Abstract
On solid growth media with limiting nitrogen source, diploid budding-yeast cells differentiate from the yeast form to a filamentous, adhesive, and invasive form. Genomic profiles of mRNA levels in Saccharomyces cerevisiae yeast-form and filamentous-form cells were compared. Disparate data types, including genes implicated by expression change, filamentation genes known previously through a phenotype, protein–protein interaction data, and protein–metabolite interaction data were integrated as the nodes and edges of a filamentation-network graph. Application of a network-clustering method revealed 47 clusters in the data. The correspondence of the clusters to modules is supported by significant coordinated expression change among cluster co-member genes, and the quantitative identification of collective functions controlling cell properties. The modular abstraction of the filamentation network enables the association of filamentous-form cell properties with the activation or repression of specific biological processes, and suggests hypotheses. A module-derived hypothesis was tested. It was found that the 26S proteasome regulates filamentous-form growth.
The availability and maturation of high-throughput biotechnologies is transforming the way cellular systems are studied. Our expanding genome-scale understanding suggests a hierarchical view of the cell in which groups of interacting molecules form biological modules, and biological modules interact in complex networks that control the properties of a cell. By traversing this hierarchy, biologists can discover properties of cells responding to perturbations or stimuli, and formulate molecular hypotheses on the control of cell properties such as metabolic capabilities, cell-cycle progression, and cell morphology. A key intermediate level in the organizational hierarchy is the module. Biological modules are loose associations of preferred molecular interaction partners that interact to perform a collective function (Hartwell et al. 1999). These loose molecular associations can be identified based on structural characteristics such as their closely connected members (Ravasz et al. 2002; Rives and Galitski 2003) and interfaces to other modules (Girvan and Newman 2002). In addition, there is evidence that modules are evolutionarily conserved (Snel et al. 2002), and that module co-members tend to be coordinately expressed (Ihmels et al. 2002; Segal et al. 2003). Applying module-level network analysis to several integrated genome-scale data sets, we studied a complex network controlling the differentiation of budding-yeast cells into a filamentous form. This approach implicated numerous module-associated biological processes in filamentous-form growth.
Various fungi, including major pathogens, can transform from a cellular yeast form to an invasive filamentous form in a morphogenetic program initiated by environmental stimuli (for review, see in Lengeler et al. 2000). For budding-yeast MATa/α diploid cells, low availability of ammonium and a solid growth substrate trigger the dimorphic switch to filamentous-form growth, characterized by cell elongation, unipolar distal budding, adhesion, and invasion (Gimeno et al. 1992; Kron et al. 1994). This transformation is often referred to as pseudohyphal development. Here it is referred to as filamentous-form growth. Several conserved signaling pathways that regulate this process have been intensively studied. Most prominent among these are the cAMP-dependent protein–kinase-A pathway, and the filamentation mitogen-activated protein–kinase pathway (fMAPK; for review, see Gancedo 2001). In addition, as in the yeast-form mitotic cell cycle (Lew and Reed 1995), Cdc28 kinase activity controls cell-cycle progression and morphogenesis during filamentous-form growth (for review, see Rua et al. 2001). The G1 cyclin Cln1 is required for polarized growth and is transcriptionally induced by enhanced fMAPK activity (Loeb et al. 1999; Madhani et al. 1999). In addition, the stability of the Cln1 protein is controlled by phosphorylation-dependent ubiquitination by the SCF ubiquitin-ligase (Skowyra et al. 1999). The present work implicates the subsequent step, ubiquitin-dependent protein degradation by the 26S proteasome, in the control of filamentous-form growth.
RESULTS
Identification of Filamentation Genes
Current knowledge of yeast filamentous-form growth is based mainly on molecular genetics and studies of expression profiles. Previous expression-profiling studies of filamentous-form growth have compared transcriptional profiles of key mutants grown in liquid culture (Madhani et al. 1999; Pan and Heitman 2000; Roberts et al. 2000; Robertson et al. 2000), in which wild-type strains normally do not form filaments.
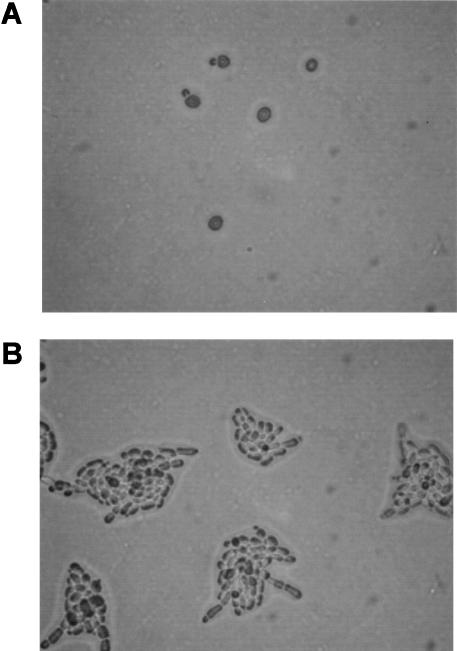
Our experimental design for expression analysis was motivated by the fact that both low availability of nitrogen and a solid growth substrate are required to induce diploid filamentous-form growth. Either of these two stimuli alone induces some (but not all) filamentation-related changes in gene expression and cell properties (e.g., Madhani et al. 1999). The connections between nutrient limitation, solid-surface growth, and dimorphism may extend to the situation faced by fungal pathogens colonizing and invading a host. The genetic links among the determinants of cell elongation, invasion, surface colonization, adhesion, and metabolic adaptation to an inhospitable host environment support this view (Gimeno et al. 1992; Kron et al. 1994; Roberts and Fink 1994; Lorenz and Fink 2001; Reynolds and Fink 2001; Rubin-Bejerano et al. 2003). Accordingly, we compared gene expression in yeast-form MATa/α diploid yeast cells (Fig. 1A) grown in liquid SHAD, a synthetic medium with plentiful glucose and ammonium, and filamentous-form cells (Fig. 1B) grown on SLAD (low ammonium) agar plates.
Figure 1.
Filamentous growth response of yeast cells. (A) Wild-type yeast-form cells grown in SHAD liquid medium. (B) Wild-type filamentous-form cells grown for 10 h on SLAD agar medium.
Experimental conditions for the induction and collection of filamentous-form cells were optimized (data not shown). High density of plating on SLAD results in inefficient switching to the filamentous form. The density of plating was optimized to 106 cells per 150-mm plate. The first buds are of the filamentous form. At about 10 h of incubation on SLAD, microcolonies of cell filaments have formed, and these begin to invade the agar. Cells were collected from the plates by washing with a rubber policeman (Methods). High efficiency and robustness of differentiation to the filamentous form were confirmed (Supplemental Data available online at www.genome.org).
Hundreds of genes were newly implicated in filamentous growth by the observation of significant expression difference in yeast-form and filamentous-form cells. In quadruplicate expression-analysis experiments in which filamentous-form cells were collected hourly from 1 to 10 h of filamentous-form growth, yeast-form targets were cohybridized with filamentous-form targets on whole-genome 70-mer oligonucleotide-probe microarrays. Using the maximum-likelihood methods of Ideker et al. (2000), a total of 873 differentially expressed genes were identified, with an estimated false-positive frequency of 2%. There were no striking differences among expression patterns at early and later times in the nascent filaments. This observation suggests that the switch to a filamentous-form expression pattern occurs rapidly and persists. The observation of filamentous-form first buds (e.g., Suppl. Fig. 1A) supports this suggestion. The mean ratio of the 10 hourly comparisons was used as a representative filamentous-form/yeast-form expression ratio for all subsequent analyses. The 873 differentially expressed genes and their expression ratios can be found in Supplemental Table 1.
Previous studies have implicated numerous genes in filamentous-form growth. A query, as in Rives and Galitski (2003), of the Incyte Yeast Protein Database (https://www.incyte.com/control/tools/proteome; Costanzo et al. 2001) identified 181 genes implicated in filamentous growth by virtue of their mutant phenotype. The 181 genes associated with a filamentation phenotype (a phenotype-implicated gene class) combine with the 873 genes showing significant differential expression (an expression-implicated gene class) for a total of 1026 genes, referred to henceforth as “filamentation genes.” Among the filamentation genes, only 28 genes (less than 3%) appear in both the phenotype-implicated and expression-implicated classes. Similar results have been reported for several other yeast responses to environmental perturbation (Giaever et al. 2002).
Assembly of an Integrated Yeast Filamentation Network
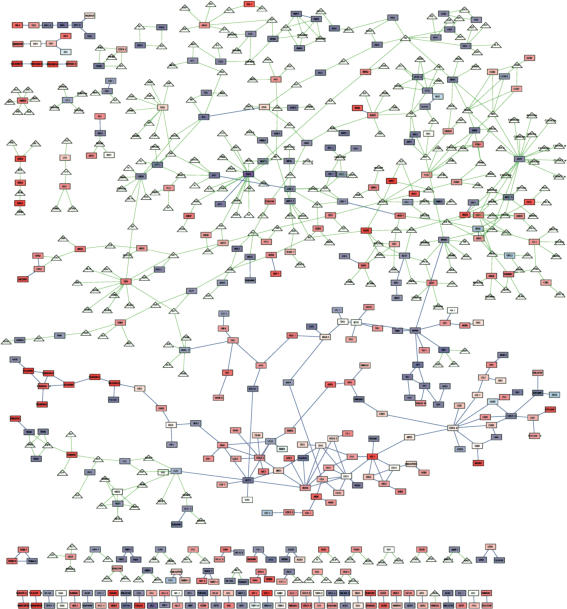
Physical interaction data linking filamentation proteins (encoded by the filamentation genes above) were integrated to form a “filamentation network.” The interactions between filamentation proteins were extracted from a global yeast protein–protein interaction network (Suppl. Data) and a global metabolic-reaction network (Suppl. Data; Forster et al. 2003). There were 231 protein–protein interactions between pairs of filamentation proteins. Metabolic reactions were represented as interactions between protein–enzymes and small-molecule metabolites. A protein–metabolite interaction indicates that the metabolite is a substrate or product of the protein–enzyme. Only protein–metabolite interactions (and the associated metabolites) involving a filamentation protein were included in the filamentation network. This integration added 295 metabolites and 500 protein–metabolite interactions to the filamentation network. In total, the integrated filamentation network includes 731 interactions. Figure 2 shows the filamentation network; filamentation proteins with no interaction with another filamentation protein are not shown. Attributes (molecule type, interaction type, and expression ratio) of the molecules and interactions are displayed visually.
Figure 2.
Integrated filamentation network. The filamentation network includes proteins (rectangular nodes) implicated in filamentous growth by expression profiling or known phenotypes, and metabolites (triangular nodes) that are either substrates or products of filamentation-protein enzymes. Not shown are filamentation proteins with neither a protein–metabolite interaction nor a protein–protein interaction with another filamentation protein. Blue edges indicate protein–protein interactions. Green edges indicate protein–metabolite interactions. Each gene node is colored based on its expression log-ratio. Shades of red indicate higher expression in the filamentous form relative to the yeast form; shades of blue indicate the opposite response; white indicates no difference.
Network Clustering
The integrated filamentation network is restricted to the filamentation proteins and their interactions with each other. This restriction applied to network clustering, which is based on similarity of network shortest-path distance profiles. Alternatively, it was possible to cluster the filamentation network based on global network interactions and paths. However, not all molecules, modules, and information-flow paths are available and active in a specific biological response. In addition, the global protein–protein interaction network has a high frequency of false interactions (von Mering et al. 2002). Our approach is advantageous because molecular interactions mediating the filamentous-growth response are likely to be reinforced by data integration in a restricted network, whereas inactive or false interactions are less likely to be reinforced by other data. For example, each protein–protein interaction in the filamentation network is present due to at least two data of at least two types: interaction assay data (e.g., a two-hybrid test) and the data resulting in the inclusion of both interacting proteins among the filamentation proteins (expression and/or phenotype data). This view presumes that there is a nonrandom tendency of filamentation proteins to interact with each other. This presumption was confirmed (Suppl. Data and Suppl. Table 2).
The filamentation network embodies several integrated data types. The formalism of the graph enables the algorithmic analysis of these disparate data. At many localities in the network there are clusters of tightly connected graph elements. The exploration of these clusters is of interest because they are likely to represent cellular modules (Ravasz et al. 2002; Rives and Galitski 2003) whose collective functions combine to determine the dimorphic properties of yeast cells.
The network-clustering method of Rives and Galitski (2003) was modified, extended, and implemented in the form of a program, Biomodules, that functions as a plug-in for Cytoscape (Shannon et al. 2003), a Java-based, open-source, biological network visualization and analysis platform. The unsupervised method is detailed in the Supplemental Data. One can obtain Biomodules source code, or use Biomodules without any upload or download of files, at http://labs.systemsbiology.net/galitski.
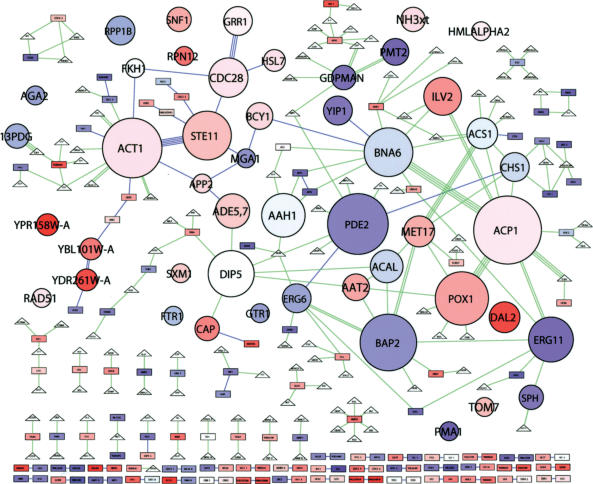
Network clustering allows the modular abstraction of the filamentation network, that is, a representation as a network of interacting modular units. The clustering algorithm was applied and the results displayed using Biomodules (Fig. 3). A total of 47 clusters, each represented as a round node, emerged in the filamentation network. Visual characteristics of the module nodes convey information on the number of member nodes and their expression ratios. Each module-node label (in bold all-caps when used in the text) is from the member protein or metabolite with the most connections to co-members, the “module-organizer” (Rives and Galitski 2003). Supplemental Table 3 lists the molecular members of each cluster. Expected clusters appear, for example, ACT1, the actin cytoskeleton, STE11, the filamentation MAP-kinase cascade, and CDC28, the cell-cycle oscillator, among others. In addition, the emergence of many other clusters, as well as their molecular members and connections among clusters, provides an intuitive, information-dense, mostly nonverbal, global view of the organization of the network.
Figure 3.
Modular abstraction of the filamentation network. Network clusters are abstracted as circular “module nodes.” Otherwise, colors and shapes represent the same as in Figure 2. The area of each module node is proportional to the number of member molecules. The color of each module node reflects the average expression log-ratio of member genes. Each module node is automatically assigned the name of the member node of highest intracluster degree (the highest number of interactions with cluster co-members); most are proteins, some are metabolites. See Supplemental Table 3 for a list of cluster members.
Coordinated Expression Change Within Clusters
The color of each module node represents the mean expression log-ratio of member genes. In Figure 3 many module nodes have intense color, suggesting that cluster co-members show coordinated expression change in the direction indicated by the color. This suggestion was tested. All genes in clusters were classified in one of three groups: induced, repressed, and not changed. We formulated a null hypothesis stating that these three gene classes are distributed randomly, according to their respective proportions, among the members of the various clusters. Using a likelihood-ratio test and gene randomizations, the null hypothesis was evaluated and rejected (Suppl. Data). The expression response of cluster co-members shows significant coordination (P = 10–11). Note that even the genes without significant expression change tend to occur together in clusters (Suppl. Tables 3,4). Because genes without significant expression change are of the phenotype-implicated class of genes, one may consider clusters in which they predominate to be a phenotype-implicated class of clusters.
Collective Functions of Network Clusters
If clusters in an integrated network represent biological modules, the clusters should have collective functions in specific biological processes (Hartwell et al. 1999). Specific biological-process gene annotations are overrepresented in specific filamentation-network clusters (Table 1). Biological-process gene annotations were obtained from the Gene Ontology Consortium Database (Ashburner et al. 2000), and were attached to their respective genes in the filamentation network. Clusters were computationally annotated using Biomodules. For all instances in which two or more genes within a cluster shared an annotation, a P-value was calculated for overrepresentation of the annotation among genes in the cluster compared to random expectation. For each cluster, all annotations with a -log10 P-value greater than 1.3 (P < 0.05) were identified. Table 1 lists the annotation of highest significance for each cluster. Most of the clusters (39 of 47) have computationally assigned annotations of high significance. For six of the remaining eight clusters, inspection of cluster members allowed the manual assignment of obvious collective functions (Table 1), for example, Ty element genes. The significance of the data in Table 1 was confirmed in tests in which genes were randomly reassigned to clusters (Suppl. Data). The finding of collective function supports the conclusion that clusters in the integrated network represent functional network modules.
Table 1.
Overrepresented Gene Annotations in Network Clusters
| Cluster | Top overrepresented gene annotationa | -log10Pb |
|---|---|---|
| ACT1 | cytoskeleton organization and biogenesis | 12 |
| CDC28 | cell cycle | 8.9 |
| STE11 | development | 7.7 |
| PDE2 | phospholipid metabolism | 7.4 |
| TOM7 | mitochondrial translocation | 6.5 |
| ERG11 | ergosterol biosynthesis | 6.0 |
| CAP | arginine biosynthesis | 5.9 |
| NH3xt | ammonium transport | 5.9 |
| SPH | sphingolipid metabolism | 5.9 |
| ACP1 | energy derivation by oxidation of organic compounds | 5.6 |
| BNA6 | nucleocytoplasmic transport | 5.5 |
| PMT2 | glycoprotein biosynthesis | 5.5 |
| 13PDG | glycolysis | 5.3 |
| PMA1 | cation transport | 4.7 |
| RPP1B | translational elongation | 4.3 |
| RAD51 | nuclear division | 4.1 |
| AGA2 | cell adhesion | 4.0 |
| FTR1 | high-affinity iron transport | 4.0 |
| GDPMAN | N-linked glycosylation | 4.0 |
| HMLα2 | regulation of transcription, mating-type specific | 4.0 |
| DAL2 | allantoin catabolism | 3.6 |
| SNF1 | regulation of transcription from Pol II promoter | 3.5 |
| BCY1 | pseudohyphal growth | 3.4 |
| ACAL | pyruvate dehydrogenase bypass | 3.3 |
| DIP5 | aspartate family amino acid catabolism | 3.3 |
| YIP1 | ER to Golgi transport | 3.2 |
| ERG6 | methionine metabolism | 3.1 |
| POX1 | glyoxylate cycle | 3.0 |
| RPN12 | ubiquitin-dependent protein catabolism | 2.9 |
| BAP2 | amino acid transport | 2.7 |
| FKH1 | regulation of transcription, DNA-dependent | 2.2 |
| HSL7 | G2/M transition of mitotic cell cycle | 2.1 |
| GRR1 | ubiquitin-dependent protein catabolism | 2.0 |
| SXM1 | protein biosynthesis | 2.0 |
| APP2 | protein transport | 2.0 |
| ACS1 | carboxylic acid metabolism | 1.8 |
| ILV2 | amino acid biosynthesis | 1.8 |
| AAT2 | amino acid and derivative metabolism | 1.6 |
| MET17 | carboxylic acid metabolism | 1.4 |
| YBL101W-A | [Ty element] | N/A |
| YDR261W-A | [Ty element] | N/A |
| YPR158W-A | [Ty element] | N/A |
| CHS1 | [chitin synthesis] | N/A |
| GTR1 | [phosphate metabolism] | N/A |
| AAH1 | [nucleotide base catabolism] | N/A |
| ADE5,7 | N/A | N/A |
| MGA1 | N/A | N/A |
Biological-process gene annotations were from the Gene Ontology Consortium database version 1.608. Each listed annotation applied to at least two cluster member genes, and showed significant (-log10p > 1.3; P < 0.05) overrepresentation in the cluster compared to random expectation. For most clusters, several annotations were significantly over-represented. In these cases, annotations with the highest significance are listed. Some clusters showed no annotations meeting the criteria and were annotated manually (indicated by brackets).
Significance (-log10 cumulative probability of the observed data and all more extreme possibilities) was calculated using the hypergeometric distribution and: (1) the total number of proteins in all clusters, 259; (2) the number of proteins in all clusters with the annotation; (3) the number of proteins in the cluster; and (4) the number of proteins with the annotation in the cluster.
Complex Cell Properties and Extended Network Context
The clustered filamentation network reflects functional “guilt-by-association” at the intramodular and intermodular levels. Molecules and modules that function together in specific cellular processes, to determine specific cell properties, can be found together in the network. For example, genes with prominent cell-elongation phenotypes (e.g., STE11, KSS1, CLB2, CLN1, CDC28, HSL7, SWE1, GRR1) are members of the STE11, CDC28, HSL7, and GRR1 modules. These modules are contiguous in the modular network abstraction (Fig. 3). As another example, a module involved in the generation of activated mannose (GDPMAN) interacts with another module involved in protein mannosylation (PMT2). In general, the modular network abstraction can be viewed as a network of collective-function units whose activities and interactions specify complex cell properties.
Hypothesis Generation
The network abstraction suggests hypothetical metabolic and cell biological properties of filamentous-form yeast cells. For example, the ERG11, PDE2, and SPH modules (Fig. 3) were annotated with functions in related pathways; ergosterol, phospholipid, and sphingolipid metabolism, respectively (Table 1). As reflected in the blue color of these module nodes, enzymes in these pathways are repressed in filamentous-form cells. It is intriguing to speculate that fungal pathogens alter their membrane composition in contact with host tissue. In support of this hypothesis, filamentous-form yeast cells are more resistant to the sterol-binding membrane-pore-forming polyene antibiotic nystatin (S. Prinz, unpubl.).
As another example, the DAL2 and POX1 modules (Fig. 3) contain enzymes associated with allantoin catabolism, fatty-acid β-oxidation, and the glyoxylate cycle (Table 1, Suppl. Table 3). These enzymes are highly induced during filamentation. The Dal pathway (degradation of alantoin) utilizes purines as a source of urea (reduced nitrogen), which is limiting in filamentation-inducing conditions. Aside from urea, another product of the Dal pathway is glyoxylate, which can be metabolized in the glyoxylate cycle. The glyoxylate cycle has been implicated in microbial virulence and pathogen survival in the host (Lorenz and Fink 2001). Many enzymes of these metabolic pathways are housed in peroxisomes. Indeed, peroxisomes proliferate in filamenting cells (Suppl. Data). The modular filamentation network, and the observed proliferation of peroxisomes, suggests that filamentous-form fungi utilize degradation of purines (via the Dal pathway and the glyoxylate cycle) and fatty acids (via β-oxidation and the glyoxylate cycle) as strategies to assimilate nitrogen and carbon. It may be that for dimorphic fungal pathogens, these nutrient sources are available from host cells.
The foregoing examples demonstrate that the automated graphical structure/function analysis of complex networks can be used to immediately suggest intriguing unknown cell biological properties of cells responding to perturbations or stimuli. In addition, the modular organization of the network associates cell-biological insights with the molecules of specific modules. This association can serve as a basis for molecular hypotheses.
RPN4 of the RPN12 Module Regulates Filamentous Growth
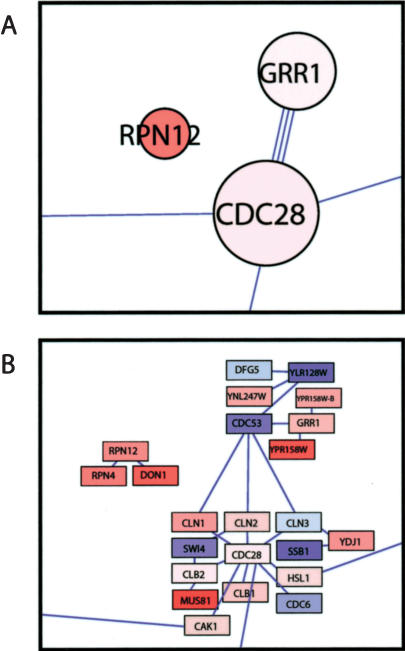
The emergence of the RPN12 module (Fig. 3, Table 1) implicates ubiquitin-dependent proteolysis, and specifically the 19S regulatory particle of the 26S proteasome, in the regulation of filamentous growth. The 19S regulatory particle caps the cylindrical 20S core particle and regulates protein binding, unfolding, deubiquitination, and translocation into the lumen of the core where proteolysis occurs (for review, see Glickman and Ciechanover 2002). The RPN12 module contains three proteins, Rpn12, Rpn4, and Don1 (Fig. 4). The expression of their respective genes is induced in filamentous-form cells. Rpn12, the module-organizer protein, is a subunit of the 19S regulatory particle (Kominami et al. 1995). The Rpn4 protein is loosely bound to the 26S proteasome (Fujimuro et al. 1998) and is a transcriptional activator of many 19S-cap and other proteasomal subunits (Mannhaupt et al. 1999; Xie and Varshavsky 2001). The Don1 protein has a CUE domain that might be involved in binding ubiquitin-conjugating enzymes (Shih et al. 2003). Moreover, two more genes encoding subunits of the 19S regulatory particle, RPN7 and RPN8, are significantly induced in filamentous-form cells (Suppl. Table 1). Both proteins interact with Rpn12 (Glickman et al. 1998). These proteins do not appear in the RPN12 module because interaction-data sources from which the filamentation network was derived are incomplete.
Figure 4.
RPN12, GRR1, and CDC28 modules and their components. Modules (A), and their respective components (B) with collective functions in cell-cycle control and ubiquitin-dependent proteolysis are shown. Graphic representations are as in Figures 2 and 3.
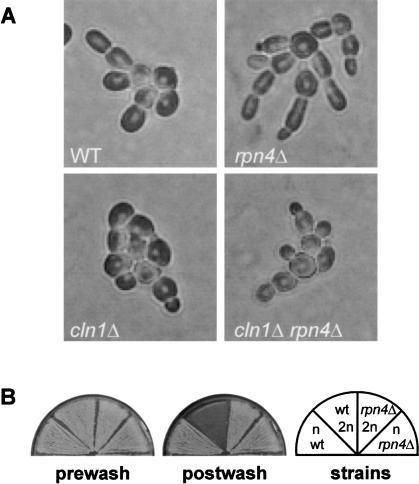
To test the possibility of a role for the RPN12 module, filamentation phenotypes of an RPN mutant were assessed. Of the four RPN genes implicated in filamentous growth by geneexpression data, RPN4 was an attractive focus for study because it is required for wild-type ubiquitin-mediated proteolysis (Johnson et al. 1995), and yet is not an essential gene like the others. The RPN4 gene was deleted from a filamentation-competent strain. The resulting rpn4Δ mutant is viable but grows somewhat slower than wild type. On SLAD plates the rpn4Δ mutant forms hyperelongated cells and shows enhanced unipolar distal budding (Fig. 5A). After continued growth on SLAD, the mutant forms colonies with a greater profusion of extruding cell chains compared to wild type (data not shown). The hyperelongated morphology of rpn4Δ mutants is observed even in liquid SHAD medium with plentiful ammonium (data not shown). In addition, rpn4Δ mutants show agar adhesion independent of their cell type. In wash assays of agar adhesion, rpn4Δ diploids adhere to agar as avidly as rpn4Δ haploids, whereas in wild-type strains haploids adhere but diploids do not (Fig. 5B). Thus, Rpn4, controlling more than one dimorphic cell property, is a regulator of filamentous-form growth.
Figure 5.
rpn4Δ mutants show Cln1-dependent hyperelongation, and cell type-independent agar adhesion. (A) Diploid wild-type, rpn4Δ, cln1Δ, and rpn4Δ cln1Δ strains were grown on SLAD agar plates and photographed after 9 h. (B) Patches of strains of the indicated cell types and genotypes were subjected to a wash-off assay of adhesion. The plate was imaged before and after washing with water.
Modules Controlling the Filamentous-Form Cell Cycle and Proteolysis
Though the RPN12 module has no physical connections to other modules shown in Figure 3, it shares the biological process annotation `ubiquitin-dependent proteolysis' with the GRR1 module (Table 1). Thus, there is a functional link between the RPN12 and GRR1 modules. Other functional links are suggested by the functional analysis of modules in Table 1. For example, there are several modules with functions in amino-acid metabolism. These examples illustrate the extension of the principle of guilt-by-association among modules from shared physical interactions to shared or related collective functions.
Grr1 is an F-box protein of the SCF ubiquitin ligase required for the ubiquitination of the G1 cyclins (Li and Johnston 1997). This information is captured in the modular network abstraction by the connections between cyclin proteins in the CDC28 module and the GRR1 module (Fig. 4B). The role of CDC28-module components in determining filamentous-form cell properties is complex. The cyclin-dependent kinase, Cdc28, plays either a positive or a negative role in cell elongation, depending on the class of cyclins with which it is associated (for review, see Rua et al. 2001). Cln1-Cdc28 promotes apical growth. Deletion of GRR1 results in constitutive filamentation (Blacketer et al. 1995; Loeb et al. 1999).
The occurrence of the RPN12, GRR1, and CDC28 modules in the filamentation network, combined with the molecular biology of these modules and the phenotype of rpn4Δ mutants, suggests the hypothesis that filamentous-form cell elongation is regulated at the level of the 26S proteasome through controlled degradation of the Cln1 cyclin.
Genetic Interaction Between CLN1 and RPN4
The hypothesis proposed above predicts a genetic interaction between CLN1 and RPN4 mutant genes. The rpn4Δ mutant is hyperelongated, whereas a cln1Δ mutant is unable to filament and forms round cells. If rpn4Δ hyperelongation is due to stabilization of Cln1 protein, a cln1Δ rpn4Δ double mutant should have the round-cell phenotype of a cln1Δ mutant, because the stabilization of Cln1 protein by rpn4Δ is not possible when Cln1 is absent. As shown in Figure 5A, the cln1Δ rpn4Δ double mutant, like the cln1Δ single mutant, forms round cells as predicted by the model. This genetic interaction occurs on SLAD plates (Fig. 5A) and in liquid SHAD medium (data not shown). Note that the cln1Δ rpn4Δ double mutant exhibits unipolar budding (Fig. 5A) and cell type-independent agar adhesion (data not shown). These observations suggest that the CLN1-RPN4 genetic interaction is specific for the cell-elongation phenotype, and that there are other genes with which rpn4Δ interacts to cause the full spectrum of its phenotypes.
Rpn4 Regulates Filamentous-Form mRNA Levels of RPN12, but not CLN1
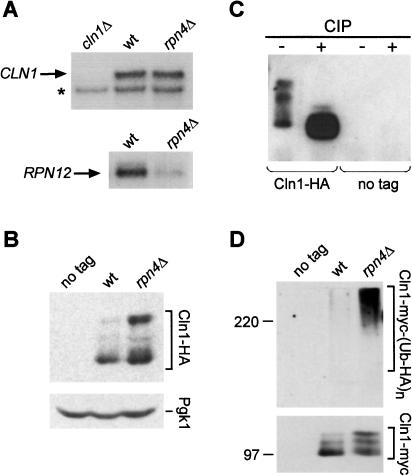
Rpn4 is known to be a transcriptional regulator. This fact raises the alternative hypothesis that deletion of the RPN4 gene results in enhanced Cln1 activity through an increase in CLN1 transcript levels. This possibility was excluded. Northern blot analysis of CLN1 mRNA levels in filamenting wild-type and rpn4Δ cells showed that CLN1 mRNA levels are not affected by rpn4Δ (Fig. 6A). This observation supports the hypothesis that the genetic interaction between the RPN4 and CLN1 genes involves posttranscriptional stabilization of Cln1 protein. Rpn4 activates the expression of proteasomal subunits (Mannhaupt et al. 1999). To confirm that this activation is occurring in filamentous-form cells, the expression of RPN12 was compared in wild-type and rpn4Δ strains grown on SLAD plates. In rpn4Δ mutants, RPN12 transcript levels are substantially lower than in wild-type filamentous-form cells (Fig. 6A). Filamentous-form regulation of proteasomal gene expression suggests that the molecular basis of the observed genetic interaction is the stabilization of Cln1 at the level of the 26S proteasome.
Figure 6.
Stabilization of Cln1 protein in rpn4Δ mutants. (A) Northern blot analysis of total RNA from wild-type and rpn4Δ strains grown on SLAD plates, and a cln1Δ strain. The blot was probed consecutively with probes for CLN1 and RPN12. The asterisk in the CLN1 blot indicates a cross-hybridizing band that also serves as a loading control. (B) Western blot analysis of Cln1 protein in diploid wild-type and rpn4Δ strains carrying HA-tagged CLN1, and a no-tag wild-type control strain. Protein extracts were prepared from cells grown for 10 h on SLAD agar plates. Pgk1 protein levels served as a loading control. (C) Cln1-HA protein was immunoprecipitated from an rpn4Δ strain. Aliquots of the immunoprecipitate were incubated with calf-intestine phosphatase (CIP), or without CIP, and analyzed by Western blotting. (D) myc-tagged Cln1 protein was immunoprecipitated in diploid wild-type and rpn4Δ strains, and a no-tag control strain. All strains had a multicopy plasmid expressing HA-tagged ubiquitin. Immunoprecipitates were analyzed by gel electrophoresis and immunoblotting with anti-HA antibody to detect ubiquitin conjugates. The blot membrane was stripped and reprobed with anti-myc antibodies to detect the immunoprecipitated Cln1.
Stabilization of Cln1 Protein by Deletion of RPN4
To test the hypothesis of Cln1 protein stabilization by rpn4Δ, Cln1 protein levels were monitored directly. Endogenous Cln1 protein was tagged at the C-terminus with a triple-HA epitope (Longtine et al. 1998). The tagged protein is functional; homozygous CLN1-HA diploid cells are filamentation-proficient (data not shown). Cln1 protein levels were compared in wild type and an rpn4Δ mutant grown on SLAD agar. Western blot analysis showed that Cln1 protein levels are elevated in the rpn4Δ mutant (Fig. 6B). In particular, the most striking difference in abundance was observed with lower-mobility forms of Cln1. Because Cln1 protein phosphorylation is a prelude to rapid ubiquitination by SCF and degradation by the proteasome (Lanker et al. 1996), it was possible that the low-mobility bands were phosphorylated forms of Cln1. Cln1 protein was immunoprecipitated and either subjected to treatment with calf intestinal alkaline phosphatase (CIP) or a mock reaction lacking CIP. As shown in Figure 6C, phosphatase treatment eliminates the low-mobility species of Cln1. These results support the model of post-phosphorylation stabilization of Cln1 protein by rpn4Δ. The positive regulation of the expression of proteasomal subunits by Rpn4 (Fig. 6A; Mannhaupt et al. 1999) suggests that altered 26S proteasome activity is responsible for increased levels of Cln1 in rpn4Δ cells. This model predicts the accumulation of polyubiquitinated Cln1 in rpn4Δ cells. To test this prediction, Cln1 tagged at the C-terminus with a 13-myc epitope (Longtine et al. 1998) was immunoprecipitated from wild-type and rpn4Δ strains expressing HA-tagged ubiquitin (Methods). Figure 6D shows that polyubiquitinated forms of Cln1 accumulate in the rpn4Δ mutant but not in wild type, though a substantial amount of Cln1 protein was immunoprecipitated from both. In addition, in a control experiment, accumulation of polyubiquitinated Cln1 was observed when the epitope tags on Cln1 and ubiquitin were switched (data not shown).
Combined genetic (Fig. 5) and molecular (Fig. 6) lines of evidence strongly suggest that in filamentous-form yeast cells, Rpn4 plays a negative role in cell elongation by controlling Cln1 protein levels through regulated proteasome activity, possibly by controlling 19S regulatory particle composition or abundance. The molecular effect of rpn4 mutation on Cln1 is to stabilize the ubiquitinated form and phosphorylated precursors of the ubiquitinated form. Genetic interaction data support the functional significance of this molecular effect for the cell-elongation phenotype. Thus not only the attachment of polyubiquitin to substrates, but also the 26S proteasome itself is a target of regulation controlling filamentous growth.
DISCUSSION
Modular Network Abstraction and Biological Insights
Scattered facts, when integrated, displayed, and analyzed in a global module-level context, can lead researchers to specific hypotheses on cell properties and their molecular determination. Cell-biological hypothesis generation is an inherently integrative process in which insight is extracted from connections that are apparent at the level of collective functions, that is, modules (Hartwell et al. 1999). As shown in the present work, network cluster analysis directly identifies relevant modules, module collective functions, changes in module activities, and connections (both physical and functional) among modules.
The evidence validating the modular network inferred here (and for other biological-response networks; data not shown) takes several forms: (1) In an integrated network, data on molecules and interactions shows clustered organization that can be identified quantitatively. (2) Cluster co-member genes show significant coordination of expression change, as expected for genes involved in a collective function. (3) Cluster co-member genes show significant overrepresentation of biological-process annotations, indicating collective function. (4) The modular network abstraction intuitively stimulates testable biological insights on complex biological properties.
Regulation of Filamentous Growth by the 26S Proteasome
The modular abstraction of the filamentation network, and its assigned collective functions, implicated the 19S regulatory cap of the 26S proteasome in filamentous growth. It seems likely that the 26S proteasome is not a passive receptacle that degrades any polyubiquitinated protein. Rather, the proteasome may be a dynamic complex whose composition and activities are subjected to regulation. There is evidence that different subunits of the regulatory particle are required for selective degradation of proteins (e.g., Bailly and Reed 1999). In addition, it was recently shown that 26S proteasome activity is subject to regulation during stationary phase (Bajorek et al. 2003). The present work suggests that the 26S proteasome controls filamentous-form cell properties through regulation by Rpn4. The genetics and molecular biology of CLN1 and RPN4 can account for the hyperelongation of rpn4Δ mutants. The enhanced unipolar budding and cell type-independent adhesion of the rpn4Δ mutant suggests that additional filamentous-form substrates of the 26S proteasome await identification. These findings add another layer of complexity to ubiquitin-dependent proteolysis, where much of the focus has been on the regulation of attachment of polyubiquitin chains by ubiquitin ligases.
It remains to be determined whether 26S proteasome abundance is controlled, or whether the induction of specific proteasomal subunits in the filamentous form results in proteasome complexes of different composition. One can speculate that proteasome substrate recognition or affinity might differ in the filamentous growth mode. The data suggest that ubiquitin-dependent proteolysis continuously inactivates proteins that promote filamentous-cell properties. Proteolytic clearing of these key proteins may be part of a complex global regulatory mechanism to stabilize the state of the network; alternatively, it may allow for prompt response when environmental conditions change. Also, Gonzalez et al. (2002) have shown that the 19S cap complex (without the 20S proteasome core) is involved in transcription. This raises the possibility that Rpn4 acts not solely through proteolysis, but also through 19S-dependent transcription.
Prospects
The examples of peroxisome proliferation and proteasomal control of filamentous growth illustrate the value of integrative unsupervised analysis of module-level biological organization. In addition, there is a wealth of information in the modular filamentation network that has not yet been explored. Furthermore, the present work suggests several possible extensions of modular network analysis. The integration of genetic interaction data may uncover hidden interactions between modules. For example, the epistatic relationship of cln1Δ to rpn4Δ (Fig. 5) leads to the recognition of the molecular interaction between their respective modules. In addition, ontological associations between modules (e.g., RPN12 and GRR1) may suggest molecular interactions. Furthermore, because module co-member genes tend to show coordinated expression, series of expression-profiling experiments may allow the association of specific constellations of modules with genetic and environmental perturbations.
METHODS
Strains and Plasmids
All yeast strains were derivatives of a Σ1278b ura3Δ0, his3Δ0::hisG strain from the Sigma2000 collection of Microbia, Inc. Strain G85, a MATa/α homozygous diploid, served as the wild type. Deletion derivatives of G85 were made using a PCR-based strategy in which KanMX4 “barcode” deletion alleles (Winzeler et al. 1999) were amplified, transformed into G85, and verified by PCR and tetrad dissection. The Cln1 protein was tagged at its C-terminus with either the triple-HA epitope or the 13-Myc epitope using pFA6a-3HA-kanMX6 or pFA6a-13Myc-kanMX6 (Longtine et al. 1998) as a PCR template. Tags were confirmed by PCR and Western blot analysis. Plasmid pRG147 (kindly provided by R. Gardner and D. Gottschling, Fred Hutchinson Cancer Research Center, Seattle, WA) is a 2-micron derivative of pRH988 (Gardner and Hampton 1999) and encodes triple-HA-tagged ubiquitin under the control of the constitutive TDH3 promoter.
Growth Conditions
For filamentous-form growth, cells were grown at 30° on SLAD agar plates. SLAD is synthetic low-ammonium (50 μM) dextrose medium (Gimeno et al. 1992). For yeast-form growth, cells were grown at 30° in liquid SHAD medium (same as SLAD but high ammonium, 32 mM). Synthetic media were supplemented with uracil and histidine as appropriate.
Comparison of Yeast-Form and Filamentous-Form Gene Expression
In four independent experiments, exponential-phase (OD600 = 0.8) yeast-form cells were collected. Half of these cells were retained for preparation of yeast-form RNA as the reference state for expression profiling. The remaining cells were washed and diluted in liquid SLAD medium and plated immediately on SLAD agar. Aliquots of 106 cells in 1 mL were spread evenly onto multiple 150-mm plates and incubated. Every hour for 10 h, a plate was microscopically imaged, then collected by washing twice with SLAD liquid and a sterile rubber policeman. Efficient collection of cells was verified microscopically (data not shown). Total RNA was extracted. PolyA+ RNA was converted to cDNA and in vitro transcribed to yield Cy-dye-labeled targets. The methods of Wodicka et al. (1997) were used except that Cy-labeled ribonucleotides were used in the in vitro transcription. Labeled targets were hybridized to microarrays of oligonucleotide probes for all yeast genes using methods detailed in Smith et al. (2002). To generate control data for error estimation, the four yeast-form replicates were cohybridized in all possible pairs with both dye orientations. Expression data have been deposited at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE679.
Network Clustering
See the Supplemental data. One can obtain Biomodules source code, or use Biomodules without any upload or download of files, at http://labs.systemsbiology.net/galitski.
Tests of Filamentation and Adhesion
To test filamentation phenotypes, strains were streaked on SLAD plates and incubated at 30° for about 10 h. Cells were microscopically imaged using a Nikon Coolpix 990 digital camera mounted on a Nikon TS100 inverted microscope with a 40X objective. To test adhesion, strains were patched on YPD agar plates and incubated at 30° for 4 d. The plates were washed with water and mild agitation until negative-control patches (diploid wild-type) washed off (Roberts and Fink 1994). Plates were imaged before and after washing using a Nikon Coolpix 990 digital camera.
RNA Preparation and Northern Blot Analysis
Cells were harvested after 10 h on SLAD agar as described above. Total RNA was extracted with hot acid phenol. Ten μg of total RNA were separated in a formaldehyde agarose gel and transferred to a nylon membrane. The CLN1 probe was created by PCR-amplifying the last 941 bp of the CLN1 ORF; the RPN12 ORF was amplified using GenePair primers (Research Genetics). Probes were labeled with [32P]dCTP using PrimeIt (Stratagene).
Protein Extracts and Western Blot Analysis
Filamentous-form cells from SLAD plates were precipitated using a trichloroacetic-acid method. Extracts were prepared from cell pellets using lysis buffer (50 mM Tris pH 7.5, 1 mM EDTA, 50 mM DTT, 1 mM PMSF, 1 mM benzamidine, aprotinin [10 μg/mL], leupeptin [1 μg/mL], pepstatin [1 μg/mL], 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, and 100 μM sodium orthovanadate), acid-washed glass beads, and a Vibrax-VXR (IKA) at 4°. Protein-sample buffer was added and extracts were immediately boiled for 5 min and clarified by centrifugation. For Western blot analysis, extracts were separated on an 8% polyacrylamide-SDS gel. The HA epitope and the Pgk1 protein were detected using mouse monoclonal antibody HA.11 (Covance) and mouse monoclonal 22C5 (Molecular Probes), respectively. Immunodetection was done with horseradish-peroxidase-linked secondary antibodies (Amersham) and enhanced chemiluminescence using SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Immunoprecipitation
To obtain sufficient cell material, cells were harvested from exponential-phase cultures in liquid SHAD. Immunoprecipitation of Cln1 was performed as described in Moll et al. 1991, using mouse monoclonal antibody HA.11 (anti-HA) or mouse monoclonal antibody 9E10 (anti-Myc) from Covance and ImmunoPure Immobilized Protein G beads (Pierce). For phosphatase treatment, washed beads were incubated in phosphatase buffer either with or without calf-intestine phosphatase (Roche). For detection of ubiquitin conjugates, proteins were separated on a 6% polyacrylamide-SDS gel and blotted without methanol to facilitate the transfer of high-molecular-weight proteins.
Acknowledgments
We thank J. Aitchison, A. Amon, R. Christmas, E. Deutsch, P. Edlefsen, G. Fink, R. Gardner, A. Golden, D. Gottschling, M. Johnson, G. Lake, A. Markiel, B. Marzolf, A. Rives, B. Schwikowski, P. Shannon, V. Thorsson, R. Visintin, M. Zahler, and E. Zitzler for their contributions. This work was funded by Merck & Co., Inc. and by NIH grant 1P30DA01562501. A.F. Siegel holds the Grant I. Butterbaugh Professorship at the University of Washington. T. Galitski is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2020604.
Footnotes
[Supplemental material is available online at www.genome.org. Software is available at http://labs.systemsbiology.net/galitski. The gene expression data from this study have been submitted to Gene Expression Omnibus database under accession no. GSE679.]
References
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, E. and Reed, S.I. 1999. Functional characterization of rpn3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol. Cell. Biol. 19: 6872–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek, M., Finley, D., and Glickman, M.H. 2003. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 13: 1140–1144. [DOI] [PubMed] [Google Scholar]
- Blacketer, M.J., Madaule, P., and Myers, A.M. 1995. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics 140: 1259–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo, M.C., Crawford, M.E., Hirschman, J.E., Kranz, J.E., Olsen, P., Robertson, L.S., Skrzypek, M.S., Braun, B.R., Hopkins, K.L., Kondu, P., et al. 2001. YPD, PombePD and WormPD: Model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29: 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, J., Famili, I., Fu, P., Palsson, B.O., and Nielsen, J. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro, M., Tanaka, K., Yokosawa, H., and Toh-e, A. 1998. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 423: 149–154. [DOI] [PubMed] [Google Scholar]
- Gancedo, J.M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 107–123. [DOI] [PubMed] [Google Scholar]
- Gardner, R.G. and Hampton, R.Y. 1999. A “distributed degron” allows regulated entry into the ER degradation pathway. EMBO J. 18: 5994–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gimeno, C.J., Ljungdahl, P.O., Styles, C.A., and Fink, G.R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 68: 1077–1090. [DOI] [PubMed] [Google Scholar]
- Girvan, M. and Newman, M.E. 2002. Community structure in social and biological networks. Proc. Natl. Acad. Sci. 99: 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H. and Ciechanover, A. 2002. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82: 373–428. [DOI] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623. [DOI] [PubMed] [Google Scholar]
- Gonzalez, F., Delahodde, A., Kodadek, T., and Johnston, S.A. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296: 548–550. [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H., Hopfield, J.J., Leibler, S., and Murray, A.W. 1999. From molecular to modular cell biology. Nature 402: C47–52. [DOI] [PubMed] [Google Scholar]
- Ideker, T., Thorsson, V., Siegel, A.F., and Hood, L.E. 2000. Testing for differentially-expressed genes by maximum-likelihood analysis of microarray data. J. Comput. Biol. 7: 805–817. [DOI] [PubMed] [Google Scholar]
- Ihmels, J., Friedlander, G., Bergmann, S., Sarig, O., Ziv, Y., and Barkai, N. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31: 370–377. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., Ma, P.C., Ota, I.M., and Varshavsky, A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270: 17442–17456. [DOI] [PubMed] [Google Scholar]
- Kominami, K., DeMartino, G.N., Moomaw, C.R., Slaughter, C.A., Shimbara, N., Fujimuro, M., Yokosawa, H., Hisamatsu, H., Tanahashi, N., Shimizu, Y., et al. 1995. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 14: 3105–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron, S.J., Styles, C.A., and Fink, G.R. 1994. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 5: 1003–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker, S., Valdivieso, M.H., and Wittenberg, C. 1996. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 271: 1597–1601. [DOI] [PubMed] [Google Scholar]
- Lengeler, K.B., Davidson, R.C., D'Souza, C., Harashima, T., Shen, W.C., Wang, P., Pan, X., Waugh, M., and Heitman, J. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D.J. and Reed, S.I. 1995. Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev. 5: 17–23. [DOI] [PubMed] [Google Scholar]
- Li, F.N. and Johnston, M. 1997. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16: 5629–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J.D., Kerentseva, T.A., Pan, T., Sepulveda-Becerra, M., and Liu, H. 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153: 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.C. and Fink, G.R. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412: 83–86. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Galitski, T., Lander, E.S., and Fink, G.R. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. 96: 12530–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhaupt, G., Schnall, R., Karpov, V., Vetter, I., and Feldmann, H. 1999. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450: 27–34. [DOI] [PubMed] [Google Scholar]
- Moll, T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66: 743–758. [DOI] [PubMed] [Google Scholar]
- Pan, X. and Heitman, J. 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 20: 8364–8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz, E., Somera, A.L., Mongru, D.A., Oltvai, Z.N., and Barabasi, A.L. 2002. Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555. [DOI] [PubMed] [Google Scholar]
- Reynolds, T.B. and Fink, G.R. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291: 878–881. [DOI] [PubMed] [Google Scholar]
- Rives, A.W. and Galitski, T. 2003. Modular organization of cellular networks. Proc. Natl. Acad. Sci. 100: 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., Nelson, B., Marton, M.J., Stoughton, R., Meyer, M.R., Bennett, H.A., He, Y.D., Dai, H., Walker, W.L., Hughes, T.R., et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873–880. [DOI] [PubMed] [Google Scholar]
- Roberts, R.L. and Fink, G.R. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes & Dev. 8: 2974–2985. [DOI] [PubMed] [Google Scholar]
- Robertson, L.S., Causton, H.C., Young, R.A., and Fink, G.R. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. 97: 5984–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua, D., Tobe, B.T., and Kron, S.J. 2001. Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4: 720–727. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano, I., Fraser, I., Grisafi, P., and Fink, G.R. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, E., Shapira, M., Regev, A., Pe'er, D., Botstein, D., Koller, D., and Friedman, N. 2003. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34: 166–176. [DOI] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, S.C., Prag, G., Francis, S.A., Sutanto, M.A., Hurley, J.H., and Hicke, L. 2003. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 22: 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Koepp, D.M., Kamura, T., Conrad, M.N., Conaway, R.C., Conaway, J.W., Elledge, S.J., and Harper, J.W. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284: 662–665. [DOI] [PubMed] [Google Scholar]
- Smith, J.J., Marelli, M., Christmas, R.H., Vizeacoumar, F.J., Dilworth, D.J., Ideker, T., Galitski, T., Dimitrov, K., Rachubinski, R.A., and Aitchison, J.D. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell. Biol. 158: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M.A. 2002. The identification of functional modules from the genomic association of genes. Proc. Natl. Acad. Sci. 99: 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417: 399–403. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wodicka, L., Dong, H., Mittmann, M., Ho, M.H., and Lockhart, D.J. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15: 1359–1367. [DOI] [PubMed] [Google Scholar]
- Xie, Y. and Varshavsky, A. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc. Natl. Acad. Sci. 98: 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.incyte.com/control/tools/proteome; Incyte Yeast Protein Database.
- http://labs.systemsbiology.net/galitski; Galitski Lab home page.
- http://www.ncbi.nlm.nih.gov/geo/; Gene Expression Omnibus database.