Abstract
The epithelial-to-mesenchymal transition (EMT) is an important event converting compact and ordered epithelial cells into migratory mesenchymal cells. Given the molecular and cellular similarities between pathological and developmental EMTs, studying this event during neural crest development offers and excellent in vivo model for understanding the mechanisms underlying this process. Here, we review new and old insight into neural crest EMT in search of commonalities with cancer progression that might aid in the design of specific therapeutic prevention.
Keywords: Neural crest, EMT, Development
1. Introduction
The neural crest (NC) is a transient embryonic cell population characterized by its multipotency and migratory ability. Induced at the boundary between neural and non-neural ectoderm, the NC is specified via a well-orchestrated transcriptional program, now referred to “gene regulatory network” [1–6]. As neurulation proceeds, NC precursors are restricted to the dorsal aspect of the neural fold and neural tube, where they can be distinguished by the expression of “neural crest specifier genes”. Subsequently, they undergo an epithelial-to-mesenchymal transition (EMT) to become migratory NC cells that migrate extensively to diverse locations. At their destinations, some undergo a reaggregation process via a mesenchymal-to-epithelial transition (MET) and further differentiate into many types of cells, ranging from neurons and glia of sensory, autonomic and enteric ganglia, to adrenomedullary secretory cells, smooth muscle cells, melanocytes, and bone and cartilage cells.
NC cells have attracted the attention of embryologists for over a century as a model for studying embryonic induction, specification, migratory potential and differentiation. In fact, perturbation experiments yield very different anomalies depending upon the phase of NC cell development that is disrupted (e.g., migration versus differentiation), with disruption in NC EMT generally causing the most severe phenotypes. Interestingly, common signaling pathways appear to occur during NC EMT as in other developmental EMTs such as those occurring during gastrulation in the primitive streak, somite decondensation, cardiac valve formation, etc. [7]. Notably, malignant cells also appear to use the same mechanisms to delaminate from an epithelial tumor as those used by embryonic epithelial cells to delaminate and migrate during development. This highlights the importance of understanding the normal mechanisms of NC EMT as these might provide important clues regarding the mistakes that lead to abnormal development or loss of the differentiated state.
In this review, we focus on new and old insights into NC formation as one of the best studied developmental examples of EMT, highlighting their importance during embryogenesis as well as a model for understanding cancer cells and tumor progression.
2. The EMT process in NC cells
A variety of in vivo and in vitro analyses in chick and Xenopus along with genetic studies in the mouse and zebrafish have identified some of the cellular and molecular mechanisms underlying EMT during NC cell delamination as well as some of the signaling cascades that trigger these events (Fig. 1). After their specification, premigratory NC precursors from the dorsal neural tube undergo an EMT process that can be parsed into several, sometimes overlapping, steps that ultimately allow the precursors to leave the neural tube, becoming bona fide NC cells that migrate through the extracellular matrix. The process of NC EMT events requires: i) the coordinated activity of transcription factors and molecular signaling pathways, ii) changes in cell junctions and polarity, iii) changes in adhesion properties, and iv) changes in the extracellular matrix.
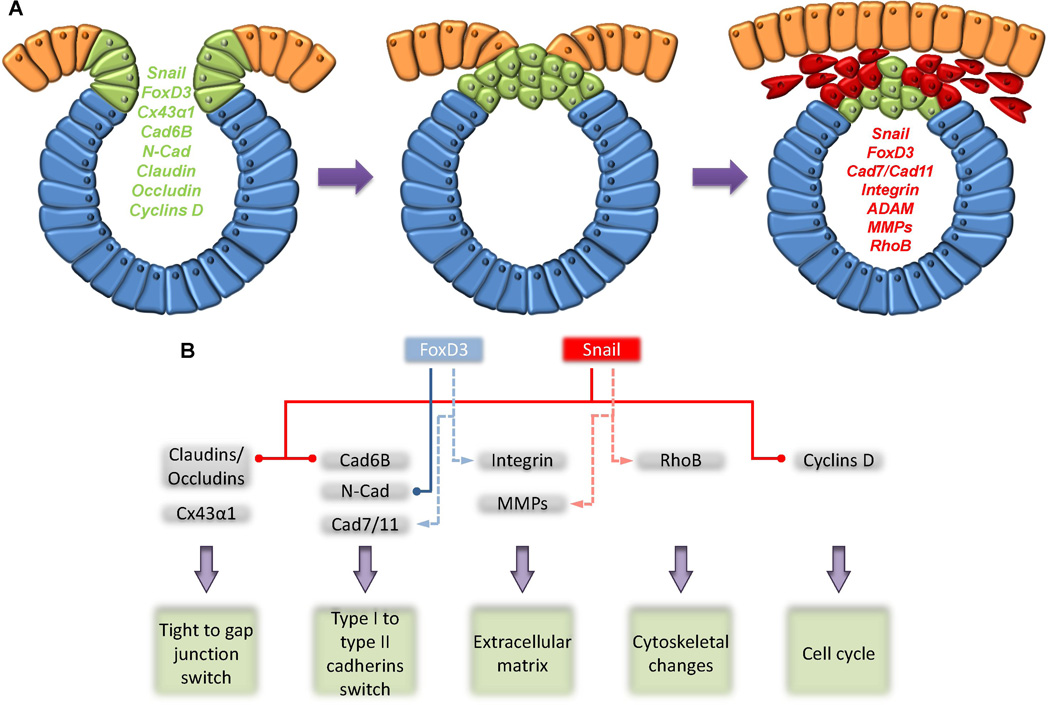
Figure 1.
(A) Schematic representation of the genes expressed on neural crest cells prior (green) and after (red) undergoes epithelial-to-mesenchymal transition. (B) Neural crest epithelial-to-mesenchymal transition regulation. NC specifiers, FoxD3 and Snail down-regulate expression of molecules that are associated with epithelial static cell populations, such as N-Cad and E-Cad (or Cad6B in chick and mouse), respectively, to give place to the up-regulation of mesenchymal migratory proteins, such as Cad7. Similarly, Snail down-regulates tight junction claudins/occludins to permit the upregulation of gap junction protein connexin-43α1 (Cx43α1), which may also depend on Snail expression. Gene regulation in which the repressors Snail or FoxD3 up-regulate the expression of matrix metalloproteases (MMPs), integrins, Cad7 or RhoB may denote indirect regulatory interactions, possibly mediated by other repressors (represented by dotted lines).
After EMT, the migratory ability of NC cells starts either prior to or soon after fusion of the neural folds depending upon the vertebrate species [8]. Concomitantly, NC cells acquire mesenchymal characteristics, as they express the intermediate filament vimentin and possess a flattened morphology with filopodia and lamellipodia, facilitating their spreading [9–11]. Migratory NC cells follow stereotypical pathways depending upon their axial level of origin. Cranial NC cells invade the surrounding cranial mesenchyme and ultimately condense to contribute to various cranial ganglia and craniofacial cartilage and bones. Migratory NC cells in the trunk that follow the ventral pathway differentiate into components of the peripheral nervous system, while those migrating dorsolaterally become melanocytes [8].
2.1. Transcription factors
The signaling pathways utilized during EMT in the NC are similar to those that are active in other developmental EMT process. Indeed, NC EMT is triggered by the integration of extracellular signals, which include components of the extracellular matrix as well as a number of secreted ligands including members of TGFβ, Wnt and FGF families. This initial event is necessary to convert neuroepithelial precursors into migratory NC cells through activation of a number of transcriptional regulators, including the zinc finger transcription factors, Snail1 and Snail2 (formerly known as Slug), and the winged-helix transcription factor FoxD3, which are critical factors that coordinate the cellular changes occurring during EMT [5].
Snail
Snail promotes NC EMT by directly mediating transitions in cell-junction assembly, motility and adhesion [5]. In chicks, NC cells express Snail2 whereas Snail1 is expressed in the mouse and both factors coexist in Xenopus [12–14]. Loss of function experiments in chick and Xenopus result in a strong abrogation of cranial NC cell migration [12, 15, 16]. Conversely, gain-of-function experiments reveal that Snail1 in Xenopus and Snail2 in chick are sufficient to induce expansion of the cranial NC territory and production of a greater number of migrating NC cells. However, numerous observations suggest that Snail genes may be neither sufficient nor necessary for NC cell specification and delamination, and may play different roles at different axial level [12, 17, 18]. These results indicated that Snail-expressing cells must either receive additional inputs or express other transcriptional regulators at different axial levels to achieve specification and execute the EMT program.
Until recently, it was not possible to discriminate whether Snail genes functioned during specification, delamination or both. However, several recent studies indicate that Snail can regulate target genes, such as E-cadherin [19], as well as genes encoding structural proteins that constitute the junction’s backbone such as claudin-3, claudin-4, claudin-7, and occludin [20], by directly binding to E-box sequences within their promoters. In mouse and chick embryos, a non-overlapping complementary mRNA expression pattern between Snail1/E-Cadherin and Snail2/Cadherin-6B, respectively, has been observed at the boundary of the ectoderm and the neural tube in the head region [19, 21]. Moreover, it has been demonstrated in chick that Snail2 directly binds to Cad6B regulatory sequence and represses it expression to allow NC cell delamination [21]. Conversely, Cad6B expression persists in migrating NC in mouse and the trunk of chick embryos after cessation of NC cell delamination [22–24]. Taken together, these results suggest that it is likely that Snail is not the sole regulator of Cad6B expression in NC cells.
Snail2 and Sox5, a transcription factor belonging to the SoxD family, are able to upregulate RhoB, a member of the Rho family of small GTPases [25]. RhoB exhibits a very dynamic pattern in prospective NC cells prior to and during early migration [26] and is a well-known regulator of events that change cell morphology necessary for NC delamination [27, 28]. These cellular changes involve actin cytoskeletal rearrangements as well as the formation of focal adhesions and stress fibers [26]. Studies in chick and zebrafish have established that Rho is necessary for cranial NC EMT [29, 30]. However, at trunk levels, it acts as a negative modulator, downregulating N-cadherin [30].
Foxd3
In all species analyzed to date, Foxd3 is expressed in both premigratory and migratory NC cells at all axial levels [31–36]. Gain and loss of function experiments in different vertebrates have ascribed multiple functions to Foxd3 in NC cells, possibly through recruitment of different partners at defined steps of development. Overexpression of Foxd3 promotes a massive EMT accompanied by a decrease in N-cadherin expression, alteration in cell polarity, and upregulation of Cad7 [31, 37]. However, this effect is observed only two days after overexpression, suggesting that the mechanism may be complex and involve many secondary interactions, possibly including epigenetic modifications.
Although such loss and gain of function experiments are a good start toward understanding the transcriptional network controlling NC cell EMT, the analysis is far from complete. Several recent studies utilizing enhancer regulatory analysis, together with chromatic immunoprecipitation (ChIP), DNA-pull down, and gel-shift assays have been able to demonstrate direct regulatory interactions between transcription factors and key genes related with EMT [38–40]. Further work along these lines is necessary to understand the direct regulatory inputs controlling NC EMT.
2.2. Cadherins
Cadherins are a large family of calcium-dependent cell-adhesion molecules that play an essential role in cell-cell interactions during NC EMT [22, 41]. In particular, the transition from the expression of type I cadherins (E-cadherin, N-cadherin, P-cadherin, R-cadherin), which are usually associated with epithelial cells, to the expression of type II cadherins (Cad5 and beyond), which characterize mesenchymal cells, are a hallmark of EMT. This transition leads to much lower adhesiveness of NC cells and allows them to increase their motility [42]. However, the repertoire of cadherins varies from one species to another, suggesting that what is important in the progress of NC EMT is the switch between different types of cadherin rather than the type itself. Moreover, this switch is quite complex as, in different species and at different axial levels, premigratory NC cells express more than one cadherin, and many others have been described in migrating cells [43]. Indeed, in zebrafish, mouse and birds, premigratory NC cells co-express both N-cadherin and Cad6 or its variant, Cad6B [22, 44, 45]. The significance of the co-expression of several cadherins and the observed axial level differences in their regulation in prospective NC cells are largely not understood and require further elucidation. During NC EMT, N-cadherin and Cad6b are downregulated in the prospective NC within the closing dorsal neural tube through FoxD3 and Snail2 activity, respectively [21, 37]. As NC cells exit the neural tube, the expression of Cad7 (in birds) and Cad11 (in mouse and Xenopus) are up-regulated, possibly by the combined action of the NC specifiers FoxD3 and Sox10 [22, 23, 37, 46, 47].
2.3. Junctions
The apical zones of premigratory NC cells are joined by intercellular junctions until fusion of the neural folds begins. Starting at this time, the junctions progressively disappear and the cells lose their apico-basal polarity [9, 48]. Occludin and claudin, which are the major components of tight junctions, are downregulated from the neural tube and a de-epithelization process takes place before the onset of NC cells migration [49, 50]. The dissolution of tight junctions seems to be directly regulated by Snail [20]. Similar to the transition that occurs in cadherins, there is a transition from tight junctions to gap junctions in the prospective NC cells prior to EMT [For review see 5]. For example, the gap junction protein connexin-43α1 (Cx43α1) is expressed in migrating NC cells at all axial levels [51] and its knockout produces defects in NC derivatives, possibly via dynamic regulation of the locomotory apparatus [52].
2.4. Extracelular matrix
The extracellular matrix (ECM) is not a passive structure that allows the movement of NC cells. Rather, the ECM is comprised of a dynamic milieu of collagens, fibronectin, laminins, vitronectin and proteoglycans, that together with integrin molecules on NC cells themselves play an active role during the EMT process [53]. In addition to switching cadherin expression, digestion of the extracellular matrix in the basement membrane that overlies the dorsal neural tube is a necessary step to achieve full NC EMT [54]. This job is carried out by membrane-bound and/or secreted forms of proteins called matrix metalloproteases (MMPs). MMP2 is one of the most common metalloproteases that plays an important role in NC migration. Indeed, specific morpholino knockdown of MMP2 expression in the dorsal neural tube disturbs NC cell EMT [55]. Consistent with this, the Ets1 transcription factor is known to induce the expression of MMPs and is upregulated in cranial NC cells precisely at the time of NC cell delamination in the chick and frog [56, 57]. Under normal condition, MMP2 is rapidly downregulated in migrating NC by tissue inhibitors of MMPs (TIMPs), such as TIMP-2, and has an important role on cardiac migratory NC cells [55, 58, 59]. In addition to the NC, MMPs are also expressed in various human cancer cells [60–63].
NC cells also express several other MMPs, including members of the novel transmembrane metalloprotease family ADAM [64, 65], such as ADAM-10 in the chick [66] and ADAM13 in the frog [67–69]. These are membrane-bound enzymes that cleave the extracellular portion of transmembrane proteins, thereby affecting cell-cell adhesion. ADAM10, expressed in the dorsal neural tube [66], is able to cleave the extracellular domain of N-cadherin resulting in the deterioration of the intercellular junctions [70]. Moreover, the remaining part of N-cadherin molecule is further cleaved off from the membrane and its CTF2 intracellular peptide acts as a transcriptional regulator [71]. This demonstrates a dual function for the proteolytic degradation of N-cadherin, which acts as a critical trigger for EMT. On the other hand, ADAM13 has been shown to cleave fibronectin and proposed to facilitate cell migration by locally reorganizing the fibronectin network [67].
NC cells express different integrins receptors that alter their contacts with the ECM following EMT. Loss-of-function studies have shown that integrins play an essential role in NC migration [72–76].
2.5. Epigenetic factors
Many studies in a variety of different vertebrate models have revealed critical roles for transcription factors in establishing the “gene regulatory network” that controls NC development [2]. Although each NC cell generally shares the same genetic information with every other, there are intrinsic properties giving them particular characteristics in terms of migration and differentiation capacities. In this context, a growing body of evidence has emerged demonstrating that epigenetic mechanisms also play a critical role in NC development [77–80]. Moreover, in this regulation not only the “writing” of the “epigenetic code”, but also the “reader”, play an important role in the fine tuning of gene expression and specific biological outcomes.
We recently reported that a dynamic histone modification is necessary for the proper temporal control of NC gene expression in vivo [80]. Accordingly, we described that the demethylation of H3K9me3 by JmjD2A is required for activation of several key NC specifier genes in the chick embryo. Expression of JmjD2A was found in the forming dorsal neural tube, and loss of JmjD2A function causes dramatic downregulation of several NC specifier genes, such as Sox9, Sox10, FoxD3, and Snail2. Importantly, in vivo chromatin immunoprecipitation reveals direct stage-specific interactions of JmjD2A with regulatory regions of Sox10 and associated temporal modifications in the methylation states of lysine residues directly affected by JmjD2A activity. Our findings show that chromatin modifications directly control the setting up of a developmental program for NC specification in vertebrate embryos via modulating histone methylation.
There are now diverse examples of the effects of different epigenetic modifications on NC cells or their derivatives. For example, histone acetylation, regulated by the histone deacetylase HDAC8, seems to have a unique role in cranial NCCs differentiation into facial skeleton [77]. It was further demonstrated that HDAC8 is required for the suppression of Otx2, Lhx1, and other homeobox transcription factors in cranial NC cells. Intriguingly, this affected skull elements, but no other NC derived tissues [77]. Differentiation of human wild-type ES cells into migratory neural crest-like cells (hNCLCs) was significantly inhibited by the depletion of the ATP-dependent chromatin remodeling chromodomain Helicase DNA-binding protein 7 (CHD7) [81]. Similarly, depletion of the CHD7 in Xenopus embryos resulted in drastic downregulation of three NC specifiers (Slug/Snail, Twist, and Sox9), due to a regulation of distal enhancer elements, which in turn regulate cranial NC cell migration in the developing pharyngeal arches [82].
Finally, it has been recently demonstrated that Aebp2, a potential targeting protein for the mammalian Polycomb Repression Complex 2 (PRC2), is upstream of several NC genes involved in migration and development processes [78]. In addition, many heterozygotes mouse display a set of phenotypes, such as enlarged colon and hypopigmentation, similar to those observed in human patients with Hirschsprung’s disease and Waardenburg syndrome. These phenotypes are usually caused by the absence of the NC-derived ganglia in hindguts and melanocytes [78].
Taken together, it is becoming clear that epigenetic mechanisms play a major role in NC development. However, there role in the EMT process is still poorly understood. It will be of great interest to map out the epigenetic regulation network during NCC development using highthroughput analysis such as ChIP-seq.
3. Cranial versus trunk NC cells EMT
Although it is tempting to try to present a single, unifying model to describe the molecular cascade leading to NC cell EMT, it has become increasingly evident that the process is heterogeneous. This is largely due to the heterogeneity of the NC population at different axis levels. Thus, NC cell EMT involves distinct cellular and molecular mechanisms that vary along the neural axis. Cranial NC cell delamination involves the cooperation of multiple regulators recruited simultaneously in a short period of time concomitant with the neurulation process. On the other hand, trunk NC cell delamination is more progressive, involving a restricted number of transcription factor and is comparatively delayed with respect to the neurulation. Moreover, it has been extensively demonstrated that the role the EMT players vary at different axial levels. For example, the expression of Snail genes varies in an axis dependent manner [12–14]. Particularly in chick, Snail2 is generally induced very precociously in cranial NC, long before neural tube closure, and persists during delamination as well as in early migrating NC cells to gradually disappear as they reach the ventral portion of the embryo. However, in the chick trunk, Snail2 expression appears soon after neural tube closure, and becomes repressed once cells start the migration [83]. In contrast to Snail2, Sox9 is required for the trunk NC development but not for cranial [37].
There are also cranial and trunk differences related to cadherin regulation. For example, in chick, Cad6B is expressed only in premigratory cranial NC and appears to prevent their delamination from the neural tube [84]. Later on, Snail2 directly downregulates Cad6B expression to induce the NC EMT [21]. On the other hand, Cad6B expression persists in early migrating trunk NC just after they have undergone an EMT and emigrated from the neural tube [24]. Similarly, in mouse, Cad6 expression persists in a subpopulation of early migrating NC [44]. Likewise, in the head both N-Cad transcripts and proteins are absent from the entire prospective NC cell population at the time of fold fusion, delineating cells undergoing EMT [8, 41, 85]. However, in the trunk, N-Cad transcripts are maintained in NC cell progenitors until their completely segregate from the neural tube [86].
As mentioned above, RhoB has different functions in cranial versus trunk NC cells. In the head, RhoB is responsible for mediating changes in the cell morphology necessary for NC delamination, but in the trunk acts as a negative modulator downregulating N-Cad [30].
There are also head and trunk differences in term of cell-cycle control. Studies show that premigratory NC cells in the trunk are arrested in the G1 phase and synchronously enter the S phase upon delamination. Snail, expressed in premigratory cells, represses the expression of cyclin D1 and D2 preventing the progression of G1-S. Upon delamination, Snail expression decreases, which allows the cyclin levels to rise and the cell-cycle to progress [87]. However, in the head the dynamics of the delamination process vary considerably. In contrast to the trunk, cranial NC cells delaminate en masse from the dorsal neural tube under the influence of the Ets1 transcription factor, which activity abolishes the necessity of the G1-S transition [57].
4. Conclusion
Here, we review the molecular mechanisms involved in NC EMT. As a general trend, the NC EMT process involves a myriad of molecular cascades resulting in a variety of cellular events that ultimately control cell-cell and cell-matrix adhesion, cell proliferation, and cell survival. To date, there is no single, simple mechanism that can describe the process of NC EMT and delamination. Rather, there appear to be a variety of mechanisms that lead to the same goal, depending upon the axial level of NC origin. It seems very likely that malignant cells use similar mechanisms to delaminate from an epithelial tumor as those used by embryonic NC cell. In this context, in-depth understanding of the EMT process occurring during normal embryonic development may contribute to the specific design of therapeutic treatment to stop the metastatic cascade and tumor progression.
Acknowledgements
Some of the work reviewed here is based on studies supported by the National Institutes of Health under Award No. R03DE022521 to MEB and PS-M, HD037105 and The California Institute of Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare there are no conflicts of interest.
References
- 1.Aybar MJ, Mayor R. Early induction of neural crest cells: lessons learned from frog, fish and chick. Curr Opin Genet Dev. 2002;12:452–458. doi: 10.1016/s0959-437x(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 2.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayor R, Young R, Vargas A. Development of neural crest in Xenopus. Curr Top Dev Biol. 1999;43:85–113. doi: 10.1016/s0070-2153(08)60379-8. [DOI] [PubMed] [Google Scholar]
- 4.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 6.Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Semin Cell Dev Biol. 2005;16:647–654. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Douarin NM, Kalcheim C. The neural crest. Second edition. Cambridge, Massachusetts, USA: Cambridge University Press; 1999. [Google Scholar]
- 9.Newgreen D, Gibbins I. Factors controlling the time of onset of the migration of neural crest cells in the fowl embryo. Cell Tissue Res. 1982;224:145–160. doi: 10.1007/BF00217274. [DOI] [PubMed] [Google Scholar]
- 10.Nichols DH. Ultrastructure of neural crest formation in the midbrain/rostral hindbrain and preotic hindbrain regions of the mouse embryo. Am J Anat. 1987;179:143–154. doi: 10.1002/aja.1001790207. [DOI] [PubMed] [Google Scholar]
- 11.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–923. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 12.Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- 13.Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- 14.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 15.LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 16.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 17.del Barrio MG, Nieto MA. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129:1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- 18.Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc Natl Acad Sci U S A. 2006;103:10300–10304. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 20.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 21.Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa S, Takeichi M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development. 1998;125:2963–2971. doi: 10.1242/dev.125.15.2963. [DOI] [PubMed] [Google Scholar]
- 24.Park KS, Gumbiner BM. Cadherin 6B induces BMP signaling and de-epithelialization during the epithelial mesenchymal transition of the neural crest. Development. 2010;137:2691–2701. doi: 10.1242/dev.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- 26.Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development. 1998;125:5055–5067. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- 27.Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- 28.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 29.Berndt JD, Clay MR, Langenberg T, Halloran MC. Rho-kinase and myosin II affect dynamic neural crest cell behaviors during epithelial to mesenchymal transition in vivo. Dev Biol. 2008;324:236–244. doi: 10.1016/j.ydbio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groysman M, Shoval I, Kalcheim C. A negative modulatory role for rho and rho-associated kinase signaling in delamination of neural crest cells. Neural Dev. 2008;3:27. doi: 10.1186/1749-8104-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- 32.Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- 33.Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 34.Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- 35.Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Mundell NA, Labosky PA. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138:641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–397. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci U S A. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J Biol Chem. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- 41.Hatta K, Takagi S, Fujisawa H, Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev Biol. 1987;120:215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- 42.Chu YS, Eder O, Thomas WA, Simcha I, Pincet F, Ben-Ze'ev A, Perez E, Thiery JP, Dufour S. Prototypical type I E-cadherin and type II cadherin-7 mediate very distinct adhesiveness through their extracellular domains. J Biol Chem. 2006;281:2901–2910. doi: 10.1074/jbc.M506185200. [DOI] [PubMed] [Google Scholar]
- 43.Taneyhill LA. To adhere or not to adhere: the role of Cadherins in neural crest development. Cell Adh Migr. 2008;2:223–230. doi: 10.4161/cam.2.4.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue T, Chisaka O, Matsunami H, Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev Biol. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, Liu B, Wilson AL, Rostedt J. cadherin-6 message expression in the nervous system of developing zebrafish. Dev Dyn. 2006;235:272–278. doi: 10.1002/dvdy.20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hadeball B, Borchers A, Wedlich D. Xenopus cadherin-11 (Xcadherin-11) expression requires the Wg/Wnt signal. Mech Dev. 1998;72:101–113. doi: 10.1016/s0925-4773(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 47.Vallin J, Girault JM, Thiery JP, Broders F. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mech Dev. 1998;75:171–174. doi: 10.1016/s0925-4773(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 48.Newgreen DF. Adhesion to extracellular materials by neural crest cells at the stage of initial migration. Cell Tissue Res. 1982;227:297–317. doi: 10.1007/BF00210888. [DOI] [PubMed] [Google Scholar]
- 49.Aaku-Saraste E, Hellwig A, Huttner WB. Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure--remodeling of the neuroepithelium prior to neurogenesis. Dev Biol. 1996;180:664–679. doi: 10.1006/dbio.1996.0336. [DOI] [PubMed] [Google Scholar]
- 50.Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol. 2006;22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- 51.Lo CW, Cohen MF, Huang GY, Lazatin BO, Patel N, Sullivan R, Pauken C, Park SM. Cx43 gap junction gene expression and gap junctional communication in mouse neural crest cells. Dev Genet. 1997;20:119–132. doi: 10.1002/(SICI)1520-6408(1997)20:2<119::AID-DVG5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 52.Xu X, Francis R, Wei CJ, Linask KL, Lo CW. Connexin 43-mediated modulation of polarized cell movement and the directional migration of cardiac neural crest cells. Development. 2006;133:3629–3639. doi: 10.1242/dev.02543. [DOI] [PubMed] [Google Scholar]
- 53.Perris R, Perissinotto D. Role of the extracellular matrix during neural crest cell migration. Mech Dev. 2000;95:3–21. doi: 10.1016/s0925-4773(00)00365-8. [DOI] [PubMed] [Google Scholar]
- 54.Erickson CA, Perris R. The role of cell-cell and cell-matrix interactions in the morphogenesis of the neural crest. Dev Biol. 1993;159:60–74. doi: 10.1006/dbio.1993.1221. [DOI] [PubMed] [Google Scholar]
- 55.Duong TD, Erickson CA. MMP-2 plays an essential role in producing epithelial-mesenchymal transformations in the avian embryo. Dev Dyn. 2004;229:42–53. doi: 10.1002/dvdy.10465. [DOI] [PubMed] [Google Scholar]
- 56.Tahtakran SA, Selleck MA. Ets-1 expression is associated with cranial neural crest migration and vasculogenesis in the chick embryo. Gene Expr Patterns. 2003;3:455–458. doi: 10.1016/s1567-133x(03)00065-6. [DOI] [PubMed] [Google Scholar]
- 57.Theveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai DH, Vollberg TM, Sr, Hahn-Dantona E, Quigley JP, Brauer PR. MMP-2 expression during early avian cardiac and neural crest morphogenesis. Anat Rec. 2000;259:168–179. doi: 10.1002/(SICI)1097-0185(20000601)259:2<168::AID-AR7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 59.Cantemir V, Cai DH, Reedy MV, Brauer PR. Tissue inhibitor of metalloproteinase-2 (TIMP-2) expression during cardiac neural crest cell migration and its role in proMMP-2 activation. Dev Dyn. 2004;231:709–719. doi: 10.1002/dvdy.20171. [DOI] [PubMed] [Google Scholar]
- 60.Nomura H, Sato H, Seiki M, Mai M, Okada Y. Expression of membrane-type matrix metalloproteinase in human gastric carcinomas. Cancer Res. 1995;55:3263–3266. [PubMed] [Google Scholar]
- 61.Pulyaeva H, Bueno J, Polette M, Birembaut P, Sato H, Seiki M, Thompson EW. MT1-MMP correlates with MMP-2 activation potential seen after epithelial to mesenchymal transition in human breast carcinoma cells. Clin Exp Metastasis. 1997;15:111–120. doi: 10.1023/a:1018444609098. [DOI] [PubMed] [Google Scholar]
- 62.Springman EB, Angleton EL, Birkedal-Hansen H, Van Wart HE. Multiple modes of activation of latent human fibroblast collagenase: evidence for the role of a Cys73 active-site zinc complex in latency and a "cysteine switch" mechanism for activation. Proc Natl Acad Sci U S A. 1990;87:364–368. doi: 10.1073/pnas.87.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stetler-Stevenson WG, Krutzsch HC, Liotta LA. Tissue inhibitor of metalloproteinase (TIMP-2). A new member of the metalloproteinase inhibitor family. J Biol Chem. 1989;264:17374–17378. [PubMed] [Google Scholar]
- 64.Smith KM, Gaultier A, Cousin H, Alfandari D, White JM, DeSimone DW. The cysteine-rich domain regulates ADAM protease function in vivo. J Cell Biol. 2002;159:893–902. doi: 10.1083/jcb.200206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei S, Whittaker CA, Xu G, Bridges LC, Shah A, White JM, Desimone DW. Conservation and divergence of ADAM family proteins in the Xenopus genome. BMC Evol Biol. 2010;10:211. doi: 10.1186/1471-2148-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall RJ, Erickson CA. ADAM 10: an active metalloprotease expressed during avian epithelial morphogenesis. Dev Biol. 2003;256:146–159. doi: 10.1016/s0012-1606(02)00133-1. [DOI] [PubMed] [Google Scholar]
- 67.Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- 68.Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- 69.Cousin H, Abbruzzese G, Kerdavid E, Gaultier A, Alfandari D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for Calpain8-a expression and cranial neural crest cell migration. Dev Cell. 2011;20:256–263. doi: 10.1016/j.devcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, Robakis NK. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 72.Alfandari D, Cousin H, Gaultier A, Hoffstrom BG, DeSimone DW. Integrin alpha5beta1 supports the migration of Xenopus cranial neural crest on fibronectin. Dev Biol. 2003;260:449–464. doi: 10.1016/s0012-1606(03)00277-x. [DOI] [PubMed] [Google Scholar]
- 73.Burstyn-Cohen T, Kalcheim C. Association between the cell cycle and neural crest delamination through specific regulation of G1/S transition. Dev Cell. 2002;3:383–395. doi: 10.1016/s1534-5807(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 74.Kil SH, Lallier T, Bronner-Fraser M. Inhibition of cranial neural crest adhesion in vitro and migration in vivo using integrin antisense oligonucleotides. Dev Biol. 1996;179:91–101. doi: 10.1006/dbio.1996.0243. [DOI] [PubMed] [Google Scholar]
- 75.Strachan LR, Condic ML. Cranial neural crest recycle surface integrins in a substratum-dependent manner to promote rapid motility. J Cell Biol. 2004;167:545–554. doi: 10.1083/jcb.200405024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bronner-Fraser M. An antibody to a receptor for fibronectin and laminin perturbs cranial neural crest development in vivo. Dev Biol. 1986;117:528–536. doi: 10.1016/0012-1606(86)90320-9. [DOI] [PubMed] [Google Scholar]
- 77.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H, Kang K, Ekram MB, Roh TY, Kim J. Aebp2 as an epigenetic regulator for neural crest cells. PLoS One. 2011;6:e25174. doi: 10.1371/journal.pone.0025174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y, Xiao A. Epigenetic regulation in neural crest development. Birth Defects Res A Clin Mol Teratol. 2011;91:788–796. doi: 10.1002/bdra.20797. [DOI] [PubMed] [Google Scholar]
- 80.Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell. 2010;19:460–468. doi: 10.1016/j.devcel.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 82.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sela-Donenfeld D, Kalcheim C. Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development. 1999;126:4749–4762. doi: 10.1242/dev.126.21.4749. [DOI] [PubMed] [Google Scholar]
- 84.Coles EG, Taneyhill LA, Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev Biol. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akitaya T, Bronner-Fraser M. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Dev Dyn. 1992;194:12–20. doi: 10.1002/aja.1001940103. [DOI] [PubMed] [Google Scholar]
- 86.Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- 87.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]