Abstract
Systematic environmental surveillance for poliovirus circulation has been conducted in Egypt since 2000. The surveillance has revealed three independent importations of wild-type poliovirus. In addition, several vaccine-derived polioviruses have been detected in various locations in Egypt. In addition to acute flaccid paralysis (AFP) surveillance, environmental surveillance can be used to monitor the wild poliovirus and vaccine-derived poliovirus circulation in populations in support of polio eradication initiatives.
TEXT
Systematic environmental surveillance was set up in Egypt in 2000 and has since been used as a supplement to acute flaccid paralysis (AFP)-based surveillance for polioviruses (PV). The first years of the environmental surveillance revealed widespread circulation of the indigenous wild-type poliovirus 1 (WPV1) of the Northeast African (NEAF) genotype in different Egyptian communities. At the same time, the poliomyelitis cases were already rare or completely missing (6, 7). As also verified in several other countries, environmental surveillance was very sensitive in detecting silent circulation of WPV (8). The recognition of WPV1 from sewage in distinct communities in Egypt guided the oral poliovirus vaccine (OPV) campaigns to the most affected areas and, finally, led to the successful eradication of the NEAF genotype in the beginning of 2005.
The intensive environmental surveillance in Egypt includes annual processing of an average of about 450 sewage specimens collected from regularly evaluated collection sites as previously described (6, 7). At present, the surveillance encompasses 19 provinces (Fig. 1), most of which have more than one specimen collection site. Hurghada, in the Red Sea province, has been followed since 2007, and the other provinces indicated in Fig. 1 have been followed for longer.
Fig 1.
Map of Egypt indicating the provinces in which the specimens have been collected for environmental surveillance for polioviruses.
The sewage specimens are concentrated at the VACSERA Laboratory, Cairo, using a two-phase polyethylene glycol (PEG)-dextran separation method (11). The concentrates are inoculated into RD and L20B cell lines, and the virus isolates are serotyped by PV antisera according to the instructions in the Polio Laboratory Manual (16). In order to increase the sensitivity of the virus isolation, selected concentrates are analyzed in parallel at the THL Virology Unit, Helsinki, Finland (7). About 1,500 PV strains are isolated annually and assayed for wild-type or vaccine origin by using standardized antigenic and/or genetic intratypic differentiation (ITD) assays (14, 16). The antigenic ITD assay, an enzyme immunoassay (EIA), utilizes cross-absorbed polyclonal antisera to Sabin and non-Sabin-like PV strains. Genetic ITD assays include Sabin strain-specific probe hybridization (VACSERA), analysis of restriction fragment length polymorphisms (RFLPs) of reverse transcription-PCR (RT-PCR) amplicons (THL) (1), and, launched more recently, at both VACSERA and THL, a PV diagnostic real-time RT-PCR assay exploiting group-, serotype-, and Sabin-specific primers and probes (9). If any of the ITD assays suggest that the virus is non-Sabin-like, the complete VP1 capsid protein-encoding region is sequenced at THL, Helsinki, Finland, as described in reference 6. The VP1 sequences are subsequently used to classify the viruses according to the WHO-approved criteria. Phylogenetic relationships between the newly produced WPV sequences and currently circulating WPV strains are used to genotype the WPV isolate and trace the transmission chains (WHO Polio Laboratory Network and Centers for Disease Control and Prevention, Atlanta, GA). PV isolates that originate from the Sabin strains used in the OPV vaccine are classified according to differences between their VP1 sequence and those of the parental Sabin strains. One-percent nucleotide divergence or more in the VP1 sequence classifies the types 1 and 3 Sabin-derived strains as vaccine-derived PV (VDPV), and differences of 6 or more nucleotides (0.6%) classify type 2 viruses as VDPV. These amounts of mutations suggest that the replication of the OPV-derived virus has occurred long enough to give the virus wild-type transmission and pathogenic properties.
The main objective of the environmental surveillance in Egypt has been the early detection of the potential emergence of WPV transmission. Importation of WPV1 has been detected three times since the eradication of the indigenous NEAF type in 2005 (Table 1). First, WPV1 was found in Al Haram in the Giza province in September 2008. The virus belonged to the West Africa B (WEAF-B) genotype, which is still endemic in Nigeria and has reemerged in several previously polio-free African countries. WEAF-B-type WPV1 was found again in December 2010 from the Aswan province in Upper Egypt. The virus was genetically related to the WPV1 strains that caused poliomyelitis in Sudan in 2008 and 2009. A representative of the second still-existing WPV1 genotype, South Asia (SOAS), was detected in Al Hagana in the Cairo province. The positive specimen was collected on 30 December 2008. The phylogenetic analysis revealed that the strain was genetically linked to polioviruses originating in India. Fortunately, no WPV-caused poliomyelitis cases have occurred in Egypt since 2005. Wider transmission of WPV1 has also been unlikely, as each of the imported WPV1 strains was identified only once from a single sewage specimen.
Table 1.
Imported wild-type poliovirus strains detected in Egypt by environmental surveillance from 2005 to 2010
| Specimen code | Collection date (day-mo-yr) | Sampling site | WPV genotype |
|---|---|---|---|
| EGY08/E310/V1 | 29-09-2008 | Al Haram, Giza | WEAF-B |
| EGY08/E444/V4 | 30-12-2008 | Al Hagana, Cairo | SOAS |
| EGY10/E405/K71797 | 21-12-2010 | Aswan | WEAF-B |
OPV is used as the primary vaccine against poliomyelitis in Egypt. The Sabin strains of OPV replicate in the intestine of vaccine recipients for several weeks and, like all RNA viruses, undergo rapid genetic changes upon replication. In rare cases, reverted Sabin strains may cause vaccine-associated poliomyelitis. The Sabin strains with higher levels of drift regain transmission and pathogenic properties that are like those of wild PVs. Circulating VDPVs (cVDPVs) have caused poliomyelitis outbreaks in populations in which vaccine coverage is low and the herd immunity has diminished below the critical low limit (10). In Egypt, widespread circulation of VDPV-2 between 1983 and 1993 was documented retrospectively by studying PV archives stored at VACSERA. VDPV-2 caused infections and paralysis in low-OPV-coverage areas (18).
Environmental surveillance conducted in Egypt has also effectively recovered VDPVs (3, 4). Altogether, 13 VDPV strains have been isolated from sewage between the years 2004 and 2010 (Table 2). Originally, 11 of these PV strains gave aberrant restriction patterns in the ITD-RFLP assay and two reacted exceptionally (double reactive) in the ITD-EIA. The VDPV definition was made according to the VP1 sequences. All but one of the VDPVs were of serotype 2, which has globally been the most frequent VDPV type in cVDPV outbreaks (10). The three VDPV-2 strains with the highest levels of drift (VP1 nucleotide divergence from that of PV2/Sabin ranging from 1.22% to 1.66%) were isolated from the city of Damanhour in the Beheira province in the years 2005, 2007, and 2008. The strains had reciprocal VP1 nucleotide differences of up to 2.4% to 2.8%, results which were suggestive of three independent emergences of VDPVs rather than continuous circulation of the same VDPV lineage in the given population (Fig. 2). The most recent environmental VDPV strain, isolated in February 2010 in Altebeen, Helwan, close to Cairo, was exceptionally of serotype 1.
Table 2.
Vaccine-derived poliovirus strains detected in Egypt by environmental surveillance from 2004 to 2010
| Specimen code | Collection date (day-mo-yr) | Sampling site | Serotype | % nt divergence from Sabin straina |
|---|---|---|---|---|
| EGY04/E428/V2 | 23-11-2004 | Menoufia | 2 | 0.66 |
| EGY05/E136/V1 | 14-04-2005 | Damanhour, Beheira | 2 | 1.22 |
| EGY05/E483/K218 | 17-12-2005 | Aleshreen, Giza | 2 | 0.66 |
| EGY06/E223/VR1 | 14-06-2006 | Fayoum | 2 | 0.66 |
| EGY07/E124/V1 | 30-03-2007 | Al Hagana, Cairo | 2 | 0.66 |
| EGY07/E276/V2 | 21-07-2007 | Aswan | 2 | 0.78 |
| EGY07/E277/VR2 | 23-07-2007 | Hurghada, Red Sea | 2 | 0.78 |
| EGY07/E310/V2 | 21-08-2007 | Aswan | 2 | 0.66 |
| EGY07/E432/K44466 | 06-12-2007 | Damanhour, Beheira | 2 | 1.55 |
| EGY08/E110/V1 | 07-04-2008 | Damanhour, Beheira | 2 | 1.66 |
| EGY08/E422/K626401 | 16-12-2008 | Alsaf, Cairo | 2 | 0.66 |
| EGY09/E360/V3 | 15-09-2009 | Al Hagana, Cairo | 2 | 0.66 |
| EGY10/E42/V1 | 07-02-2010 | Helwan, Altebeen, Cairo | 1 | 1.10 |
nt, nucleotide.
Fig 2.
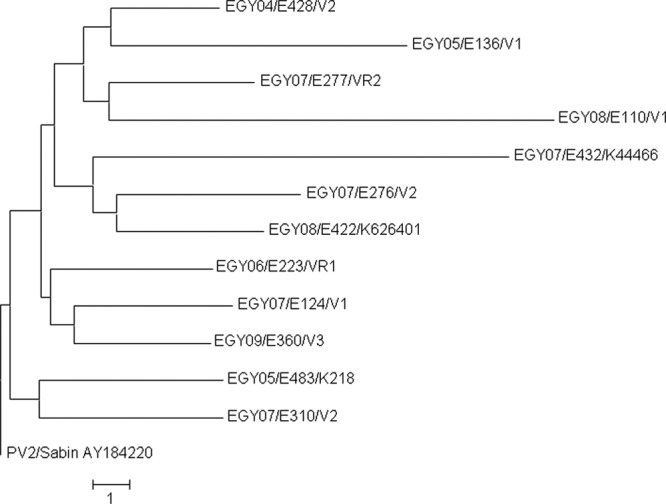
The evolutionary history of the VP1 nucleotide sequences of type 2 vaccine-derived polioviruses inferred by using the neighbor-joining method in MEGA5 (13). The distances are based on the numbers of base differences per sequence. The sequence of PV2/Sabin is used as a reference.
Our results highlight the importance of continuous environmental surveillance as a support to the clinically based AFP surveillance. Presently, while WPV transmission is globally restricted but not yet interrupted, there exists still a continuous possibility of spreading of WPV to polio-free countries. It is well documented that imported WPV1 frequently causes poliomyelitis outbreaks in susceptible populations (2, 5). The surveillance for emerging VDPV circulation is equally important. The risk for cVDPV transmission and outbreaks is increased in populations with locally lowered vaccine coverage (17). In both cases, the early detection of circulation is essential for the prevention and limitation of the possible poliomyelitis outbreaks. More widespread and regular use of environmental surveillance in risk populations is desirable for the rapid detection of WPV importations and the emergence of cVDPV outbreaks. The environmental surveillance has provided vital information in outbreaks in Finland in 1984 (11), in Israel in 1988 (12), and in The Netherlands in 1992 (15) about the extent of circulation of WPV in the community and has complemented the reported clinical cases. Targeted environmental surveillance can help to identify the reservoir communities and high-risk areas, as it helped in Pakistan to isolate WPVs (especially WPV3) in a Pashtun community in a high-risk area in Karachi, corresponding very closely with virus isolation in clinical cases (H. Asghar, personal communication).
In addition to AFP surveillance, environmental surveillance can be used to monitor the WPV and VDPV circulation in populations to support polio eradication initiatives (6). Knowing the proven value of environmental surveillance, the World Health Organization has included environmental surveillance in its Global Polio Emergency Action plan for 2012 to 2013 to be used extensively where gaps persist in surveillance performance, supplementing AFP surveillance (16a).
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences determined in this study are JQ613982 to JQ613997.
ACKNOWLEDGMENTS
This work was supported by World Health Organization Technical Services Agreements with the Holding Company for Biological and Vaccine Production (VACSERA), Cairo, Egypt, and with the National Institute for Health and Welfare (THL), Helsinki, Finland.
We are grateful to Päivi Hirttiö, Alena Kaijalainen, Marja-Liisa Ollonen, Eija Penttilä, and Johanna Rintamäki at THL and Tamer Hassan and Dina Maher at VACSERA laboratories for excellent technical assistance.
Footnotes
Published ahead of print 11 May 2012
REFERENCES
- 1. Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology 184:645–654 [DOI] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention (CDC) 2006. Resurgence of wild poliovirus type 1 transmission and consequences of importation—21 countries, 2002–2005. MMWR Morb. Mortal. Wkly. Rep. 55:145–150 [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention (CDC) 2009. Update on vaccine-derived polioviruses—worldwide, January 2008–June 2009. MMWR Morb. Mortal. Wkly. Rep. 58:1002–1006 [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention (CDC) 2011. Update on vaccine-derived polioviruses—worldwide, July 2009–March 2011. MMWR Morb. Mortal. Wkly. Rep. 60:846–850 [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention (CDC) 2009. Wild poliovirus type 1 and type 3 importations—15 countries, Africa, 2008-2009. MMWR Morb. Mortal. Wkly. Rep. 58:357–362 [PubMed] [Google Scholar]
- 6. El Bassioni L, et al. 2003. Prolonged detection of indigenous wild polioviruses in sewage from communities in Egypt. Am. J. Epidemiol. 158:807–815 [DOI] [PubMed] [Google Scholar]
- 7. Hovi T, et al. 2005. Environmental surveillance of wild poliovirus circulation in Egypt—balancing between detection sensitivity and workload. J. Virol. Methods 126:127–134 [DOI] [PubMed] [Google Scholar]
- 8. Hovi T, et al. 2012. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 140:1–13 [DOI] [PubMed] [Google Scholar]
- 9. Kilpatrick DR, et al. 2009. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J. Clin. Microbiol. 47:1939–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nathanson N, Kew OM. 2010. From emergence to eradication: the epidemiology of poliomyelitis deconstructed. Am. J. Epidemiol. 172:1213–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poyry T, Stenvik M, Hovi T. 1988. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 54:371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shulman LM, et al. 2000. Resolution of the pathways of poliovirus type 1 transmission during an outbreak. J. Clin. Microbiol. 38:945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Avoort HG, et al. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van der Avoort HG, Reimerink JH, Ras A, Mulders MN, van Loon AM. 1995. Isolation of epidemic poliovirus from sewage during the 1992-3 type 3 outbreak in The Netherlands. Epidemiol. Infect. 114:481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. WHO 2004. Polio laboratory manual. World Health Organization, Geneva, Switzerland [Google Scholar]
- 16a. WHO 2012. Global polio emergency action plan 2012–2013. World Health Organization, Geneva, Switzerland: http://www.polioeradication.org/Portals/0/Document/Resources/StrategyWork/EAP_201205.pdf [Google Scholar]
- 17. Wringe A, Fine PE, Sutter RW, Kew OM. 2008. Estimating the extent of vaccine-derived poliovirus infection. PLoS One 3:e3433 doi:10.1371/journal.pone.0003433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang CF, et al. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366–8377 [DOI] [PMC free article] [PubMed] [Google Scholar]