of RIPK2 significantly impaired NF-κB activation, and knockdown of firefly luciferase served as internal control (Fig. S1B). Each transcript was targeted using three different siRNAs, resulting in a total number of 23,349 assays for NF-κB activation. Genes with normalized fold-induction values higher than 1.5 or lower than −1.5 were considered as candidate genes and were further filtered according to the scores of the individual siRNA (Fig. S1C and SI Materials and Methods). Bioinformatics analysis demonstrated significantly enriched gene ontology (GO) terms such as signal transduction, NF-κB signaling, proteolysis, and inhibition of apoptosis among 261 candidate genes from the first screen (Fig. 1A). The 261 candidate genes and a network analysis of their connection to NOD2 are illustrated in Fig. S1D. To narrow down the number of candidate genes, the 261 genes were rescreened using the same setup. The remaining 69 candidate genes were validated in a third screen changing the application of individual siRNAs to a pool of four siRNAs per transcript, which differed in their respective target sequences. A network score for the identified hits versus randomly selected genes was calculated based on information from a protein-interaction database (STRING 8.3) (21). Candidate genes were scored (9, 4, 1, or 0 points) according to their distance to NOD2 in the precalculated matrix (Fig. S1E, Upper), and the ratio of aggregated scores of candidate genes to aggregated scores of randomly picked genes and a false discovery rate (FDR) from 50,000 random simulations (out of the druggable genome pool) were calculated.
Fig. 1.
Systematic siRNA screening identifies modulators of NOD2 signaling. (A) Enrichment of candidate genes in different functional groups (GO terms) after the primary screen. (B) Screening procedure and number of candidate genes at different screening stages. Primary and secondary screens were performed with three individual siRNAs per gene. Pools of four siRNAs were used for the third screen. Candidate genes with an enhancing or inhibitory effect on NF-κB activation are indicated in blue or red, respectively.
This procedure finally generated a set of 20 positive NF-κB regulators (Fig. 1B). We found a continuous increase in ratio from the primary to the tertiary screen, implying a stepwise accumulation of hits that map closer to known NOD2 interactors and a decrease in FDR, indicating relevance of the remaining 20 candidate genes for the NOD2 pathway (Fig. S1E, Lower). Because initiation of NOD2 signaling activates the canonical NF-κB pathway, we sought to distinguish between general NF-κB modulators and those specific for NOD2 signaling. To this end, we rescreened the set of 20 candidate genes using TNF-α stimulation (“counterscreen”; Fig. S1F). Ten out of 20 candidate genes showed no effect upon TNF-α stimulation (blue font) and were thus considered to be specific modulators within the NOD2-signaling pathway. Normalized NF-κB activation of the final 20 candidate genes from all screens is summarized in Table S1.
FRMPD2 Represents a Positive Regulator of NOD2-Mediated NF-κB Activation.
To identify genes relevant for the intestinal epithelium, we rescreened the 20 candidate genes using Caco-2 cells and found 14 of them beyond the cutoff level (bold in Fig. 2A). We next mapped functional relationships of candidate proteins including known NOD2-signaling components based on interaction data from STRING 8.3 (highest confidence; P > 0.9) and BioGRID (http://thebiogrid.org/) (Fig. 2B). We found that 16 out of 20 candidate genes are either directly or by a short distance connected to NOD2, whereas a subset (including the gene FRMPD2) has not yet been associated with immunological functions or NF-κB signal transduction. Next, we selected genes for further follow-up studies. We included genes from the TNF-α counterscreen that were MDP-responsive only (blue in Fig. 2B), and from these we selected genes confirmed as regulators from the Caco-2 screen (bold in Fig. 2B). This assortment method resulted in six genes: RIPK2, EP400, BIRC4, PSMB4, FRMPD2, and EEF1E1. Because RIPK2 and BIRC4 are known modulators of NOD2 signaling (3, 8), only RIPK2 was included as positive control. Moreover, karyopherin subunit β 1 (KPNB1) was integrated in the follow-up as a “canonical” control. Expression of the selected candidate genes was confirmed in different cell lines of epithelial and myeloid origin (Fig. S2A). Next, we measured IL-8 chemokine release of selected candidate genes, which is induced upon MDP-mediated NF-κB activation (22, 23). Knockdown of all six genes significantly reduced IL-8 release in response to MDP stimulation in NOD2-transfected HEK293 cells (Fig. 2C) and in the monocytic cell line THP-1, which endogenously expresses NOD2 (Fig. S2B). Because NOD2 signaling is linked to antimicrobial mechanisms (10, 22, 24), we also analyzed bacterial cytoinvasion using fluorescence-labeled Listeria monocytogenes. Knockdown of all six genes significantly altered bacterial entry and (except for KPNB1) led to an increased intracellular bacterial load (Fig. S2C). For further functional studies, we chose FRMPD2 because of its function related to basolateral membrane targeting in polarized epithelial cells (20). We found that silencing of Frmpd2 in murine IECs significantly decreased keratinocyte-derived cytokine (KC) release from WT cells but not in Nod2 KO cells (Fig. 2D). Viability of cells was assured using the MTS viability assay (Fig. S3). Although other mechanisms could also account for the observed change in intracellular bacteria, we could show using real-time PCR that induction of mRNA levels of antimicrobial peptides/proteins [e.g., human β-defensin 1 (BD-1), phospholipase A2 (PLA2), and human α-defensin 6 (HD6)] was significantly decreased after infection with L. monocytogenes upon FRMPD2 silencing (Fig. S2D).
Fig. 2.
Confirming FRMPD2 as a positive regulator of NOD2-mediated NF-κB activation. (A) Validation of 20 candidate genes in Caco-2 cells using a luciferase assay. Cells were transfected and stimulated with MDP according to previous assay procedures. (B) Functional interaction network of candidate proteins including members of the NOD2-signaling pathway (gray boxes) based on data from STRING 8.3 (highest confidence; P > 0.9) and BioGRID. Dashed lines represent manually added interactions based on publications not yet integrated into databases. Nodes represent candidate genes. Colored in blue are candidate genes from the TNF-α counterscreen that did not share canonical NF-κB activation. Out of these candidates, those whose fold inductions fell below the cutoff level in the Caco-2 screen are in bold. (C) IL-8 release of MDP-stimulated HEK293 cells transfected with siRNA of the indicated genes and NOD2 plasmids. IL-8 concentration was normalized against Renilla luciferase. (D) KC release of Frmpd2 and control. siRNA-transfected and MDP-stimulated IECs from WT and Nod2 KO mice (n = 3 each group) cultured in a 3D collagen matrix. (E) Western blot (WB) analysis of MDP-mediated IκB-α degradation in HEK293 cells transfected with control and FRMPD2 siRNAs together with plasmids encoding NOD2. (F) MDP-induced luciferase activity in HEK293 cells at 48 h after transfection with negative control or RIPK2 siRNAs, plasmids encoding NOD2, FRMPD2, or mock control, and luciferase reporter constructs. (G) Fluorescence microscopy of polarized Caco-2 cells transfected with plasmids encoding FRMPD2-DsRed and ERBB2IP-myc [stained with antibody to myc (green); Upper] or with NOD2-GFP and FRMPD2-DsRed constructs (Lower). (Scale bars, 10 µm.) Arrowheads indicate colocalization. (H) Coimmunoprecipitation (Co-IP) of FLAG- and GFP-tagged proteins from whole-cell lysates of HEK293 cells with or without MDP stimulation (10 µg/mL for 90 min), followed by WB analysis of NOD2 (anti-FLAG), FRMPD2 (anti-GFP), and ERBB2IP (anti-ERBB2IP). (I) Co-IP of endogenous FRMPD2 with overexpressed NOD2-FLAG from Caco-2 cells followed by WB analysis with antibody to FRMPD2 (anti-FRMPD2) or NOD2 (anti-FLAG). Data in A, C, D, and F show mean and SD of at least three in-plate replicates; *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, we determined the influence of FRMPD2 on the NOD2-signaling pathway. Knockdown of FRMPD2 abolished MDP-induced IκB-α degradation, supporting a function upstream of IκB-α (Fig. 2E). Further, the MDP-induced increase in NF-κB activation upon overexpression of FRMPD2 was abrogated when RIPK2 was silenced, indicating that FRMPD2 acts upstream of RIPK2 activation (Fig. 2F). Silencing of RIPK2 and FRMPD2 was confirmed at 72 h after siRNA transfection using Western blot analysis (Fig. S4). To further investigate the specificity of the FRMPD2 effect for the NOD2 pathway, we found that FRMPD2 silencing does not alter NF-κB activation induced by various Toll-like-receptor (TLR) ligands (Fig. S5A), whereas NOD1-induced NF-κB activation was significantly decreased, too (Fig. S5B).
The functional network analysis suggested that FRMPD2 and NOD2 are functionally connected through ERBB2IP (ERBIN) (Fig. 2B), a member of the LRR and PDZ domain family. ERBB2IP has been shown to interact with NOD2 at the basolateral plasma membrane (19, 25). We therefore asked whether FRMPD2 is directly linked to NOD2. Coexpression of FRMPD2 and ERBB2IP as well as FRMPD2 and NOD2 showed a colocalization at the plasma membrane of Caco-2 cells (Fig. 2G).
To assess a potential physical interaction between NOD2 and FRMPD2, we performed coimmunoprecipitation experiments. We demonstrate that FRMPD2 (both overexpressed and endogenous protein) coprecipitates with NOD2 (Fig. 2 H and I) without addition of ERBB2IP. Simultaneous overexpression of ERBB2IP clearly facilitated interaction of NOD2 and FRMPD2 (Fig. 2H). Subsequent NOD2-domain mapping showed that the C-terminal LRR domains and to a lesser extent the central nucleotide-binding domain (NBD) are required for the interaction with FRMPD2 (Fig. S6A). In a reciprocal setup, all FRMPD2 domain constructs containing PDZ domains fused to GFP were capable of precipitating with full-length NOD2, whereas interaction was observed neither with the FRMPD2-FERM domain nor with the FRMPD2-KIND domain alone (Fig. S6B). In addition, we found that FRMPD2 and ERBB2IP interact via the FERM domain of FRMPD2 independent of the presence of NOD2 (Fig. S6C). From these results, we concluded that NOD2, FRMPD2, and ERBB2IP form a tripartite interaction complex that modulates NOD2 signaling.
FRMPD2 Controls NOD2 Signaling from the Basolateral Membrane in Intestinal Epithelial Cells.
To study FRMPD2 function in the context of NOD2 signaling in polarized IECs, we stained for endogenous FRMPD2 and NOD2 in human colonic biopsy sections. We demonstrate colocalization of endogenous FRMPD2 and NOD2 at the lateral membrane and in proximity to the basal membrane of the IEC layer (Fig. 3A). To confirm this finding, we used NOD2- and FRMPD2-transfected polarized Caco-2 monolayers and found them to be colocalized with the basolateral marker Na+/K+-ATPase (Fig. 3B). It has been previously shown that membrane recruitment of NOD2 is essential for NF-κB activation (18) and that NOD2 is localized at the basolateral plasma membrane (19). We therefore asked (i) whether MDP induces NOD2 signaling in a polarized manner, and (ii) whether FRMPD2 is involved in compartmentalized NOD2 signaling. To this aim, we used small intestinal organ cultures from WT and Nod2 KO mice and disrupted epithelial junctions by calcium depletion to facilitate substance diffusion to the basolateral compartment (26, 27) or left tissue in normal media. MDP-mediated KC release was only induced in calcium-free cultured intestinal segments from WT mice, not in Nod2-deficient mice (Fig. 3C). Next, we stimulated polarized Caco-2 monolayers with MDP at either the apical or basal side. We found that only basal MDP stimulation induced BD-2 mRNA expression (Fig. 3D) and led to IL-8 cytokine secretion (Fig. S7A). Notably, when FRMPD2 was silenced, basal BD-2 and IL-8 induction was completely abrogated. These results suggest that MDP-mediated NF-κB activation and subsequent cytokine release are specifically triggered from the basolateral membrane. Because we found an impact of FRMPD2 on NOD1-mediated NF-κB activation as well, we analyzed the response of polarized Caco-2 monolayers to D-gamma-Glu-mDAP (iE-DAP) stimulation. Comparable to NOD2, we found that only basal iE-DAP stimulation induced IL-8 secretion (Fig. S7B), indicating that FRMPD2 could function as a general modulator of NLR signaling. To test whether FRMPD2 is able to restrict NOD2 to the membrane compartment, we silenced FRMPD2 in Caco-2 cells and analyzed NOD2 distribution in the cytosolic and membrane compartments after MDP stimulation. We found a reduction of NOD2 in the membrane fraction in FRMPD2-silenced cells both at the endogenous level (Fig. 3E) and when NOD2 was transfected (Fig. S7C). In addition, using fluorescence microscopy, we observed diminished NOD2 levels at the plasma membrane when cells were treated with FRMPD2 siRNA (Fig. S7D). To examine the function of FRMPD2 as a scaffold for NOD2 signaling, we checked whether other components of the NOD2 signalosome are recruited to the membrane in an FRMPD2-dependent manner. Notably, we found RIPK2 recruited to the plasma membrane when FRMPD2 and NOD2 are coexpressed in RIPK2-transfected polarized Caco-2 cells (Fig. 3F). As demonstrated previously (28), endogenous RIPK2 was recruited to the membrane in Caco-2 cells in an MDP-dependent fashion (Fig. S8A) in the presence of NOD2. Silencing of FRMPD2 resulted in diminished recruitment of RIPK2 after MDP stimulation (Fig. 3E). Interestingly, we were not able to detect a strong direct interaction of FRMPD2 and RIPK2 (Fig. S8B).
Fig. 3.
FRMPD2 controls NOD2 signaling from the basolateral membrane in intestinal epithelial cells. (A) Fluorescence microscopy of paraffin sections from colonic epithelial biopsy samples stained using anti-NOD2 (green) and anti-FRMPD2 (red) antibodies (Upper). Secondary Abs only were used as control (Lower). Images are representative of at least three independent experiments or n = 5 individuals. (Scale bar, 10 µm.) (B) Cross-sectional xy, xz, and yz images acquired by confocal fluorescence microscopy of FRMPD2-dsRED– and NOD2-GFP–transfected Caco-2 cells grown on transwell inserts for induction of polarized monolayers. Cells were fixed and stained with antibody to the basolateral marker Na+/K+-ATPase (purple). Arrowheads indicate sites of lateral colocalization. (Scale bar, 10 µm.) (C) KC ELISA of small intestinal mucosal explants of culture supernatants. Intestinal segments from WT and Nod2 KO mice were incubated in either calcium-free and EDTA-containing or normal RPMI 1640 media and stimulated with MDP (10 µg/mL, 24 h). (D) Real-time SYBR Green PCR on BD-1, BD-2, lysozyme, and IL-8 transcripts normalized to GAPDH from polarized Caco-2 cells treated with control or FRMPD2 siRNA in response to either apical or basal MDP stimulation (10 µg/mL, 12 h). (E) WB analysis of cellular distribution of NOD2 and RIPK2 protein in response to MDP stimulation in polarized Caco-2 monolayers at 72 h after transfection with FRMPD2 siRNA or control. (F) Fluorescence microscopy of RIPK2-, FRMPD2-dsRED–, and NOD2-GFP–transfected Caco-2 cells grown on transwell inserts for induction of polarized monolayers. Cells were fixed and stained with antibody to RIPK2 (purple). (Scale bar, 10 µm.) Data in C and D show mean and SD of at least three in-plate replicates; *P < 0.05.
FERM-PDZ2 Domain of FRMPD2 Serves as a Membrane Scaffold and Binds to the LRR of NOD2.
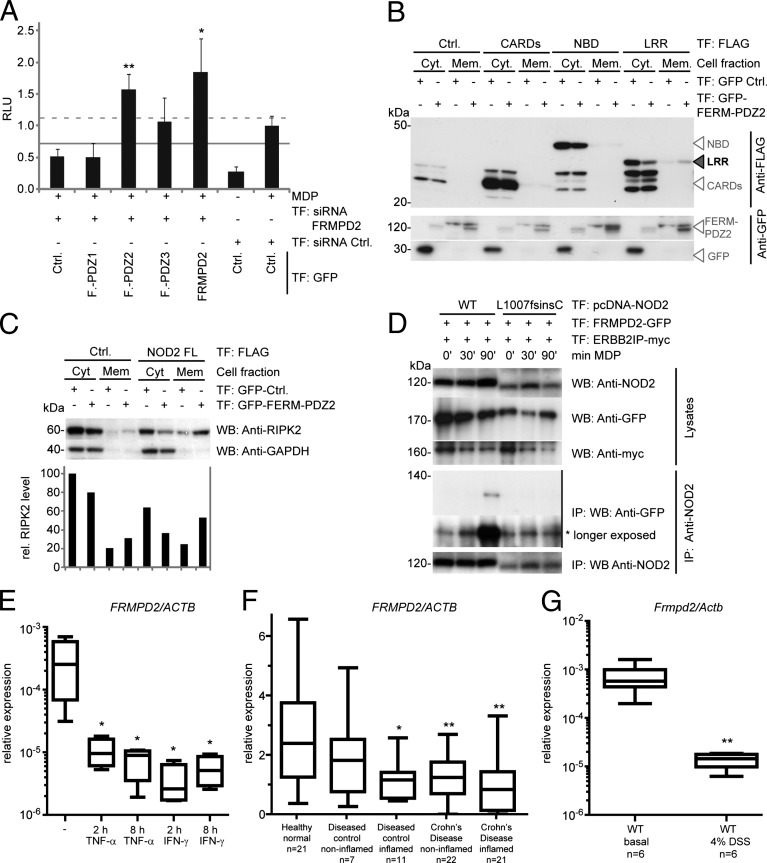
To further dissect the molecular basis of the FRMPD2-scaffolding complex, we studied the localization and function of FRMPD2 and NOD2 domains. First, we analyzed whether a specific FERM-PDZ domain of FRMPD2 is sufficient to restore NF-κB activation after FRMPD2 silencing. As shown in Fig. 4A, overexpression of FERM-PDZ2 increased MDP-induced NF-κB activation significantly, whereas FERM-PDZ1 and FERM-PDZ3 had no or little effect. We next analyzed whether the FERM-PDZ2 domain influences localization of CARD, NBD, and LRR domains of NOD2 and found the LRR to be redistributed from the cytosolic to the membrane fraction when coexpressed with the FERM-PDZ2 domain (Fig. 4B). In addition, we demonstrated that coexpression of the FERM-PDZ2 domain and NOD2 increases RIPK2 membrane localization (Fig. 4C). We observed that the LRR of NOD2 is recruited to the membrane already at low levels of overexpressed PDZ2 domain (Fig. S8C).
Fig. 4.
The FERM-PDZ2 domain of FRMPD2 serves as a membrane scaffold for the LRR of NOD2 and recruits RIPK2 to the membrane. (A) Luciferase assay of MDP-induced NF-κB activation in HEK293 cells after transfection with FRMPD2 siRNAs or negative control, plasmids encoding NOD2, GFP-FERM-PDZ domains, GFP-FRMPD2-full length, or GFP-control, and luciferase reporter constructs. (B) WB analysis of cellular distribution of NOD2 CARD, NBD, or LRR domains dependent on GFP-control or GFP-FERM-PDZ2 cotransfection in HEK293 cells. The black arrowhead indicates the shift of the LRR domain from the cytosolic to the membrane fraction when coexpressed with FERM-PDZ2. (C) WB analysis of cellular distribution of RIPK2 in HEK293 cells cotransfected with NOD2 or empty vector, GFP-control, or GFP-FERM-PDZ2. (D) Co-IP of WT and mutant (L1007fsinsC) NOD2 with endogenous RIPK2 in HEK293 cells transfected with FLAG-NOD2 or empty vectors. (E) Real-time TaqMan PCR on FRMPD2 transcripts normalized to ACTB from Caco-2 cells treated with 25 ng/mL TNF-α or IFN-γ for 2 or 8 h. (F) Real-time TaqMan PCR on FRMPD2 transcripts normalized to ACTB from sigmoid colon biopsies from healthy individuals (n = 21), normal and inflamed intestinal mucosa of patients with CD (n = 22 and n = 21), and diseased controls (n = 7 and n = 11). (G) Real-time SYBR Green PCR on Frmpd2 transcripts normalized to Actb from colon biopsies of untreated C57BL/6J WT mice (n = 6) and DSS-treated mice (n = 6, 4% DSS for 6 d). Data in A and E–G show mean and SD of at least three in-plate replicates; *P < 0.05 and **P < 0.01.
Interaction with FRMPD2 Is Abrogated in the Crohn Disease-Associated NOD2 Variant L1007fsinsC.
We next asked (i) whether the spatially defined interaction of NOD2 and FRMPD2 is altered in the CD-associated NOD2 variant L1007fsinsC, which affects the LRR domain of NOD2, and (ii) whether this could in part explain the molecular dysfunction of the variant protein. The NOD2 L1007 frameshift mutation encodes a C-terminal truncated NOD2 protein, thereby abrogating its MDP-mediated NF-κB activation ability (4) and, interestingly, its membrane recruitment as well (18, 28). Notably, wild-type NOD2 immunoprecipitated with FRMPD2, reaching the highest interaction level at 90 min after MDP stimulation, whereas NOD2 L1007fsinsC had a very weak interaction consistently throughout the time-course stimulation study (Fig. 4D).
To further analyze whether FRMPD2 expression is differentially regulated under proinflammatory conditions, we stimulated Caco-2 cells for 2 and 8 h with TNF-α or IFN-γ and found that FRMPD2 mRNA levels were diminished compared with unstimulated cells (Fig. 4E). Next, we determined the FRMPD2 expression profile in normal and inflamed human intestinal mucosa using sigmoid colon samples from healthy individuals, patients with CD, and diseased controls (sigma diverticulitis patients). We found that in comparison with healthy individuals, FRMPD2 was significantly down-regulated in the inflamed intestinal mucosa of diseased individuals and in the noninflamed tissues as well as in the inflamed intestinal mucosa from patients with CD (Fig. 4F). Accordingly, we found that in a mouse model of intestinal inflammation [dextran sodium sulfate (DSS)-induced colitis], Frmpd2 expression was significantly decreased (Fig. 4G). This result remained consistent when relative expression was calculated using Villin as reference gene (Fig. S9) to correct for a potential loss of epithelial cells.
Discussion
Defining the mechanisms of physiological NOD2 activation is pivotal not only to our understanding of how innate immune responses are initiated but also to identifying molecular components relevant for NOD2 dysfunction in intestinal inflammation and Crohn disease.
In this study, we describe the regulatome of NOD2-dependent NF-κB signaling using a stringent high-throughput siRNA screen targeting 7,783 human transcripts. We confirmed known pathway members RIPK2, RELA, and BIRC4 (XIAP) among the top 20 validated NOD2 modulators. Strikingly, our data revealed the FERM and PDZ domain-containing 2 membrane protein as a NOD2 interactor and spatial organizer of NOD2-mediated NF-κB activation in IECs. FRMPD2 is an example of a basolateral membrane protein that is necessary for effective NOD2 downstream signaling. Although membrane recruitment of NOD2 has been shown to be essential for NF-κB activation (18), hitherto identified NOD2-interacting membrane proteins ERBB2IP and CD147 (19, 25, 29) exert a negative regulatory function on NOD2-mediated NF-κB activation.
FRMPD2 contains PDZ domains, which are known to direct proteins to the basolateral membrane (20) and to facilitate assembly of large signaling complexes (30). We found that FRMPD2 critically directs NOD2-mediated MDP recognition to the basolateral membrane of IECs. NOD2 colocalizes and physically interacts with FRMPD2, and the presence of ERBB2IP further enhances this interaction. Notably, a previous study showed that also the adaptor molecule RIPK2 is recruited to the plasma membrane, thereby enhancing NF-κB transcriptional activity (28). We show that silencing of FRMPD2 clearly diminishes membrane recruitment of RIPK2, yet we are not able to demonstrate a strong physical interaction of FRMPD2 and RIPK2 by immunoprecipitation. Therefore, our data suggest that upon basolateral MDP stimulation, FRMPD2 acts as a scaffold for NOD2 and ERBB2IP that may overcome the previously noted inhibitory influence of ERBB2IP and enables subsequent RIPK2 recruitment, leading to NF-κB activation and induction of IL-8 and BD-2 (illustrated in Fig. 5). Because FRMPD2-silenced IECs are impaired in their NF-κB response to MDP and iE-DAP, but not to TNF-α and TLR ligands, FRMPD2 can be considered as a specific modulator of the NOD2 and presumably also NOD1 pathways. In light of these findings, it will be interesting to further elucidate the exact influence of FRMPD2 on signaling events induced by NOD1 [which also localizes to the membrane (31) but does not interact with ERBB2IP (25) and other NLR-family members]. Of note, silencing of FRMPD2 also diminished NF-κB activation upon MDP stimulation in nonpolarized monocytic THP-1 cells, pointing to a broader role of FRMPD2 in membrane-associated effective NOD2 signaling in nonpolarized cell populations.
Fig. 5.
Spatial specificity of MDP-induced NOD2 signaling in polarized IECs. Upon basolateral MDP stimulation, NOD2 is recruited to the plasma membrane by FRMPD2, which is facilitated by ERBB2IP. Subsequently, RIPK2 joins the complex and enables NF-κB activation and induction of the proinflammatory chemokine IL-8 and antimicrobial peptide BD-2 (Left). The CD-associated genetic variant of NOD2 (L1007fsinsC) leads to a truncated LRR of NOD2. In response to MDP stimulation, the interaction of FRMPD2 with NOD2 L1007fsins variant C is impaired. This potentially leads to a disturbed compartmentalization of NOD2 and failure of NF-κB activation and target gene induction (Right).
As noted earlier, the most prevalent CD-associated genetic variant of NOD2 (L1007fsinsC) leads to a truncation of the last 33 amino acids in the LRR of NOD2. Cells expressing this variant are impaired in their NF-κB response upon MDP stimulation (14, 17), and NOD2 membrane localization is completely abrogated (18). Interestingly, other known NOD2 membrane-interacting proteins identified so far, namely ERBB2IP and CD147, interact with the CARD effector-binding domains that remain structurally unaltered in the L1007fsinsC form of NOD2. Thus, the question why the mutated NOD2 form loses its propensity to shuttle to the cellular membrane (19, 25, 29) has remained unsolved so far. We show that the PDZ2 domain of FRMPD2 recruits the LRR of NOD2 to the membrane and that this direct interaction of the LRR of NOD2 with FRMPD2 is drastically impaired in the L1007fsinsC variant. This interaction sheds light on the potential mechanism of how genetic truncation of the LRR leads to disturbed intracellular compartmentalization of NOD2. As a consequence, the spatial specificity of MDP-induced NOD2 signaling is lost and IL-8 and BD-2 expression is abrogated (illustrated in Fig. 5). Our findings that FRMPD2 is down-regulated in intestinal inflammation and in the mucosa of CD patients may account for dysregulated NF-κB activation and disturbed barrier integrity in diseased individuals. Although we did not find a specific PDZ-binding motif in the LRR of NOD2, which usually occurs at the C-terminal end of a protein, mutations of the last two leucines in the C terminus of NOD2 have previously been shown to abolish membrane localization (18) and could potentially represent the PDZ-binding site that is deleted in the L1007fsinsC NOD2 variant.
In conclusion, we identify FRMPD2 as a scaffold molecule that spatially organizes the assembly of the critical NOD2-signalosome components of NOD2 and RIPK2 at the basolateral membrane of epithelial cells, which is required for effective NF-κB activation. We hypothesize that further insights into the regulation of this molecular landing pad of NOD2 at the membrane might provide novel therapeutic strategies modulating innate immune responses in acute and chronic inflammatory diseases.
Materials and Methods
First and second siRNA screens were performed with Silencer Human Druggable Genome siRNA Library V3 (Ambion). The third siRNA screen and the TNF-α and Caco-2 screens were performed using siRNA pools (siGENOME; Dharmacon). siRNAs were transfected using siPORT Amine (Ambion) in a 96-well format. Plasmids for NOD2 and luciferases were transfected using FuGENE 6 (Roche). Cell lysates were analyzed on a 96-well microplate reader (Tecan). Please refer to SI Materials and Methods for detailed explanations of experimental procedures. Polarized Caco-2 monolayers were grown for 14–21 d on transwell microplates (Corning). Coimmunoprecipitations and Western blotting were performed on cells transfected with respective plasmids or on endogenous proteins (see SI Materials and Methods for full details). Precipitates were washed, eluted, and separated by SDS/PAGE. After transfer onto PVDF membranes (Millipore), Western blotting was performed as described (29). Fluorescence images were captured using a Zeiss AxioImager.Z1 apotome fluorescence microscope or Leica TCS SP5 AOBS confocal microscope (see SI Materials and Methods for full details). Analysis of the biological processes was conducted as previously published (32); gene ontology terms (category: biological process) were retrieved from the Gene Ontology Consortium database (www.geneontology.org). Statistically significant differences between samples were determined using the Student’s t test. A P value <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Thomas Kufer for the gift of ERBB2IP-myc, and D. Quitzau, T. Kaacksteen, K. Goebel, D. Oelsner, and M. Reffelmann for their technical assistance. This work was supported by grants from the National Genome Research Network (NGFN) Chronic Inflammatory Barrier Diseases, German Research Foundation (DFG) Individual Grant RO2994/5-1, DFG Clusters of Excellence “Inflammation at Interfaces” and “Future Ocean,” and Interreg IV Project HIT-ID.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.P.-Y.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209673109/-/DCSupplemental.
See Commentary on page 21188.
References
- 1.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol. 2011;12(9):817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 2.Lange C, et al. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. 2011;28(5):1687–1702. doi: 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- 3.Ogura Y, et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276(7):4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 4.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 5.Mo J, et al. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J Biol Chem. 2012;287(27):23057–23067. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inohara N, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275(36):27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 7.Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14(24):2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 8.Krieg A, et al. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci USA. 2009;106(34):14524–14529. doi: 10.1073/pnas.0907131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, et al. Regulation of IL-8 and IL-1beta expression in Crohn’s disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13(16):1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 10.Voss E, et al. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem. 2006;281(4):2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- 11.Lipinski MM, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci USA. 2010;107(32):14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6(5):376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 13.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 14.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 15.Hampe J, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357(9272):1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 16.Bonen DK, et al. Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124(1):140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 17.Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, Nalps and Naip: Intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 2003;5(9):581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- 18.Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-kappaB activation in muramyl dipeptide recognition. J Cell Biol. 2005;170(1):21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald C, et al. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280(48):40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 20.Stenzel N, Fetzer CP, Heumann R, Erdmann KS. PDZ-domain-directed basolateral targeting of the peripheral membrane protein FRMPD2 in epithelial cells. J Cell Sci. 2009;122(Pt 18):3374–3384. doi: 10.1242/jcs.046854. [DOI] [PubMed] [Google Scholar]
- 21.Jensen LJ, et al. STRING 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hisamatsu T, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124(4):993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 23.Rosenstiel P, et al. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology. 2003;124(4):1001–1009. doi: 10.1053/gast.2003.50157. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski S, et al. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci. 2009;122(Pt 19):3522–3530. doi: 10.1242/jcs.050690. [DOI] [PubMed] [Google Scholar]
- 25.Kufer TA, Kremmer E, Banks DJ, Philpott DJ. Role for Erbin in bacterial activation of Nod2. Infect Immun. 2006;74(6):3115–3124. doi: 10.1128/IAI.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sedar AW, Forte JG. Effects of calcium depletion on the junctional complex between oxyntic cells of gastric glands. J Cell Biol. 1964;22(1):173–188. doi: 10.1083/jcb.22.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov AI, et al. Microtubules regulate disassembly of epithelial apical junctions. BMC Cell Biol. 2006;7:12. doi: 10.1186/1471-2121-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lécine P, et al. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282(20):15197–15207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- 29.Till A, et al. A role for membrane-bound CD147 in NOD2-mediated recognition of bacterial cytoinvasion. J Cell Sci. 2008;121(Pt 4):487–495. doi: 10.1242/jcs.016980. [DOI] [PubMed] [Google Scholar]
- 30.Nourry C, Grant SG, Borg JP. PDZ domain proteins: Plug and play! Sci STKE. 2003;2003(179):re7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 31.Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008;10(2):477–486. doi: 10.1111/j.1462-5822.2007.01062.x. [DOI] [PubMed] [Google Scholar]
- 32.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22(3):281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.