Summary
DNA double strand breaks are repaired by two main pathways: non-homologous end joining (NHEJ) and homologous recombination (HR). The choice between these pathways depends on cell cycle phase; however the continuous effect of cell cycle on the balance between them is still unclear. We used live cell imaging and fluorescent reporters for 53BP1, Rad52 and cell cycle to quantify the relative contribution of NHEJ and HR at different points of the cell cycle in single cells. We found that NHEJ is the dominant repair pathway in G1 and G2 even when both repair pathways are functional. The shift from NHEJ to HR is gradual, with the highest proportion of breaks repaired by HR in mid-S, where amount of DNA replication is highest. Higher proportions of HR also strongly correlate with slower rates of repair. Our study shows that the choice of repair mechanism is continuously adjusted throughout the cell cycle and suggests that the extent of active replication, rather than the presence of a sister chromatid influences the balance between the two repair pathways in human cells.
Introduction
DNA double strand breaks (DSBs) are potentially cytotoxic lesions generated during normal cell metabolism or by ionizing radiation and chemotherapeutic drugs. Their repair is critical for the successful maintenance and propagation of genetic information. In mammalian cells, two distinct pathways promote repair of DSBs, non-homologous end joining (NHEJ) and homologous recombination (HR). In NHEJ, the broken DNA ends are aligned and ligated together without requiring long sequence complementarities (Lieber et al., 2003). HR requires an intact homologous sequence located on a sister chromatid, or elsewhere in the genome. It is initiated by resection of DNA at the break site to generate 3′ single-stranded DNA overhangs that invade the DNA double helix of the undamaged, homologous partner and copy information back to the break site (West, 2003). Both NHEJ and HR are essential for genome maintenance, and defects in either pathway are linked to immunodeficiency, cancer predisposition and other diseases.
A critical question is how the choice of DNA repair pathway is regulated. The current view is that cell cycle is the key regulatory factor that guides the decision between pathways (Shrivastav et al., 2008). NHEJ functions throughout the cell cycle, but is assumed to be most important in G0/G1 (Lieber et al., 2003). HR was suggested to be active only in the post-replicative stages of the cell cycle, S and G2, during which time the preferred homologous template – the sister chromatid - is available (Aylon et al., 2004; Johnson and Jasin, 2000; Kadyk and Hartwell, 1992; West, 2003). This cell cycle dependent competency for HR was suggested to be regulated by the activity of cyclin dependent kinases (CDKs), which control DSB resection, a prerequisite for HR (Aylon et al., 2004; Huertas et al., 2008; Huertas and Jackson, 2009; Ira et al., 2004; Yun and Hiom, 2009). Interestingly, the physical presence of replicated DNA did not affect the choice of repair mechanism in yeast (Aylon et al., 2004). It is unclear whether this is also the case in mammalian cells. In addition, it is unclear how individual mammalian cells transition between the two mechanisms with cell cycle progression (Figure 1A), whether all breaks in a cell are repaired exclusively by one mechanism, and is the choice of repair mechanism fixed at the time of damage or changed during the course of repair. Most of our knowledge about the relationship between cell cycle and the choice of repair pathway comes from measurements of fixed cells at specific times post damage. Such measurements allow estimation of cell cycle phase based on one fixed snapshot of a cell, followed by grouping of cells into three major phases G1, S and G2. However, each group includes cells that enter that phase at different times, potentially leading to large heterogeneity within each group. Direct connection between cell cycle phase and the choice of repair mechanism therefore requires quantification of these events over time in the same cell.
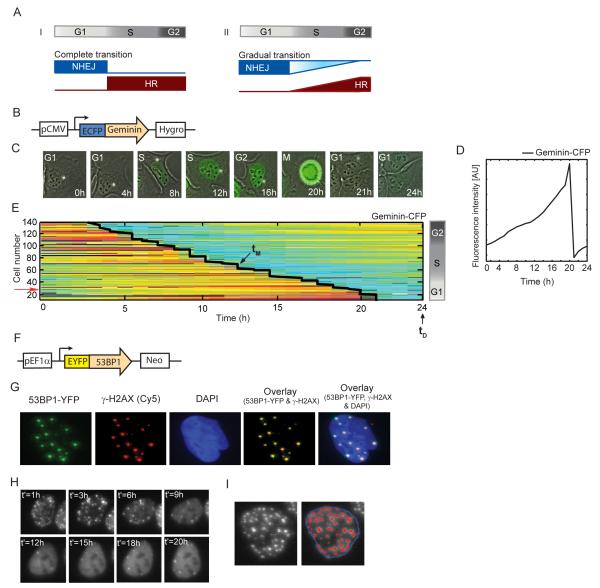
Figure 1. Experimental system for quantifying DSBs and cell cycle phase in single, living cells.
(A) Potential models of the transition between NHEJ and HR with cell cycle progression. The transition can be switch like (i) where cells completely shift to HR on entering S phase; or a more gradual change (ii) where cells utilize more HR with greater progress in S and G2 as the amount of replicated homologous substrate accrues. (B) Schematic drawing of the Geminin reporter. (C) Time lapse images of a freely cycling U2OS cell expressing the Geminin-CFP reporter. Images are overlays of the phase and CFP channels. (D) Quantification of the average nuclear Geminin-CFP intensity in a freely cycling cell over one cell division (indicated by the sharp drop in intensity). (E) Heat map of Geminin-CFP intensities in individual cells over time. Each horizontal line represents a single cell; blue represents low Geminin intensity; red represents high intensity. Cells were clustered according to their time of mitosis (tM, diagonal black line). Damage was applied at the 24h time point (tD). Cells at the top were in G2 when damage was applied, while cells in G1 are at the bottom. The red arrow indicates the trajectory of the cell shown in (C). (F) Schematic drawing of the 53BP1 reporter. (G) Cells expressing 53BP1-YFP were fixed and stained with anti γ-H2AX antibody after damage. The overlaid image shows co-localization between 53BP1 and γ-H2AX foci (see additional examples and quantification in Figure S1C-E). (H) Time-lapse images of a cell expressing 53BP1-YFP after damage. t’ is the time elapsed from the initiation of DNA damage. Images are maximum projections of z-stacks through the nucleus (see Experimental Procedures) in the YFP channel. (I) Example of the automated segmentation for the enumeration of 53BP1-YFP foci in a cell. Image processing was performed using the Ensemble Thresher software package developed in our lab (see experimental procedures for algorithmic details and additional examples in Figure S4). See also Figure S1.
Here, we use long-term, time-lapse microscopy and fluorescent reporters to measure DSBs, HR and cell cycle phase in asynchronous, individual living cells, and accurately determine the relationship between cell cycle state, choice of repair mechanism and kinetics of repair. Our results show that the choice of repair pathway is not fixed at the time of damage but rather is adjusted during the course of repair. NHEJ is the exclusive repair pathway in G1; however cells damaged in late G1 show low levels of HR as they progress into S. Once HR is activated, it does not capture all breaks, and the balance between NHEJ and HR changes gradually with cell cycle progression. Specifically, S and G2 cells exhibit both HR and NHEJ with maximal use of HR in mid-S phase; during which time repair is the slowest and the amount of DNA replication is highest. Our data argue against the idea that the presence of replicated DNA determines the choice of repair, and instead suggest a direct link between the extent of active replication and HR.
Results
1. Quantifying DSB repair and cell cycle in individual living cells
We developed a fluorescent reporter system that allows quantification of cell cycle phase and DNA DSBs in individual living cells. To monitor cell cycle phase we expressed a CFP fusion of the N-terminal domain of Geminin (Figure 1B); which was previously shown to faithfully report APC inactivation (Sakaue-Sawano et al., 2008). Geminin-CFP slowly accumulates in cells post division; reaches maximal levels as cells enter mitosis, and then rapidly degrades during cytokinesis (Figure 1C, D). We imaged freely cycling cells for 24 hours prior to inducing DNA damage. During this time most cells divided at least once and their time of division was identified computationally based on the rapid drop in Geminin-CFP levels. Based on each cell’s division time, we determined its cell cycle phase at the time of damage and the time that had passed since it entered that phase(see experimental procedures, Figure 1E, Figure S1A, B).
We quantify DNA DSBs in single cells using 53BP1 fused to yellow fluorescent protein (YFP; Figure 1F). 53BP1 is a mediator protein in the DNA damage response. It localizes to DSB sites within minutes after damage and forms sub-nuclear compartments (foci) on chromatin regions adjacent to the break (Anderson et al., 2001; Bekker-Jensen et al., 2005; Schultz et al., 2000). As previously reported, we found that the tagged 53BP1 protein forms distinct foci that co-localize with the canonical marker for DSBs, γ-H2AX (Figures 1G, S1C-E, Lobrich et al., 2010). Foci formed by DNA damage response proteins such as 53BP1 and γ-H2AX provide an indirect measurement of DSBs in cells. For example, γ-H2AX and 53BP1 foci may be affected by factors that alter the signal persistence rather than the actual breaks. In addition, multiple breaks may cluster into a single focus. Despite these limitations, measurements of foci provide a sensitive, high-resolution quantification of breaks in individual cells and have been shown to faithfully reproduce results obtained from populations of fixed cells by more direct measurements of DNA breaks such as pulsed-field gel electrophoresis (Rothkamm and Lobrich, 2003). Our analysis showed that the number of 53BP1-YFP foci in a cell decreases with time (Figure 1H, S1C). Note that the number of 53BP1 foci we measure represents a balance between repair and generation of new breaks by internal cellular events such as replication. We were able to distinguish the large, high intensity 53BP1 foci, which were recently shown to form at sites of mitotic chromosomal lesions in non-damaged G1 cells (Lukas et al., 2011), from the numerous, smaller 53BP1 foci generated after damage induction by imaging the same cell before and after inducing damage (Fig. S1F). We also confirmed that the decay in the number of foci post damage represents repair (and not decay of the fluorescent signal due to photo bleaching), by showing that the distribution of foci at 24 hours post damage is similar between cells that were imaged frequently (every hour) and cells that were imaged only at 24 hours post damage (Figure S1G, p-value 0.99, Kolmogorov-Smirnov test). We generated a double reporter cell line expressing both Geminin-CFP and 53BP1-YFP and confirmed that these reporters do not alter cell cycle distribution of asynchronously growing cells (Figure S1H).
To induce DSBs, we used the radiomimetic drug neocarzinostatin (NCS). NCS creates a burst of DSBs and has been shown to act solely within five minutes following addition of the drug to the cell culture medium (Shiloh et al., 1983). We chose to use NCS instead of irradiating cells because this allowed us to add the drug directly to cells on the microscope and follow the same cells before and after damage without disturbing the imaging setup and without a significant time delay in image acquisition. We verified that the kinetics of DSB repair post NCS treatment are similar to those obtained in irradiated cells (compare Figure S2A to Figure 2B). To minimize photo bleaching, we limited the imaging of cells to hourly intervals post NCS treatment. The mobility of foci and cells does not permit tracking individual foci between time points. Our measurements therefore provide the total numbers and intensity of foci in each cell over time.
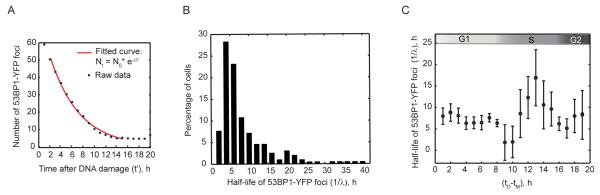
Figure 2. Cell cycle position at the time of damage affects kinetics of DSB repair.
(A) Enumerated 53BP1-YFP foci (black dots) and the exponential fit to the raw data (red line) for the cell shown in Figure 1H). (B) The distribution of half-lives of 53BP1-YFP foci in an unsynchronized population (n>220 cells) and as a function of cell cycle progression (C). In (C), the average half-life of 53BP1-YFP foci is plotted for cells binned according to their cell cycle position at the time of damage. The plot was calculated with a sliding window of bin size W = 2 hours. Bars represent mean + SEM for a total population of > 220 cells. See also Figure S2.
2. Cell cycle phase at the time of damage affects the rate of DSB repair
We first asked if cells damaged in different stages of the cell cycle repair DSBs uniformly or if they vary in their kinetics of repair. Previous studies in fixed cells have suggested that DSBs are repaired with bi-phasic kinetics comprising a fast repair process (half-life of 0.5h - 2h) and a slower repair process (half-life of 12h - 24h). Since our measurements were collected at hourly intervals, the sampling of the early, rapid decay was limited to two data points. This resolution was insufficient to faithfully characterize and separate the fast repair kinetics from the overall repair trajectory, and we were able to observe clear, biphasic repair only in 12% of the cells. We therefore focused on the slow phase of repair and fit the enumerated 53BP1 foci from 2 hours post damage to an exponential decay until the net decay approached zero (Figure 2A). Our analysis revealed a large variation in the half-life of 53BP1 foci across cells with a peak around 4-5 hours (Figure 2B). To test whether these variations relate to cell cycle phase, we binned cells according to their position in cell cycle at the time of damage and plotted the half-life of 53BP1 foci for cells in each bin (Figure 2C). Our results indicate a significant association (p<0.01, one-way ANOVA) between cell cycle phase and the rates of DSB repair. We observed that cells damaged in G1 and G2 have comparable half-lives of DSBs. In contrast, cells damaged in S phase show a large variation in their kinetics of repair. Cells at the G1/S transition attain the shortest half-lives of DSBs amongst all cells. Half-lives then gradually increase as cells enter S phase and peak in mid-S; followed by an acceleration of repair as presented by shorter half-lives in late-S and G2.
The rapid kinetics of repair at the G1/S boundary may result from changes in chromatin structure in preparation for DNA replication, promoting accessibility of repair factors to break sites. Additionally, there may be increased availability of key substrates such as dNTPs, and DNA processing enzymes that allow rapid DNA repair during this time. The accelerated kinetics of repair towards the end of the cell cycle may be important to address DNA damage in a timely manner as cells progress towards mitosis.
To ensure that the observed effect of cell cycle on repair kinetics is not merely due to differences in the initial number of DSBs, we plotted the rate of repair as a function of the initial number of breaks. We found no correlation between the initial number of breaks and the rate of repair (Figure S2B). At present, we cannot rule out the possibility that clustering of multiple repair sites into a single focus leads to the appearance of longer repair times.
Our results show that the dynamics of DSB repair differ significantly between cells in different cell cycle phases and even more strikingly, repair rates are not uniform for S phase cells, but rather are determined by the exact time a cell has spent in S phase at the time of damage.
3. NHEJ is the dominant repair mechanism in both G1 and G2 cells
Several factors, including the degree of chromatin compaction, level of CDK activity, and availability of nucleotide substrates could affect the rates of DSB repair through the cell cycle. Another potential factor is the balance between NHEJ and HR. It was recently suggested that NHEJ repairs breaks with faster kinetics than HR (Shibata et al., 2011). To quantify the extent of activation of the alternative repair pathways and the cell cycle dependent balance between them, we added a fluorescent reporter of Rad52 (Rad52-mCherry) to cells expressing 53BP1-YFP and Geminin-CFP (Figure 3A). Rad52 is a recombinase mediator protein that forms foci at DSB sites that are committed to homology dependent repair (Essers et al., 2002). Evidence in mammalian cells suggests that Rad52 functions primarily in single strand annealing (Stark et al., 2004). An in vitro study of the human Rad52 protein indicates that it functions to catalyze the capture of the second DSB end prior to D-loop dissociation in canonical HR reactions (McIlwraith and West, 2008). More recently, it was demonstrated that Rad52 is synthetically lethal with the recombinase mediator BRCA2, and depletion of Rad52 in BRCA2 deficient cells impairs Rad51 foci formation, (Feng et al., 2011), suggesting that Rad52 functions in an independent, alternate pathway that supports Rad51 mediated classical HR repair.
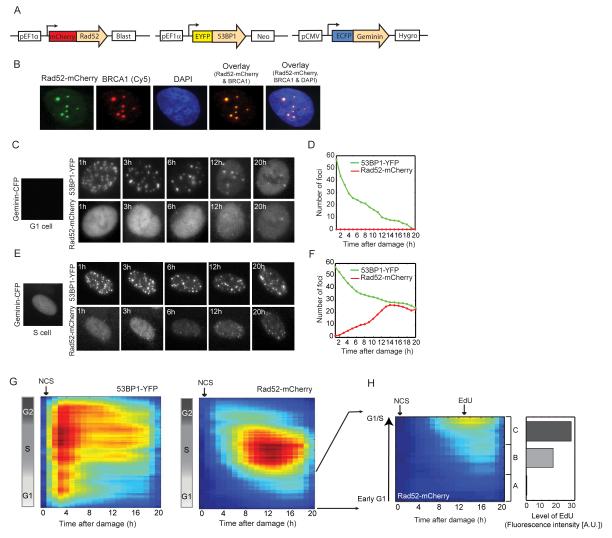
Figure 3. NHEJ dominates the repair of DSBs in G1 and G2 cells.
(A) Schematic drawing of the Rad52, 53BP1 and Geminin reporters for quantifying HR, total DSBs and cell cycle phase respectively in individual cells. (B) U2OS cells expressing the Rad52-mCherry reporter were fixed and stained with anti-BRCA1 antibody after damage. The overlaid image shows co-localization of the Rad52-mCherry and BRCA1 foci (see additional examples and quantification in Figure S3A-C). (C, E) Time-lapse images of U2OS cells expressing the reporters in (A) that were damaged in the G1 (C) or S (E) cell cycle phases. The 53BP1-YFP and Rad52-mCherry images are maximum projections of z-stacks through the nucleus (see Experimental Procedures). (D, F) Quantification of the number of 53BP1-YFP (green) and Rad52-mCherry (red) foci in the cells shown in (C) and (E) respectively. (G) Heat maps of 53BP1-YFP and Rad52-mCherry foci as a function of time after damage (x-axis) and cell cycle progression (y-axis). Cells were binned to 20% full interval on both axes. Blue represents low foci numbers and red represents high foci numbers in a range of 0-120 (53BP1-YFP) or 0-100 (Rad52-mCherry) foci. Number of cells >140 (H) Heat map of Rad52-mCherry foci zoomed in on cells damaged in G1. The gray bars on the right indicate the average EdU content (total nuclear intensity) for cells binned into three groups A, B and C (from early to late G1) based on their cell cycle stage at the time of damage. Number of cells >40. See also Figure S3.
Homology dependent repair comprises many sub-pathways, which precludes the development of a single protein reporter that can capture all homology dependent events. We chose Rad52 to report on HR since it is not a key factor for mammalian HR (Rijkers et al., 1998; van Veelen et al., 2005) and we were concerned that high expression of core HR proteins (such as Rad51 and BRCA1) may disturb the natural balance between NHEJ and HR. However, one limitation of using Rad52 as an HR marker is that it may not accompany all HR reactions in mammalian cells. To evaluate if our Rad52 reporter captures all homology dependent repair events or a specific subset of HR reactions, we performed immunofluorescence based comparisons of Rad52-mCherry foci with BRCA1, a protein that functions in DNA resection, an early, essential step for all homology dependent events. (Figure 3B, Figure S3A). We found that 93.1% of Rad52-mCherry foci in a cell had a corresponding, co-localized BRCA1 focus (Figure S3B). This may result from BRCA1 leaving the break before Rad52 is loaded. Interestingly, only 72% of the BRCA1 foci in a cell had a corresponding, co-localized Rad52-mCherry focus, indicating that 28% of breaks accompanied by BRCA1 are repaired by Rad52 independent HR mechanisms. Thus the Rad52 reporter underrepresents the total number of foci repaired by homology dependent repair in our experimental system. However, this underrepresentation was found to be systematic and did not depend on cell cycle phase (Figure S3C). This demonstrates that the Rad52 reporter is reliable for studying the effect of cell cycle on all HR reactions that utilize BRCA1 mediated DNA resection. Lastly, since fluorescent reporters may provide a risk of altering the natural balance of proteins and cellular responses, we confirmed that insertion of the triple reporters does not alter the kinetics of repair (Figure S3D).
While all cells treated with NCS showed 53BP1 foci, formation of Rad52 foci was highly dependent on cell cycle phase, in agreement with previous studies in fixed cells (Bekker-Jensen et al., 2006; Jazayeri et al., 2006; Lisby et al., 2004; Tashiro et al., 2000). S phase cells show higher numbers of Rad52 foci, while early/mid G1 cells show no Rad52 foci (Figure 3C-F). Interestingly, late G1 cells show low numbers of Rad52 foci at 12-16 hours post damage (Figure 3G). To determine if this activation of HR results from G1 cells progressing into S phase during the course of the repair, we exposed damaged G1 cells to a 40 minute pulse of the thymine analogue EdU at the time of HR activation (13 hours post damage) and analyzed their EdU content. We observed that early G1 cells that did not develop Rad52 foci also did not incorporate EdU (Figure 3H). Cells damaged in late G1 and at the G1/S transition incorporated higher amounts of EdU and developed Rad52 foci, indicating that they had progressed into S phase when HR was activated. This shows that the decision to activate HR in G1 cells is not fixed at the time of damage; cells in early G1 exclusively activate NHEJ for repair, however mid/late G1 cells first repair exclusively by NHEJ, but then progress into S phase and activate HR. Since NCS also leads to a small proportion of single strand breaks (SSBs, Shiloh et al., 1983), it is possible that as cells undergo the G1/S transition, SSBs may cause replication forks to collapse and generate one-ended DSBs that initiate HR repair.
Based on current literature, the precise role of HR in G2 is somewhat controversial. On one hand, studies in HR deficient cells suggested HR as the dominant repair pathway in both S and G2 (Rothkamm et al., 2003; Takata et al., 1998). On the other hand, a more recent study shows that HR deficient cells can repair up to 85% of DSBs by NHEJ in G2 (Beucher et al., 2009). It is unclear if this reflects the behavior in wild-type cells or results from the ability of NHEJ to compensate for the absence of functional HR. Our system allowed us to determine the contribution of HR and NHEJ when both mechanisms are intact in wild-type cells. We found that cells damaged in G2 show low levels of HR, comparable to the levels seen in late G1 cells (Figure 3G). However this activation was much earlier (4h post damage in G2 cells compared to 12h in late G1 cells). Late G2 cells show very little or no induction of HR (Figure 3G). Importantly, G2 cells that divided after damage were eliminated from this analysis to ensure that the predominant NHEJ observed in this phase did not result from G2 cells progressing into G1 phase during the course of repair. Thus NHEJ is the dominant repair mechanism in both G1 and G2 cells even in the presence of a functional HR pathway.
4. The balance between HR and NHEJ changes gradually with highest activation of HR in mid-S
What fraction of DSBs is repaired by HR in S and G2 cells? To answer this we divided the number of Rad52 foci by the number of 53BP1 foci induced post damage (Figure 4A). We observed that the proportion of HR gradually increases as cells progress from early S towards mid-S phase, followed by a decrease as cells progress to late-S and G2 (Figure 4A). The same pattern was observed when the maximum proportion of HR in individual cells (calculated as the ratio between the maximum number of Rad52 foci to the total number of 53BP1 foci induced post damage), and the rates of Rad52 foci accumulation were plotted against cell cycle position (Figure 4B, C). Our data demonstrate that cells do not show an immediate and complete activation of HR on entering S phase. Instead, the level of HR increases gradually as cells progress from early to mid-S phase, followed by a gradual decrease when they progress to late-S and G2. We confirmed that the relative position of cells in S (early, mid, late) at the time of damage had not changed when they showed maximal activation of HR (11h post damage; Figure S3E).
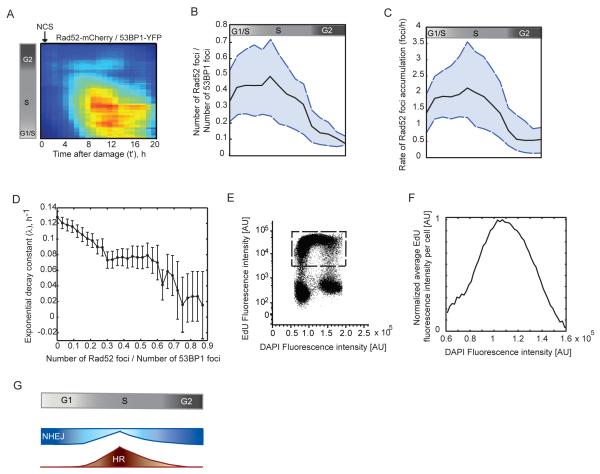
Figure 4. Contribution of HR to DSB repair changes gradually with cell cycle progression and is highest in mid-S.
(A) Heat map showing the proportion of DSBs channeled to the HR repair pathway over time post damage, calculated as the ratio between Rad52-mCherry foci to 53BP1-YFP foci. The ratio is shown as a function of the time elapsed from the induction of DSBs and cell cycle progression (indicated by the reference bar on the left). Cells were binned to 20% full interval on both axes. Blue represents low ratios and red indicates a higher proportion of HR. (B) The maximum proportion of HR in individual cells post damage is plotted against their cell cycle progression at the time of damage indicated by the reference bar on top. The median (black line), 25th and 75th percentile (dashed blue lines) of the population (n> 220 cells) are shown. (C) The rate at which Rad52-mCherry foci accumulate in individual cells post damage is plotted against their cell cycle progression at the time of damage. The median (black line), 25th and 75th percentile (dashed blue lines) of the population (n> 220 cells) are shown. (D) The rate of repair as a function of HR usage is plotted for cells binned according to their maximum Rad52-mcherry/53BP1-YFP foci ratio. Cells are binned according to a bin size of 0.03. Bars represent mean + SEM. Population of n>220 cells. (E, F) The amount of DNA replication as a function of S phase progression was measured by pulse labeling cells with EdU. Levels of EdU fluorescence are shown as a function of the DAPI fluorescence (E) for a non-synchronized population of cells. The level of DNA replication is quantified as the average EdU intensity per cell. To avoid bias from non-replicating cells, (F) was calculated from cells in the window shown in (E). (G) A new model for the transition between NHEJ and HR with cell cycle progression. Cells in G1 repair DSBs exclusively by NHEJ. Cells then increase their use of HR gradually as they progress from G1 to early S. Following a peak in mid-S, HR decreases gradually as cells move towards late S and G2, with late G2 cells repairing DSBs almost entirely by NHEJ.
The observation that mid-S phase cells attain the greatest proportions of HR (Figure 4A, B) and exhibit the longest half-lives of DSBs (Figure 2C) suggests a potential correlation between the choice of repair pathway and the kinetics of repair. To investigate this further, we plotted the average decay constant obtained from exponential fits to the enumerated 53BP1-YFP foci for cells binned according to their proportions of HR post damage (Figure 4D). Indeed, a strong correlation was observed; the rates of repair declined with increasing contribution of HR to total DSB repair, supporting the idea that HR mediated DSB joining proceeds at a slower rate than NHEJ mediated repair. We also calculated the half-lives of Rad52 foci in cells damaged in S and G2, and found no significant difference (Figure S3F, p-value 0.2379, t-test), suggesting that the 5 – 10 hour variation in the half-life of DSBs observed between S and G2 results mainly from the choice of repair mechanism and not from differences in the kinetics of HR. Interestingly, there were several regimes in which an increase in HR did not produce a proportional change in repair rates (Figure 4D), supporting that, in addition to the extent of HR, other factors such as chromatin compaction and cyclin-CDK activity affect the kinetics of DSB repair.
5. Increased levels of HR in mid-S correlate with high levels of DNA replication
When damaged cells replicate their DNA, SSBs and blocking lesions on the DNA may cause active replication forks to stall and collapse, resulting in one-ended DSBs. One-ended DSBs breaks are repaired exclusively by HR (Helleday et al., 2007), as NHEJ requires two free DNA duplex ends for repair. The proportion of HR in S phase therefore, might depend on the level of active DNA replication. DNA does not replicate uniformly throughout S phase, rather different regions of the genome are replicated at distinct rates and times during S phase. In yeast, the highest replication origin firings occur near mid-S (Raghuraman et al., 2001). Even though replication timing is less well characterized in human cells, studies indicate that replication rates are highest around mid-S (Woodfine et al., 2004). To measure the level of DNA replication, we exposed an asynchronously growing population to a 20 minute pulse of EdU and quantified its content by flow cytometry. Within such a short time interval, the number of cells entering or exiting S phase is negligible and EdU fluorescence per cell is indicative mainly of level of EdU incorporation (i.e. amount of DNA replication). We found that cells in mid-S amassed the highest quantities of EdU (Figure 4E, F). This indicates that DNA replication was greatest in mid-S, correlating with the high proportions of HR observed during this time. This suggests that in mammalian cells, HR is most important for repair of DSBs created during active DNA replication.
Discussion
With recent advances in imaging techniques and single cell analyses, it has become clear that variability in internal states leads to remarkable heterogeneity in the behavior of isogenic cells exposed to a uniform stimulus (Snijder and Pelkmans, 2011). Studying how basal states affect individual cellular behavior is crucial for our ability to understand and predict cellular responses and for developing efficient drugs. In this study, we used fluorescent reporters for 53BP1, Geminin and Rad52 to assess how variations in cell cycle state impact the kinetics of DSB repair and the balance between alternate repair pathways in individual cells. Although an analysis of the rates of repair has been previously undertaken in populations of fixed cells (Shibata et al., 2011), our live cell system allowed us to develop a more comprehensive picture of how the kinetics of repair vary throughout the cell cycle and within each phase. We show that not only do the rates of repair differ between cells damaged in different cell cycle phases, but individual cells damaged in the same phase also vary significantly in the kinetics of repair. This heterogeneity is most pronounced for S phase cells; cells damaged closer to the G1/S transition attain the highest rates of repair amongst all cells, following which the rates decrease as cells progress to mid-S and then increase towards late S and G2. Based on these findings, we argue that the rates of DSB repair are fine tuned according to the exact time each cell has spent in a phase.
Next, we show that the rate of repair strongly correlates with the contribution of HR to DSB repair, which also varies continuously with cell cycle progression. HR is absent in cells damaged in early G1, following which it increases gradually and peaks at mid-S then declines towards late S and G2 (Fig. 4G). This shows that cells do not initiate a maximal use of HR immediately on entering S phase, nor do they show maximal HR at the end of S and in G2 when replication is complete. Our data therefore do not favor the idea that the presence of sister chromatids control the level of HR in the post replicative phases in mammalian cells.
Our analysis also provided a measure of the variation between cells. We found that the proportion and rates of active HR vary widely even between individual cells damaged at the exact cell cycle position. For example, the proportion of breaks repaired by HR in cells at the G1/S transition (~ 8 h post division) vary from approximately 20% to 65% (Figure 4B). This suggests that the choice between NHEJ and HR is influenced by additional factors such as the nature of the break (one-ended or two-ended); chromatin complexity; or dose of DNA damage (Beucher et al., 2009; Shibata et al., 2011).
One explanation for the high proportions of HR observed in mid-S is that NHEJ is unable to compete with HR for break sites during this phase. It is known that HR plays an exclusive role in the repair of one-ended DSBs that arise when replication forks stall at nicks in the DNA template and collapse in S phase (Helleday et al., 2007). When cells are damaged during peak DNA replication, nicks created on the DNA template may cause collapse of several active forks and create substrate that can only be addressed by HR mediated repair. We observe that in our cells; the highest amount of DNA replication occurs in mid-S, correlating well with increased proportions of HR observed during this time. It is further tempting to speculate that a close association between replication machinery and HR factors leads to the gradual transition between NHEJ and HR as cells enter or exit S phase. The interaction of CtIP, a protein essential for DNA resection in the HR pathway, with PCNA, provides some evidence for this hypothesis (Gu and Chen, 2009). G2 cells demonstrate a reduced efficiency of DNA resection compared to S phase cells that may further allow an increased channeling of DSBs into NHEJ as cells progress into G2 (Zierhut and Diffley, 2008). Additionally, while both cyclins A and B can activate the resection machinery, differences in their individual efficiencies could lead to a decline in HR with a gradual decrease in cyclin A activity as cells progress towards mitosis. Future studies employing conditional replication defective cell lines and perturbations that uncouple DNA replication from cyclin activity will help consolidate the relationship between active DNA replication, cyclin levels and HR repair.
In this study we used U2OS cells, which lack a stable G1/S checkpoint. This allows cells damaged in G1 to progress into S phase and activate HR during the course of repair (Figure 3H). As in the case of U2OS, many cancers arise due to mutations in key regulators of the G1/S checkpoint and hence understanding how cells with disabled checkpoints repair DNA damage in response to chemotherapy is of clinical relevance. In addition, recent work has uncovered substantial limitations to the G1/S checkpoint even in damaged normal cells (Deckbar et al., 2010). First, the G1/S checkpoint is not fully initiated until several hours post damage, during which time many G1 cells enter S phase with unrepaired breaks. Second, at high doses of damage most cells undergo a permanent G1/S arrest, but a small fraction of cells escape arrest and enter S phase with DSBs. It would be important to determine the choice and kinetics of repair in additional cancer and primary cells.
The quantitative analysis presented here was made possible due to the use of fluorescent reporters in live cells. Previous studies that have used similar approaches provided important insights about the complex mechanisms that function to preserve genomic integrity in response to DNA damage. For example, fluorescent tagged 53BP1, Mdc1 and NBS1 provided a detailed understanding of the spatiotemporal sequence of events initiated on DSB generation and the causal relationship between them (Bekker-Jensen et al., 2005; Lukas et al., 2004). In addition, using fluorescent H2B and probes that bind to an ISce1 induced break led to the discovery that free DNA ends at DSB sites have limited local movement and that damaged chromatin does not undergo large-scale movements (Kruhlak et al., 2006; Soutoglou et al., 2007). Therefore, fluorescent reporters that bind DSBs present a powerful tool for dissecting signaling kinetics and cellular decisions in individual cells.
Ultimately, measurements using DNA damage and repair reporters in live cells will enable us to address additional long-standing, fundamental questions in this field. For example, reporters for NHEJ and HR in altered cellular states can teach us about their ability to compensate for each other under selective drug action. Reporters for the different sub-pathways of homology dependent repair can provide insights into their interplay and balance at different stages of repair. Such reporters can also help determine the timing at which commitment to a specific repair pathway occurs and the factors leading to these decisions. Live cell reporters also enable the observation of time separated events in the same cells such as repair, cell cycle checkpoints and activation of tumor suppressor proteins. Such analyses in cellular backgrounds where key DNA response proteins are mutated have the promise of providing a comprehensive understanding of how specific mutations or polymorphisms lead to carcinogenesis.
Experimental procedures
Cell culture
U2OS cells were grown in McCoy’s 5A medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, 100 mg/mL streptomycin and 250 ng/mL fungizone (Gemini Bio-Products). When required, the medium was supplemented with selective antibiotics (400 μg/mL G418, 5 μg/mL blasticidin, 50 μg/mL hygromycin). When indicated, medium was replaced with fresh medium supplemented with 200 ng/mL Neocarzinostatin (National Cancer Institute) during experiments. Cell cycle distributions were analyzed by flow cytometry using propidium iodide or DAPI staining as indicated. DNA replication was measured by incorporation of EdU using the Click-iT EdU kit (Invitrogen).
Cell Line Construction
The original pCMV-EGFP-53BP1 construct was kindly provided by Prof. Yasuhisa Adachi (Jullien et al., 2002). We generated our pEF1α-EYFP-53BP1 plasmid by replacing GFP with YFP and combining this fluorescent protein-cDNA fragment with the EF1α promoter in a vector harboring a neomycin resistance cassette using standard molecular biology techniques. This plasmid was stably transfected into U2OS cells using FuGENE6 (Roche), which were maintained in selective media and sorted into single cells using fluorescence activated cell sorting to generate a clonal population.
Our pEF1α-mCherry-Rad52 plasmid was generated similarly from the original pCMV-EGFP-Rad52 construct kindly provided by Prof. Roland Kanaar (Essers et al., 2002). The GFP tag was replaced with mCherry and the fluorescent protein-cDNA fragment was combined with the EF1α promoter in a vector harboring a blasticidin resistance cassette. Stable, clonal cell lines were established as described above.
The pCMV-ECFP-Geminin construct was generated by PCR amplification of the sequence coding for the 110 amino acid N-terminus of Geminin from genomic DNA isolated from human cells. The PCR product was combined with the CMV promoter and CFP tag in a lentiviral vector harboring a hygromycin resistance cassette by Multisite Gateway cloning (Invitrogen). This plasmid was transfected into 293T cells to generate replication-defective viral particles using standard protocols, which were used to stably infect our engineered U2OS cell line.
In silico mapping of cell cycle progression in individual cells
We first established the average cell cycle duration of our cell line by imaging undamaged, freely cycling cells for 48h. During this time most cells underwent at least two successive divisions, which were identified from the Geminin-CFP reporter. The time between successive divisions was measured and averaged for ~100 cells to establish the average cell cycle. For in silico mapping of cell cycle progression of individual cells at the time of damage, we imaged cells for 24h prior to addition of NCS (Figure S1B). This allowed most cells to divide at least once prior to damage; and we isolated trajectories where the time of division could be clearly determined. To relate elapsed time after division to cell cycle phase and progression, we measured the distribution of DNA content in an asynchronously growing culture by flow cytometry using propidium iodide staining. These distributions were fit using a modification of the Dean-Jett model (Dean and Jett, 1974) to determine the amount of cells in G1, S and G2 phases; and were subsequently translated to the time spent in various cell cycle phases using a previously published model (Toettcher et al., 2009). These durations were then mapped to the time since last cell division before damage was applied for individual cells to establish their cell cycle progression at the time of damage.
Time-Lapse Microscopy
24h prior to microscopy, cells were plated in RMPI lacking riboflavin and phenol red in poly-D-lysine coated glass-bottom plates (MatTek Corporation). The medium was supplemented with 10% fetal calf serum, 100 U/mL penicillin, 100 mg/mL streptomycin, 250 ng/mL fungizone (Gemini Bio-Products) and 10 mM HEPES. Cells were imaged on a Nikon Eclipse Ti inverted microscope with a 40X plan apo objective (NA 0.95), Hamamatsu Orca ER camera and a Perfect Focus System. The microscope was surrounded by a custom enclosure to maintain constant temperature and atmosphere. The filter sets used were CFP - 436/20 nm; 455 nm; 480/40 nm (excitation; beam splitter; emission filter), YFP - 500/20 nm; 515 nm; 535/30 nm; and mCherry - 560/40 nm; 585 nm; 630/75 nm (Chroma). Images were acquired every 20 mins in the phase and CFP channels and every 60 mins in the YFP and mCherry channels. We acquired 6 z-sections with a step size of 0.75 μm in the YFP and mCherry channels. Image acquisition was controlled by MetaMorph software (Molecular Devices).
Image Analysis
Image analysis was done by Matlab (MathWorks) based custom written software. Cell boundaries were calculated by two complementary approaches. (i) Cells were separated from background by thresholding a Top-Hat transform of the original image. Top-Hat transformation was used to remove trends that are spatially wider than cell diameters. (ii) Boundaries between adjacent, touching cells were identified by seed based watershedding. Seeds were calculated as the regional maxima of the Gaussian smoothed image. To eliminate bias to regional maxima by bright foci, images were first pre-processed by morphological closing with a structure smaller than cell diameter but larger than foci. Foci were identified by taking advantage of their small size. Images were first transformed with Top-Hat to remove all intensities that are spatially larger that 10% of cell diameter. This transformation resulted in an image with a strongly intensified foci signal. Foci were then segmented by regional thresholding followed by seed-based watershedding (Figure S4). Similar to cell boundaries, seeds were calculated as the regional maxima of the fluorescence intensity. The image analysis algorithm was separately optimized to identify 53BP1-YFP foci and Rad52-mCherry foci, which differed in size and intensity.
Supplementary Material
Highlights.
- The rate of DSB repair depends on the cell cycle stage at time of damage
- Both NHEJ and HR are used during S and G2, and higher use of HR correlates with slower repair
- NHEJ is the dominant repair mechanism in G1 and G2 even in the presence of a functional HR pathway
- The balance between NHEJ and HR changes gradually with the highest use of HR at the peak of active replication
Acknowledgements
We thank Sara Thiebaud and Michal Mekel for early contribution to this work. We thank Roy Kishony, Tim Mitchison, Dipanjan Chowdhury, Rebecca Ward and all members of the Lahav lab for comments and discussions. We thank the Nikon Imaging Center at Harvard Medical School for assistance with microscopy. This research was supported by the National Institute of Health grant GM083303, John and Virginia Kaneb, Hoffman-La Roche and fellowships from the German Research Foundation and the Charles King trust to A.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. Embo J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. Embo J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean PN, Jett JH. Mathematical analysis of DNA distributions derived from flow microfluorometry. J Cell Biol. 1974;60:523–527. doi: 10.1083/jcb.60.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D, Stiff T, Koch B, Reis C, Lobrich M, Jeggo PA. The limitations of the G1-S checkpoint. Cancer Res. 2010;70:4412–4421. doi: 10.1158/0008-5472.CAN-09-3198. [DOI] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. Embo J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Chen PL. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle. 2009;8:1409–1420. doi: 10.4161/cc.8.9.8322. [DOI] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien D, Vagnarelli P, Earnshaw WC, Adachi Y. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J Cell Sci. 2002;115:71–79. doi: 10.1242/jcs.115.1.71. [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. Embo J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. Embo J. 2011 doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, van der Schans GP, Lohman PH, Becker Y. Induction and repair of DNA damage in normal and ataxia-telangiectasia skin fibroblasts treated with neocarzinostatin. Carcinogenesis. 1983;4:917–921. doi: 10.1093/carcin/4.7.917. [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol. 2011;12:119–125. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. Embo J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Loewer A, Ostheimer GJ, Yaffe MB, Tidor B, Lahav G. Distinct mechanisms act in concert to mediate cell cycle arrest. Proc Natl Acad Sci U S A. 2009;106:785–790. doi: 10.1073/pnas.0806196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res. 2005;574:34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. Embo J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.