Abstract
Background. Respiratory syncytial virus (RSV) reinfects individuals repeatedly. The extent to which this is a consequence of RSV antigenic diversity is unclear.
Methods. Six-hundred thirty-five children from rural Kenya were closely monitored for RSV infection from birth through 3 consecutive RSV epidemics. RSV infections were identified by immunofluorescence testing of nasal washing samples collected during acute respiratory illnesses, typed into group A and B, and sequenced in the attachment (G) protein. A positive sample separated from a previous positive by ≥14 days was defined as a reinfection a priori.
Results. Phylogenetic analysis was undertaken for 325 (80%) of 409 identified infections, including 53 (64%) of 83 reinfections. Heterologous group reinfections were observed in 28 episodes, and homologous group reinfections were observed in 25 episodes; 10 involved homologous genotypes, 5 showed no amino acid changes, and 3 were separated by 21–24 days and were potentially persistent infections. The temporal distribution of genotypes among reinfections did not differ from that of single infections.
Conclusions. The vast majority of infection and reinfection pairs differed by group, genotype, or G amino acid sequence (ie, comprised distinct viruses). The extent to which this is a consequence of immune memory of infection history or prevalent diversity remains unclear.
Respiratory syncytial virus (RSV) is recognized as a major cause of severe lower respiratory tract infection in infants, young children, and vulnerable adults [1, 2]. A key unanswered question in RSV molecular and immuno-epidemiology is the mechanism underlying RSV reinfections [3, 4] that are observed to occur repeatedly throughout the lifespan of any individual [5–8]. Rational design and implementation of an RSV vaccine, which has thus far remained elusive, will benefit from a better understanding of the role of viral diversity and variation in reinfections.
It has been previously hypothesized that the antigenic diversity observed in the RSV surface proteins, the attachment glycoprotein (G) and fusion (F) glycoprotein, plays a role in the reinfection process, but direct data to support this are lacking [3, 4, 9]. RSV isolates can be classified into 2 genetically and antigenically distinct groups (A and B) [10, 11]; these differ most extensively in the G glycoprotein, which has 47% amino acid variations between the prototype strains [12], whereas F has 9% [4]. Phylogenetic analysis of the G-gene nucleotide sequences has allowed the identification of a number of genotypes and lineages in the groups [13–17], and this has helped with the current understanding of RSV molecular epidemiology and evolution [9].
Both heterologous and homologous group and genotype reinfections have been observed in previous studies [18–20]. However, there is uncertainty regarding the extent to which these patterns that were observed were influenced by the infection history of the reinfected individuals rather than being a reflection of the prevailing distribution of groups and genotypes in circulation. On one hand, the regular pattern of dominance of group A or B seen in numerous time series data [21] and the infrequent reoccurrence of dominant genotypes [13, 22] suggests the build-up of strain-specific immunity in the population favoring the transmission of heterologous variants. This would support the notion that reinfection of individuals is influenced by exposure history. On the other hand, repeated RSV infection with the same genetic strain has been demonstrated in human experimental challenges, even within a 2-month interval [23]. Thus, it is so far unknown what fraction of RSV natural reinfections are a consequence of antigenic variation in the virus.
Few previous studies have focused on explaining the nature of RSV reinfection at the molecular level (i.e., characterizing the group, genotype, and amino acid sequence of the strains involved) [9, 19, 20, 24, 25]. Furthermore, these had important limitations. First, small numbers by which patterns were determined curtail statistical power. Second, identification of reinfection through hospital-based surveillance is likely to have missed mild infections (the majority) that did not warrant a hospital visit. Third, frequently, the prevalence of the reinfecting group or genotype in the overall population of circulating viruses was not investigated [19].
The design of the present study was to closely monitor a cohort of children from birth for RSV infection until each child experienced 3 consecutive RSV epidemics, with the aim of identifying all infections and reinfections occurring in these children during their infancy and early childhood years [26, 27]. We have previously published a report on the molecular analysis of virus strains from 12 children in this cohort who were reinfected during the first year of life [28]. Here, we present results of the molecular analysis, at group, genotype, and G-gene amino acid sequence levels, of all the reinfections that we observed in the full period of follow-up and the epidemiological context in which these reinfections occurred.
METHODS
Recruitment and Follow-Up of Participants
The cohort and the population that this study involved has been described in detail elsewhere [26–28]. In brief, 635 children were recruited into a birth cohort from the Kilifi District Hospital maternity ward and the hospital's maternal child health clinic. Informed consent to participate in the study was obtained from a parent or a guardian, and both the Kenya National Ethics Review Committee and the Coventry Research Ethics Committee in UK approved the study protocol.
Recruitment was in 2 phases: from January through June 2002 (338 children) and from December 2002 through May 2003 (297 children). Follow-up was conducted through active household visits (weekly during epidemic periods, otherwise every 4 weeks) and passively through health clinic presentations until each child experienced 3 consecutive RSV epidemics during the period 2002–2005. Prospectively, when a child showed an acute respiratory illness (ARI) symptom, a nasal washing sample was taken and tested for the presence of RSV by an indirect immunoflourescence assay test (IFAT). ARI severity was categorized into upper respiratory tract infection, mild pneumonia, severe pneumonia, or very severe pneumonia in accordance with the modified World Health Organization criteria [26]. Infections were considered as separate episodes if a positive result was determined ≥14 days after a previous positive result [26].
Laboratory Methods
Typing of RSV IFAT Positives
Viral RNA was extracted from the samples positive by IFAT with use of the QIAmp viral RNA mini kit (Qiagen), in accordance with the manufacturer's protocol. The RNA was analyzed in a multiplex real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assay that distinguished the RSV-positive samples as either group A or B, using group-specific TaqMan probes [29]. Tests with negative results were repeated to confirm the result.
G-Gene Amplification and Sequencing
Only RNAs with a positive real-time RT-PCR assay result were included in the G-gene amplification and sequencing. Reverse transcription and first round amplification were performed using QIAGEN One-Step RT-PCR kit, with the forward primer AG20 (5′-GGGGCAAATGCAAACATGTCC-3′) and the reverse primer F164 (5′-GTTATGACACTGGTATACCAACC-3′) at 0.4 pmoles/μL. Thermocyling was 50°C for 30 minutes, 95°C for 15 minutes, and then 40 cycles of 94°C for 30 seconds, 54°C for 30 seconds, and 72°C for 1 minute. A final extension of 10 minutes at 72°C was allowed. Two microliters of the One-Step RT-PCR products were amplified under a nest PCR protocol with Qiagen TaqMan PCR kit mastermix and the primers BG10 (5′-GCAATGATAATCTCAACCTC-3′) and F1 (5′-CAACTCCATTGTTATTTGCC-3′) at the same concentration as the one-step reaction. Thermocycling was: 95°C for 2 minutes, followed by 30 cycles of 95°C for 45 seconds, 54°C for 45 seconds, and 72°C for 1 minute, followed by a final extension at 72°C for 5 minutes. Products from positive reactions (confirmed on a 2% agarose gel) were purified using the Illustra GFX DNA purification kit and sequenced using the BigDye terminator chemistry on the 3130xl ABI instrument. In addition to the 2 nest primers, group-specific primers were included to allow full sequencing coverage of the entire product length both in the forward and the reverse directions. For group A, these were G523F primer (5′-ATATGCAGCAACAATCCAAC-3′) and G523R primer (5′-GTTGGATTGTTGCTGCATAT-3′), and for group B, they were G533F primer (5′-TGTAGTATATGTGGCAACAA-3′) and G533R primer (5′-TTGTTGCCACATATACTACA-3′). Sequence contigs were assembled in Sequencher software, version 4.10.0.
Sequence Analysis
Sequences were aligned manually in the Se-al software, version 2.0 (http://tree.bio.ed.ac.uk/software/seal/), groups A and B separately, and trimmed to be of equal length. Identical sequences were identified in DAMBE [30]. Phylogenetic analyses were performed in MEGA-5 [31] using maximum likelihood methods. The appropriate model of evolution was determined in jmodeltest program [32]. Sequences representing previously identified RSV genotypes were downloaded from GenBank and included in the alignments and in the generation of phylogenetic trees; for RSV A, strains representing GA1-7 and SAA1 [13, 16, 33] and, for RSV B, strains representing GB1-4, SAB1-3, and BA [13, 16, 17, 33] were included. The robustness of the tree branching patterns was tested by bootstrapping with 1000 iterations. The Kilifi sequences that clustered with known genotypes on a major branch with a bootstrap support value of ≥70 were considered to be of the same genotype.
Statistical Analysis
Proportions were compared using Fisher's exact test. Association between heterologous reinfecting group and severe disease (all pneumonia syndromes) was tested by logistic regression. Age group (0–11 months, 12–23 months, and ≥24 months) was assessed as a potential confounder for RSV disease severity with use of the likelihood ratio test. Analyses were performed using Stata, version 11 (StataCorp).
GenBank Accession Numbers
The accession numbers for the nucleotide sequences determined in this study are JQ838257–JQ838584. The strain nomenclature is Country (Ken for Kenya)/unique identifier/month identified/year identified.
RESULTS
Specimen Sequencing
Over the entire study period (2002–2005), 409 infections were identified by antigen testing on the collected nasal specimens during ARI episodes [26]. Of these, 325 (79.4%) were characterized as either group A or B and their G-gene sequenced (Table 1). Forty-two of the specimens were already depleted (10.3%), and another 42 gave a negative PCR result and, thus, could not be characterized. Only 53 (63.8%) of the IFAT-identified 83 reinfections gave complete group and G-gene sequence data. These came from 52 children, 51 of whom had 2 infections and 1 with 3 infections.
Table 1.
Group and Genotype Distribution Over the 4 Epidemics in the Whole Cohort
| Variable | 2002a (%) | 2002–2003b (%) | 2004c (%) | 2004–2005d (%) | Combined (%) |
|---|---|---|---|---|---|
| Number on follow-upe | 335 | 508 | 524 | 219 | 635 |
| Samples positive by IFAT | 78 | 79 | 174 | 78 | 409 |
| Sequenced | 61 (78.2) | 75 (94.9) | 123 (70.7) | 66 (84.6) | 325 (79.5) |
| Group A | 60 (98) | 25 (33) | 69 (55) | 8 (12) | 162 (49.8) |
| GA2f | 10 (16.7) | 0 (0) | 0 (0) | 5 (62.5) | 15 (9.3) |
| GA5f | 50 (83.3) | 25 (100) | 69 (100) | 3 (37.5) | 147 (90.7) |
| Group B | 1 (2) | 50 (67) | 54 (45) | 58 (88) | 163 (50.2) |
| SAB1g | 1 (100) | 48 (96.0) | 54 (100) | 2 (3.4) | 105 (64.4) |
| BAg | 0 (0) | 2 (4.0) | 0 (0) | 56 (96.6) | 58 (35.6) |
a Epidemic was from March 2002 through September 2002.
b Epidemic was from December 2002 through August 2003.
c Epidemic was from January 2004 through May 2004.
d Epidemic was from November 2004 through April 2005.
e Refers to the number of children observed during each of the epidemics.
f Represents the genotype classification within group A.
g Represents the genotype classification within group B.
Group and Genotype Prevalence and Circulation Patterns
The monthly number of RSV A and B cases that were observed over the entire study period is shown in Figure 1. Four epidemic seasons were observed. Groups A and B varied in predominance in succeeding epidemics, resulting in a pattern of A, A/B, A/B, and B.
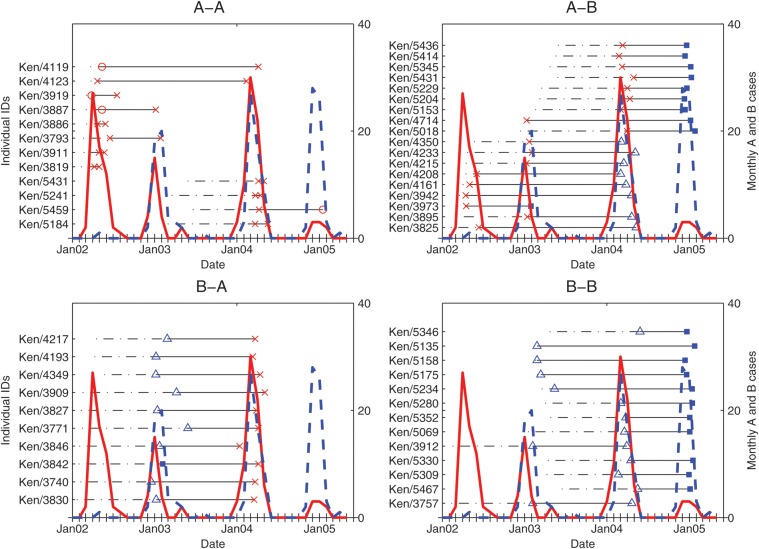
Figure 1.
The time course of 53 RSV infections in the 52 children who had repeat infection. The different subplots represent the group reinfection patterns observed. In the subplots, the dotted portions of the black horizontal straight lines represent the time from birth to the first positive sample of a reinfected participant. Each symbol between the continuous portions of the horizontal line represents a positive sample for which genotyping was completed: empty red circle, GA2; a red x symbol, GA5; empty blue triangle, SAB1 and filled blue square, BA. The continuous red curve and the dotted blue curve represent the monthly trends in the number of RSV A and RSV B cases detected, respectively, in the whole cohort, shown by the secondary y-axis.
We analyzed a G-gene fragment that was 618 nucleotides long (corresponding to nucleotides 295–912 in reference strain A2, accession number M11486) for all 162 group A positives, of which only 61 were of unique sequence. Phylogenetic analysis revealed that these sequences fell into 2 major clusters: one close to the GA2 genotype and the other close to the GA5 genotype (Supplementary Figure 1). GA5 positives were observed during all the 4 epidemics, Table 1. Identical GA5s in the analyzed region were frequently found both during and between epidemics (Supplementary Figure 1). GA2 sequences were identified only during the first and last epidemics of the study period, Table 1.
Sequences analyzed from group B positive samples (n = 163) were variable in length (either 660 or 720 nucleotides long, corresponding to nucleotides 256–915 in reference strain CH18537, accession number M17213). The variation was a result of some viruses containing a deletion of 6 nucleotides in the first hypervariable region of the G-gene together with a duplication of 60 nucleotides in the second hypervariable region. Of the 163 nucleotide sequences, 71 were unique, and phylogenetic analysis revealed that these were of 2 major types: one close to the SAB1 genotype and the other close to the BA genotype (Supplementary Figure 2). The BA-like strains had the 60-nucleotide duplication in their G-gene similar to the duplication that was first observed in Argentinean RSV B strains in 1999 [34]. The SAB1-like strains occurred in all 4 epidemics, and the BA genotype occurred in only the 2002–2003 and 2004–2005 epidemics and were the predominant RSV genotype in the latter epidemic (Table 1).
Reinfection Patterns
The details of age at first infection, age at reinfection, RSV group, and genotype of all infections identified among the 52 reinfected children are given in Figure 1. Among the reinfection pairs, 28 were heterologous at the group level (18 A-B; 10 B-A) and the remaining 25 were homologous (12 A-A; 13 B-B). The mean interval between the infection and reinfection was 365 days (range, 21–699 days). Seven of the reinfections occurred during the same epidemic as the first infection, 37 occurred in consecutive epidemics, and 11 occurred in nonconsecutive epidemics. All 7 reinfections that occurred in the same epidemic as the first infection were of homologous group A, and 6 of these had either identical or near-identical G-gene sequences (Table 2).
Table 2.
Details of the Genetic Differences in the Homologous Group Reinfections
| Subject ID | Age at Diagnosis (1st, 2nd), Days | Interval Between the Infections, Days | G Genotype (1st, 2nd) | No. Of Nucleotide Differences | No. Of Amino Acid Differences |
|---|---|---|---|---|---|
| Ken/4119 | 52, 745 | 693 | GA2, GA5 | 59 | 41 |
| Ken/4123 | 30, 691 | 661 | GA5, GA5 | 3 | 2 |
| Ken/5459 | 294, 577 | 283 | GA5, GA2 | 65 | 38 |
| Ken/3887 | 72, 310 | 238 | GA2, GA5 | 61 | 35 |
| Ken/3793 | 119, 344 | 225 | GA5, GA5 | 4 | 0 |
| Ken/3919 | 24, 137 | 113 | GA2, GA5 | 61 | 38 |
| Ken/5184a | 388, 444 | 56 | GA5, GA5 | 0 | 0 |
| Ken/3886a | 51, 86 | 35 | GA5, GA5 | 1 | 1 |
| Ken/5431a | 314, 338 | 24 | GA5, GA5 | 0 | 0 |
| Ken/3911a | 68, 92 | 24 | GA5, GA5 | 0 | 0 |
| Ken/5241a | 371, 393 | 22 | GA5, GA5 | 1 | 1 |
| Ken/3819a | 59, 80 | 21 | GA5, GA5 | 0 | 0 |
| Ken/3757 | 361, 801 | 440 | SAB1, SAB1 | 3 | 2 |
| Ken/3912 | 355, 772 | 417 | SAB1, SAB1 | 16 | 12 |
a The individuals had repeat positives with RSV A, occurring within a limited time period and with high similarity in the G sequences. We could not definitively ascertain that these were re-infections. For Group B, we only show homologous genotype reinfections (Ken/3737 and Ken/3912).
Heterologous Group Reinfections
Twenty-eight (52.8%) of the reinfections that we characterized were heterologous at group level. All occurred between epidemics (23 in consecutive epidemics, 5 in nonconsecutive epidemics), and the observed genotype patterns were 10 GA5-BA, 8 GA5-SAB1, 9 SAB1-GA5, and 1 BA-GA5.
Homologous Group A Reinfections
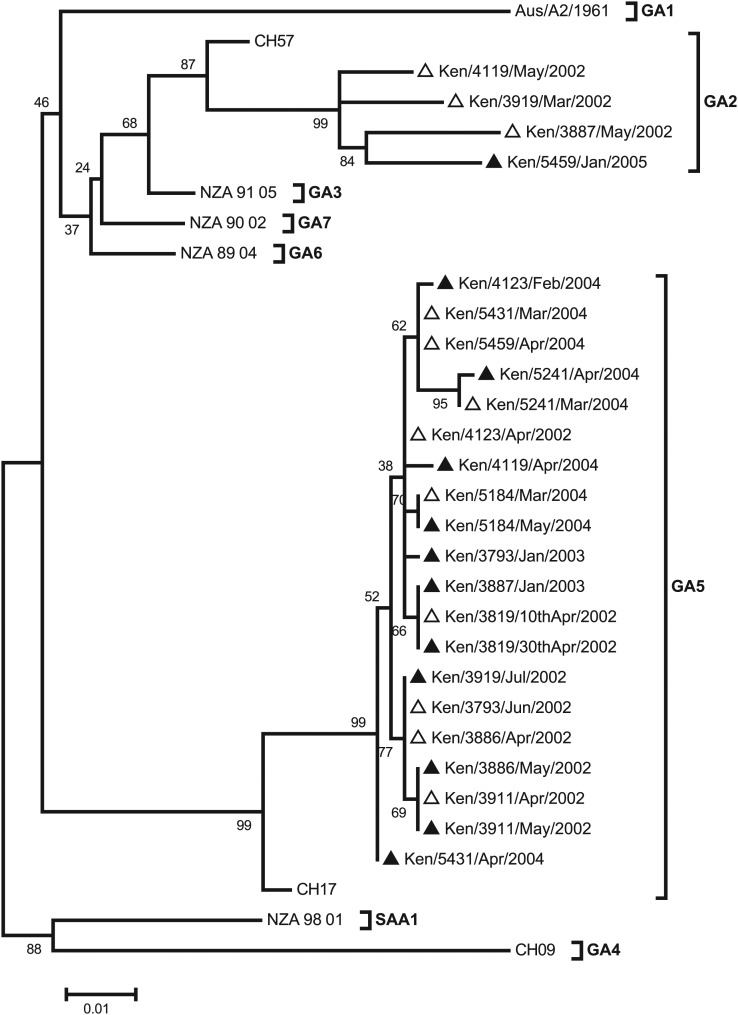
The evolutionary relationship of the pair viruses from homologous group A reinfections is shown in Figure 2. Four children were reinfected with a heterologous genotype in group A, one of them during the same epidemic (Ken/3919). Two children (Ken/4123 and Ken/3793) were infected and reinfected with the same genotype as in primary infection but in separate epidemics. The reinfecting virus of Ken/4123 had 2 nonsynonymous mutations relative to the primary infecting virus: T113A and N250S. The latter change is predicted to result in a loss of a potential N-glycosylation site (NST→SST). The viruses from Ken/3793 were identical at amino acid sequence level but with the presence of the 4 nucleotide changes.
Figure 2.
Maximum likelihood phylogenetic tree based on the sequences from the 12 subjects who experienced homologous group A reinfections. Reference sequences for genotypes GA1-7 and SAA1-3 were included to assign the strains G genotypes. The GenBank accession numbers of the reference sequences are Aus/A2/1961, M11486; CH57,AF065258 ; NZA_91_05, DQ171722; CH9, AF065254; CH17, AF065255; NZA_89_04, DQ171761, NZA_90_02, DQ171755; and NZA_98_01, DQ171792. The empty triangles represent first infections, and the dark, filled triangles represent the repeat infections.
The remaining 6 children (Ken/5184, Ken/5241, Ken/5431, Ken/3819, Ken/3911, and Ken/3886) were found to be infected and reinfected with viruses of the same genotype in group A in a single epidemic (Figure 1). In these children, the interval between the 2 identified positive samples was 21–56 days (Table 2). For patients Ken/5241 and Ken/3886, the pairs possessed single nucleotide mutations that resulted in amino acid changes L248V and T147I, respectively. However, the pairs from the other 4 children gave identical nucleotide sequences.
Homologous Group B Reinfections
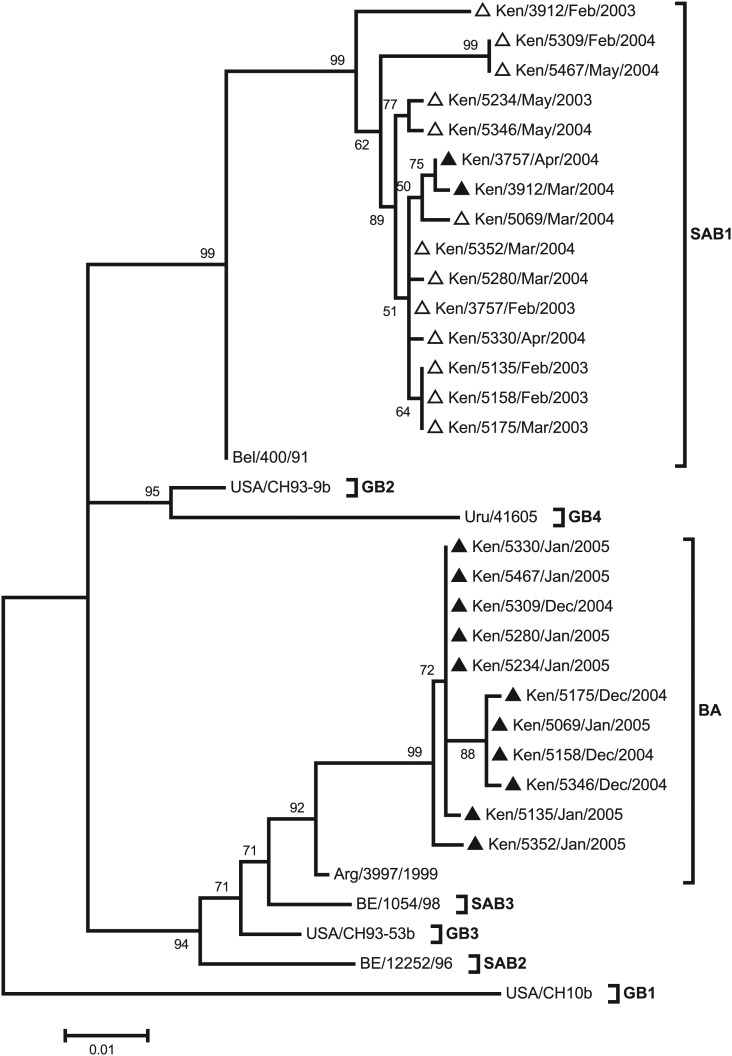
The phylogenetic relationship of the G-gene sequences from the children with homologous group B reinfection is shown in Figure 3. The sequences fell into 2 clusters: one that included virus strains that have the 60-nucleotide duplication (BA genotype) and another of virus strains that lack the duplication (SAB1 genotype). Eleven children with an initial infection with SAB1 genotype virus strains were reinfected with BA genotype virus strains during a subsequent epidemic. Two children (Ken/3757 and Ken/3912) were infected and reinfected in consecutive epidemics with SAB1 genotype virus strains that had amino acid changes. In these 2, the reinfecting strain from Ken/3757 possessed 2 nonsynonymous changes (N66H and S297L), and in Ken/3912, the reinfecting strain possessed 11 nonsynonymous changes occurring together with a mutation at a stop codon site that resulted in a changed predicted protein length (Table 2).
Figure 3.
A maximum likelihood tree showing the evolutionary relationship of strains from subjects who experienced homologous group B reinfections. The tree is based on sequences from 11 subjects and reference sequences representing genotypes GB1-4, SAB1-3 and the BA. Their GenBank accession numbers are CH10b, AF065250; CH93-9B,AF065251; CH93-53B, AF065253; BE/400/91, AY751275; BE/12252/96, AY751241; BE/1054/98,DQ985156; Uru/41605; AF251557; and Arg//3997/1999, DQ227366. The empty triangles represent first infections, and the dark, filled triangles represent the second infections.
To establish whether there were changes elsewhere in the RSV genome that would have contributed to the occurrence of homologous genotype reinfections, we sequenced a portion of the F protein gene, 310 amino acids long (from position 22, 54% of the total F length), for 4 of the homologous genotype reinfections (2 from each group) (Supplementary Figure 1). We found 100% nucleotide identity in this region between the reinfecting pairs.
Infection History Versus Genotype Prevalence
The distribution of the reinfection genotypes among the reinfected individuals mirrored the genotype prevalence among the children for whom we identified only a single infection during the whole study period. This was particularly true during the 2004 and 2004–2005 epidemics, during which we observed most of the reinfections (86.8%) (Table 3). When we examined the group level patterns, by epidemic, there was evidence for group-specific immune protection during the 2004 epidemic, notably among individuals who were reinfected after one intervening inter-epidemic period. A similar result was not observed from the data for the 2004–2005 epidemic (Table 4).
Table 3.
Comparison of the Prevalence of the Various Genotypes in the Single Infection Group and the Reinfection Group by Epidemic
| Genotype | 2002 |
2002–2003 |
2004 |
2004–2005 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st infect | % | Reinfect | % | 1st infect | % | Reinfect | % | 1st infect | % | Reinfect | % | 1st infect | % | Reinfect | % | |
| GA2 | 10 | 17.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 8.9 | 1 | 4.8 |
| GA5 | 46 | 80.7 | 4 | 100 | 23 | 31.9 | 2 | 66.7 | 64 | 59.3 | 15 | 60.0 | 3 | 6.7 | 0 | 0 |
| SAB1 | 1 | 1.8 | 0 | 0 | 47 | 65.3 | 1 | 33.3 | 44 | 40.7 | 10 | 40.0 | 2 | 4.4 | 0 | 0 |
| BA | 0 | 0 | 0 | 0 | 2 | 2.8 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 80.0 | 20 | 95.2 |
| P valuea | >.99 | .326 | >.99 | .576 | ||||||||||||
a The P values shows absence of a statistically significant difference in the distribution of genotypes in cases identified as first infection vs cases identified as reinfection following Fisher's exact test.
Table 4.
The Observed Group Reinfection Patterns Over the 4 RSV Epidemics
| Second Infection by Group and Epidemic (2–4) |
||||||||
|---|---|---|---|---|---|---|---|---|
| A2 | B2 | A3 | B3 | A4 | B4 | Total | ||
| First infection by group and epidemic (1–3) | A1 | 2 | 1 | 2 | 5 | – | – | 10 |
| B1 | 0 | 0 | 0 | 0 | – | – | 0 | |
| A2 | – | – | 0 | 4 | 0 | 1 | 5 | |
| B2 | – | – | 10 | 2 | 0 | 4 | 16 | |
| A3 | – | – | – | – | 1 | 7 | 8 | |
| B3 | – | – | – | – | 0 | 7 | 7 | |
| Total | 2 | 1 | 12 | 11 | 1 | 19 | 46 | |
Only individuals who developed a reinfection in a separate epidemic from the first infection are included. The 2 instances when a considerable number of reinfections were observed in consecutive epidemics are boxed with discontinuous line borders in bold. Fisher's exact test for the patterns observed in epidemic 3 (2004) gave a highly significant result (P = .003), but the same was not observed in epidemic 4 (ie, 2004–2005; P = .333).
Clinical Severity of Reinfections
Among the 53 reinfections, 36 (67.9%) were associated with upper respiratory tract infection, 11 (20.8%) with mild pneumonia, and 6 (11.3%) with severe pneumonia. Pneumonia occurred in 6 (24%) of 25 homologous group reinfections and 11 (39.3%) of 28 heterologous group infections. There was no statistically significant association between heterologous group reinfection and pneumonia (odds ratio, 2.05; 95% confidence interval, .623–6.74). Age was not a significant confounder (likelihood ratio test, χ2 = 3.10; P = .213).
DISCUSSION
The study presents results on RSV molecular epidemiology in a birth cohort of 635 children closely monitored over 4 RSV epidemic seasons to identify patterns of infection in relation to previous exposure. Previously, a number of population-level studies have observed that the RSV groups that predominate epidemics regularly oscillate [9, 20, 35], whereas the predominant genotypes are often replaced in consecutive seasons [13, 14, 36]. These observations have led to the hypothesis that RSV groups/genotypes create homologous herd immunity after an epidemic, favoring heterologous variants to circulate in a subsequent season [9, 21, 22].
This study investigated whether individuals previously infected with a particular RSV variant are more likely to be reinfected with a heterologous than with a homologous variant, despite the prevailing group or genotype prevalence. We characterized 53 repeat infections from an intensively monitored cohort and determined that 28 of these were of the heterologous group and 25 were of the homologous group. Among the homologous group reinfections, only 10 were with a homologous genotype, 5 of which showed limited amino acid changes in the fragment of the G-gene that we sequenced.
Therefore, overall, 80% of the reinfections that we characterized from the cohort were heterologous at group or genotype level. To understand the epidemiological context in which these reinfections occurred, we genotyped virus strains from children in whom we identified only one antigen-confirmed infection during the study period. Comparing this sample of background circulating variants with the reinfecting variants by epidemic, we found no difference in the genotype distributions (Table 3). This observation raises the question of whether the observed heterologous group/genotype reinfections were influenced by the children's previous exposures or reflected the prevailing distribution of genotypes in the community.
After examining the reinfections patterns by epidemic, the hypothesis of stronger homologous group immunity, compared with heterologous group immunity, was supported by data from the 2004 epidemic but not from the 2004–2005 epidemic (Table 4). Of note, the 2004–2005 epidemic was the first epidemic in Kilifi that was predominated by the newly recognized group B genotype virus strains (BA) that possess a 60-nucleotide duplication in the G-gene and that have replaced previously circulating group B strains worldwide [37]. Paradoxically, immunological studies in our laboratory that focused on strain-specific immunity found that infant convalescent serum samples from children infected with the SAB1 strains neutralize to the same level as the strains with the 60-nucleotide duplication [44].
Ten (∼20%) of 53 of the reinfections that we observed were due to a homologous genotype. Five of these possessed amino acid sequences differences between the pairs, which were few in number (<3), except for one that had 12 amino acid changes. The immunological significance of such limited sequence differences is uncertain, particularly in understanding whether this contributed to the reinfection process. However, some of the substitutions occurred in known antigenic epitope domains. For instance, changes affecting N-glycosylation pattern (Ken/4123) and protein length (Ken 3912) are both known to have a profound effect on antigenicity [38, 39]. However, in animal immunization studies, Sullender et al showed that recombinant G proteins that are up to 15% different in amino acid sequence give similar protection against homologous and heterologous isolates in intra-group challenge [25].
This observation of homologous group or genotype reinfection raises the question of the completeness and durability of RSV strain-specific immune responses [4]. The interval between the infections for the group B homologous genotype reinfections (Ken/3757 and Ken/3912) exceeded a year. Using data derived from this cohort, we have recently shown that, after infection, immunity to reinfection appears to last up to 6 months and is only 60%–70% effective [40]. Thus, it is possible that previous-exposure immune responses in some of these children waned to below protective levels. There was also a significant interval between the infections in Ken/4123 (21.7 months) and Ken/3793 (9.4 months) homologous genotype group A reinfections. Individual Ken/3793 provided a clear example of a reinfection with an identical virus in the G-gene occurring in a subsequent epidemic.
For the repeat positive samples of the same group (A) and genotype, separated by a short interval (<2 months; some, 21–24 days) and with limited or no genetic differences, it was not possible to be definitive on whether these were reinfections or persistent single infections. Although we did not determine whole genome sequences, the portion that we sequenced is the most variable in RSV genomes and the protein product of the G-gene is of most immunological significance together with the F protein. Partial sequencing of the F gene for some of these pairs did not find any differences (Supplementary Figure 3). Previous studies have shown that the mean duration of RSV shedding in infants and young children is 7 days (range, 1–21 days; reviewed in [41]), and prolonged shedding occurs mostly in individuals with impaired immunity [42] or cardiopulmonary disease [43]. Our examination of the records on the baseline characteristics of these children did not find presence of a predisposing condition to prolonged shedding, such as very low birth weight, malnutrition, or congenital disease.
With regard to disease severity, pneumonia occurred in 32% of the reinfections. We did not observe a statistically significant higher risk of pneumonia after heterologous group reinfection, compared with homologous group reinfection.
The sampling and testing methods of the present study undoubtedly missed some infections and reinfections in the cohort, thus presenting a possible bias [40]. Ideally, sampling should have been more frequent, irrespective of symptoms, to capture possible subclinical cases, and screening could have been done using the more sensitive molecular methods. Although our study presents data from a considerably increased sample size, compared with previous studies on RSV reinfection, the observed numbers in the individual epidemics were insufficient for conclusive interpretation.
In conclusion, our study shows that the vast majority of RSV reinfections in nature are with genetically distinct strains and that repeat infections with variants identical in the amino acid sequence of the G protein to the primary infecting variant can occur. The direct role of strain-specific immunity at individual level in reinfections remains unclear, because we observed that the distribution of the genotypes in the reinfection cases mirrored the distribution of genotypes observed at the same time in the wider study population.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the Kilifi RSV project field and clinical staff, for collecting the samples analyzed here; the guardians/parents of the children who participated in the study; and Timothy Kinyanjui, for assistance with the Matlab code that produced the reinfection graphic. The study was published with permission of the Director of KEMRI.
Financial support. This work was supported by the Wellcome Trust, UK (084663 and 084538).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–55. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falsey AR, Walsh EE. Respiratory syncytial virus infection in adults. Clin Microbiol Rev. 2000;13:371–84. doi: 10.1128/cmr.13.3.371-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011;162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullender WM. Respiratory syncytial virus genetic and antigenic diversity. Clin Microbiol Rev. 2000;13:1–15. doi: 10.1128/cmr.13.1.1-15.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson FW, Collier AM, Clyde WA, Jr, Denny FW. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N Engl J Med. 1979;300:530–4. doi: 10.1056/NEJM197903083001004. [DOI] [PubMed] [Google Scholar]
- 6.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–6. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 7.Johnson KM, Bloom HH, Mufson MA, Chanock RM. Natural reinfection of adults by respiratory syncytial virus. Possible relation to mild upper respiratory disease. N Engl J Med. 1962;267:68–72. doi: 10.1056/NEJM196207122670204. [DOI] [PubMed] [Google Scholar]
- 8.Falsey AR. Respiratory syncytial virus infection in adults. Semin Respir Crit Care Med. 2007;28:171–81. doi: 10.1055/s-2007-976489. [DOI] [PubMed] [Google Scholar]
- 9.Cane P. Molecular epidemiology and evolution of RSV. In: Cane P, editor. Respiratory syncytial virus. Vol. 1. Elsevier; 2007. pp. 89–113. [Google Scholar]
- 10.Mufson MA, Orvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66(Pt 10):2111–24. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 11.Anderson LJ, Hierholzer JC, Tsou C, et al. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–33. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 12.Johnson PR, Spriggs MK, Olmsted RA, Collins PL. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–9. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79(Pt 9):2221–9. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- 14.Cane PA, Matthews DA, Pringle CR. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J Clin Microbiol. 1994;32:1–4. doi: 10.1128/jcm.32.1.1-4.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cane PA, Pringle CR. Molecular epidemiology of respiratory syncytial virus: rapid identification of subgroup A lineages. J Virol Methods. 1992;40:297–306. doi: 10.1016/0166-0934(92)90088-u. [DOI] [PubMed] [Google Scholar]
- 16.Peret TC, Hall CB, Hammond GW, et al. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis. 2000;181:1891–6. doi: 10.1086/315508. [DOI] [PubMed] [Google Scholar]
- 17.Trento A, Viegas M, Galiano M, et al. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol. 2006;80:975–84. doi: 10.1128/JVI.80.2.975-984.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mufson MA, Belshe RB, Orvell C, Norrby E. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J Clin Microbiol. 1987;25:1535–9. doi: 10.1128/jcm.25.8.1535-1539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi M, Sano Y, Dapat IC, et al. High frequency of repeated infections due to emerging genotypes of human respiratory syncytial viruses among children during eight successive epidemic seasons in Japan. J Clin Microbiol. 2011;49:1034–40. doi: 10.1128/JCM.02132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zlateva KT, Vijgen L, Dekeersmaeker N, Naranjo C, Van Ranst M. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J Clin Microbiol. 2007;45:3022–30. doi: 10.1128/JCM.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White LJ, Waris M, Cane PA, Nokes DJ, Medley GF. The transmission dynamics of groups A and B human respiratory syncytial virus (hRSV) in England & Wales and Finland: seasonality and cross-protection. Epidemiol Infect. 2005;133:279–89. doi: 10.1017/s0950268804003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cane PA. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol. 2001;11:103–16. doi: 10.1002/rmv.305. [DOI] [PubMed] [Google Scholar]
- 23.Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis. 1991;163:693–8. doi: 10.1093/infdis/163.4.693. [DOI] [PubMed] [Google Scholar]
- 24.Parveen S, Broor S, Kapoor SK, Fowler K, Sullender WM. Genetic diversity among respiratory syncytial viruses that have caused repeated infections in children from rural India. J Med Virol. 2006;78:659–65. doi: 10.1002/jmv.20590. [DOI] [PubMed] [Google Scholar]
- 25.Sullender WM, Mufson MA, Prince GA, Anderson LJ, Wertz GW. Antigenic and genetic diversity among the attachment proteins of group A respiratory syncytial viruses that have caused repeat infections in children. J Infect Dis. 1998;178:925–32. doi: 10.1086/515697. [DOI] [PubMed] [Google Scholar]
- 26.Nokes DJ, Okiro EA, Ngama M, et al. Respiratory syncytial virus infection and disease in infants and young children observed from birth in Kilifi District, Kenya. Clin Infect Dis. 2008;46:50–7. doi: 10.1086/524019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nokes DJ, Okiro EA, Ngama M, et al. Respiratory syncytial virus epidemiology in a birth cohort from Kilifi district, Kenya: infection during the first year of life. J Infect Dis. 2004;190:1828–32. doi: 10.1086/425040. [DOI] [PubMed] [Google Scholar]
- 28.Scott PD, Ochola R, Ngama M, et al. Molecular analysis of respiratory syncytial virus reinfections in infants from coastal Kenya. J Infect Dis. 2006;193:59–67. doi: 10.1086/498246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunson RN, Collins TC, Carman WF. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J Clin Virol. 2005;33:341–4. doi: 10.1016/j.jcv.2004.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–3. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 33.Venter M, Madhi SA, Tiemessen CT, Schoub BD. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol. 2001;82:2117–24. doi: 10.1099/0022-1317-82-9-2117. [DOI] [PubMed] [Google Scholar]
- 34.Trento A, Galiano M, Videla C, et al. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84:3115–20. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- 35.Waris M. Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J Infect Dis. 1991;163:464–9. doi: 10.1093/infdis/163.3.464. [DOI] [PubMed] [Google Scholar]
- 36.Choi EH, Lee HJ. Genetic diversity and molecular epidemiology of the G protein of subgroups A and B of respiratory syncytial viruses isolated over 9 consecutive epidemics in Korea. J Infect Dis. 2000;181:1547–56. doi: 10.1086/315468. [DOI] [PubMed] [Google Scholar]
- 37.Trento A, Casas I, Calderon A, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84:7500–12. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cane PA, Thomas HM, Simpson AF, Evans JE, Hart CA, Pringle CR. Analysis of the human serological immune response to a variable region of the attachment (G) protein of respiratory syncytial virus during primary infection. J Med Virol. 1996;48:253–61. doi: 10.1002/(SICI)1096-9071(199603)48:3<253::AID-JMV7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Melero JA, Garcia-Barreno B, Martinez I, Pringle CR, Cane PA. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol. 1997;78(Pt 10):2411–8. doi: 10.1099/0022-1317-78-10-2411. [DOI] [PubMed] [Google Scholar]
- 40.Ohuma E, Okiro E, Ochola R, et al. The Natural History of Respiratory Syncytial Virus (RSV): The Influence of Age and Previous Infection on Re-infection and Disease in a Birth Cohort. Am J Epidemiol. 2012 doi: 10.1093/aje/kws257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okiro EA, White LJ, Ngama M, Cane PA, Medley GF, Nokes DJ. Duration of shedding of respiratory syncytial virus in a community study of Kenyan children. BMC Infect Dis. 2010;10:15. doi: 10.1186/1471-2334-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall CB, Powell KR, MacDonald NE, et al. Respiratory syncytial viral infection in children with compromised immune function. N Engl J Med. 1986;315:77–81. doi: 10.1056/NEJM198607103150201. [DOI] [PubMed] [Google Scholar]
- 43.Hall CB, Douglas RG, Jr, Geiman JM. Respiratory syncytial virus infections in infants: quantitation and duration of shedding. J Pediatr. 1976;89:11–5. doi: 10.1016/s0022-3476(76)80918-3. [DOI] [PubMed] [Google Scholar]
- 44.Sande CJ, Mutunga MN, Medley GF, Cane PA, Nokes DJ. Group and genotype specific neutralising antibody responses against respiratory syncytial virus (RSV) in infants and young children with severe pneumonia. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.