Abstract
For centuries ecologists have studied how the diversity and functional traits of plant and animal communities vary across biomes. In contrast, we have only just begun exploring similar questions for soil microbial communities despite soil microbes being the dominant engines of biogeochemical cycles and a major pool of living biomass in terrestrial ecosystems. We used metagenomic sequencing to compare the composition and functional attributes of 16 soil microbial communities collected from cold deserts, hot deserts, forests, grasslands, and tundra. Those communities found in plant-free cold desert soils typically had the lowest levels of functional diversity (diversity of protein-coding gene categories) and the lowest levels of phylogenetic and taxonomic diversity. Across all soils, functional beta diversity was strongly correlated with taxonomic and phylogenetic beta diversity; the desert microbial communities were clearly distinct from the nondesert communities regardless of the metric used. The desert communities had higher relative abundances of genes associated with osmoregulation and dormancy, but lower relative abundances of genes associated with nutrient cycling and the catabolism of plant-derived organic compounds. Antibiotic resistance genes were consistently threefold less abundant in the desert soils than in the nondesert soils, suggesting that abiotic conditions, not competitive interactions, are more important in shaping the desert microbial communities. As the most comprehensive survey of soil taxonomic, phylogenetic, and functional diversity to date, this study demonstrates that metagenomic approaches can be used to build a predictive understanding of how microbial diversity and function vary across terrestrial biomes.
Keywords: shotgun metagenomics, soil microbial ecology, 16S rRNA gene sequencing, biogeography
Soil microorganisms play critical roles in regulating soil fertility, plant health, and the cycling of carbon, nitrogen, and other nutrients. Every gram of soil harbors thousands of bacterial, archaeal, and eukaryotic taxa, and this taxonomic diversity is mirrored by the diversity of their protein-encoded functions, encompassing a seemingly limitless array of physiologies and life history strategies. Although these characteristics of soil microbial communities have been known for decades, the ongoing development of high-throughput molecular tools (and the tools necessary to analyze the associated flood of data) allow microbial ecologists to characterize the taxonomic, phylogenetic, and functional diversity of soil microbial communities to an extent that was unimaginable only a few years ago. We can now move beyond detailed studies of individual soils to conduct detailed comparative studies of soils across broad spatial gradients.
Perhaps the most dramatic and well-studied spatial gradients in biological diversity are those that exist across the major global terrestrial biomes. Different biomes typically harbor distinct assemblages of macrobial (plant and animal) taxa and ecologists have spent many decades describing the apparent differences in biological diversity. Although comparable research on the biogeographical patterns exhibited by microbial taxa has lagged far behind research on plant and animal communities (1), we are beginning to understand how soil microbial diversity varies across the globe and how this diversity is related to the physical, chemical, and biological characteristics of ecosystems. In particular, we now know that soil bacterial communities are strongly influenced by pH, which explains a large proportion of the variance in soil bacterial diversity and community composition at local (2, 3), regional (4–6), and continental scales (7). Soils with near-neutral pH typically have higher bacterial diversity than more acidic or more basic soils and the relative abundances of many bacterial phyla have been shown to be strongly correlated with soil pH (7). Of course, soil pH is not the only factor that can influence bacterial communities and there is evidence that other microbial taxa that are abundant in soil (including Archaea, fungi, and protists) do not necessarily exhibit the same biogeographical patterns observed for bacteria (2, 8). Changes in the types and quantities of organic carbon added to soil can have considerable influences on soil microbial communities (9, 10) and, depending on the gradients being studied or the experimental treatments imposed, other factors such as soil temperature, moisture, and nutrient availability have also been shown to influence microbial structure in soil.
Although our understanding of the phylogenetic and taxonomic biogeography of soil microbial communities continues to expand, there has been limited progress in understanding how the functional capabilities of soil microbial communities change across biomes. For individual well-studied soil microbial processes (e.g., N2 fixation) (11) or specific extracellular enzymes (12), researchers have been able to document their interbiome characteristics. Likewise, previous work has demonstrated how specific functional groups or gene categories can vary across space (e.g., ref. 13). However, we lack an integrated understanding of how the functional genes encoded in their collective genomes act to structure communities across environmental gradients. Although we might expect an overall correlation between taxonomic composition and the functional attributes of soil microbial communities, this may not always be the case as distinct taxa can share specific functional attributes and closely related taxa may have very different physiologies and environmental tolerances (14). As a result, we cannot rely entirely on our understanding of the biogeographical patterns in the taxonomic or phylogenetic structure of soil microbial communities to predict the functional attributes or the functional diversity of these communities (15). Using shotgun metagenomic sequencing, the direct sequencing of the collective genomes found in a given environmental sample, researchers have gained important insight into the potential functions of microbial communities from individual soil types (16, 17) and soils that have been experimentally manipulated in the laboratory (18). The value of shotgun metagenomic analyses is that a far more comprehensive understanding of the traits microbes may use to survive in an individual soil can be identified, traits which are often very difficult to measure using biogeochemical or culture-based approaches (19). To our knowledge, such tools have not yet been used to directly compare microbial metagenomes across soils representing a range of different biomes.
The current study was designed to test the hypothesis that the microbial communities found in desert soils are taxonomically and functionally distinct compared with those found in other biomes and that the variability across different desert sites is less than the variability between desert and nondesert biomes. This was predicated on the fact that at least three of the main factors known to shape the composition of soil microbial communities: pH, moisture availability, and inputs of plant-derived organic carbon, are often very different between desert and nondesert soils. Desert soils are drier, typically have higher pH soils than other biomes, and the paucity (or complete absence) of plant biomass reduces the inputs of organic carbon. We used shotgun metagenomic sequencing of soils from 16 sites representing a wide range of ecosystems (forests, grassland, tundra, and deserts) to determine how the functional capabilities of soil microbial communities vary across the major global terrestrial biomes and the extent to which these capabilities are predictable. We addressed three basic questions: Do deserts (both cold and hot) harbor microbial communities that are taxonomically, phylogenetically, and functionally distinct from those found in forests, grasslands, and tundra? What functional attributes distinguish desert and nondesert soil microbial communities? Can we use information on the taxonomic diversity and composition of soil microbial communities to predict their functional attributes?
Results and Discussion
General Characteristics of the Soil Microbial Communities.
Soils were collected from 16 sites: 3 from hot deserts, 6 from Antarctic cold deserts, and 7 from temperate and tropical forests, a prairie grassland, a tundra, and a boreal forest (Table S1). The sites were selected to span a wide range of ecologically distinct biomes to examine how cold desert soils compare with hot deserts, and to forests, prairie, and tundra. Using a shotgun metagenomic approach, we obtained a total of 3.9–11 million 100-bp sequences per sample (390–1,100 Mbp per sample). Only 13–23% of the sequences (688,000–1,900,000 reads per sample) could be annotated using the technique applied (Table S2), a percentage similar to that reported in previous studies that used shotgun metagenomic sequencing to characterize soil microbial communities (17, 20) and communities in other highly diverse microbial habitats (21, 22). As survey depth can affect estimation of the relative abundances of gene categories, all of the shotgun metagenomic datasets were rarefied by randomly subsampling 688,000 annotated reads per sample before downstream analyses.
The majority of the shotgun metagenomic reads were derived from bacteria, as shown by the analysis of both the large-subunit (LSU) and small-subunit (SSU) rRNA reads recovered from the metagenomic data. Between 74% and 96% of either the SSU or the LSU reads were assigned to bacterial or archaeal taxa (Table S3). Although fungi and other eukaryotes can represent a large portion of the microbial biomass contained within soils, their representation in the metagenomic data was low. A similar pattern has been observed in comparable shotgun metagenomic datasets obtained from other soils (17, 20) and is most likely a product of many eukaryotic taxa (including fungi), having a far lower ratio of rRNA gene copies per unit biomass than bacterial cells. However, the ratio of fungal:bacterial rRNA reads did vary across soils in this study, with temperate and boreal forests having the highest fungal:bacterial ratios (Table S3), a pattern similar to that noted previously (23).
An amplicon survey of a portion of the 16S rRNA gene was performed to provide a higher resolution and more in-depth analysis of the composition and diversity of the soil bacterial communities. We used barcoded primers that target the V4 region of the 16S rRNA gene from both bacteria and Archaea with the resulting amplicons sequenced on the Illumina HiSeq platform (24). All samples were compared at an equivalent sequencing depth of 118,000 randomly selected 16S rRNA gene amplicons per sample. These results show that all of the communities were dominated by Acidobacteria, Actinobacteria, Bacteroidetes, Proteobacteria, and Verrucomicrobia (Fig. S1), bacterial phyla that are known to be relatively abundant and ubiquitous in soil (25). Additional phyla including Chloroflexi, Cyanobacteria, Firmicutes, and Gemmatimonadetes were also found in nearly all soils, but their relative abundances were highly variable and typically represented less than 5% of the 16S rRNA reads in any individual soil (although Cyanobacteria were more abundant in some of the desert soils, Fig. S1). Archaea were relatively rare in all soils (0.01–6.7% of reads) but were most abundant in the three hot desert sites and one of the tropical rainforest sites (Fig. S1). The observed range in archaeal abundances and the observation that Thaumarchaeota were the dominant archaeal group in nearly all of the soils validates results reported previously (8).
Comparison of Community Structure Determined via the Amplicon and Shotgun-Metagenomic Approaches.
The 16S rRNA gene data obtained from both the amplicon and shotgun sequencing were used to directly compare the taxonomic results obtained using these two very different methods. We conducted this comparison to determine whether biases introduced by both approaches may influence the determination of bacterial community structure, as suggested previously (26). Such biases may be derived from the PCR process itself or because the 16S rRNA gene regions recovered from the metagenomic data can span the entire length of the gene, whereas the PCR-based amplicon approach only targets the V4 region. Because different regions of the 16S rRNA gene vary in the accuracy of their taxonomic assignments (27), the two approaches may not necessarily give identical results. However, this was not the case; the two methods generated nearly identical estimates of bacterial community composition. This is evident from the strong correlation between the Bray–Curtis distance matrices (Spearman r = 0.91, P < 0.001), and by directly comparing the relative abundances of the dominant taxa (Fig. S2). In addition, although the 16S rRNA amplicon dataset contained orders of magnitude more of the 16S rRNA reads than the shotgun-derived 16S rRNA dataset (118,000 and 1,884 reads per sample, respectively), the estimates of taxonomic richness for each dataset were significantly correlated (r2 = 0.81, P < 0.001). The strong concordance between these two very different approaches suggests that, at least across the wide range of soils examined here, the two methods yield nearly identical estimates of the overall differences in soil bacterial community diversity and composition.
Alpha Diversity Patterns.
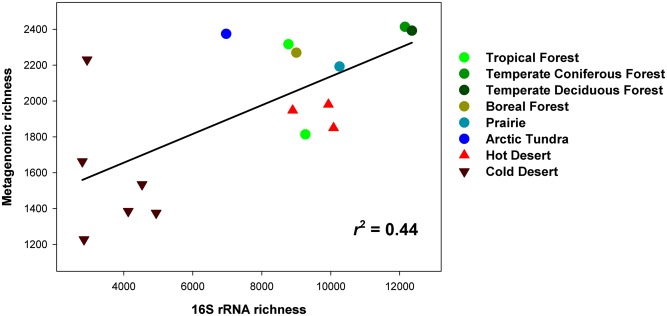
Alpha diversity, the richness and/or evenness of taxa or lineages contained within an individual community, was highly variable across the 16 soils (Fig. 1). The richness of the bacterial and archaeal communities ranged from <4,000 to >12,000 phylotypes per sample (Table S2) with all samples compared at an identical sequencing depth. The cold desert soils harbored far lower diversity than the other soils regardless of the taxonomic or phylogenetic metric used (Fig. 1 and Table S2). This trend is similar to that observed for invertebrates, with soils from the McMurdo Dry Valleys in Antarctica having very low levels of invertebrate diversity and extremely simple food webs (28). Although it has been reported that these cold desert soils harbor surprisingly high levels of bacterial diversity (29), we find that their diversity is actually far lower, on average, than that found in other biome types.
Fig. 1.
Differences in alpha diversity levels across 16 soils. X axis shows taxonomic richness of the bacterial communities (number of phylotypes out of 118,000 amplicon reads per sample). Y axis shows the functional gene richness (number of functional gene categories identified from 688,000 annotated shotgun metagenomic reads per sample). See Table S2 for additional information on diversity levels across the 16 soils.
As has been demonstrated previously (7), soil pH is a reasonably good predictor of prokaryotic diversity across the 16 soils (y = −533x2 + 6,371x −8,823; r2 = 0.6, where x = soil pH and y = phylotype richness). Soils close to neutral had the highest diversity levels, whereas soils that were either very basic (the desert soils) or acidic (the Peruvian tropical rainforest soil and the Arctic tundra soil) had lower levels of diversity. As this study only included 16 soils that differ in a wide variety of ways, we cannot use this sample set to definitively identify the edaphic or site factors responsible for the diversity patterns observed here—indeed, there are many possible reasons why these soils harbor such different levels of bacterial diversity. For example, it is possible that the low diversity of the cold desert soils is not directly related to their very high pH levels, but rather due to their high salinities, negligible plant-carbon inputs, or the extreme moisture and temperature conditions encountered at those sites (29–31).
Although functional alpha diversity is less frequently measured, it is increasingly common for both macrobial ecologists (32, 33) and microbial ecologists (15, 34) to consider the diversity and distributions of functional traits (or functional genes) across communities. Functional diversity (the richness of protein-coding gene categories identified out of 688,000 reads per metagenome, Fig. 1) was typically lowest in the cold desert soils, intermediate in the hot desert soils, and highest in the nondesert soils (a pattern unrelated to the percentage of reads that could be annotated from each soil, Table S2). However, there was notable variation within these broadly defined categories. For example, one of the cold desert soils (EB026) had far higher functional diversity than the other cold desert soils. This is likely a result of that soil having a broader array of genes associated with photosynthesis and carbon-fixation pathways than the other cold desert soils, as evidenced from both the metagenomic data (Fig. S3) and from the higher abundances of Cyanobacteria in that soil compared with the other soils (Fig. S1).
There were significant correlations between functional diversity and both the taxonomic (Fig. 1) and phylogenetic diversity of the bacterial communities (P < 0.001 in both cases), with the cold desert soils consistently harboring the lowest levels of diversity. This finding highlights that the overall diversity of functional gene categories found in a given sample is, to some degree, predictable from the taxonomic or phylogenetic diversity of the microbial communities. A similar pattern has been observed in other studies of microbial communities (16, 35, 36), demonstrating that functional redundancy at the genomic level is not so pervasive as to obscure any relationship between these very different metrics of diversity. However, the correlations between functional diversity and taxonomic or phylogenetic diversity were largely driven by the cold desert soils and were not significant when the cold desert soils were omitted from the analyses (r2 < 0.2, P > 0.1 in both cases). This suggests that functional diversity is not necessarily predictable from the taxonomic or phylogenetic diversity of communities when comparing vegetated soils. Likewise, it is worth noting that one of the samples with the highest levels of metagenomic richness (the cold desert soil EB026) had nearly the lowest level of taxonomic richness (Table S2), suggesting that the types of taxa found in a community are also important to consider when trying to predict functional diversity. Although it is often observed that macrobial communities with lower taxonomic or phylogenetic diversity have reduced functional diversity (32, 33), this paradigm does not necessarily hold true for microbial communities.
Beta Diversity Patterns—Bacterial Community Composition.
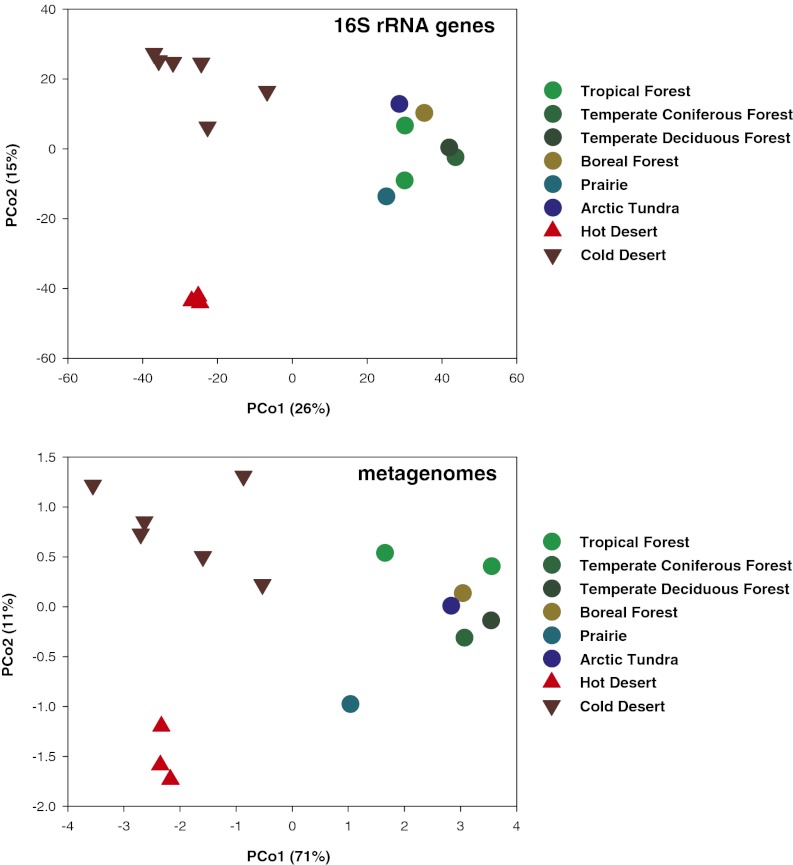
Biome-specific differences between the 16 soil communities were evident from the 16S rRNA amplicon data (Fig. S1 and Fig. 2). The desert soils harbored communities that clustered apart from the nondesert communities when community differences were measured using either a taxonomic metric (Bray–Curtis distance, Fig. 2) or a phylogenetic metric (unweighted Unifrac) with both metrics yielding nearly identical patterns. The hot desert and cold desert soils were taxonomically (Bray–Curtis analysis of similarity, ANOSIM R = 0.91 and 0.89, respectively, P < 0.002 in both cases) and phylogenetically (unweighted Unifrac ANOSIM R = 0.98 and 0.89, respectively, P < 0.005 in both cases) distinct from those found in the nondesert soils. Also, whereas the cold and hot desert soil communities were distinct (unweighted Unifrac ANOSIM R = 0.65, P = 0.01), these differences were less than the differences between the desert and nondesert soils. Although different biomes clearly harbor distinct bacterial communities (Fig. 2), the largest distinction was between the desert and nondesert biomes with the cold and hot desert soils harboring relatively similar bacterial communities.
Fig. 2.
Ordination plots derived from principal coordinates analyses of Bray–Curtis distances between bacterial community composition (Upper, based on amplicon 16S rRNA gene data) and metagenome composition (Lower, based on annotated shotgun metagenomic data).
The general taxonomic patterns evident in Fig. 2 are largely driven by differences in the abundances of major taxonomic groups. The Actinobacteria, Bacteroidetes, and Cyanobacteria phyla were generally more abundant in the desert soils than in the nondesert soils, whereas Verrucomicrobia and Acidobacteria showed the opposite pattern (Fig. S1). Overall, the composition of the desert soil communities surveyed here was similar to those reported in other studies of cold and hot desert microbial communities (30, 37). More generally, the results shown here confirm the broad-scale patterns we would expect based on pH differences; high pH soils (such as those found in the desert soils included in this study) typically have higher relative abundances of Actinobacteria and Bacteroidetes with lower abundances of Acidobacteria compared with more acidic soils (6, 7). We note that the cold desert soil EB017 has a high abundance of Acidobacteria but these Acidobacteria belong to the class Chloracidobacteria that is distinct from the acidobacterial group (Solibacteres), which dominates in low pH soils; this is a pattern we would expect based on the results reported in Jones et al. (38). Factors other than pH may also be driving the bacterial community patterns evident in Fig. S1 and Fig. 2. For example, taxa known to be tolerant of low moisture conditions, including Actinobacteria (39), were more abundant in the desert soils surveyed here, whereas those taxa commonly associated with soils receiving higher rates of organic carbon inputs (e.g., beta- and gammaproteobacteria (10), were relatively less abundant in the desert soils.
Although Cyanobacteria and Proteobacteria were typically more abundant in the hot desert soils than in the cold desert soils (Fig. S1), the hot and cold deserts harbored relatively similar bacterial communities (as noted above). Despite large differences in site and edaphic characteristics, including the complete absence of plants in the cold desert sites and very low mean annual temperatures, cold and hot desert soil communities were relatively similar. This suggests that other factors common across these desert types (such as high soil pHs and low moisture levels) are most important in structuring these communities.
Beta Diversity Patterns—Functional Genes.
The beta diversity patterns determined from the 16S rRNA gene analyses were nearly identical to the patterns determined from a comparison of functional gene abundances across the 16 soil metagenomes (Fig. 2). The Bray–Curtis distances calculated from taxon abundances and functional gene abundances were significantly correlated (Mantel r = 0.76, P < 0.001). Likewise, there was a strong correlation between unweighted Unifrac distances, a phylogenetic metric of community similarity, and the Bray–Curtis distances in functional gene abundances (Mantel r = 0.82, P < 0.001). Therefore, as with the alpha diversity patterns, the concordance in beta diversity patterns highlights that the overall functional differences between the soil microbial communities were significantly correlated with the differences in the composition of these communities. Our findings are in line with comparable studies conducted in soil (20) and other habitats that also found strong correlations between metagenome composition and taxonomic composition (34, 35, 40). Although individual functional genes may not necessarily be correlated with community structure, the overall functional attributes of soil microbial communities appear to be predictable across broad gradients in soil and biome types if one has information on the taxonomic or phylogenetic structure of the communities.
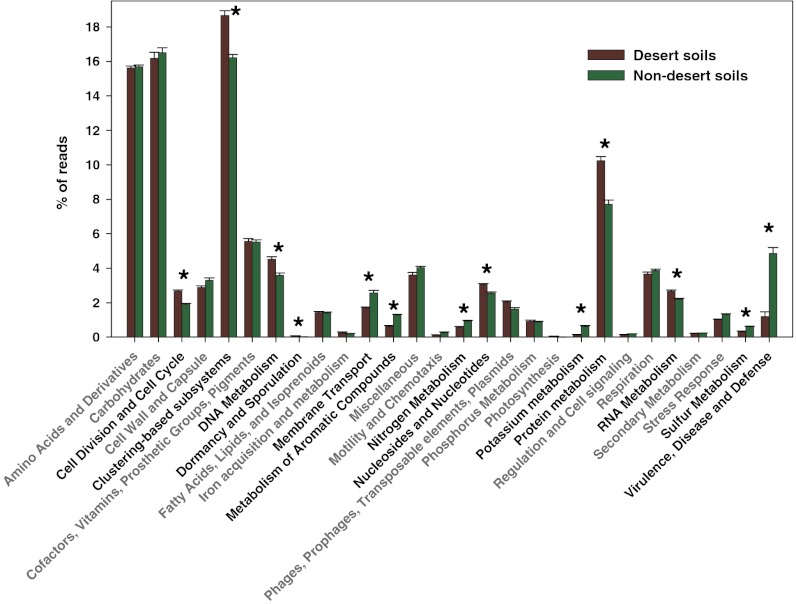
Both the cold desert soils and hot desert soils had metagenomes distinct in composition from those found in the nondesert soils (ANOSIM R = 0.97 and 0.98 respectively, P < 0.005 in both cases), a pattern clearly evident from the ordination plot (Fig. 2) and the corresponding heatmap (Fig. S3). The large differences between desert and nondesert soils were also evident from a comparison of the relative abundances of functional genes classified at the lowest level of resolution (Fig. 3). After correction for multiple comparisons, 13 of 28 major gene categories were significantly different in abundance between desert and nondesert soils (Fig. 3), patterns that were examined in more detail by identifying the 35 specific gene categories (out of 417 in total) that strongly differentiated the desert soil metagenomes from the nondesert metagenomes (Fig. S4). The cold and hot desert microbial communities also had metagenomes that were distinct in composition from one another (Fig. 2; ANOSIM R = 0.41, P = 0.03), but the differences between these desert soils were less than the differences between the desert and nondesert soils.
Fig. 3.
Relative abundances of major categories of functional genes in the shotgun metagenomes obtained from the desert soils (both cold and hot deserts) versus the other, nondesert, biomes. Asterisks and bold type indicate those categories with significantly different relative abundances in desert and nondesert soils (Bonferroni corrected P values <0.05, uncorrected P values <0.002).
Many of the gene categories that were more abundant in the desert soils than in the nondesert soils were those related to core metabolic functions (Fig. 3 and Figs. S3 and S4). Given that we were determining relative abundances, the overrepresentation of these gene categories in the desert soils may simply be a product of the desert soils having reduced diversity; lower phylogenetic or metagenomic diversity would presumably lead to an increase in the relative abundances of those core genes that are shared by nearly all cells and are required for cell survival and replication. However, some of the observed differences in functional gene abundances between the desert and nondesert soils may be more directly related to the unique conditions found in deserts, including lower moisture availability and reduced plant biomass. For example, we would expect nutrient cycling rates to be lower in desert systems than in more mesic systems due to moisture constraints (41), a pattern that was confirmed by the higher relative abundances of genes associated with nitrogen, potassium, and sulfur metabolism in the nondesert soils (Fig. 3 and Fig. S4). Likewise, exposure to frequent moisture stress may explain why the desert soils have higher relative abundances of genes associated with dormancy/sporulation, stress proteins, and amino acid metabolism (amino-acid–based solutes are commonly used by bacteria for osmoregulation) (39). The desert soils had lower relative abundances of genes associated with the degradation of complex organic compounds, including aromatics (Fig. S4), a pattern likely related to the lower levels of plant biomass found in the desert soils. Plants typically represent major sources of organic carbon to soil and these pools of organic carbon are often distinct (and more enriched in aromatics) (42) in soils supporting more plant biomass than in soils where plants are less abundant or nonexistent where we would expect microbe-derived organic carbon pools to dominate.
One of the most striking differences between desert and nondesert soil microbial communities was the differential abundance of antibiotic resistance genes and other genes likely associated with microbe–microbe competition. Genes associated with antibiotic resistance were far less abundant in the desert soils (averaging 1.5% of the annotated reads) than in the nondesert soils (averaging 4.8% of the annotated reads, Fig. S4). Likewise, murein hydrolases, which cleave bacterial peptidoglycan and are frequently associated with bacterial cell lysis (43), were consistently more abundant in nondesert soils than in the desert soils (Fig. S4). We hypothesize that these patterns reflect reduced microbial competition in the desert soils. The production of antibiotics and resistance to antibiotics are traits that are widespread among soil bacteria and fungi (44, 45), with both models and experimental data suggesting that elevated microbe–microbe competition should select for increased antibiotic production and resistance (46–48). Likewise, murein hydrolase production has been linked to antagonistic interactions between microbes and a range of antimicrobial defenses (43). In the desert soils, where conditions are less conducive to microbial growth, adaptations that enhance microbial competition may be less important than adaptations that allow for persistence of cells under adverse environmental conditions or the ability to respond rapidly to pulses in moisture availability. Although additional work is required to verify this hypothesis, our results do suggest that the intensity of competitive interactions within microbial communities varies as a function of environmental conditions, a phenomenon that has frequently been observed in plant and animal communities (49, 50).
Only three major gene categories were significantly different in abundance between the cold and hot desert communities (Fig. S5): genes associated with the metabolism of carbohydrates and aromatic compounds being relatively more abundant in the hot desert soils. This pattern is further supported by the determination of functional gene abundances at a higher level of resolution (Fig. S6) as we found genes associated with monosaccharide utilization, carbohydrate transporters, and aromatic compound catabolism to be relatively more abundant in the hot desert soils. As described above, these functional differences are likely linked to differences in the quantity or quality of plant-carbon inputs, because the hot desert soil microbial communities likely receive far more plant-derived carbon than the cold deserts where plants are absent.
Caveats.
The shotgun metagenomic results presented above should be considered carefully given that the technique has clear limitations. First, with only 688,000 annotated metagenomic reads per sample, we have not captured the full extent of the genomic diversity contained within individual samples and deeper sequencing would have allowed us to describe changes in the relative abundances of rarer (yet potentially important) genes or gene categories. Nevertheless, we were still able to detect clear differences across biomes suggesting that, for certain questions, shallower sequencing of many samples may be more useful than deeper sequencing of fewer samples (51). Second, only 13–23% of the sequence reads in this study could be annotated; the genomes of many important soil taxa have not been sequenced and even fewer have been appropriately annotated. We are invariably misannotating genes or ignoring genes that may have important functions or may account for key differences across biomes, a problem that plagues every study that uses shotgun metagenomic analyses (52). Third, even though this study represents one of the largest cross-site terrestrial metagenomic surveys conducted to date, we recognize that the sites sampled here do not necessarily represent each of the biomes in question and that even more samples are required to adequately assess intrabiome variability. However, given the strength of the patterns observed, particularly the clear separation between desert and nondesert soils, we suspect that more comprehensive analyses will further confirm the general patterns observed here. Finally, we only examined soils at a single time point per site, as it was not our goal to also quantify the temporal variability in the soil metagenomes. Although the metagenomes are unlikely to be static over time, previous work has demonstrated that the temporal variability in the composition of soil bacterial communities is typically far lower than the spatial variability (53–55) so we would expect the general patterns observed here to persist across seasons.
Conclusions.
This study represents one of the most comprehensive analyses of soil metagenomes conducted to date with >1.2 Mbp of 16S rRNA gene data and >390 Mbp shotgun metagenomic data obtained from each of 16 soils (∼20 Mbp of 16S rRNA data, and 6.2 Gbp of shotgun metagenomic data). However, even at these sequencing depths, we have not surveyed the full extent of microbial taxonomic, phylogenetic, or functional diversity found within individual soil samples and, with only 16 soils, we have not described the full range of soil microbial community types found across the globe. Nevertheless, we were still able to detect strong patterns in the datasets that highlight the predictability of soil microbial community attributes across biomes. Like plant and animal communities, the diversity and relative abundances of major soil microbial taxa and functional gene categories can be related to broad-scale gradients in biotic and abiotic characteristics. Functional diversity was significantly correlated with phylogenetic and taxonomic diversity across the 16 soils, but these patterns were driven by the very low levels of diversity observed in the soils from the cold desert sites. The microbial metagenomes obtained from the cold and hot desert soils were relatively similar to one another, suggesting that the composition and functional attributes of the microbial communities in these two desert types may be more comparable than often assumed (56). The metagenomes recovered from the nondesert soils clustered together apart from the desert soils even though they represented a wide range of biomes that included tropical forests, tundra, and a prairie.
Microbial ecology continues to lag far behind plant and animal ecology in our ability to resolve large-scale biogeographical patterns in diversity, community composition, and functional attributes. However, this work highlights how coupling metagenomic analyses with extensive cross-site sampling efforts can reduce this disparity. As sequencing capacities continue to increase and tools for analyzing the resulting data become more effective, we will soon be able to expand upon the work presented here and gain a more comprehensive understanding of how soil microbial communities vary across time and space.
Materials and Methods
Additional information on sample collection and analytical methods is provided in SI Materials and Methods.
The cold desert soils were collected from various sites with the McMurdo Dry Valleys region of Antarctica with the hot desert soils collected from sites in the southwestern United States. The seven “nondesert” soils were collected from tropical forests in Peru and Argentina, an arctic tundra in Alaska, a native tallgrass prairie in Kansas, a temperate deciduous forest in South Carolina, a temperate coniferous forest in North Carolina, and a boreal forest in Alaska (Table S1). For both the 16S rRNA gene analyses and the shotgun metagenomic analyses DNA was extracted from each soil sample using the approach described in Fierer et al. (20). To determine the diversity and composition of the bacterial communities in each of these soils, we used the PCR-based protocol described in Caporaso et al. (24) that targets the V4–V5 region of the 16S rRNA gene. Amplicon sequencing was conducted on an Illumina HiSeq2000 with processing of the reads conducted as described in Caporaso et al. (57). For all downstream analyses, we rarefied to 118,000 randomly selected reads per sample to correct for differences in sequencing depth. Reads were assigned to phylotypes at the ≥97% sequence similarity level using the open-reference phylotype picking protocol in QIIME (58). Shotgun metagenomic analyses were conducted on the soil DNA extracts following the Illumina Paired-End Prep kit protocol with sequencing performed using a 2 × 100 bp sequencing run on the Illumina GAIIx. Sequences were uploaded to MG-RAST (59) for downstream analyses and data accession numbers are provided in Table S2. Sequences were annotated to functional categories against the M5NR database using BLASTX at an e-value cutoff of 1 × 10−2 and the SEED subsystems hierarchy. Downstream analyses were performed on the metagenomes evenly sampled at random to 688,000 annotated reads per sample.
Supplementary Material
Acknowledgments
We thank Jessica Henley and Donna Berg-Lyons for their assistance with the molecular analyses. This work was funded by a Department of Agriculture Grant 2008-34158-04713) and National Science Foundation (NSF) Grant DEB-0953331 (to N.F.). Funding for the cold desert research was provided in part by the NSF McMurdo Dry Valleys Long-Term Ecological Research Program Award OPP-0423595 (to D.H.W. and B.J.A.). The Department of Energy supported J.A.G. and S.O. under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Rapid Annotation using Subsystems Technology for Metagenomes database (MG-RAST). Accession numbers are listed in Table S2.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215210110/-/DCSupplemental.
References
- 1.Fierer N, Ladau J. Predicting microbial distributions in space and time. Nat Methods. 2012;9(6):549–551. doi: 10.1038/nmeth.2041. [DOI] [PubMed] [Google Scholar]
- 2.Rousk J, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4(10):1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 3.Osborne CA, Zwart AB, Broadhurst LM, Young AG, Richardson AE. The influence of sampling strategies and spatial variation on the detected soil bacterial communities under three different land-use types. FEMS Microbiol Ecol. 2011;78(1):70–79. doi: 10.1111/j.1574-6941.2011.01105.x. [DOI] [PubMed] [Google Scholar]
- 4.Chu H, et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12(11):2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- 5.Kuramae EE, et al. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol Ecol. 2012;79(1):12–24. doi: 10.1111/j.1574-6941.2011.01192.x. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths RI, et al. The bacterial biogeography of British soils. Environ Microbiol. 2011;13(6):1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- 7.Lauber C, Knight R, Hamady M, Fierer N. Soil pH as a predictor of soil bacterial community structure at the continental scale: A pyrosequencing-based assessment. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates S, Caporaso JG, Walters WA, Knight R, Fierer N. A global-scale survey of archaeal abundance and diversity in soils. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldfarb KC, et al. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol. 2011;2:94. doi: 10.3389/fmicb.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88(6):1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 11.Cleveland CC, et al. Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Global Biogeo Cyc. 1999;13:623–645. [Google Scholar]
- 12.Sinsabaugh RL, et al. Stoichiometry of soil enzyme activity at global scale. Ecol Lett. 2008;11(11):1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- 13.Bru D, et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2011;5(3):532–542. doi: 10.1038/ismej.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philippot L, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8(7):523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 15.Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: From taxonomy to traits. Science. 2008;320(5879):1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 16.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 17.Delmont TO, et al. Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J. 2012;6(9):1677–1687. doi: 10.1038/ismej.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackelprang R, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480(7377):368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 19.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6(11):805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 20.Fierer N, et al. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6(5):1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quaiser A, Zivanovic Y, Moreira D, López-García P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J. 2011;5(2):285–304. doi: 10.1038/ismej.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452(7187):629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 23.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. Global patterns in belowground communities. Ecol Lett. 2009;12(11):1238–1249. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 24.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steven B, Gallegos-Graves LV, Starkenburg SR, Chain PS, Kuske CR. Targeted and shotgun metagenomic approaches provide different descriptions of dryland soil microbial communities in a manipulated field study. Environ Microbiol Rep. 2012;4:248–256. doi: 10.1111/j.1758-2229.2012.00328.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35(18):e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall DH, Virginia RA. Controls on soil biodiversity: Insights from extreme environments. Appl Soil Ecol. 1999;13:137–150. [Google Scholar]
- 29.Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: The microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol. 2010;8(2):129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 30.Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC. The Inter-Valley Soil Comparative Survey: The ecology of Dry Valley edaphic microbial communities. ISME J. 2012;6(5):1046–1057. doi: 10.1038/ismej.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wall D. Biodiversity and ecosystem functioning in terrestrial habitats of Antarctica. Antarct Sci. 2005;17:523–531. [Google Scholar]
- 32.Petchey OL, Gaston KJ. Functional diversity (FD), species richness and community composition. Ecol Lett. 2002;5:402–411. [Google Scholar]
- 33.Diaz S, Cabido M. Vive la difference: Plant functional diversity matters to ecosystem processes. Trends Ecol Evol. 2001;16:646–655. [Google Scholar]
- 34.Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol. 2011;7:473. doi: 10.1038/msb.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert JA, et al. The taxonomic and functional diversity of microbes at a temperate coastal site: A ‘multi-omic’ study of seasonal and diel temporal variation. PLoS ONE. 2010;5(11):e15545. doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant JA, Stewart FJ, Eppley JM, DeLong EF. Microbial community phylogenetic and trait diversity declines with depth in a marine oxygen minimum zone. Ecology. 2012;93(7):1659–1673. doi: 10.1890/11-1204.1. [DOI] [PubMed] [Google Scholar]
- 37.Pointing SB, et al. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 2009;106(47):19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones RT, et al. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009;3(4):442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris R. Effect of water potential on microbial growth and activity. In: Parr J, Gardner W, Elliott L, editors. Water Potential Relations in Soil Microbiology. Madison, WI: Soil Science Society of America; 1981. pp. 23–95. [Google Scholar]
- 40.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manzoni S, Schimel JP, Porporato A. Responses of soil microbial communities to water stress: Results from a meta-analysis. Ecology. 2012;93(4):930–938. doi: 10.1890/11-0026.1. [DOI] [PubMed] [Google Scholar]
- 42.Grandy A, Neff J. Molecular C dynamics downstream: The biochemical decomposition sequence and its impact on on soil organic matter structure and function. Sci Total Environ. 2008;404:221–446. doi: 10.1016/j.scitotenv.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32(2):259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 44.Allen HK, et al. Call of the wild: Antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8(4):251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 45.Dantas G, Sommer MOA, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320(5872):100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 46.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams ST, Vickers JC. The ecology of antibiotic production. Microb Ecol. 1986;12:43–52. doi: 10.1007/BF02153221. [DOI] [PubMed] [Google Scholar]
- 48.Czárán TL, Hoekstra RF, Pagie L. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci USA. 2002;99(2):786–790. doi: 10.1073/pnas.012399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiens JA. Competition and variable environments. Am Sci. 1977;65:590–597. [Google Scholar]
- 50.Chesson P, Huntly N. The roles of harsh and fluctuating conditions in the dynamics of ecological communities. Am Nat. 1997;150(5):519–553. doi: 10.1086/286080. [DOI] [PubMed] [Google Scholar]
- 51.Knight R, et al. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30(6):513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert JA, Meyer F, Bailey MJ. The future of microbial metagenomics (or is ignorance bliss?) ISME J. 2011;5(5):777–779. doi: 10.1038/ismej.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103(3):626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallenstein MD, McMahon S, Schimel J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol Ecol. 2007;59(2):428–435. doi: 10.1111/j.1574-6941.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 55.Krave AS, et al. Stratification and seasonal stability of diverse bacterial communities in a Pinus merkusii (pine) forest soil in central Java, Indonesia. Environ Microbiol. 2002;4(6):361–373. doi: 10.1046/j.1462-2920.2002.00304.x. [DOI] [PubMed] [Google Scholar]
- 56.Pointing SB, Belnap J. Microbial colonization and controls in dryland systems. Nat Rev Microbiol. 2012;10(8):551–562. doi: 10.1038/nrmicro2831. [DOI] [PubMed] [Google Scholar]
- 57.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.