Abstract
Signal transducer and activator of transcription (STAT) proteins perform key roles in mediating signaling by cytokines and growth factors, including platelet-derived growth factor (PDGF). In addition, Src family kinases activate STAT signaling and are required for PDGF-induced mitogenesis in normal cells. One STAT family member, Stat3, has been shown to have an essential role in cell transformation by the Src oncoprotein. However, the mechanisms by which STAT-signaling pathways contribute to mitogenesis and transformation are not fully defined. We show here that disruption of Stat3 signaling by using dominant-negative Stat3β protein in NIH 3T3 fibroblasts suppresses c-Myc expression concomitant with inhibition of v-Src-induced transformation. Ectopic expression of c-Myc is able to partially reverse this inhibition, suggesting that c-Myc is a downstream effector of Stat3 signaling in v-Src transformation. Furthermore, c-myc gene knockout fibroblasts are refractory to transformation by v-Src, consistent with a requirement for c-Myc protein in v-Src transformation. In normal NIH 3T3 cells, disruption of Stat3 signaling with dominant-negative Stat3β protein inhibits PDGF-induced mitogenesis in a manner that is reversed by ectopic c-Myc expression. Moreover, inhibition of Src family kinases with the pharmacologic agent, SU6656, blocks Stat3 activation by PDGF. These findings, combined together, delineate the signaling pathway, PDGF → Src → Stat3 → Myc, that is important in normal PDGF-induced mitogenesis and subverted in Src transformation.
Signal transducer and activator of transcription (STAT) proteins originally were discovered as key effectors of normal IFN signaling and subsequently were shown to be involved in signaling by numerous cytokines and growth factors (1, 2). Upon engagement of polypeptide ligands with their cognate cell surface receptors, tyrosine kinases induce phosphorylation of cytoplasmic STAT proteins, which then translocate to the nucleus and regulate gene expression (3). One of the growth factors that stimulates robust activation of STAT signaling under normal physiological conditions is platelet-derived growth factor (PDGF) (4, 5). In addition to cytokines and growth factors, many studies have shown that diverse oncoproteins, including the viral Src oncoprotein, v-Src, and an oncogenic form of PDGF, v-Sis, constitutively activate one STAT family member, Stat3 (6–10). Furthermore, activated Stat3 signaling has been shown to be required for cell transformation by v-Src (11, 12). Among the genes regulated by STAT proteins are those involved in controlling the fundamental cellular processes of cell proliferation and apoptosis, including cyclin D1, bcl-x, and c-myc (13–19). Stable expression of a constitutively activated mutant form of Stat3 is sufficient to induce transformation in murine fibroblasts and up-regulate expression of key mediators of cell cycle progression, including c-Myc protein (15).
Overexpression or amplification of the c-myc gene has been detected in numerous solid tumors and blood malignancies (20). In addition, earlier studies showed that c-Myc protein is required for transformation by the oncogenic tyrosine kinase, BCR-Abl (21, 22). Mounting evidence indicates that Stat3 is activated with high frequency in many blood malignancies and solid tumors (10, 23), suggesting an important role for constitutive Stat3 activation in human cancer. Elevated expression or kinase activity of c-Src protein also has been associated with the progression of certain human cancers, including carcinomas of the breast and colon as well as various blood malignancies (24). With respect to normal cellular signaling, PDGF stimulation induces expression of c-Myc (25) as well as activation of c-Src kinase (26). Furthermore, both c-Src and c-Myc have been shown to be required for PDGF-induced mitogenesis (27–30). Recent evidence indicates that Stat3 exists in a complex with PDGF receptors and c-Src and that Stat3 activation is induced rapidly upon PDGF stimulation in a c-Src kinase-dependent manner (31). Based on the collective findings described above, we considered c-Myc a probable Stat3 effector involved in normal mitogenic signaling by PDGF and oncogenic signaling by v-Src. In this study, we investigated the role of Stat3-mediated c-Myc expression in cells transformed by v-Src or stimulated with PDGF.
Methods
Focus Formation Assays.
Plasmid DNA expression vectors for v-Src, Stat3β, and c-Myc were transfected into NIH 3T3 cells along with β-galactosidase (0.2 μg for each condition) by the calcium phosphate precipitation method by using the quantity of plasmid indicated in the figure (12, 32, 33) (Fig. 1a). For each transfection condition, the transfection efficiency was determined by detection of β-galactosidase expression by using the βGal Staining Kit (Invitrogen). The average of the number of cells expressing β-galactosidase in five independent fields of vision (×200) was used for normalization. Foci were quantified after 17 days, normalized for β-galactosidase expression, and expressed relative to the number of foci induced by v-Src alone. Transfections were performed in triplicate, and data represent the average of three independent experiments. TGR1 (c-myc+/+) or HO15.19 (c-myc−/−) cells (34) were transfected with 50 ng of v-Src expression vector (Fig. 2a) in duplicate by using Fugene 6 (Roche, Gipf-Oberfrick, Switzerland), or HO15.19 cells were transfected with v-Src alone (50 ng) or together with c-Myc expression vector (5 μg) (Fig. 2b) in triplicate by using Lipofectamine Plus (GIBCO/BRL). Cells were fixed in methanol and stained with neat Karyo-Max (GIBCO/BRL). Foci were quantified after 35 days and expressed as the number of foci induced by v-Src alone (Fig. 2a) or by v-Src together with c-Myc (Fig. 2b). Values represent the mean and SD for 2–3 independent experiments.
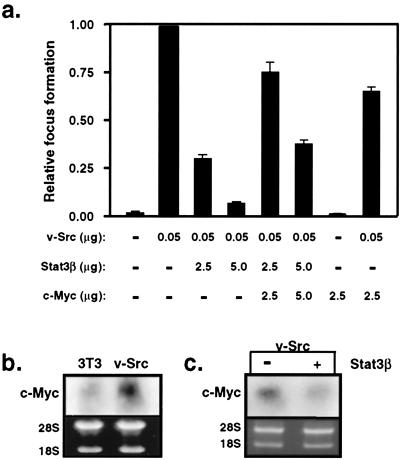
Figure 1.
Stat3-dependent c-Myc expression is required for Src transformation. (a) NIH 3T3 cells were transfected with the indicated expression vectors, and v-Src-induced focus formation in monolayer was quantified. (b) Northern blot analysis of c-myc expression in normal NIH 3T3 fibroblasts or these cells stably transformed by v-Src (NIH 3T3/v-Src). (c) Northern blot analysis of c-myc expression in FACS-selected NIH 3T3/v-Src cells transiently transfected with vectors encoding GFP alone (−) or together with Stat3β.
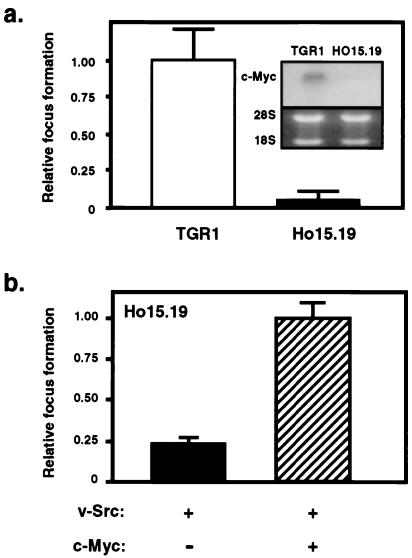
Figure 2.
Fibroblasts lacking the c-myc gene are refractory to transformation by v-Src. (a) Parental rat-1 (TGR1) and c-myc knockout cells (HO15.19) were transfected with v-Src, and monolayer focus formation assays were performed. (Inset) Northern blot, demonstrating lack of c-myc mRNA expression in HO15.19 cells. (b) HO15.19 cells were transfected with v-Src vector alone or together with an expression vector encoding human c-Myc protein and subjected to focus formation assays.
Northern Blots.
For Northern blot analysis, total mRNA from exponentially growing NIH 3T3 or NIH 3T3/v-Src cells (Fig. 1b) or NIH 3T3/v-Src cells expressing either green fluorescent protein (GFP) alone or GFP together with Stat3β (Fig. 1c) was isolated by the guanidine isothiocyanate method. A total of 15 μg of RNA was resolved on a 1% agarose-formaldehyde gel, transferred to a nylon membrane, and probed with a 32P-labeled probe corresponding to the 1.4-kb XhoI fragment of murine c-myc cDNA. Detection of 28S and 18S ribosomal RNA by ethidium bromide staining confirmed equalization and integrity of samples.
Flow Cytometry.
NIH 3T3 or NIH 3T3/v-Src cells were seeded at a density of 5 × 105/60-mm dish in DMEM/5% bovine calf serum/antibiotics 24 h before transfection. Cells were transfected with a total of 6 μg of EGFP-C3 (CLONTECH), encoding green fluorescent protein (16), or a 5:1 molar ratio of pSG5hStat3β, encoding human Stat3β (32), to EGFP-C3 by using Lipofectamine Plus. For cell cycle analysis, cells were transfected followed by treatment with nocodazole (50 ng/ml) as described (35). Cells were harvested 20 h later by trypsinization and fixed with 0.5% formaldehyde for 10 min on ice, followed by fixation in 70% ethanol for 30 min. Fixed cells were harvested by centrifugation, resuspended in PBS containing RNase A (1 mg/ml), and incubated at 37°C for 30 min. Cells were collected by centrifugation and resuspended in PBS containing propidium iodide (50 μg/ml) (Sigma). GFP-positive cells (5,000–10,000) were analyzed for DNA content by FACScan (Becton-Dickinson). Values represent the mean and SD for three independent experiments (Fig. 3a). Alternatively, cells were sorted 48 h posttransfection by FACStar and 5 × 105 GFP-positive cells were collected per sample. To measure apoptosis levels (16), cells were harvested 24 h postsort by trypsinization and 3–5 × 105 cells were collected by centrifugation in a 5-ml polystyrene, round-bottom tube, washed with PBS, and resuspended in 1× Annexin V binding buffer (PharMingen) at 1 × 106 cells/ml. A 100-μl aliquot was incubated with 5 μl of Annexin V-PE (PharMingen) for 15 min at room temperature in the dark. Samples were diluted with 400 μl of 1× Annexin V binding buffer and analyzed by FACScan. Values represent the difference in Annexin V binding between GFP-positive cells and mock-transfected GFP-negative cells, expressed as percentage of cells binding Annexin V. The mean and SD for two independent experiments is shown (Fig. 3b).
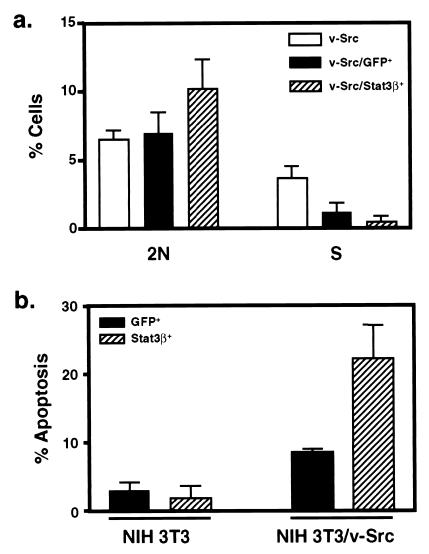
Figure 3.
Stat3 signaling contributes to Src oncogenesis by stimulating cell cycle progression and preventing apoptosis. (a) NIH 3T3/v-Src cells were mock-transfected or transfected with vectors encoding GFP alone or together with Stat3β followed by treatment with nocodazole. Cell cycle distribution of mock-transfected or GFP-positive cells with or without Stat3β expression was determined by staining with propidium iodide and FACS analysis. (b) NIH 3T3 or NIH 3T3/v-Src cells were mock-transfected or transfected with vectors encoding GFP alone or together with Stat3β. GFP-positive cells with or without Stat3β expression were harvested by FACS and analyzed for apoptosis with Annexin V staining.
Microinjections and Western Blot Analysis.
Quiescent NIH 3T3 cells were injected with expression vectors encoding Stat3β (50 μg/ml), c-Myc (20 μg/ml), or c-Fos (20 μg/ml) by using an automated microinjection system (Zeiss). After microinjection, fibroblasts (200 per construct) were stimulated with PDGF-BB (20 ng/ml) 6 h postinjection or not stimulated, and PDGF-induced DNA synthesis was measured by BrdUrd incorporation 18 h later. The data are shown as the percentage of total microinjected cells incorporating BrdUrd, and values represent the mean and SD for three independent experiments (Fig. 4a). For analysis of Stat3 tyrosine phosphorylation (Fig. 4b), cells were treated with the Src family kinase selective inhibitor SU6656 (1 μM) for 1 h before stimulation with PDGF-BB for 10 min. Stat3 protein was immunoprecipitated from cell lysates with anti-Stat3 antibody (Transduction Laboratories, Lexington, KY) and subjected to SDS/PAGE followed by transfer to nitrocellulose membranes. Activated Stat3 was detected by antibodies to p-Tyr705-Stat3 (New England Biolabs), and the membrane was stripped and reprobed with antibodies to total Stat3 (Transduction Laboratories).
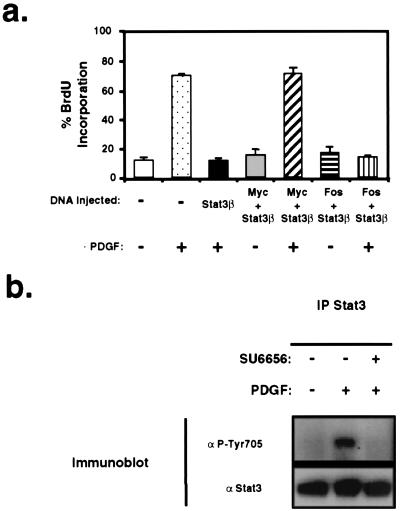
Figure 4.
PDGF-induced mitogenesis depends on Stat3 signaling and c-Myc expression. (a) Quiescent NIH 3T3 cells were microinjected with expression vectors encoding the indicated cDNAs and stimulated with PDGF or not stimulated, and DNA synthesis was measured by BrdUrd incorporation. (b) NIH 3T3 cells were treated with DMSO or the Src family kinase-selective inhibitor SU6656 (1 μM) for 1 h before stimulation with PDGF. Stat3 proteins were immunoprecipitated from cell lysates and analyzed by Western blot by using antibodies to activated phosphotyrosine-Stat3 or total Stat3 protein.
Results
To define the role of c-Myc in Src-induced transformation, expression vectors encoding v-Src, Stat3β, or c-Myc were transfected into NIH 3T3 fibroblasts alone or in various combinations, and the ability of v-Src to induce foci in monolayer focus formation assays was quantified (Fig. 1a). Stat3β is a naturally occurring isoform of Stat3 lacking the C-terminal transcriptional activation domain and, consequently, blocks Stat3 signaling in a dominant-negative fashion (12, 32, 33). In this assay of v-Src transformation, no additional selection was applied to the transfected cells to minimize any potential bias that may arise from selection or clonal variation. Because this approach precludes measurement of protein expression levels because stable clones are not selected, lacZ expression vector was included as an internal control for transfection efficiency, and the focus formation data were normalized to β-galactosidase expression. Consistent with previous studies (11, 12), increasing amounts of dominant-negative Stat3 protein expression inhibited Src-induced focus formation in a dose-dependent manner, verifying that Stat3 signaling is required for Src transformation. Importantly, ectopic expression of c-Myc was able to rescue the defect elicited by coexpression of Stat3β and partially restore v-Src-induced transformation, even at the highest amount of Stat3β expression. This partial restoration may reflect other effects of Stat3β or the possibility that Stat3 regulates genes in addition to c-myc that contribute to cell transformation. These results suggest that the c-myc gene is a downstream target of Stat3 signaling in Src transformation.
In NIH 3T3 cells stably transformed by v-Src, elevated endogenous c-myc mRNA expression was detected compared with parental, nontransformed cells (Fig. 1b). To determine the contribution of Stat3 signaling to c-myc expression, cells stably transformed by v-Src were transiently transfected with GFP expression vector alone or together with Stat3β expression vector, followed by isolation of GFP-positive cells by FACS. Consistent with a role for Stat3 in the regulation of the c-myc gene (15, 17, 18), disruption of Stat3 signaling by expression of dominant-negative Stat3β protein significantly reduced endogenous c-myc mRNA levels in Src-transformed cells (Fig. 1c). These results demonstrate that induction of c-myc expression depends on v-Src-mediated activation of Stat3 signaling.
The requirement for c-Myc expression in Src oncogenesis was evaluated further by using Rat-1 fibroblasts lacking c-Myc protein expression because of homozygous deletion of both c-myc alleles (34). Fig. 2a Inset confirms the absence of any detectable c-myc mRNA expression in these cells. Moreover, these cells lack detectable expression of L-myc or N-myc (34). Results from monolayer focus formation assays demonstrate that c-myc-null cells (HO15.19) are refractory to transformation by the Src oncoprotein compared with the parental cell line (TGR1, Fig. 2a). Because the c-myc-null cells proliferate more slowly than wild-type cells (34), incubation of the focus formation assay was doubled from 17 to 35 days for the c-myc-null cells. Although wild-type cells yielded large foci within 17 days, there were few detectable foci (even very small ones) after 35 days in the case of c-myc-null cells. To determine whether the lack of transformation by v-Src was due to loss of c-Myc expression rather than differences in transfection efficiency between cell lines or independent events associated with gene targeting, v-Src expression vector was transfected alone or together with an expression vector encoding c-Myc into HO15.19 cells. Restoring c-Myc expression, in turn, partially restored transformation by the v-Src oncoprotein, demonstrating that c-Myc is required for efficient Src transformation (Fig. 2b). This restoration of Src transformation cannot be attributed simply to enhanced transfection efficiency in the presence of c-Myc, because ectopic c-Myc expression was observed to reduce transfection efficiency in the HO15.19 cells (data not shown). Taken together, these findings indicate that Stat3-dependent expression of c-Myc is required for Src transformation.
The biological mechanism by which Stat3 signaling contributes to Src oncogenesis was assessed by transfection of v-Src-transformed NIH 3T3 cells with expression vectors encoding GFP alone or together with Stat3β. To detect perturbations in cell cycle progression, cells were treated with nocodazole as described (35), and GFP-positive cells were analyzed for their DNA content by FACS. Inhibition of Stat3 signaling in v-Src transformed cells resulted in a small but reproducible arrest of cells in G1 with a 2N DNA content (Fig. 3a). The nocodazole block approach (35) strengthens the conclusions that the increase in the G1 population is due to arrest of cells in G1 by Stat3, whereas the effect of GFP on S phase primarily is a result of nonspecific inhibition of DNA synthesis rather than G1 arrest. To assess further the contribution of Stat3 signaling to Src oncogenesis, GFP-positive cells were isolated by FACS and analyzed for apoptosis by using Annexin V staining. Inhibition of Stat3 signaling induces a marked increase in apoptosis of v-Src-transformed but not parental NIH 3T3 cells (Fig. 3b). These results are consistent with the findings that, in human multiple myeloma and squamous cell carcinoma cell lines, Stat3 signaling is required for survival (16, 36). Furthermore, gene therapy with dominant-negative Stat3 induces apoptosis of melanoma cells in a mouse model (37). Together, these findings provide evidence that constitutive activation of Stat3 contributes to oncogenesis through the combined effects of preventing apoptosis and stimulating cell cycle progression.
Previous studies have demonstrated that c-Src kinase activity is required for normal cell proliferation in the context of PDGF signaling (27–30). Moreover, ectopic c-Myc expression rescues a block in PDGF-induced mitogenesis caused by kinase-inactive c-Src (29). Using a similar strategy used to characterize the role of c-Src in the context of PDGF signaling, we analyzed the contribution of Stat3 signaling to PDGF-induced mitogenesis. Stat3β expression vector was microinjected into quiescent, normal NIH 3T3 cells or coinjected with c-Myc or c-Fos expression vectors followed by stimulation with PDGF (Fig. 4a). Expression of Stat3β was detected by immunofluorescent Stat3 antibodies and was in excess of endogenous Stat3 protein (data not shown). Expression of Stat3β alone effectively blocked PDGF-induced DNA synthesis as measured by BrdUrd incorporation. Ectopic expression of c-Myc, but not c-Fos, alleviated the Stat3β-mediated block in PDGF signaling and restored mitogenesis. The amount of c-Myc or c-Fos plasmid was shown previously to be sufficient to rescue the block in PDGF-induced mitogenesis resulting from expression of dominant-negative Src kinase or dominant-negative N17 Ras, respectively (29). These data provide evidence that Stat3 signaling is required for normal quiescent fibroblasts to progress from G1 to S phase in response to PDGF. To determine the role of c-Src in PDGF-induced Stat3 activation and c-myc expression, we used the Src family kinase selective inhibitor, SU6656 (38). Consistent with recent studies demonstrating a requirement for c-Src in PDGF-induced activation of Stat3 DNA-binding activity (31), the Src inhibitor blocked tyrosine phosphorylation of Stat3 (Fig. 4b) and repressed induction of c-myc mRNA in response to PDGF (38).
Discussion
Our findings delineate a complete signal transduction pathway from the cell surface to the nucleus in which activation of PDGF receptor results in signaling through c-Src kinase to Stat3, which, in turn, induces the expression of c-myc (Fig. 5). In normal fibroblasts stimulated with PDGF, we show that Stat3-mediated c-Myc induction is required for cell cycle progression from G1 to S phase. In v-Src transformed cells, this signaling pathway is subverted in the absence of PDGF, resulting in constitutive activation of Stat3 and elevated expression of c-Myc protein that is required for cell transformation. Unlike normal cells, however, only a relatively small proportion of Src-transformed cells were arrested in G1 by Stat3β expression. Instead, blocking Stat3 signaling in Src-transformed cells induced predominantly apoptosis. Recent evidence indicates that v-Src is capable of generating proapoptotic stimuli that are suppressed by various antiapoptotic-signaling pathways downstream of Src (39, 40). In particular, blocking phosphatidylinositol 3-kinase or Ras-mediated antiapoptotic-signaling pathways allows manifestation of the proapoptotic pathways activated by Src (39, 40). Because of the antiapoptotic activity of Stat3 (13, 16, 36), it is likely that Stat3 represents another survival pathway in Src transformation. Thus, blocking any one of these critical survival signaling pathways, including Stat3, may result in the activation of proapoptotic effectors and cell death induced by Src. Our results are completely consistent with an earlier report demonstrating that Stat3-mediated induction of c-Myc contributes to cytokine-induced cell cycle progression and survival in mouse pre-B cells (18). However, it remains to be determined whether Stat3 directly or indirectly regulates the c-myc promoter during Src transformation and in response to PDGF stimulation.
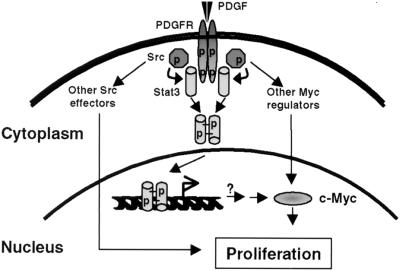
Figure 5.
Model for normal mitogenic PDGF signal transduction through Stat3 that is subverted by Src oncogenesis. In this model, PDGF-induced receptor dimerization leads to activation of receptor-associated c-Src kinase activity, which, in turn, phosphorylates Stat3 monomers on tyrosine. Phosphorylated Stat3 forms activated dimers that translocate to the nucleus and induce c-myc gene expression that is required for normal PDGF-stimulated mitogenesis. In cells transformed by the Src oncoprotein, constitutive activation of Stat3 leads to continuous induction of c-myc expression, thereby short-circuiting PDGF receptor signaling and contributing to oncogenesis. It remains to be determined whether Stat3 regulates the c-myc promoter directly or indirectly in this model. In addition, other signaling pathways independent of Stat3 regulate c-myc expression and cell proliferation.
Our results show that expression of c-Myc is necessary but not sufficient for mitogenesis and cell transformation, consistent with the findings that Stat3 regulates multiple genes involved in cell cycle progression and survival, including cyclin D1 and Bcl-x (3, 10, 15, 16). A requirement for other Stat3-regulated genes may explain our observation that c-Myc only partially rescues the Stat3β-mediated block in Src transformation. In this regard, it could be relevant that v-Src also induces cyclin D1 expression in a Stat3-dependent manner (41). In addition, our findings reported here do not contradict earlier evidence that induction of c-myc by v-Src depends on the Raf/MEK/MAPK pathway (42), because a dependence on Stat3 does not exclude a role for other signaling pathways in the regulation of c-myc by v-Src. Furthermore, recent studies demonstrated that c-myc expression is required for cell transformation by other oncoproteins, including Ras and Raf (43). Our findings are compatible with these earlier studies and establish a requirement for c-Myc protein in cell transformation by v-Src. Moreover, our results demonstrate that Stat3 participates in c-Myc induction in response to PDGF- and Src-induced signaling. In contrast to our findings reported here and an earlier study (31), another recent study (44) reported that Src is not required for PDGF-induced Stat3 activation or c-myc expression. It is probable that this discrepancy results from the different approaches used in these studies. In particular, one study used PDGF receptor mutants that do not bind Src (44), whereas the present and another earlier study (31) used two different pharmacologic inhibitors (SU6656 and PD180970) that likely block all Src family kinases, including both receptor-bound and nonreceptor-bound Src family kinases (38, 45). Finally, it is notable that the requirement for c-Src in PDGF signaling is abrogated in p53 null fibroblasts or by expression of simian virus 40 large T antigen, suggesting that the Stat3-dependent pathway delineated here can be bypassed by alternative pathways (46).
It has long been postulated that the Src oncoprotein induces cell transformation by subverting normal receptor tyrosine kinase-signaling pathways (47, 48). Although there are likely to be multiple pathways involved, our findings delineate one such pathway from the cell surface to the nucleus that is essential for normal mitogenesis induced by PDGF receptor and subverted in Src oncogenesis. In addition, the results presented here suggest that c-Myc target genes participate in Stat3-mediated signaling downstream of PDGF receptor and Src. A recent study using microarray gene expression profiling of human fibroblast cells identified numerous potential Myc target genes involved in controlling cell proliferation (49). Additional c-Myc targets that participate in regulating cell cycle progression are the cdk4 and p27kip1 genes (refs. 50–52; A. Obaya, B. O'Connell, and J.M.S., unpublished data). Future studies using methods such as gene-expression profiling should provide further insight into the target genes of Stat3 and c-Myc proteins that contribute to mitogenesis and cell survival induced by Src-mediated signaling.
Acknowledgments
We thank members of our laboratories for stimulating discussions, Robert Eisenman for the human c-Myc expression plasmid, Eric Caldenhoven and Rolf de Groot for the Stat3β cDNA, and Jodi Kroeger and Matt Morrow of the Moffitt Cancer Center Flow Cytometry Core for flow cytometric analyses. This work was supported in part by National Cancer Institute Grants CA55652 (to R.J.) and CA67360 (to W.J.P.), American Cancer Society Grant RPG99099 (to T.J.Y.), and the Cortner–Couch Endowed Chair for Cancer Research (W.J.P.).
Abbreviations
- STAT
signal transducer and activator of transcription
- PDGF
platelet-derived growth factor
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg J, Darnell J E., Jr Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 4.Wagner B J, Hayes T E, Hoban C J, Cochran B H. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silvennoinen O, Schindler C, Schlessinger J, Levy D E. Science. 1993;261:1736–1739. doi: 10.1126/science.8378775. [DOI] [PubMed] [Google Scholar]
- 6.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 7.Danial N N, Pernis A, Rothman P B. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 8.Garcia R, Yu C L, Hudnall A, Catlett R, Nelson K L, Smithgall T, Fujita D J, Ethier S P, Jove R. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 9.Besser D, Bromberg J F, Darnell J E, Jr, Hanafusa H. Mol Cell Biol. 1999;19:1401–1409. doi: 10.1128/mcb.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman T, Garcia R, Turkson J, Jove R. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot R P, Jove R. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 14.Fukada T, Ohtani T, Yoshida Y, Shirogane T, Nishida K, Nakajima K, Hibi M, Hirano T. EMBO J. 1998;17:6670–6677. doi: 10.1093/emboj/17.22.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bromberg J F, Wrzeszczynska M H, Devgan G, Zhao Y, Pestell R G, Albanese C, Darnell J E., Jr Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 16.Catlett-Falcone R, Landowski T H, Oshiro M M, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna J L, Nunez G, et al. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 17.Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, Hibi M, Hirano T. J Exp Med. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirogane T, Fukada T, Muller J M, Shima D T, Hibi M, Hirano T. Immunity. 1999;11:709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 19.Socolovsky M, Fallon A E, Wang S, Brugnara C, Lodish H F. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 20.Dang C V, Resar L M, Emison E, Kim S, Li Q, Prescott J E, Wonsey D, Zeller K. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 21.Sawyers C L, Callahan W, Witte O N. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 22.Afar D E, Goga A, McLaughlin J, Witte O N, Sawyers C L. Science. 1994;264:424–426. doi: 10.1126/science.8153630. [DOI] [PubMed] [Google Scholar]
- 23.Catlett-Falcone R, Dalton W S, Jove R. Curr Opin Oncol. 1999;11:490–496. doi: 10.1097/00001622-199911000-00010. [DOI] [PubMed] [Google Scholar]
- 24.Biscardi J S, Tice D A, Parsons S J. Adv Cancer Res. 1999;76:61–119. doi: 10.1016/s0065-230x(08)60774-5. [DOI] [PubMed] [Google Scholar]
- 25.Kelly K, Cochran B H, Stiles C D, Leder P. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 26.Gould K L, Hunter T. Mol Cell Biol. 1988;8:3345–3356. doi: 10.1128/mcb.8.8.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twamley-Stein G M, Pepperkok R, Ansorge W, Courtneidge S A. Proc Natl Acad Sci USA. 1993;90:7696–7700. doi: 10.1073/pnas.90.16.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche S, Koegl M, Barone M V, Roussel M F, Courtneidge S A. Mol Cell Biol. 1995;15:1102–1109. doi: 10.1128/mcb.15.2.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barone M V, Courtneidge S A. Nature (London) 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 30.Broome M A, Hunter T. J Biol Chem. 1996;271:16798–16806. doi: 10.1074/jbc.271.28.16798. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y Z, Wharton W, Garcia R, Kraker A, Jove R, Pledger W J. Oncogene. 2000;19:2075–2085. doi: 10.1038/sj.onc.1203548. [DOI] [PubMed] [Google Scholar]
- 32.Caldenhoven E, van Dijk T B, Solari R, Armstrong J, Raaijmakers J A M, Lammers J W J, Koenderman L, de Groot R P. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 33.Turkson J, Bowman T, Adnane J, Zhang Y, Djeu J Y, Sekharam M, Frank D A, Holzman L B, Wu J, Sebti S, et al. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 35.Wu C L, Classon M, Dyson N, Harlow E. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandis J R, Drenning S D, Zeng Q, Watkins S C, Melhem M F, Endo S, Johnson D E, Huang L, He Y, Kim J D. Proc Natl Acad Sci USA. 2000;97:4227–4232. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, Jove R, Yu H. Cancer Res. 1999;59:5059–5063. [PubMed] [Google Scholar]
- 38.Blake R A, Broome M A, Liu X, Wu J, Gishizky M, Sun L, Courtneidge S A. Mol Cell Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson D, Agochiya M, Samejima K, Earnshaw W, Frame M, Wyke J. Cell Death Differ. 2000;7:685–696. doi: 10.1038/sj.cdd.4400700. [DOI] [PubMed] [Google Scholar]
- 40.Webb B L, Jimenez E, Martin G S. Mol Cell Biol. 2000;20:9271–9280. doi: 10.1128/mcb.20.24.9271-9280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinibaldi D, Wharton W, Turkson J, Bowman T, Pledger W J, Jove R. Oncogene. 2000;19:5419–5427. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 42.Aziz N, Cherwinski H, McMahon M. Mol Cell Biol. 1999;19:1101–1115. doi: 10.1128/mcb.19.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bazarov A V, Adachi S, Li S F, Mateyak M K, Wei S, Sedivy J M. Cancer Res. 2001;61:1178–1186. [PubMed] [Google Scholar]
- 44.Sachsenmaier C, Sadowski H B, Cooper J A. Oncogene. 1999;18:3582–3592. doi: 10.1038/sj.onc.1202694. [DOI] [PubMed] [Google Scholar]
- 45.Kraker A J, Hartl B G, Amar A M, Barvian M R, Showalter H D, Moore C W. Biochem Pharmacol. 2000;60:885–898. doi: 10.1016/s0006-2952(00)00405-6. [DOI] [PubMed] [Google Scholar]
- 46.Broome M A, Courtneidge S A. Oncogene. 2000;19:2867–2869. doi: 10.1038/sj.onc.1203608. [DOI] [PubMed] [Google Scholar]
- 47.Hunter T, Cooper J A. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 48.Bishop J M. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- 49.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mateyak M K, Obaya A J, Sedivy J M. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Obaya A J, Mateyak M K, Sedivy J M. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 52.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett J F, Obaya A J, O'Connell B, Mateyak M K, Tam W, Kohlhuber F, et al. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. . (First Published February 25, 2000; 10.1073/pnas.050586197) [DOI] [PMC free article] [PubMed] [Google Scholar]