Abstract
Small heat shock proteins (sHsps) are molecular chaperones that prevent the aggregation of nonnative proteins. The sHsps investigated to date mostly form large, oligomeric complexes. The typical bacterial scenario seemed to be a two-component sHsps system of two homologous sHsps, such as the Escherichia coli sHsps IbpA and IbpB. With a view to expand our knowledge on bacterial sHsps, we analyzed the sHsp system of the bacterium Deinococcus radiodurans, which is resistant against various stress conditions. D. radiodurans encodes two sHsps, termed Hsp17.7 and Hsp20.2. Surprisingly, Hsp17.7 forms only chaperone active dimers, although its crystal structure reveals the typical α-crystallin fold. In contrast, Hsp20.2 is predominantly a 36mer that dissociates into smaller oligomeric assemblies that bind substrate proteins stably. Whereas Hsp20.2 cooperates with the ATP-dependent bacterial chaperones in their refolding, Hsp17.7 keeps substrates in a refolding-competent state by transient interactions. In summary, we show that these two sHsps are strikingly different in their quaternary structures and chaperone properties, defining a second type of bacterial two-component sHsp system.
Keywords: stress response, chaperone function, protein aggregation, heat stress, chaperone evolution
In the cellular stress response, small heat shock proteins (sHsps) represent a central component for balancing protein homeostasis. They act as promiscuous, ATP-independent molecular chaperones that prevent the aggregation of proteins upon stress conditions (1–4). In contrast to ATP-dependent chaperones, sHsps lack refolding activity and serve as storage for unfolded proteins (2, 5). Studies in different organisms revealed the cooperation of sHsps with members of the Hsp100 and Hsp70 chaperone families in the refolding of the bound substrate proteins (3, 6–10).
These proteins are found in most species except a few, mostly pathogenic bacteria (11, 12). All known sHsps consist of a conserved α-crystallin domain of ∼90 amino acids, flanked by variable amino- and carboxyl-terminal extensions (12). The α-crystallin domain, named after the respective eye lens protein (13), constitutes the structural hallmark of sHsps that forms a compact β-sheet sandwich fold (14–18). The dimer is the basic building block of higher oligomers. Available crystal structures and electron microscopic studies revealed that sHsps usually assemble into hollow, ball-, or barrel-like oligomers (14, 15, 17, 19–21). In the case of Hsp16.5 from Methanocaldococcus jannaschi, the structure consisting of 24 subunits forms a ball-like shell (15), whereas wheat Hsp16.9 assembles into two hexameric double disks resulting in a barrel-shaped structure (17).
In Escherichia coli, the two members of the sHsp family, IbpA and IbpB, have been found to be associated with inclusion bodies (22) and aggregated proteins after heat stress (23). The Ibp proteins share 48% sequence identity and form large multimeric assemblies with sizes of up to 3 MDa (24), with IbpA even adopting short fibrillar structures (25–27). IbpA and IbpB are present under physiological conditions and are both overexpressed by a factor of ∼300 upon heat stress (28). They physically interact with each other (24), and for substrate reactivation they cooperate with the Hsp70/40 and the Hsp100 chaperone system (10, 24) in a synergistic manner (29). Whereas IbpA reduces the size of substrate complexes but inhibits their further processing by disaggregases, IbpB facilitates substrate transfer to the Hsp70/40 and the Hsp100 chaperone machinery but does not alter the morphology of model substrate aggregates (29).
Besides the E. coli system, no other sHsp system in prokaryotes has been studied in detail to date, and it is therefore an open question whether the findings obtained for the E. coli system can be generalized for the prevailing bacterial two-component sHsp systems. In these bacterial two-component systems, the sHsps are either near relatives (located in the same branches of the phylogenetic tree, e.g., E. coli; Fig. S1A) or they are rather distantly related (located in different branches) (12). To study the latter type of two-component sHsp systems, we choose the sHsp system of Deinococcus radiodurans, an extremotolerant, Gram-positive bacterium (30). D. radiodurans has an optimal growth temperature of 30–32 °C. Above, the organism shows a heat stress response and cell division is slowed. Prolonged incubation at 46–52 °C results in a total arrest of cell division and lethality (31, 32). In its genome, nine common chaperones and cochaperones are found among a total of 67 stress-induced genes (32). The two-component sHsp system comprises Hsp20.2 (gene name ibpA, DR1114, gi number 15806134) and Hsp17.7 (gene name ibpB, DR1691, gi number 15806694) (Fig. S1A). Our analysis reveals that the two potential sHsps differ substantially in their quaternary structure and their functional role within the chaperone network, expanding our view on bacterial sHsp systems.
Results
Two sHsps of D. radiodurans Are Active as Molecular Chaperones in Vitro.
Hsp17.7 and Hsp20.2 were expressed in E. coli and purified to homogeneity. To ensure that both tag-free proteins were properly folded, their secondary structures and stabilities against thermal unfolding were monitored by CD spectroscopy (Fig. S1B). Similar to other sHsps (33), Hsp17.7 and Hsp20.2 show CD spectra typical for β-sheet proteins and melting temperatures of 67.3 °C and 57.8 °C, respectively (Fig. S1B). The structural integrity of the proteins is not influenced at temperatures below 48 °C.
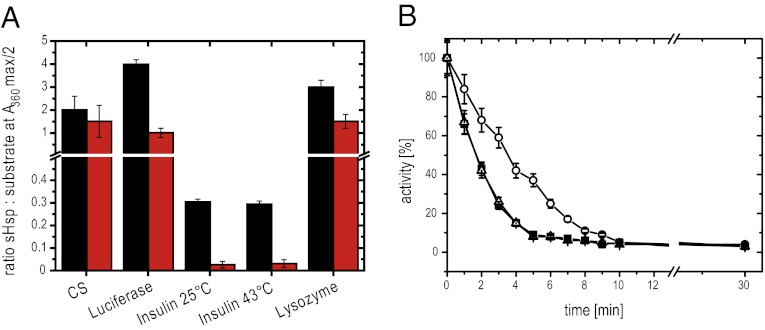
To assess the chaperone activity of Hsp20.2 and Hsp17.7, we first tested their ability to suppress protein aggregation at elevated temperatures, a key trait of sHsps (4, 8, 34). We used citrate synthase (CS, 48 kDa), firefly luciferase (61 kDa), insulin (3 kDa), and lysozyme (14 kDa) as model substrates. Hsp17.7 was able to suppress the thermal aggregation of both CS and luciferase in a concentration-dependent manner, as well as the aggregation of reduced lysozyme (Fig. 1A and Fig. S2). Additionally, the reduction-induced aggregation of the insulin B chain at 25 °C and at 43 °C was suppressed, showing similar ratios of sHsp: substrate needed for half-maximum suppression at both temperatures (Fig. 1A and Fig. S2 E and G). Thus, it seems that the chaperone activity of Hsp17.7 is not affected by temperature. Because the amount of substrate molecules to sHsps is dependent on the size of the substrate, for peptide substrates like insulin, usually substoichiometric concentrations of sHsps are sufficient (35, 36). The ratio of half-maximum aggregation suppression, however, does not correlate with the binding stoichiometry in this type of assay, because the decrease of the concentration of aggregation-prone insulin is sufficient to inhibit aggregation even if not the majority of insulin molecules is captured by the sHsp. In comparison with Hsp17.7, Hsp20.2 is the more potent chaperone because lower amounts of Hsps20.2 were sufficient to fully suppress the heat- or reduction-induced aggregation of the substrate proteins (Fig. 1A and Fig. S2).
Fig. 1.
Comparison of chaperone activity of Hsp17.7 and Hsp20.2. (A) Influence of Hsp17.7 (black) and Hsp20.2 (red) on the thermal aggregation of different substrate proteins. The ratio of sHsp to substrate at half-maximum suppression of aggregation is displayed. Error bars represent the SD in three independent assays. (B) Influence of Hsp17.7 and Hsp20.2 on the thermal inactivation of CS. Aliquots from a thermostated solution (43 °C) of 75 nM CS (filled squares) and 75 nM CS in the presence of 600 nM Hsp17.7 (open circles) or 600 nM Hsp20.2 (open triangles) were extracted at the indicated time points, and the enzymatic activity was determined (37). Mean values of three independent assays and the respective SD are indicated.
In the case of CS, monitoring the kinetics of thermal inactivation allows conclusions on the interaction of a molecular chaperone with unfolding intermediates and on the stability of the chaperone–substrate complex (37). For Hsp17.7, we observed a deceleration of CS inactivation (Fig. 1B), indicating a transient interaction between Hsp17.7 and CS unfolding intermediates, as was described for Hsp26 and Hsp42 (38). In contrast, the thermal inactivation of CS was not affected by the presence of Hsp20.2, pointing toward a more stable complex between Hsp20.2 and its substrate.
Quaternary Structures of Hsp17.7 and Hsp20.2 Are Strikingly Different.
To determine the size of the Hsp17.7 oligomer, we used size exclusion chromatography (SEC). The resulting single peak had an apparent molecular mass of 150 kDa, which could correspond to ∼8 subunits (Fig. 2A). Next we investigated the influence of heat stress on the oligomeric size of Hsp17.7, because several sHsps dissociate at elevated temperatures (39, 40). At 43 °C, Hsp17.7 eluted with an identical apparent mass as observed at 25 °C (Fig. 2A), demonstrating that the quaternary structure of Hsp17.7 is unaffected by changes in temperature. A more detailed analysis of the quaternary structure of Hsp17.7 by analytical ultracentrifugation (aUC) sedimentation velocity experiments showed a surprisingly low S20w value of 2.1 S, corresponding to a molecular mass of ∼40 kDa and a frictional coefficient (f/f0) of 1.78, independent of the protein concentration used (Fig. 2C and Fig. S3A). Thus, the aUC experiments suggest that Hsp17.7 is an elongated dimer. Further refinement of the aUC analysis using a 2D grid analysis (2-DSA) and Monte Carlo simulations revealed a single species with an S20w value of 2.08 S, a frictional coefficient of 1.78, and a molecular mass of 37 kDa (Fig. S3B), confirming that Hsp17.7 is an elongated dimer in solution that does not change its size at elevated temperatures. This sets Hsp17.7 apart from most other sHsps studied to date.
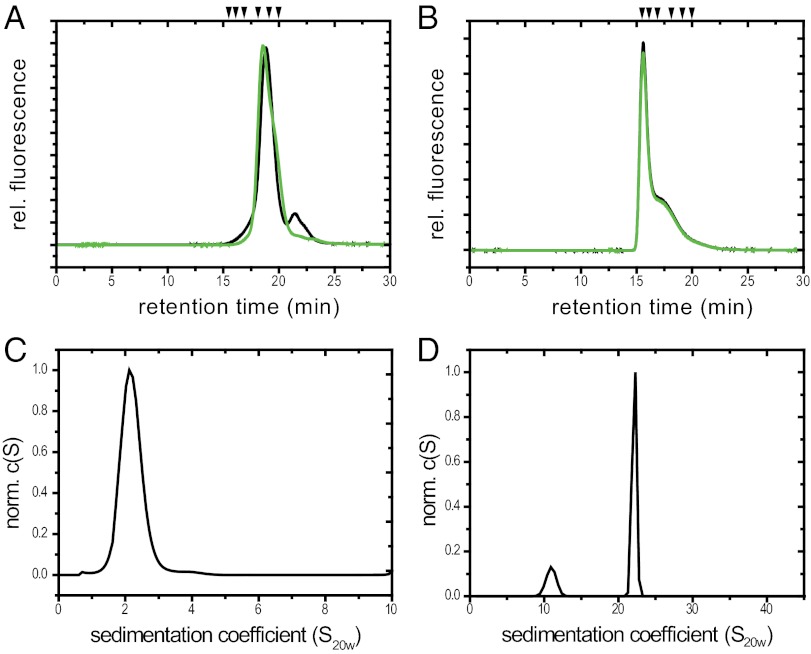
Fig. 2.
Quaternary structure of Hsp17.7 and Hsp 20.2. (A and B) SEC-HPLC was performed using a TosoHaas TSK 4000 PW column as described in Materials and Methods. Hsp17.7 (A) and Hsp20.2 (B) at a concentration of 1.5 mg/mL were separated at 25 °C (green) and at 43 °C (black). Arrows indicate calibration markers as described in Materials and Methods. (C and D) Analytical ultracentrifugation sedimentation analysis. Normalized c(S) distribution of Hsp17.7 at 1.0 mg/mL (C) and Hsp20.2 at 1.5 mg/mL (D).
In contrast, Hsp20.2 exhibited one main peak in SEC (Fig. 2B), corresponding to a large oligomeric complex of 600–800 kDa, corresponding to roughly 36 subunits and a peak shoulder, indicative of an 18mer. No temperature-dependent shift to smaller oligomeric species was observed at 43 °C. aUC experiments yielded a sedimentation coefficient of 22 S for the high molecular weight oligomer and 11 S for the smaller oligomer (Fig. 2D and Fig. S3C). Using interference optics we were able to fit the data to a frictional coefficient of 1.23 and a molecular mass of ∼700 kDa and ∼350 kDa, in accordance with a 36mer and an 18mer.
To investigate whether the assembly state of the two sHsps is similar in vivo, their separation behavior in D. radiodurans extracts was analyzed by native PAGE. Both proteins were detected at the same position as the purified proteins (Fig. S3D). For Hsp20.2, the two visible bands correlated well with the observed two main species. For Hsp17.7, a prominent dimer band was detected. When purified Hsp20.2 was added to cell lysate of D. radiodurans at physiological conditions (25 °C), an additional high molecular mass band appeared, most likely indicating the formation of substrate complexes with unfolding proteins (see below). Furthermore, immunoblot analysis suggests that the two proteins do not form mixed complexes, and aUC experiments with mixtures of the two proteins support this finding (Fig. S3E).
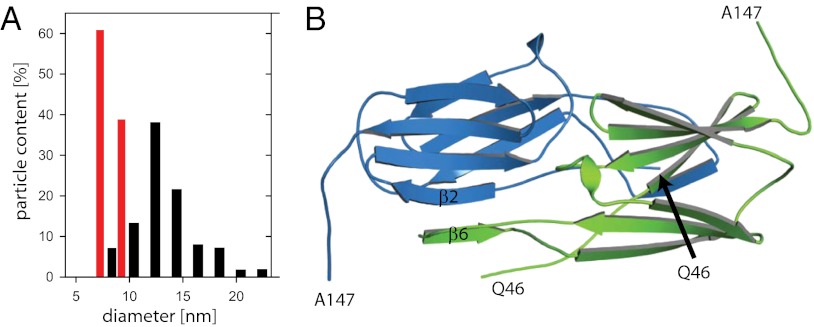
To elucidate the quaternary structure of the oligomers in more detail, the samples were visualized by negative stain electron microscopy (Fig. S4 A and B). We could confirm the small size of Hsp17.7, with dimensions in agreement with the postulated dimers (Fig. 3A and Fig. S4B). Hsp20.2 formed heterogeneous, mostly spherical oligomers with diameters between 9 and 18 nm (Fig. 3A and Fig. S4B). The main population had a diameter of ∼13 nm.
Fig. 3.
Structure of Hsp20.2 and Hsp17.7. (A) Size distributions (circumscribing diameters, in nm) of Hsp20.2 oligomers (black) and Hsp17.7 (red), based on electron microscopic images from negatively stained particles. (B) Crystal structure of Hsp17.7 (Protein Data Bank ID code 4FEI). Ribbon diagram of the Hsp17.7 dimer. The monomers are colored in green and blue. The N-terminal 45 and the C-terminal 19 amino acids are not shown as they are structurally distorted.
For Hsp17.7, crystallization conditions could be determined that resulted in a crystal diffracting to 2.4-Å resolution (Table S1). The atomic structure obtained after molecular replacement shows that Hsp17.7 crystallized as a dimer. Similar to other structures of sHsps, the N-terminal region (amino acids 1–45) and the C-terminal extension (amino acids 148–166) of Hsp17.7 are flexible and therefore lack defined electron density. The monomers adopt the characteristic α-crystallin core fold, consisting of two antiparallel β-sheets, which are packed in parallel layers (Fig. 3B and Fig. S4). In agreement with other family members (1, 16), the peripheral β6-strand in the extended loop stabilizes the dimer of Hsp17.7 by interacting with the β2-strand of the partner monomer. A backbone superposition (Fig. S4E) of sHsps from different organisms showing this typical strand-exchanging dimer interface revealed structural divergence for the extended loop. Interestingly, the backbone of the extended loop of Hsp17.7 structurally matches best with HspA from the plant pathogenic bacterium Xanthomonas sp. (XAC1151) (41, 42). However, XaHspA assembles to a 36mer in vitro. In comparison with XaHspA, MjHsp16.5, and TaHsp16.9, the extended loop of Hsp17.7 adopts the narrowest geometry (Fig. S4E), caused by the sequence gap between the β5- and β6-strands (Fig. S4C). The Hsp17.7 dimers are packed in a helical, line-up order (Fig. S4D). Interestingly, the C-terminal IXI/V motif still forms a crystal contact to the neighboring dimer, which resembles the oligomerization interface also found in the structures of MjHsp16.5 and TaHsp16.9 (4). However, the β10-strand formed by the IXI/V motif is oriented parallel to the β4-strand, which is opposed to its orientation in MjHsp16.5 and TaHsp16.9 oligomers (Fig. S4F). In this context it should also be noted that the C-terminal extension of Hsp17.7 is peculiarly short, showing a sequence gap between the α-crystallin domain and the IXI/V motif (Fig. S4C).
Substrate Complexes of Hsp20.2 and Hsp17.7.
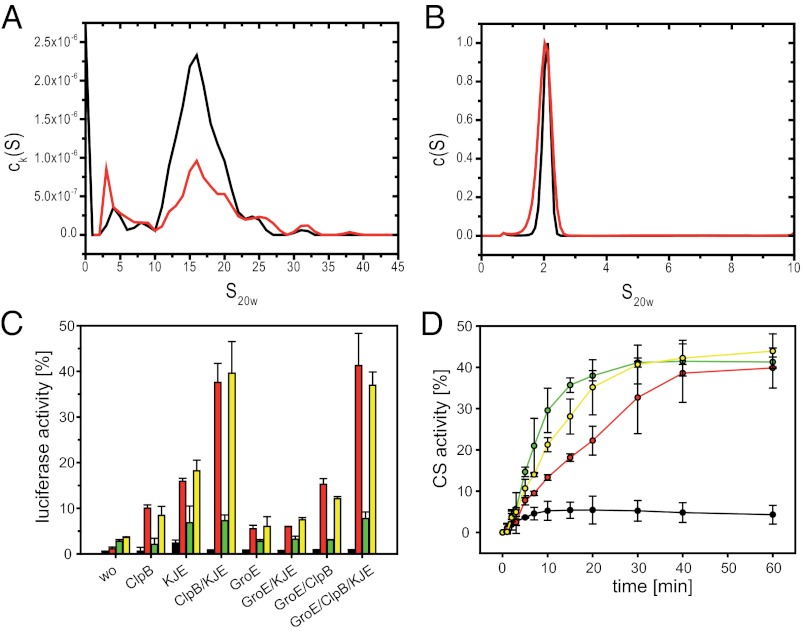
To determine the active chaperone species and the stoichiometry of the substrate interaction, aUC experiments were performed. We exploited the tremendous spectral differences between Hsp20.2 (A280 = 4,470 M−1⋅cm−1) and lysozyme (38,000 M−1⋅cm−1) and conducted a multisignal sedimentation velocity analysis (43). A 16-S complex was detected, which did not change when the Hsp20.2 concentration was varied (Fig. 4A). Intriguingly, the observed size of the Hsp20.2–lysozyme complex lies between the Hsp20.2 18mer and the 36mer, indicating that the 18mer is the substrate binding species. Collecting aUC data with absorbance and interference optics in parallel, we were able to spectrally resolve the complex. It showed a molecular mass of approximately 550 kDa and a stoichiometry of 3:1 Hsp20.2 to lysozyme. Thus, the complex most likely consists of 18 subunits Hsp20.2 and six molecules of bound substrate.
Fig. 4.
Analysis of substrate binding and release of Hsp17.7 and Hsp20.2. (A) Multisignal sedimentation velocity analysis of substrate binding to Hsp20.2. ck(S) distribution of 10 µM lysozyme + 1 mM Tris(2-carboxyethyl)phosphine + 20 µM Hsp20.2 calculated from parallel scans with absorbance and interference optics. The contribution of Hsp20.2 (black) and lysozyme (red) to the total signal was deconvoluted using the multiwavelength discrete/continuous distribution analysis of SEDPHAT. (B) c(S) distribution of 1 µM Hsp17.7-Alexa 488 (black) and 10 µM Hsp17.7-Alexa 488 + 100 µM Hsp20.2 (red). (C) Effects of Hsp17.7 and Hsp20.2 on the refolding efficiency of heat-denatured luciferase by the KJE, ClpB, and GroE chaperone machineries. Luciferase (80 nM) was incubated at 43 °C for 10 min in the absence (black) of sHsps or in the presence of 600 nM Hsp20.2 (red), 600 nM Hsp17.7 (green), or 600 nM of Hsp20.2 and Hsp17.7 (yellow). After complete inactivation, the samples were shifted to 25 °C and subsequently DnaK/DnaJ/GrpE (KJE, 0.6, 1.2, and 0.6 µM), ClpB (600 nM), and GroE (600 nM GroEL and GroES) were added in different combinations as indicated. Luciferase activity was assayed after 60 min in comparison with 80 nM untreated luciferase (100% value). Error bars represent the SD in three independent assays. (D) CS was denatured for 1 h in 6 M GdmCl and diluted 200-fold to a final concentration of 300 nM in a solution containing DnaK/DnaJ/GrpE (2, 2, and 0.5 µM), 2 mM ATP, and 3 mM MgCl2 (black) supplemented with 1.2 µM Hsp17.7 (green), 1.2 µM Hsp20.2 (red), or a mixture of 0.6 µM Hsp17.7 and 0.6 µM Hsp20.2 (yellow). Aliquots were withdrawn at the indicated time points, and the enzymatic activity was determined (37). Mean values of three independent assays and the respective SDs are indicated.
To analyze the interaction of Hsp17.7 with lysozyme, fluorescently labeled Hsp17.7 was incubated with lysozyme, and complex formation was analyzed by aUC. In the presence of reduced lysozyme, the sedimentation behavior of Hsp17.7 did not change compared with Hsp17.7 alone (Fig. 4B). Thus, no stable Hsp17.7–substrate complex could be detected consistent with the results obtained for CS inactivation (Fig. 1B).
The depletion of IbpA/B in E. coli leads to enhanced aggregation of proteins (7) and to a reduction of maximum cell numbers in the stationary phase. The plasmid-based overexpression of IbpA/B in the respective strain is able to complement both phenotypes (Fig. S5 C and D). When Hsp17.7 or Hsp20.2 was overexpressed no complementation of the IbpA/B phenotype was achieved (Fig. S5 C and D), indicating that both systems are not comparable to each other and that the D. radiodurans sHsps cannot even compensate partially for the E. coli Ibps.
To investigate the chaperone activity of Hsp20.2 and Hsp17.7 in vivo, we analyzed aggregated protein in stressed and unstressed D. radiodurans lysates (Fig. S6A). Interestingly, only Hsp20.2 was associated with aggregates, whereas Hsp17.7 stayed in the soluble fraction. Additionally, when D. radiodurans lysates were separated by native PAGE after 30 min at 43 °C (Fig. S6B), Hsp20.2 was found in the high molecular weight aggregates. To determine which substrates are bound by Hsp20.2, the high molecular aggregates were analyzed by mass spectrometry (see below and Table S2). This analysis further confirmed the presence of Hsp20.2 and the absence of Hsp17.7 in these complexes, and thus differences in substrate binding. When we isolated sHsp–substrate complexes by immunoprecipitation, only for Hsp20.2 substantial protein amounts were coisolated from D. radiodiurans lysates (Fig. S6C). Subsequent mass spectrometry analysis allowed us to identify ∼100 potential substrate proteins bound by Hsp20.2 (Table S2), including a number of metabolic enzymes, ribosomal proteins, and elongation factors. These aggregation-prone proteins seem to be the basic substrates of sHsps in D. radiodurans. Similar to other sHsps (38, 44), the molecular mass distribution of the substrates of Hsp20.2 is very broad, ranging from 15 to more than 100 kDa. Interestingly, besides these potential substrates, a number of S-layer proteins including the gene DR1115 (NP 294839.1), which is encoded directly upstream of hsp20.2, were detected. Usually these proteins are extracellular, but they might be recognized as substrates on their secretion pathway. Within the identified proteins, DnaK and GroEL were also detected, indicating a potential cooperation of these chaperone systems in substrate release similar to observations for E. coli (7, 10). To investigate potential synergistic effects with other molecular chaperones, we analyzed the ability of the D. radiodurans sHsps to cooperate with the E. coli Hsp70 (DnaK/DnaJ/GrpE; KJE), Hsp100 (ClpB), and Hsp60 (GroEL/GroES) chaperone systems. We determined the refolding of substrates from preformed sHsp complexes with thermally unfolded firefly luciferase (Fig. 4C and Fig. S7) and chemically denatured CS (Fig. 4D). A few percent of luciferase were reactivated spontaneously (below 2%) or in the presence of the sHsps only (below 5%). However, when Hsp20.2 was present during the inactivation reaction, 10–15% of the luciferase activity was regained in presence of GroEL or ClpB and 15–20% when KJE was added. The combination of the ClpB and the KJE systems resulted in the refolding of nearly 40% of luciferase. Similar to results for E. coli IbpB (7), the combination of all three ATP-dependent chaperone systems indicated that the GroE system is able to compensate for the role of KJE to some extent, but no additive or synergistic effects on luciferase refolding were achieved when both GroE and KJE were present. In the presence of Hsp17.7, on the other hand, only minor levels of luciferase activity were regained, independent of the combination of chaperone systems added. Hsp17.7 seems to interact only transiently, releasing its substrate spontaneously without the help of other chaperone systems. When both D. radiodurans sHsps were present in the reaction, no additive or synergistic effects were observed, indicating that the two sHsps work independently of each other. Interestingly, in experiments with chemically denatured CS, which was diluted in a solution containing KJE and Hsp17.7 or Hsp20.2, respectively, we detected a slight acceleration of the refolding kinetics in the presence of Hsp17.7, whereas the total amount of recovered activity was similar, independent of the sHsp present (Fig. 4D). Again, the two sHsps did not show any synergistic effects in this assay. The finding that Hsp20.2 and Hsp17.7 act independently of each other is in clear contrast to studies on E. coli IbpA/B, which work together synergistically in refolding with the help of other, ATP-dependent chaperone systems (24, 29).
Discussion
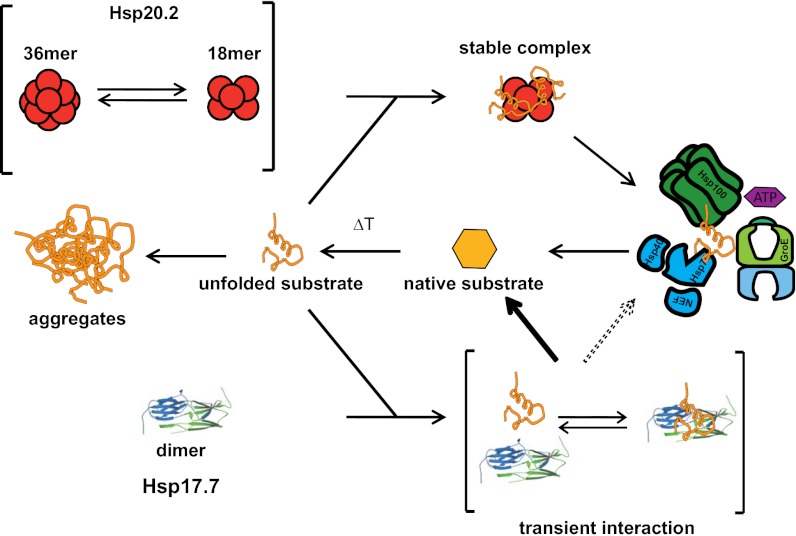
sHsps can be seen as the guardians of the proteome, especially upon sudden proteotoxic stresses, which result in the accumulation of aggregation-prone nonnative proteins. In bacteria, mainly one or two sHsps are found per species (12). For the two-component systems, our analysis of the D. radiodurans sHsps revealed a strikingly different organization and behavior compared with the well-studied E. coli system and hence defines a unique paradigm. Whereas in the E. coli system, two highly homologous sHsps (IbpA and IbpB) cooperate, the D. radiodurans sHsp system consists of two proteins that are far apart in the phylogenetic tree, with different structures and different chaperone function. Hsp20.2 belongs to the large spherical homooligomeric sHsps, which are constitutively active and form stable substrate complexes. In contrast and unexpectedly, Hsp17.7 is a dimer that is also active as a chaperone but forms only transient complexes with the substrate proteins tested. Presumably, stable substrate interactions are favored by multiple interactions in oligomeric sHsp complexes.
Two different assembly forms of the α-crystallin dimers have been reported to date (1, 4, 18). In the first variant, the β6/7-strands of the two monomers are part of a compact β-sheet, whereas in the second variant, the peripheral β6-strand is located in the extended loop and stabilizes the dimer by interacting with the β2-strand of the partner monomer. Hsp17.7 adopts the second variant of assembly. However, because of a sequence gap between the β5- and β6-strands, the loop geometry is rather narrow in Hsp17.7. Previous studies demonstrated that the C-terminal extension of sHsp monomers crucially contributes to the assembly of the α-crystallin dimers and oligomers by binding to the β4–β8 hydrophobic groove of a neighboring monomer via the IXI/V motif (15, 17, 45, 46). In Hsp17.7 the short length of the C-terminal extension and the resulting close location of IXI/V motif to the α-crystallin domain, together with an alternate extended loop conformation, might hinder the formation of oligomers. Intriguingly, the IXI/V motif of the short C-terminal extension forms a crystal contact to the β4–β8 edge of a symmetry mate by adopting an ordered β-strand conformation, indicating that the principal contact site is conserved in Hsp17.7. However, in the crystal, the β10-strand is orientated opposed to the common direction within the β4–β8 groove in MjHsp16.5 or TaHsp16.9 oligomers.
It is remarkable that Hsp17.7 is active as a dimer in suppressing the aggregation of diverse substrates. Only under certain buffer conditions, a few sHsps like HspB6 (Hsp20) (47, 48) and HspB1 (Hsp27) (49) could be shown to be active as dimers. However, in these cases, the sHsps usually form large oligomers.
The atypical quaternary structure of Hsp17.7 goes along with remarkable differences in its mode of substrate interaction compared with other sHsps. Although Hsp17.7 is able to suppress the aggregation of different model substrates efficiently, we could not detect any stable substrate complexes. Together with its ability to affect the kinetics of CS thermo-inactivation and the KJE-mediated refolding of chemically denatured CS, this supports the view of a transient interaction of Hsp17.7 with unfolded polypeptide chains. Free Hsp17.7 and its substrate-bound state may exist in a dynamic equilibrium, preventing the formation of large aggregates (Fig. 5). Thus, Hsp17.7 is an atypical sHsps that might act as a rather generic low-affinity chaperone or might be necessary for some specific substrates in D. radiodurans.
Fig. 5.
Model of the D. radiodurans two-component sHsp system. Dimeric Hsp17.7 and the ensemble of 36 and 18meric Hsp20.2 form the D. radiodurans two-component sHsp system. Stress conditions, like heat shock, lead to the unfolding of substrate proteins (orange). Unfolding proteins are stably trapped by Hsp20.2 18mers or transiently bound by Hsp17.7. Hsp104 and Hsp70 or GroE can reactivate the Hsp20.2-bound substrate proteins in an energy-dependent reaction or refold the substrate proteins spontaneously released from Hsp17.7. This indicates that Hsp20.2 and Hsp17.7 work in parallel and independently of each other.
In contrast to Hsp17.7, Hsp20.2 is a typical sHsp forming dynamic oligomers with seemingly two main species, a 36mer and an 18mer. Compared with archaeal and eukaryotic sHsps, which have been described as 12mers or 24mers (15, 17, 20, 21, 45), the Hsp20.2 assembly seems to be more closely related to Xanthomonas HspA, which has also been reported to be a 36mer (45, 46). Interestingly, Xanthomonas HspA and Hsp20.2 are found in the same branch of the phylogenetic tree (12).
Concerning chaperone activity, Hsp20.2 suppresses the aggregation of substrate proteins with a binding capacity of ∼6 substrate molecules per Hsp20.2 18mer. This also indicates that the 36mer observed in the absence of substrate proteins might represent a storage form (Fig. 5).
sHsps have been shown to cooperate with Hsp100 proteins and the Hsp70 system in refolding the sHsp-bound substrates (7–9, 29). For Hsp20.2, our studies indicate a similar functional network. Hsp17.7, however, does not seem to be integrated in this chaperone cascade. Because of its transient binding behavior, it is active without the need of other ATP-dependent chaperones.
Taken together, on the systemic level, our results show that the D. radiodurans sHsp system is structurally and functionally strikingly distinct from the two-component sHsp system of E. coli. Intriguingly, the two sHsps of D. radiodurans, neither of which has been acquired through horizontal gene transfer (according to Alien Hunter, Sanger Institute, Cambridge, United Kingdom), seem to function independently of each other. This view is supported by in vivo experiments in which we substituted the E. coli ibp deletion strain by the D. radiodurans sHsps and could not detect complementation. Overall, this indicates that the diversity of the bacterial sHsp systems might be higher than expected. Besides the basic classification of bacteria encoding one or two sHsp members (12), the two-member systems need to be subdivided into two types: type I with two highly homologous sHsps that (closely) cooperate (E. coli type) and type II consisting of two sHsps that are structurally different and act independently of each other (D. radiodurans type).
Materials and Methods
Materials.
E.coli DnaK, DnaJ, GrpE, and pig heart CS were purified as described elsewhere (8, 50). Yeast SUMO (small ubiquitin-like modifier) protease representing the C-terminal fragment (403–621) of ubiquitin-like-protease 1 (51) was amplified by PCR from cDNA and cloned into pET28b (Invitrogen), yielding N-terminally His6-tagged protein. The protein was purified by Ni2+-chelating chromatography (GE Healthcare) according to the manufacturer’s protocol. Bovine insulin and hen egg lysozyme were obtained from Sigma.
Expression and Purification of sHsps.
For purification of Hsp17.7 and Hsp20.2, N-terminal 6× His plus SUMO fusions were constructed. The proteins were expressed in E. coli BL21 and purified by Ni2+ chelating and SEC according to the manufacturer's protocols (GE Healthcare). Details are provided in SI Materials and Methods.
Analysis of Chaperone Activity.
Thermal aggregation of model substrates, inactivation, and refolding of CS was performed as described elsewhere (8). For refolding of chemically denatured CS by the DnaK chaperone machinery, 2 µM DnaK, 2µM DnaJ, and 0.5 µM GrpE were used in the presence of 1 µM Hsp17.7 or Hsp20.2.
Inactivation and reactivation of firefly luciferase (Photinus pyralis; Promega) was assayed as described elsewhere (8). For each measurement, native luciferase was diluted to a final concentration of 4 µg/mL (80 nM) and incubated for 10 min at 43 °C in the presence or absence of sHsps. Then different combinations of chaperones were added, and the reactions were shifted to 25 °C for 65 min for reactivation. The activity of luciferase was compared with the activity of untreated, native luciferase, which was set to 100%. Luciferase activity was measured using a thermostated Tecan GENios Microplate Reader.
Analytical SEC.
SEC was performed as described previously (35). Buffer conditions were 40 mM Hepes/KOH (pH 7.4) and 150 mM NaCl, and a flow rate of 0.5 mL/min was used. The TSK 4000 pw column (TosoHaas) was calibrated using the Sigma gel filtration molecular weight marker kit (Sigma) including carbonic anhydrase, 29 kDa; albumin, 66 kDa; alcohol dehydrogenase, 150 kDa; amylase, 200 kDa; apoferritin, 443 kDa; and thyroglobulin, 669 kDa.
aUC Sedimentation Velocity Experiments.
Analytical ultracentrifugation was carried out with a ProteomLab XL-I (Beckman) supplied with absorbance and interference optics. Sample sedimentation was monitored at 280 nm or 230 nm, continuously scanning with a radial resolution of 30 microns. Data analysis was carried out with the Ultrascan and the USLIMS platform (52). Details are provided in SI Materials and Methods.
Protein Crystallization and Structure Determination.
Crystals of sHsp17.7 were grown at 20 °C by using the sitting drop vapor diffusion method. The drops contained equal volumes of protein (12 mg/mL) and reservoir solution [0.2 M ammoniumacetate, 0.1 M bis·Tris (pH 6.5), 25% PEG 3350]. The dataset was collected on a Bruker Microstar/X8 Proteum (Bruker AXS) with a Cu rotating anode (λ = 1.54 Å) and processed with the Proteum software suite (Bruker AXS). Hsp17.7 crystallized in the trigonal space group P3121 with cell parameters of a,b = 51.3 Å, c = 80.5 Å. For structure determination, molecular replacement was performed in Phaser (53) using the coordinates of HspA from Xanthomonas sp. (XAC1151; PDB-ID: 3GLA) as a starting model. Details are provided in SI Materials and Methods.
Electron Microscopy and Image Processing.
For negative stain electron microscopy, 5 µL of the protein sample (0.05 mg/mL protein in 50 mM Hepes/KOH, 75 mM NaCl, pH 7.4) were adsorbed for 2 min onto carbon-coated grids that were glow discharged in air. Samples were negatively stained with uranyl acetate [1.5% (wt/vol) at pH 4.5], and electron micrographs were recorded at a calibrated magnification of 58,000 using a JEOL JEM 100CX electron microscope operated at 100 kV. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Katrin Heinz (University of Konstanz) for the kind gift of the Deinococcus radiodurans strain R1, Dr. Axel Mogk for the kind gift ibp deletion strains and expression plasmids, and Bianca Ludwig for excellent experimental assistance. This research was supported by Deutsche Forschungsgemeinschaft Grants SFB 594 A5 and SFB 1035 A06; and in part by the National Science Foundation through TeraGrid resources provided by Texas Advanced Computing Center Grant TG-MCB070040N.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [Research Collaboratory for Structural Bioinformatics ID code rcsb072800 and PDB ID code 4FEI (Hsp17.7 from Deinococcus radiodurans)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209565109/-/DCSupplemental.
References
- 1.McHaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: Chaperone activity of small heat shock proteins. Biochemistry. 2009;48(18):3828–3837. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12(10):842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 3.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27(2):328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basha E, O'Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37(3):106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter K, Haslbeck M, Buchner J. The heat shock response: Life on the verge of death. Mol Cell. 2010;40(2):253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Kampinga HH, Brunsting JF, Stege GJ, Konings AW, Landry J. Cells overexpressing Hsp27 show accelerated recovery from heat-induced nuclear protein aggregation. Biochem Biophys Res Commun. 1994;204(3):1170–1177. doi: 10.1006/bbrc.1994.2586. [DOI] [PubMed] [Google Scholar]
- 7.Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50(2):585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 8.Haslbeck M, Miess A. Stromer T. Walter S. Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005;280(25):23861–23868. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 9.Cashikar AG, Duennwald ML, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280(25):23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273(18):11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- 11.Kappé G, Leunissen JA, de Jong WW. Evolution and diversity of prokaryotic small heat shock proteins. Prog Mol Subcell Biol. 2002;28:1–17. doi: 10.1007/978-3-642-56348-5_1. [DOI] [PubMed] [Google Scholar]
- 12.Kriehuber T, et al. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24(10):3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- 13.Horwitz J. Alpha-crystallin. Exp Eye Res. 2003;76(2):145–153. doi: 10.1016/s0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 14.Stamler R, Kappé G, Boelens W, Slingsby C. Wrapping the alpha-crystallin domain fold in a chaperone assembly. J Mol Biol. 2005;353(1):68–79. doi: 10.1016/j.jmb.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394(6693):595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 16.Bagnéris C, et al. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392(5):1242–1252. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 17.van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8(12):1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 18.Laganowsky A, et al. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19(5):1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun N, et al. Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc Natl Acad Sci USA. 2011;108(51):20491–20496. doi: 10.1073/pnas.1111014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haslbeck M, Kastenmüller A, Buchner J, Weinkauf S, Braun N. Structural dynamics of archaeal small heat shock proteins. J Mol Biol. 2008;378(2):362–374. doi: 10.1016/j.jmb.2008.01.095. [DOI] [PubMed] [Google Scholar]
- 21.White HE, et al. Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26. Structure. 2006;14(7):1197–1204. doi: 10.1016/j.str.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Laskowska E, Wawrzynów A, Taylor A. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie. 1996;78(2):117–122. doi: 10.1016/0300-9084(96)82643-5. [DOI] [PubMed] [Google Scholar]
- 23.Allen SP, Polazzi JO, Gierse JK, Easton AM. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174(21):6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matuszewska M, Kuczyńska-Wiśnik D, Laskowska E, Liberek K. The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J Biol Chem. 2005;280(13):12292–12298. doi: 10.1074/jbc.M412706200. [DOI] [PubMed] [Google Scholar]
- 25.Kitagawa M, Miyakawa M, Matsumura Y, Tsuchido T. Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur J Biochem. 2002;269(12):2907–2917. doi: 10.1046/j.1432-1033.2002.02958.x. [DOI] [PubMed] [Google Scholar]
- 26.Ratajczak E, et al. IbpA the small heat shock protein from Escherichia coli forms fibrils in the absence of its cochaperone IbpB. FEBS Lett. 2010;584(11):2253–2257. doi: 10.1016/j.febslet.2010.04.060. [DOI] [PubMed] [Google Scholar]
- 27.Shearstone JR, Baneyx F. Biochemical characterization of the small heat shock protein IbpB from Escherichia coli. J Biol Chem. 1999;274(15):9937–9945. doi: 10.1074/jbc.274.15.9937. [DOI] [PubMed] [Google Scholar]
- 28.Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27(19):3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratajczak E, Zietkiewicz S, Liberek K. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J Mol Biol. 2009;386(1):178–189. doi: 10.1016/j.jmb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Heinz K, Marx A. Lesion bypass activity of DNA polymerase A from the extremely radioresistant organism Deinococcus radiodurans. J Biol Chem. 2007;282(15):10908–10914. doi: 10.1074/jbc.M611404200. [DOI] [PubMed] [Google Scholar]
- 31.Battista JR. Against all odds: The survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 32.Airo A, Chan SL, Martinez Z, Platt MO, Trent JD. Heat shock and cold shock in Deinococcus radiodurans. Cell Biochem Biophys. 2004;40(3):277–288. doi: 10.1385/CBB:40:3:277. [DOI] [PubMed] [Google Scholar]
- 33.Stromer T, Fischer E, Richter K, Haslbeck M, Buchner J. Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation: The N-terminal domail is important for oligomer assembly and the binding of unfolding proteins. J Biol Chem. 2004;279(12):11222–11228. doi: 10.1074/jbc.M310149200. [DOI] [PubMed] [Google Scholar]
- 34.Friedrich KL, Giese KC, Buan NR, Vierling E. Interactions between small heat shock protein subunits and substrate in small heat shock protein-substrate complexes. J Biol Chem. 2004;279(2):1080–1089. doi: 10.1074/jbc.M311104200. [DOI] [PubMed] [Google Scholar]
- 35.Haslbeck M, et al. Hsp26: A temperature-regulated chaperone. EMBO J. 1999;18(23):6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stromer T, Ehrnsperger M, Gaestel M, Buchner J. Analysis of the interaction of small heat shock proteins with unfolding proteins. J Biol Chem. 2003;278(20):18015–18021. doi: 10.1074/jbc.M301640200. [DOI] [PubMed] [Google Scholar]
- 37.Buchner J, Grallert H, Jakob U. Analysis of chaperone function using citrate synthase as nonnative substrate protein. Methods Enzymol. 1998;290:323–338. doi: 10.1016/s0076-6879(98)90029-5. [DOI] [PubMed] [Google Scholar]
- 38.Haslbeck M, et al. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23(3):638–649. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy GB, Das KP, Petrash JM, Surewicz WK. Temperature-dependent chaperone activity and structural properties of human alphaA- and alphaB-crystallins. J Biol Chem. 2000;275(7):4565–4570. doi: 10.1074/jbc.275.7.4565. [DOI] [PubMed] [Google Scholar]
- 40.Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270(18):10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- 41.Hilario E, Martin FJ, Bertolini MC, Fan L. Crystal structures of Xanthomonas small heat shock protein provide a structural basis for an active molecular chaperone oligomer. J Mol Biol. 2011;408(1):74–86. doi: 10.1016/j.jmb.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Lin CH, et al. Characterization of Xanthomonas campestris pv. campestris heat shock protein A (HspA), which possesses an intrinsic ability to reactivate inactivated proteins. Appl Microbiol Biotechnol. 2010;88(3):699–709. doi: 10.1007/s00253-010-2776-z. [DOI] [PubMed] [Google Scholar]
- 43.Padrick SB, et al. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal Biochem. 2010;407(1):89–103. doi: 10.1016/j.ab.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basha E, et al. The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem. 2004;279(9):7566–7575. doi: 10.1074/jbc.M310684200. [DOI] [PubMed] [Google Scholar]
- 45.Takeda K, et al. Dimer structure and conformational variability in the N-terminal region of an archaeal small heat shock protein, StHsp14.0. J Struct Biol. 2011;174(1):92–99. doi: 10.1016/j.jsb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Saji H, et al. Role of the IXI/V motif in oligomer assembly and function of StHsp14.0, a small heat shock protein from the acidothermophilic archaeon, Sulfolobus tokodaii strain 7. Proteins. 2008;71(2):771–782. doi: 10.1002/prot.21762. [DOI] [PubMed] [Google Scholar]
- 47.Bukach OV, Glukhova AE, Seit-Nebi AS, Gusev NB. Heterooligomeric complexes formed by human small heat shock proteins HspB1 (Hsp27) and HspB6 (Hsp20) Biochim Biophys Acta. 2009;1794(3):486–495. doi: 10.1016/j.bbapap.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Bukach OV, Seit-Nebi AS, Marston SB, Gusev NB. Some properties of human small heat shock protein Hsp20 (HspB6) Eur J Biochem. 2004;271(2):291–302. doi: 10.1046/j.1432-1033.2003.03928.x. [DOI] [PubMed] [Google Scholar]
- 49.Hayes D, Napoli V, Mazurkie A, Stafford WF, Graceffa P. Phosphorylation dependence of hsp27 multimeric size and molecular chaperone function. J Biol Chem. 2009;284(28):18801–18807. doi: 10.1074/jbc.M109.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haslbeck M, Schuster I, Grallert H. GroE-dependent expression and purification of pig heart mitochondrial citrate synthase in Escherichia coli. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;786(1-2):127–136. doi: 10.1016/s1570-0232(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 51.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5(5):865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 52.Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39(3):405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.