Background: Nucleotide binding and oligomerization domain-containing protein 2 (NOD2) is a protein involved in the recognition of bacterial pathogens through detection of muramyl dipeptide.
Results: Purified recombinant NOD2 was found to bind ATP and muramyl dipeptide.
Conclusion: NOD2 is an intracellular signaling receptor for muramyl dipeptide.
Significance: These results help to define the molecular events involved in NOD2 signaling.
Keywords: ATP, Cellular Immune Response, Inflammation, Innate Immunity, Nod-like Receptors (NLR), Pathogen-associated Molecular Pattern (PAMP), Signal Transduction, Nucleotide Binding and Oligomerization Domain-containing Protein 2, Muramyl Dipeptide, Adenosine Triphosphate
Abstract
Nucleotide binding and oligomerization domain-containing protein 2 (NOD2/Card15) is an intracellular protein that is involved in the recognition of bacterial cell wall-derived muramyl dipeptide. Mutations in the gene encoding NOD2 are associated with inherited inflammatory disorders, including Crohn disease and Blau syndrome. NOD2 is a member of the nucleotide-binding domain and leucine-rich repeat-containing protein gene (NLR) family. Nucleotide binding is thought to play a critical role in signaling by NLR family members. However, the molecular mechanisms underlying signal transduction by these proteins remain largely unknown. Mutations in the nucleotide-binding domain of NOD2 have been shown to alter its signal transduction properties in response to muramyl dipeptide in cellular assays. Using purified recombinant protein, we now demonstrate that NOD2 binds and hydrolyzes ATP. Additionally, we have found that the purified recombinant protein is able to bind directly to muramyl dipeptide and can associate with known NOD2-interacting proteins in vitro. Binding of NOD2 to muramyl dipeptide and homo-oligomerization of NOD2 are enhanced by ATP binding, suggesting a model of the molecular mechanism for signal transduction that involves binding of nucleotide followed by binding of muramyl dipeptide and oligomerization of NOD2 into a signaling complex. These findings set the stage for further studies into the molecular mechanisms that underlie detection of muramyl dipeptide and assembly of NOD2-containing signaling complexes.
Introduction
Nucleotide-binding domain and leucine-rich region-containing proteins (NLRs)4 are a family of intracellular proteins involved in innate immune signaling (1–3). These proteins share structural similarities to a subfamily of proteins encoded by plant disease resistance genes (R genes) and the mammalian apoptotic protease-activating factor-1 (APAF-1). These proteins all contain a central, presumed nucleotide-binding domain with homology to the AAA-ATPase superfamily, which is believed to regulate the signaling activity of these proteins. Additionally, members of this family of proteins contain a variable number of C-terminal leucine-rich repeats and an N-terminal effector domain (usually a pyrin domain or caspase activation and recruitment domain (CARD)), which are thought to mediate additional protein-protein interactions. Mutations in several NLR protein-encoding genes are associated with human immunologic disorders (4). For example, mutations in nucleotide oligomerization domain-containing protein 2 (NOD2), a member of the NLR family, are strongly associated with inherited inflammatory diseases, particularly Blau syndrome and Crohn disease (5–9).
NOD2 is involved in detection of muramyl dipeptide (MDP), a component of bacterial cell walls. Cytoplasmic exposure to muramyl dipeptide results in NOD2-dependent activation of NF-κB (10, 11). This signaling event is mediated by the association of NOD2 and the kinase, RIP2K (12, 13). In addition to activation of the transcription factor NF-κB, it has recently been recognized that stimulation of NOD2 by intracytoplasmic bacterial pathogens induces formation of autophagosomes through interactions between NOD2 and the autophagy protein ATG16L1 (14–16). Mutations in NOD2 associated with Blau syndrome appear to cause constitutive, dysregulated NF-κB activation (17). Mutations associated with Crohn disease result in reduced NF-κB activity and are less responsive to muramyl dipeptide (10, 11, 18). Crohn disease-associated mutant NOD2 proteins also fail to initiate MDP-induced autophagy (14–16). The molecular mechanisms by which MDP exposure controls NOD2-mediated signaling and by which missense mutations lead to dysregulated signaling in human diseases have not been discerned.
Analysis of NOD2 function in cells after transfection with mutant cDNA-expressing constructs suggests that nucleotide binding and hydrolysis are important in the activation and deactivation of signaling by NOD2 (19, 20). Molecular modeling based on homology of NOD2 with the NLR related protein APAF-1 indicates that mutations predicted to disrupt ATP binding prevent activation of NOD2 signaling (21, 22). Further, mutations that modeling predicts may be involved in ATP hydrolysis result in constitutive activation of NOD2 signaling, suggesting that ATP hydrolysis may drive inactivation of NOD2 (20). A similar mechanism of activation has been noted in a plant R protein involved in pathogen resistance, tomato I-2, which has a domain structure similar to that of mammalian NLR proteins. Mutations that abolish biochemical ATP binding do not signal in vivo, and mutations that allow ATP binding but abolish ATP hydrolysis signal constitutively (23, 24). Although these published studies strongly support a role for nucleotide binding by NOD2 in controlling signal transduction, formal evidence for NOD2 nucleotide binding has remained elusive. Six NLR proteins have been shown to bind nucleotides. Full-length NLRP3, full-length NOD1, and isolated recombinant nucleotide-binding domains from NLRP12 and NLRC4 all bind to ATP (25–28). The MHC class II transcriptional activator (CIITA), which was the first described member of the NLR family, binds to GTP (29). Interestingly, NLRP1 has been shown to bind all nucleotide triphosphates with similar affinities (30). Additionally, NLRP1 has been shown to assemble into an inflammasome, a macromolecular complex containing NLRP1 and procaspase-1, in vitro in response to MDP and triphosphate nucleotide (30). This in vitro inflammasome formation by NLRP1 bears remarkable semblance to the assembly of the caspase-3-activating apoptosome structure by APAF-1, which requires the APAF-1 ligands cytochrome c and ATP (31). NLRP3 and NLRC4 are both thought to form similar inflammasome structures in a manner dependent on ATP binding by the NLR protein. GTP binding by CIITA is required for nuclear translocation and localization to the MHC class II promoter but not for assembly with the transcriptional activation factors required for CIITA-induced transcription (32).
We now show the successful isolation of recombinant NOD2 from eukaryotic cells. This full-length protein binds and hydrolyzes ATP. The purified recombinant protein can also bind to a biotinylated muramyl dipeptide, suggesting that NOD2 is indeed a MDP receptor. Recombinant NOD2 also homo-oligomerizes and hetero-oligomerizes with RIP2K. Finally, the homo-oligomerization, hetero-oligomerization, and MDP binding are enhanced by ATP binding. These data further illuminate the NOD2 signaling pathway and provide a path for further studies into the molecular dysfunction of NOD2 in human disease processes.
EXPERIMENTAL PROCEDURES
Construction of NOD2 Expression Plasmids
Standard techniques of DNA manipulation were utilized (33). The NOD2-encoding cDNA was amplified with high fidelity Pfx DNA polymerase (Invitrogen) with primers that generated flanking PmeI and SgfI sites (for primer sequences, see supplemental Table 1). The resulting DNA fragment was digested by PmeI and SgfI and ligated with pFN21A vector (Promega), creating a gene encoding a NOD2 fusion protein with an N-terminal HaloTag. We designated this plasmid as pFN21A-NOD2. The following primers were used to amplify the HaloTag-NOD2 DNA fragment by PCR: 5′-TCGAATCTAGAATGGCAGAAATCGGTACTGG-3′ and 5′-AAGCAAGAGTCTGGTGTCCCT-3′. HaloTag-NOD2-His6 DNA fragment was then ligated with pFastBac/CT-TOPO vector (Invitrogen), which generated a C-terminal hexahistidine tag. DNA sequencing was performed to confirm that there was not mutation in the Halo-NOD2-His coding sequence. pFastBac-Halo-NOD2 plasmid was used to transform DH10Bac competent cell to generate recombinant bacmid virus DNA. The NOD2-encoding cDNA and truncations of the NOD2-encoding cDNA were inserted in the mammalian expression vector, pcDNATM3.1/V5-His TOPO® (Invitrogen) or pFN21A (Promega) for HaloTag fusion proteins, using the manufacturers' recommendations. As described above, DNA fragments were amplified with high fidelity Pfx DNA polymerase (Invitrogen), and all plasmids and cloning intermediates were confirmed using both restriction digest and sequencing. The primer sequences used to generate these mammalian expression plasmids are noted in supplemental Table 1.
Baculovirus and Insect Cell Culture
Recombinant baculovirus expressing Halo-NOD2-His6 was generated using the Bac to Bac system (Invitrogen). Insect cell culture and baculovirus infection were performed according to the manufacturer's protocols.
Purification of Recombinant NOD2, NOD2 CARDs, and GST
Sf9 cells infected with Halo-NOD2-His6-expressing baculovirus were collected by centrifugation and washed with phosphate-buffered saline (PBS). The cell pellet was then resuspended in binding buffer (50 mm sodium phosphate, 300 mm NaCl, pH 8.0, supplemented with 0.1% CHAPS and CompleteTM proteinase inhibitor mixture (Roche Applied Science)), and homogenized using one pass through a French press. The solubilized lysate was prepared by centrifugation at 15,000 × g for 30 min and applied to an immobilized nickel column (Sepharose 6 Fast Flow, GE Healthcare). The column was washed with 5 column volumes of binding buffer, and the recombinant protein was eluted with binding buffer supplemented with 500 mm imidazole. The eluted recombinant protein was applied to HaloLink (Promega), washed, and eluted with His6-tagged tobacco etch virus (TEV) protease. The proteins eluted from HaloLink resin were subsequently applied to immobilized nickel resin to remove His6-tagged TEV protease, and the flow-through was collected.
NOD2 CARDs (residues Met28–Arg227) were inserted using a Gateway cloning system into the vector pDest-HisMBP. The recombinant protein was expressed in Rosetta 2 Escherichia coli cells as an N-terminal His-MBP and C-terminal FLAG fusion. The protein was purified via nickel-NTA affinity chromatography, followed by removal of the N-terminal fusion tags with TEV protease. The protein was passed over a 1-ml HisTrap column (GE Healthcare) to remove the cleaved tag, residual full-length protein, and His-tagged TEV protease.
GST was expressed in BL21 E. coli cells from the plasmid pGEX-4T1(GE Healthcare). The protein was purified using glutathione-Sepharose 4B (GE Healthcare) as per the manufacturer's instructions.
ATPγS Binding and ATP Hydrolysis Assays
Nucleotide binding was assayed by a nitrocellulose filter binding assay (34). Nucleotide hydrolysis assays were carried out utilizing a modified acidified charcoal precipitation (35).
Cell Culture and Transfection
THP-1 and HEK293 (ATCC) were cultured under the recommended conditions. Fugene 6 (Promega) was used for transfection of HEK293 cells according to the manufacturer's protocol.
Immunoprecipitation and Immunoblot Analysis
Immunoprecipitations were carried out in 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm MgCl2, and 0.5% Nonidet P-40 supplemented with CompleteTM protease inhibitor mixture (Roche Applied Science) and 1 mm ATP. Lysates from 5 × 107 cells were incubated for 30 min at 30 °C prior to immunoprecipitation with agarose-conjugated anti-FLAG (M2) (Sigma-Aldrich) followed by SDS-PAGE. Anti-V5 antibody was from Invitrogen. Anti-hexahistidine antibody (GenScript, Piscataway, NJ) and anti-NOD2 (NOD2-15) were from Biolegend (San Diego, CA). Electrophoresis was carried out using the NuPAGE gel system (Invitrogen), and separated proteins were transferred to nitrocellulose using the iBlot dry blotting system (Invitrogen). Immunoblots were imaged with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and SuperSignal West chemiluminescent substrate products (Pierce) using the FluorChem E CCD-based imaging system (Protein Simple).
Biotinylated MDP Binding Assay
Purified NOD2 was incubated with biotinylated MDP at 24 °C for 1 h in 50 mm Tris-HCl (pH 7.0), 150 mm NaCl, and 0.1% CHAPS in the absence or presence of 10 μm ATP or ATPγS. 100 μl of Streptavidin microbeads (Miltenyi Biotec, Auburn, CA) was added to the reaction mix after a 1-h incubation. To assess MDP binding by NOD2 and truncated NOD2 proteins in cellular extracts, HEK293T cells were plated in 12-well culture plates and transfected with vectors expressing the indicated V5-tagged NOD2 proteins. After 24 h, cells were washed with PBS and lysed with radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Igepal Co-630, 0.5% deoxycholate, and 0.1% SDS) containing proteinase inhibitors. Lysates were centrifuged to remove cell debris. Biotinylated MDP was added to lysates and rotated at 4 °C overnight. After 1-h or overnight incubation, 100 μl of Streptavidin microbeads was mixed with the proteins. MDP-bound protein was isolated by magnetic isolation using μ columns (Miltenyi Biotec, Auburn, CA) according to the manufacturer's protocol. The eluate was subjected to SDS-PAGE followed by immunoblot blot analysis using anti-NOD2 antibody or anti-V5 antibody, as indicated.
Surface Plasmon Resonance
Surface plasmon resonance experiments were performed on a Biacore T100 at 25 °C. Full-length NOD2 was diluted 10-fold in pH 4.0 acetate buffer and immobilized onto the surface of a CM5 sensor chip to a level of ∼3,500 response units. Recombinant NOD2 CARDs and GST proteins were buffer-exchanged into 10 mm sodium phosphate, pH 6.0, 150 mm NaCl, 50 μm EDTA, 0.01% Tween 20 and passed over the chip surface at a flow rate of 15 μl/min and concentrations of 0, 6.25, 12.5, 25, 50, and 100 μm. Background subtraction from a protein-free reference surface was performed, and the data were analyzed using BiaEvaluation software and GraphPad Prism 5.0.
RESULTS
Recombinant NOD2 Can Be Purified to Homogeneity by Sequential Affinity Chromatography
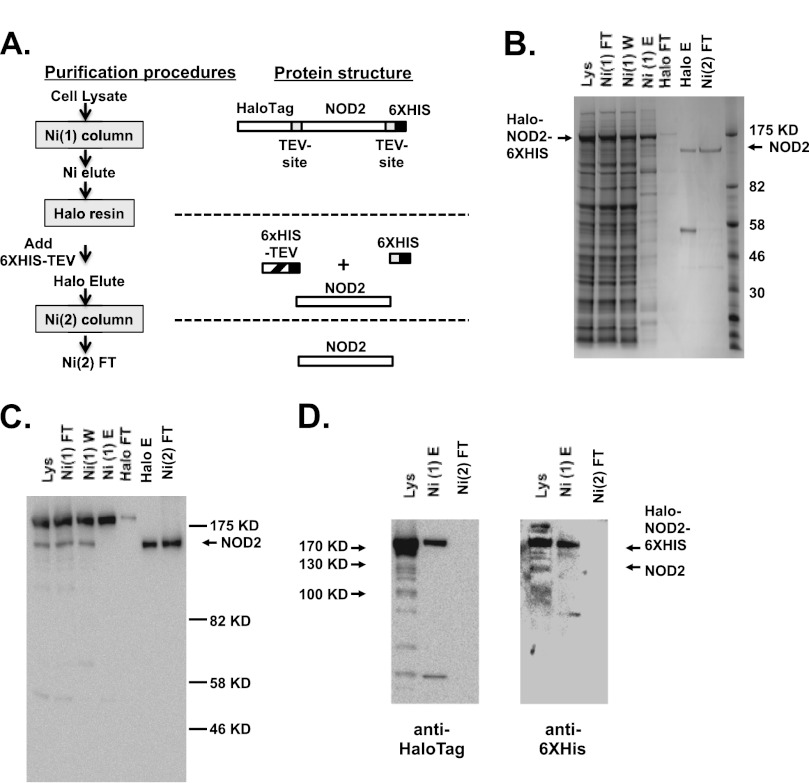
Biochemical characterization of the NLR protein functions remains largely unstudied more than 10 years after the discovery of these proteins. Generation of functional purified protein from recombinant sources has been a major challenge in this field. Portions of both NLRP12 and NLRC4 containing the central nucleotide-binding domain have been expressed, purified, and characterized (27, 28). To date, only NLRP1, NOD1, and NLRP3 have been characterized as full-length proteins (25, 26, 30). We have previously expressed and purified a dually tagged recombinant NLRP3 with an N-terminal hexahistidine tag and a C-terminal FLAGTM tag expressed using recombinant baculovirus in insect cells (26). This strategy did not yield purified recombinant NOD2 in a quantify sufficient for biochemical analysis (data not shown). We evaluated numerous additional expression systems and fusion protein partners in an attempt to isolate purified NOD2. Ultimately, we generated a recombinant baculovirus expressing an N-terminal HaloTagTM and C-terminal hexahistidine-tagged form of NOD2 in which both tags could be removed using recombinant TEV protease (Fig. 1A). The majority of NOD2 expressed in Sf9 cells infected with this recombinant baculovirus can be solubilized in CHAPS detergent (Fig. 1B). However, most of this solubilized protein fails to bind to immobilized nickel. Immunoblot analysis with antibodies directed to the hexahistidine tag indicates that the NOD2 protein that fails to bind nickel-containing chromatography resins does indeed contain the hexahistidine tag. This is probably the result of the protein folding (or misfolding) in a way that prevents access of this tag to the resin. The recombinant NOD2 protein that can be eluted from immobilized nickel can be purified to near homogeneity using HaloTag-ligand chromatography, removal of the tags using hexahistidine-tagged TEV protease, and subsequent reapplication to immobilized nickel (Fig. 1C). The final protein is immunoreactive with a monoclonal antibody directed against NOD2 and is not reactive with antibodies against the HaloTagTM or hexahistidine tag (Fig. 1D). This procedure yields between 100 and 300 μg of purified NOD2 for each liter of Sf9 culture.
FIGURE 1.
Recombinant NOD2 expressed in insect cell culture can be purified to near homogeneity. A, the purification strategy for recombinant Halo-NOD2-His6 protein is detailed with the linear structure of the protein at each step provided to the right of the step. B, recombinant NOD2 was purified from Sf9 insect cells infected with Halo-NOD2-His6-expressing baculovirus, as indicated under “Experimental Procedures” (and outlined in A). Proteins from the indicated stages of purification were separated by SDS-PAGE electrophoresis and visualized using staining with Coomassie Blue. Lanes 1–3, 20 μg of protein; lane 4, 5 μg of protein; lane 5, 1 μg of protein; lanes 6 and 7, 700 ng of protein. C, proteins from the Halo-NOD2-His6 purification strategy were separated by SDS-PAGE as in B, the proteins were transferred to nitrocellulose, and NOD2 was visualized using immunoblot with antibodies directed against NOD2. D, proteins from the lysate (Lys; 5 μg), first nickel column elution (Ni(1)E; 1.2 μg), and second nickel column flow-through (Ni(2)FT; 100 ng) were separated using SDS-PAGE, transferred to nitrocellulose, and detected using antibodies directed against HaloTag (left) and His6 tag (right).
NOD2 Binds Non-hydrolyzable ATP Analogs and Exhibits ATPase Activity
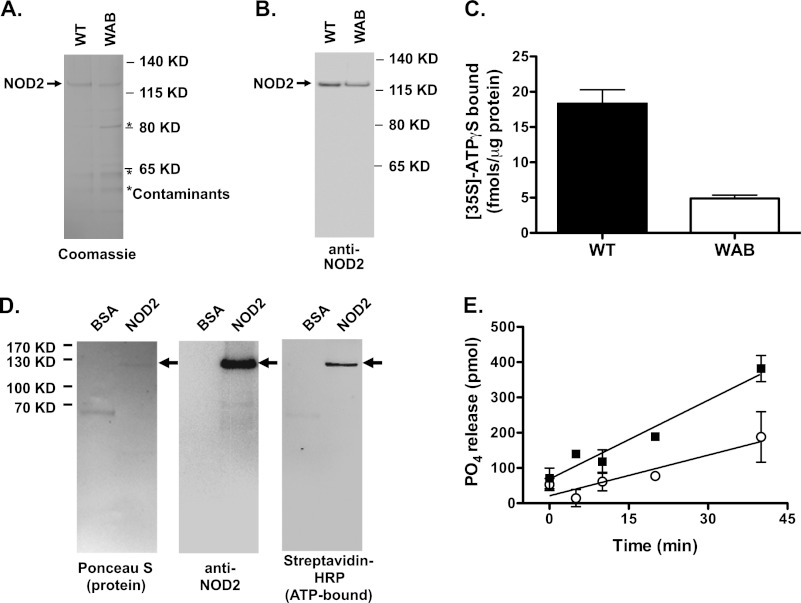
Purified NOD2 protein exhibited binding activity toward ATPγS, a non-hydrolyzable ATP analog. NLR proteins contain conserved putative nucleotide-binding domains that fall into the structural superfamily known as AAA-ATPases (2). The Walker A motif, which contains the conserved sequence, GK(S/T), is involved in coordinating the γ-phosphate of bound ATP in many proteins with this motif (36). The Walker B motif, which contains the sequence ψψψD, is predicted to be a critical element for nucleotide binding and γ-phosphate hydrolysis. A mutant NOD2-encoding cDNA was constructed in which the Walker A motif amino acids Gly304, Lys305, and Ser306 (GKS → AAA) and Walker B motif amino acids Thr377, Phe378, and Asp379 (TFD → AAA), were mutated to alanine. This mutant NOD2 designated (NOD2 WAB) protein was purified to near homogeneity, as described for the wild type protein, but contained an additional protein contaminant (Fig. 2, A and B). WT and WAB mutant NOD2 were assayed for ATPγS binding; the mutant NOD2 had diminished binding compared with WT protein (Fig. 2C). Because contaminating proteins were more abundant in the WAB mutant NOD2 protein preparation than in the WT NOD2 protein preparation, these data indicate that the ATP binding in the preparation is attributable to the recombinant NOD2 and not other contaminants in the preparations. To further demonstrate that NOD2 was responsible for ATPγS binding activity in the protein preparations, the preparations were incubated with a nonhydrolyzable, biotin-labeled, photoactivatable ATP analog (8-azido-ATPγ-biotinpentylamine) and exposed to UV radiation. Analysis of ATP analog-linked proteins using SDS-PAGE and detection of biotin with streptavidin HRP indicated that virtually all of the detectable ATP-bound protein appears to be NOD2 (Fig. 2D). Purified NOD2 also exhibited ATPase activity, as shown by the release of [32P]PO4 from [γ-32P]ATP incubated with purified NOD2 (Fig. 2E). Partially purified NOD2 WAB had diminished ATPase activity when compared with the wild type protein (Fig. 2E). The NOD2 WAB protein was found to label with 8-azido-ATPγ-biotinpentylamine, although with less intensity than WT NOD2 (supplemental Fig. S1A). Consistent with markedly diminished, but not completely extinguished, ATP binding and hydrolysis in vitro, cells transfected with cDNA encoding the NOD2 WAB mutant had minimally increased NF-κB-dependent reporter gene activity that was significantly less than that observed in cells transfected with cDNA encoding WT NOD2 and was not responsive to MDP stimulation (supplemental Fig. S1, B–D). Hence, the NOD2 WAB mutant protein retains some residual ATPγS binding activity, ATPase activity, and biological activity. Thus highly purified, recombinant NOD2 has both ATP binding and hydrolysis activities that depend on intact Walker A and Walker B motifs for full activity.
FIGURE 2.
Recombinant NOD2 binds non-hydrolyzable ATP analogs. A and B, wild-type and WAB (Walker A/Walker B mutant) NOD2 were purified as described in the legend to Fig. 1. The proteins (2 μg/lane) were run on SDS-PAGE as indicated and visualized using Coomassie Blue staining (A) and immunoblot with anti-NOD2 antibodies (B). The position of NOD2 is indicated with an arrow. The position of a contaminating protein that co-isolated with the WAB mutant of NOD2 is indicated by an asterisk. C, ATPγS binding activity of wild type and WAB mutant NOD2 was assessed. 700 ng of each protein was incubated with 1 μm [35S]ATPγS (21,000 cpm/pmol) at 30 °C for 1 h and captured on a nitrocellulose filter, as described under “Experimental Procedures.” The protein-bound nucleotide was measured by scintillation counting of nitrocellulose-bound proteins. D, purified wild type NOD2 and albumin (as a negative control) were incubated with 8-azido-ATPγ-biotinpentylamine for 1 h, and covalent linkage to nucleotide analog-associated proteins was induced by UV illumination. The proteins were separated using SDS-PAGE and transferred to nitrocellulose. The proteins were visualized using Ponceau S (left). NOD2 was identified using immunoblot with NOD2-directed antibodies (middle). The analog-bound proteins were visualized using streptavidin HRP with luminescent substrate (right). E, wild type (■) and WAB mutant (○) NOD2 (1 μg) were incubated with 1 μm [γ-32P]ATP (2000 cpm/pmol). At the indicated time points, the reaction was stopped by the addition acidified charcoal, and released [32P]orthophosphate was measured as described under “Experimental Procedures.” Data points represent the mean, and error bars represent the S.D. of triplicate measurements. The plots and images are representative of at least three experiments.
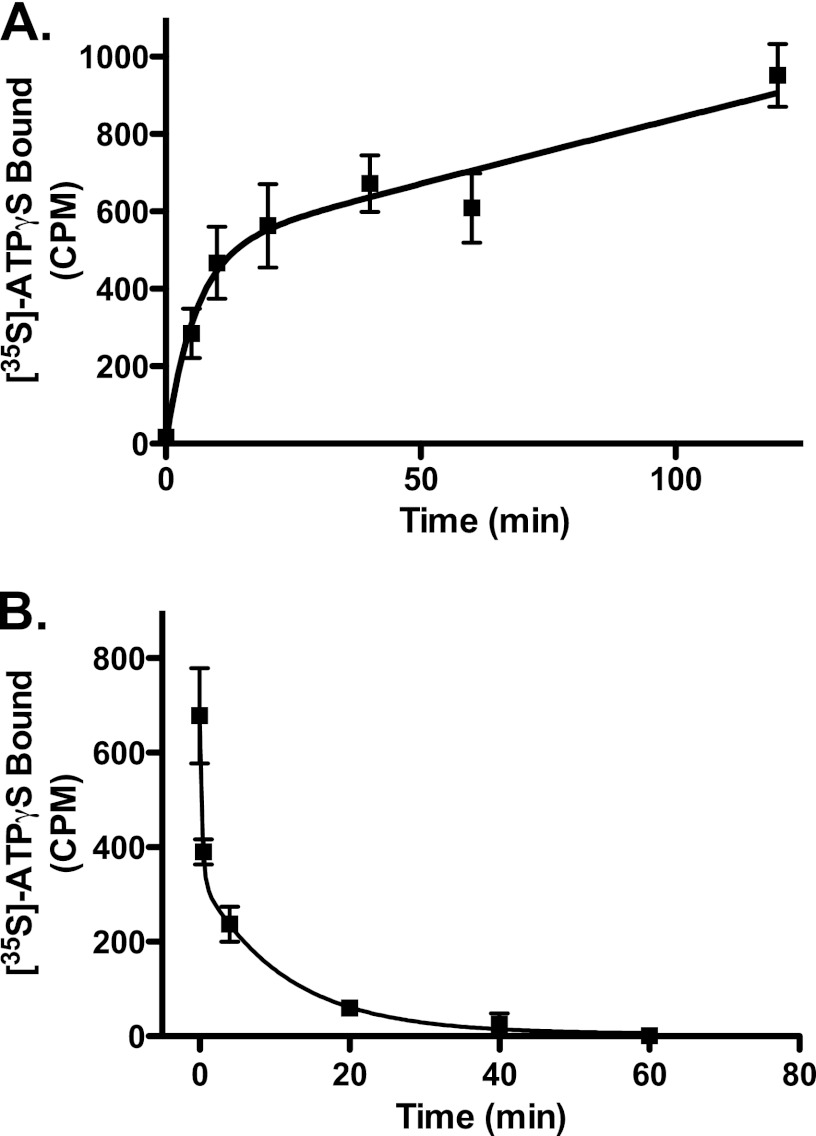
The rates of ATPγS binding and dissociation were also measured in preparations of wild type NOD2. The association of ATPγS was measured over time and demonstrated a rapid phase with a T½ of ∼4 min and a second prolonged phase. The second phase did not reach saturation in a measurable time frame (Fig. 3A). The rate of dissociation of ATPγS was also measured by loading the protein with [35S]ATPγS and then incubating in a 200-fold excess of unlabeled ATPγS and measuring the decay of bound radiolabel. The disassociation rate of nucleotide has not been documented for other NLR proteins. As with ATPγS association, nucleotide disassociation appeared to occur in two phases, one with a T½ of <1 min and the second with a T½ of ∼8 min (Fig. 3B). Approximately half of the bound counts disassociated in the first phase and half in the second. Both disassociation rates were surprisingly rapid when compared with guanine nucleotide binding signaling proteins, which bind almost irreversibly to non-hydrolyzable GTP analogs.
FIGURE 3.
Recombinant NOD2 associates and disassociates with ATPγS with rapid kinetics. A, recombinant NOD2 was incubated with 1 μm [35S]ATPγS (7700 cpm/pmol) at 30 °C, aliquots of the reaction were removed at the indicated times, and [35S]ATPγS binding by NOD2 was measured by filtration over nitrocellulose as described under “Experimental Procedures.” B, NOD2 protein was incubated with 5 nm [35S]ATPγS (17,000 cpm/pmol) for 1 h at 30 °C. A 1000-fold excess of unlabeled ATPγS (5 μm) was added to the binding reaction, aliquots were withdrawn at the indicated times, and remaining NOD2-bound [35S]ATPγS was determined as described under “Experimental Procedures.” Error bars, S.D. of ATPγs binding assays measured in triplicate. Curves represent best fitting to two-phase binding (y = Ymax1(1 − e−k1x) + Ymax2(1 − e−k2x)) or two-phase disassociation (y = Span1(e−k1x) + Span2(e−k2x)). A representative experiment of four is shown.
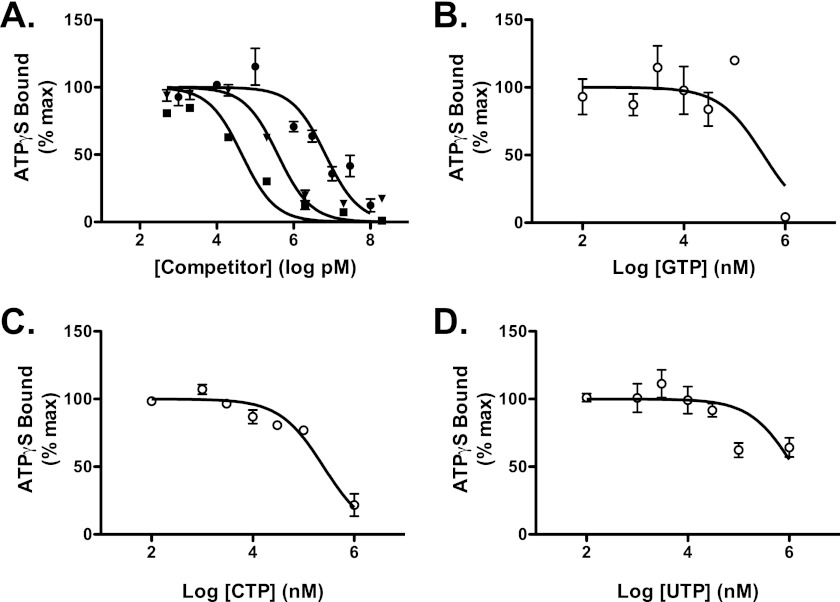
A competition assay for NOD2 ATPγS binding activity was used to assess the nucleotide binding specificity of NOD2. Unlabeled ATP and dATP competed for [35S]ATPγS binding by NOD2 with an IC50 of 57 and 277 nm, respectively. The IC50 for ADP competition with ATPγS was 2.8 μm (Fig. 4A). These data show NOD2 has a strong binding preference for triphosphate containing adenine nucleotides. Competition with non-adenine nucleotides did not fit one-site competition models well; however, estimates of the IC50 were generated using this model. The calculated IC50 for UTP, CTP, or GTP ranged from 47 μm to 24 mm, indicating at least a 1000-fold higher affinity for adenine-containing triphosphate nucleotides (Fig. 4B). Combined, these studies conclusively demonstrate that NOD2 is an adenine nucleotide-binding protein.
FIGURE 4.
NOD2 demonstrates a binding preference for ATP over other nucleotides. A, NOD2 was incubated with 5 nm [35S]ATPγS (17,000 cpm/pmol) and the indicated concentration of unlabeled competitor nucleotides (■, ATP; ▾, dATP; ●, ADP) at 30 °C for 30 min. B–D, NOD2 was incubated with 5 nm [35S]ATPγS (17,000 cpm/pmol) and the indicated concentrations of unlabeled competitor nucleotides: GTP (B), CTP (C), or UTP (D), at 30 °C for 30 min. Bound ATPγS was then assessed as described in Fig. 2. Data points show mean and S.D. (error bars) of triplicate or quadruplicate measurements. Plots are representative of at least three experiments. Curves represent best nonlinear regression fit to a single-site competition model (y = 100/(1 + 10(x−log IC50))).
Purified Recombinant NOD2 Binds to MDP
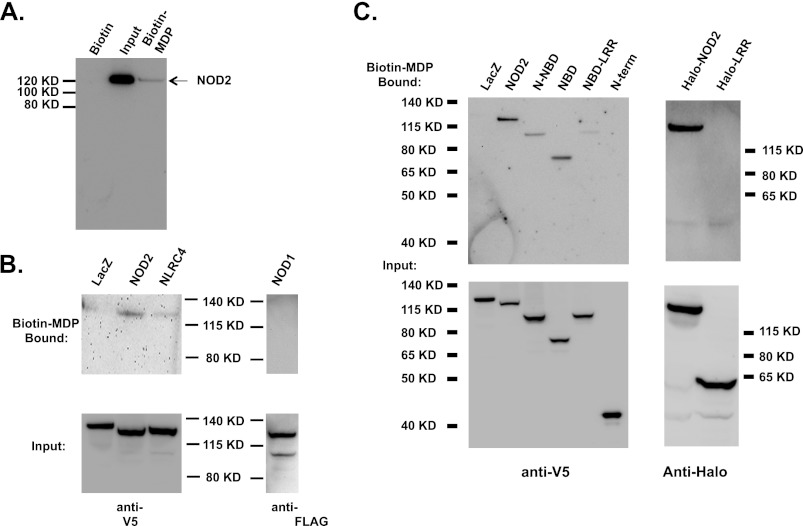
Although NOD2 signaling is clearly activated by MDP in vivo and NOD2 is commonly referred to as a “receptor” for this bacterial PAMP, binding of this or other bacterial ligands to NOD2 has not been demonstrated. We sought to determine whether NOD2 was able to directly associate with MDP. Purified NOD2 was incubated with biotinylated MDP or biotin (as a negative control). The biotin-associated material was captured using streptavidin-carrying paramagnetic beads, and the associated proteins were analyzed by immunoblot. Purified NOD2 was found to associate with biotinylated MDP but not biotin alone (Fig. 5A). In this assay, only a small portion (∼3%) of the purified NOD2 was recovered from the streptavidin beads, which may result form poor affinity of NOD2 for the biotinylated MDP or steric hindrance limiting simultaneous binding of NOD2 and streptavidin to biotinylated MDP. Further experiments using isotope-labeled MDP would allow us to further address this, but these reagents are not commercially available at this time. The binding of MDP by NOD2 was also confirmed using lysates from cells transfected with cDNA encoding NOD (Fig. 5B). Lysates prepared from the transfected cells were exposed to biotinylated MDP. Subsequently, the MDP-associated proteins were isolated using streptavidin paramagnetic isolation. NOD2 was found to associate with biotinylated MDP, whereas other control proteins expressed after transfection, β-galactosidase, NLRC4, and NOD1, did not (Fig. 5B). To determine the domain(s) of NOD2 that is required for MDP binding, a series of NOD2 deletion mutants were expressed in HEK293 cells (Fig. 5C). Interestingly, all NOD2-derived proteins containing the central NBD were found to associate with MDP (Fig. 5C). Because the leucine-rich repeat-containing C terminus of NOD2 had previously been implicated in MDP-responsive signaling, we sought to further test whether this domain might also associate with MDP (37). Because the LRR domain failed to express as an isolated V5-tagged fusion protein at reasonable levels, full-length NOD2 and the LRR domain of NOD2 were expressed as HaloTag fusion proteins in transfected HEK293 cells. Full-length NOD2 fused to HaloTag associated with biotinylated MDP, as had been observed with other recombinant full-length NOD2 proteins. However, the HaloTag-NOD2 LRR fusion protein did not associate with MDP (Fig. 5C). Combined, these data suggest that NOD2 is a bona fide intracellular MDP receptor and that the nucleotide-binding domain containing amino acids 216–821 is sufficient to associate with MDP.
FIGURE 5.
NOD2 directly associates with muramyl dipeptide. A, purified NOD2 was mixed with biotin or biotinylated MDP and then isolated using μMACS streptavidin beads as detailed under “Experimental Procedures.” The MDP-bound proteins were analyzed by SDS-PAGE and immunoblot analysis with anti-NOD2 antibodies. The lanes were loaded as follows. Control, μMACS streptavidin bead-associated NOD2 after incubation of NOD2 with biotin; Input, the amount of NOD2 incubated with μMACS streptavidin beads; Biotin-MDP, μMACS streptavidin bead-associated NOD2 after incubation of NOD2 with biotinylated MDP. B, HEK293T cells were transfected with plasmids encoding V5-tagged β-galactosidase, NOD2, NLRC4, or FLAG-tagged NOD1. Cell lysates were prepared from the transfected cells, and MDP-binding proteins were isolated using biotinylated MDP and μMACS streptavidin beads as described in A. The MDP-bound proteins were resolved using SDS-polyacrylamide gel and immunoblot analysis with anti-V5 antibody or anti-FLAG antibody as indicated. C, cells were transfected with plasmids encoding full-length NOD2 or the indicated NOD2 truncation tagged with V5 epitope or HaloTag as indicated. Cell lysates were prepared from the transfected cells, and MDP-binding proteins were isolated using biotinylated MDP and detected as described in B. Cell lysates were prepared from the transfected cells, and MDP-binding proteins were isolated using biotinylated MDP and detected as described in B. The protein and domain truncations are indicated as follows. LacZ, E. coli β-galactosidase; NOD2, full-length NOD2 (amino acids 1–1040); N-NBD, NOD2 amino acids 1–821; NBD, NOD2 amino acids 216–821; NBD-LRR, NOD2 amino acids 216–1040; N-term, NOD2 amino acids 1–215; Halo-NOD2, full-length NOD2 (NOD2 amino acids 1–215); Halo-LRR, NOD2 amino acids 705–1040.
Recombinant NOD2 Homo-oligomerizes and Binds RIP2K
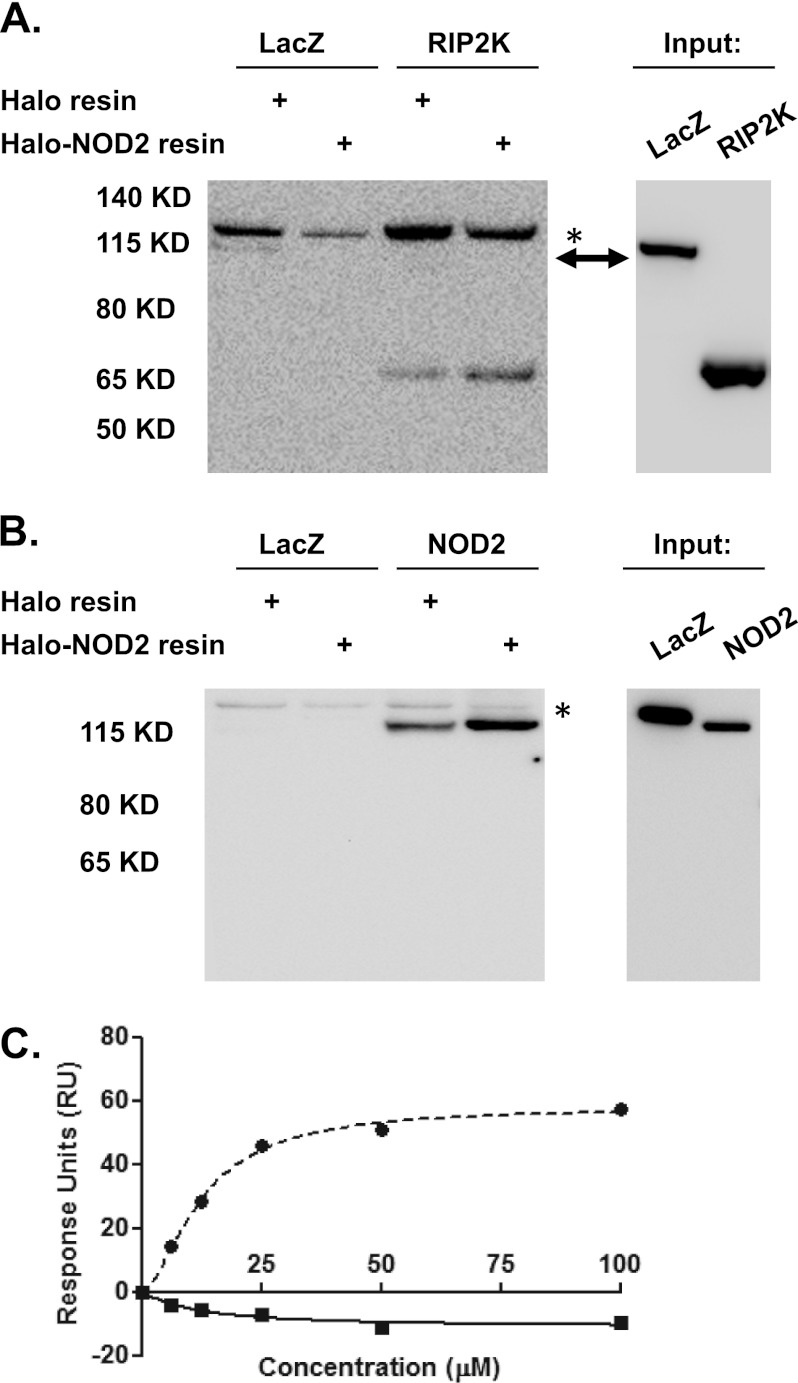
The activation of NOD2 signaling is thought to depend on the homo-oligomerization of the protein that facilitates the formation of a signaling complex that includes the kinase RIP2K (12, 13). We sought to determine whether recombinant purified NOD2 was able to form this signaling complex. Recombinant Halo-NOD2-His6, enriched by immobilized nickel chromatography, was immobilized by incubation with HaloLink resin. Lysates from cells transfected with plasmids that express either V5-tagged NOD2 or RIP2K were incubated with immobilized NOD2 or control resin in which NOD2 was removed using TEV protease. The association of NOD2 or RIP2K from the transfected cell lysates with immobilized Halo-NOD2 and control resin was assayed by immunoblot analysis. Both NOD2 and RIP2K were found to associate with Halo-NOD2-His6, whereas V5-tagged β-galactosidase did not (Fig. 6, A and B). Weak association of NOD2 and RIP2K with control resin was noted and may be the result of incomplete removal of NOD2 or nonspecific interactions with the resin (Fig. 6, A and B). Yeast two-hybrid studies have previously shown the CARDs of NOD2 to undergo homotypic interactions (38). To further assess the functionality of the full-length NOD2, we sought to repeat these observations using surface plasmon resonance. Full-length NOD2 was immobilized, and increasing concentrations of recombinant NOD2 CARDs or GST were used as analyte. Specific binding was only observed to the NOD2 CARDs (Fig. 6C) with a measurable Kd of 12.37 ± 0.58 μm. Non-linear regression using a specific binding model with Hill slope produced a value of the Hill slope of 1.6 ± 0.13, indicative of multiple binding sites and potential cooperativity consistent with a tandem CARD interaction process (Fig. 6C). These data suggest that recombinant purified NOD2 not only binds ATP and MDP but also is capable of supporting protein-protein interactions important in in vivo NOD2 signaling.
FIGURE 6.
Recombinant NOD2 binds to NOD2 and RIP2K. A, HEK293T cells were transfected with β-galactosidase (LacZ) or RIP2K, as indicated. Immobilized NOD2 was generated by incubation of Halo-NOD2-His6 expressed in Sf9 cells (as described in the legend to Fig. 1) with HaloLink resin. Control resin was prepared by removal of the NOD2 fusion protein using TEV protease cleavage prior to incubation with the cellular lysates. Lysates from the transfected cells were incubated with immobilized NOD2 or the control resin as indicated. The bound proteins were subjected to SDS-PAGE and immunoblot analysis with anti-V5 antibody. B, HEK293T cells were transfected with β-galactosidase or NOD2, as indicated, and NOD2 binding of transfected proteins was determined as described in A. C, recombinant NOD2 binds to NOD2 CARD domain. Recombinant NOD2 CARDs and GST proteins at concentrations of 0, 6.25, 12.5, 25, 50, and 100 μm were passed over full-length NOD2 immobilized on a CM5 chip. Background subtraction from a protein-free reference surface was performed, and the data were analyzed using BiaEvaluation software and GraphPad Prism 5.0.
ATP Binding Modulates MDP Binding and Homo-oligomerization by Recombinant NOD2
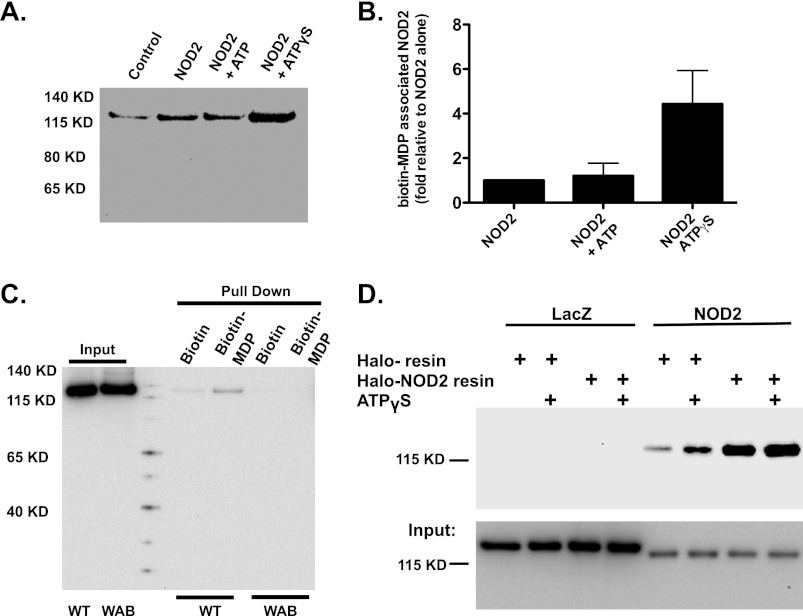
We sought to determine whether ATP binding by NOD2 could alter biologically relevant activities of the recombinant protein. The association of purified NOD2 with MDP was assessed as described above after NOD2 was incubated with ATP, ATPγS, or no nucleotide. MDP binding by NOD2 was enhanced by incubation with non-hydrolyzable ATP analog but not with ATP (Fig. 7, A and B). Interestingly, the NOD2 WAB protein binding to biotinylated MDP was dramatically reduced when compared with WT NOD2 (Fig. 7C), further supporting the concept that MDP binding by NOD2 is modulated by association of the protein with ATP. The homo-oligomerization of NOD2 was also assessed by assaying interaction between V5-tagged NOD2 in transfected cell lysates and HaloLink, resin-bound, recombinant Halo-NOD2-His6 incubated with ATP, ATPγS, or no nucleotide. As with association between NOD2 and MDP, NOD2 homo-oligomerization was increased by binding to ATPγS (Fig. 7D).
FIGURE 7.
Recombinant NOD2 MDP binding and homo-oligomerization is regulated by ATP binding. A, recombinant NOD2 was mixed with biotinylated MDP in the absence or presence of ATP and ATPγS (10 μm). MDP-bound protein was separated by magnetic isolation and analyzed with SDS-PAGE and immunoblot analysis with anti-NOD2 antibody. Each panel represents an independent experiment. B, band intensity from the immunoblots in A was determined using AlphaView® image analysis software. The quantity of NOD2 associated with immobilized Halo-NOD2-His6 in the presence of ATP or ATPγS was calculated relative to quantity associated in the absence of nucleotide after subtraction of the background (control resin pull-down) for each individual experiment. The relative quantities of recovered NOD2 are plotted as mean ± S.E. (error bars). C, purified WT and WAB NOD2 (as indicated) was mixed with biotin or biotin-MDP, isolated using μMACS streptavidin beads, and analyzed by SDS-PAGE and immunoblot analysis with anti-NOD2 antibodies as described in the legend to Fig. 5A. Lanes were loaded with samples as indicated. Input, the amount of NOD2 (WT or WAB, as indicated) incubated with μMACS streptavidin beads; Biotin, μMACS streptavidin bead-associated NOD2 (WT or WAB, as indicated) after incubation of NOD2 with biotin; Biotin-MDP, μMACS streptavidin bead-associated NOD2 (WT or WAB, as indicated) after incubation of NOD2 with biotinylated MDP. D, lysates from HEK293T cells transfected with V5-tagged NOD2-expressing plasmid were incubated with immobilized Halo-NOD2-His6 or control resin in the presence of ATP or ATPγS (10 μm), as described in the legend to Fig. 6B. The binding of V5-tagged NOD2 to Halo-NOD2-His6 was assessed by immunoblot analysis as described in the legend to Fig. 6.
DISCUSSION
This work shows that highly purified, recombinant full-length NOD2 binds to and hydrolyzes ATP. Similar to related proteins NOD1, NLRP3, and NLRP12, NOD2 favors ATP binding over other nucleotides (25–28). Unlike purified NOD1, which does not bind dATP, NOD2 appears to have some affinity for dATP (25). NLRP1 and CIITA do not share this nucleotide preference, and it is not clear what selective pressures may have led to the alterations in nucleotide specificity by different NLR proteins (29, 30). Some intracellular bacteria produce ecto-ATPases and ecto-NTPDases that might affect host intracellular nucleotide triphosphate levels (39). Additionally, physiologic stressors, such as starvation or hypoxia, may also alter the intracellular nucleotide triphosphate pools. It may be that signaling activities of some NLR family members are beneficial to the host under these conditions, whereas others are not.
The rapid disassociation of ATPγS from purified NOD2 suggests that the ATP-bound enzyme is likely to associate with other cellular factors that slow the rate of ATP disassociation during signaling or, alternatively, that ATP hydrolysis and not ATP binding alone is important in the signaling activities of NOD2. Our observation that non-hydrolyzable ATP analogs seem to promote NOD2 activation, as measured by homo-oligomerization, in vitro suggests that NOD2-ATP may be stabilized during signal transduction. In addition to the effector pathways of RIP2K and ATG16L, numerous proteins have been found to bind to NOD2 and regulate MDP-induced signaling in cell-based assays of NOD2 signaling. These include 1) proteins that regulate small GTP-binding protein function (ERBIN and GEF-H1); 2) CARD domain-containing proteins (CARD8 and caspase-12); and 3) proteins involved in ubiquitination of signaling proteins (TRAF4, A20, and SGT1) (40–47). It is possible that these proteins or post-translational modifications resulting from the activity of these may act to inhibit ATP disassociation or slow ATP hydrolysis in order to promote NOD2 signaling. In studies of transfected cells, NOD2 E383K has a high level of NF-κB activation in the absence of MDP stimulation that is not further activated by MDP exposure (20). Structural predictions suggest that this mutant might have intact ATP binding but impaired ATP hydrolysis, which supports our in vitro findings with non-hydrolyzable ATPγS. Other mutations in the Walker A or Walker B motif that are predicted to disrupt ATP binding lead to decreased signaling base line and MDP-stimulated NF-κB activation. Thus, analysis of the in vivo function of rationally designed mutant NOD2 and our biochemical analysis of purified NOD2 suggest that ATP binding is important for assembly of the signaling complex and that hydrolysis is required for deactivation. In vivo analysis of similar mutations in NOD1 indicates that ATP binding and hydrolysis are important in NOD1 signaling (20). However, analysis of ATP binding and hydrolysis in vitro by NOD1 and mutant NOD1 needs to be explored to confirm this apparent difference in signaling mechanisms by these related proteins. Additionally, dysregulated NOD2 signaling, resulting from missense mutations, is present in hereditary inflammatory disorders, including Crohn disease and Blau syndrome. Many of these mutations actually fall within the nucleotide-binding domain of NOD2 (5–9). Further studies of NOD2 nucleotide binding in the setting of these mutations are needed to better understand how this activity of the protein regulates inflammatory signaling.
Our understanding of the role of NOD1 and NOD2 as peptidoglycan component receptors has evolved over the past decade. Early studies by Nuñez and colleagues (48) suggested that NOD1 and NOD2 conferred responsiveness to bacterial LPS. These studies were coupled with data indicating that radiolabeled LPS could co-immunoprecipitate with NOD1 when added to lysates generated from cells overexpressing NOD1. This work led to an early hypothesis that NOD1 and NOD2 were the physical receptors for cytoplasmic bacterial products. Later, NOD2 and NOD1 were found to signal in response to peptidoglycan-derived molecules, muramyl dipeptide and l-alanyl-γ-d-glutamyl-meso-diaminopimelic acid (tri-DAP), respectively (10, 11, 49, 50). It remains unclear why LPS was found in association with NOD1 in these early studies. It is possible that the observed interaction between NOD1 and LPS was mediated by cellular proteins that bind LPS and associate with NOD1 or that the preparations of labeled LPS were also contaminated with labeled peptidoglycan fragments that bind to NOD1. Although NOD2 has been clearly and repeatedly implicated as a member of the MDP-sensing signal transduction pathway, demonstration of its role as the receptor molecule for MDP has remained elusive; in one study of proteins that bound immobilized muramyl dipeptide, NOD2 did not associate with MDP, whereas calreticulin did (51, 52). Our current work clearly demonstrates that highly purified NOD2 is able to associate with MDP in vitro. Surprisingly, in cellular lysates with conditions that are permissive for MDP binding by NOD2, we now show that the binding to MDP is dependent on the nucleotide-binding domain of NOD2 rather than the leucine-rich repeat domain, where binding had previously been hypothesized to take place. Mutations in the NOD2 LRR domain that are associated with Crohn disease predisposition and numerous mutations made in the LRR resulting from systematic mutation of 60 residues predicted to be solvent-exposed confer diminished MDP responsiveness (10, 11, 19). Additionally, the substitution of the NOD2 LRR for the NOD1 LRR results in an MDP-responsive NOD1 molecule in cell-based assays (37). These data support the hypothesis that MDP-mediated activation of NOD2 requires the LRR domain of the protein. Cell-based analysis of mutant NOD1 molecules combined with molecular modeling of the LRR domain strongly suggests that the NOD1 activator, tri-DAP, associates with the LRR of NOD1 (37). Additional evidence of a direct interaction between NOD1 and tri-DAP is provided by the demonstration that full-length recombinant NOD1 purified from lentivirus-transduced mammalian cells forms homo-oligomers when incubated with ATP and tri-DAP (25). A recent study further demonstrated that full-length recombinant NOD1 covalently coupled to a gold chip associated with tri-DAP, whereas a truncated NOD1 lacking the NBD and LRR did not (53). However, this study lacks refined truncations required to differentiate binding of tri-DAP to the NBD or LRR domains. We cannot rule out the possibility that the isolated NOD2 LRR protein in our transfected cells is misfolded when expressed without the remainder of the protein or that the LRR and nucleotide-binding domain may both contribute MDP binding properties to the full-length NOD2 protein. Our data raise the possibility that intramolecular interactions between the NOD2 NBD and NOD2 LRR may control signaling activity of the protein and may be modulated by binding of MDP to the NBD or an interface of the NBD and LRR. We have not yet been able to purify the individual domains of NOD2, particularly the nucleotide-binding domain alone, to confirm that NBD domain directly interacts with MDP, and this remains an important question to be pursued.
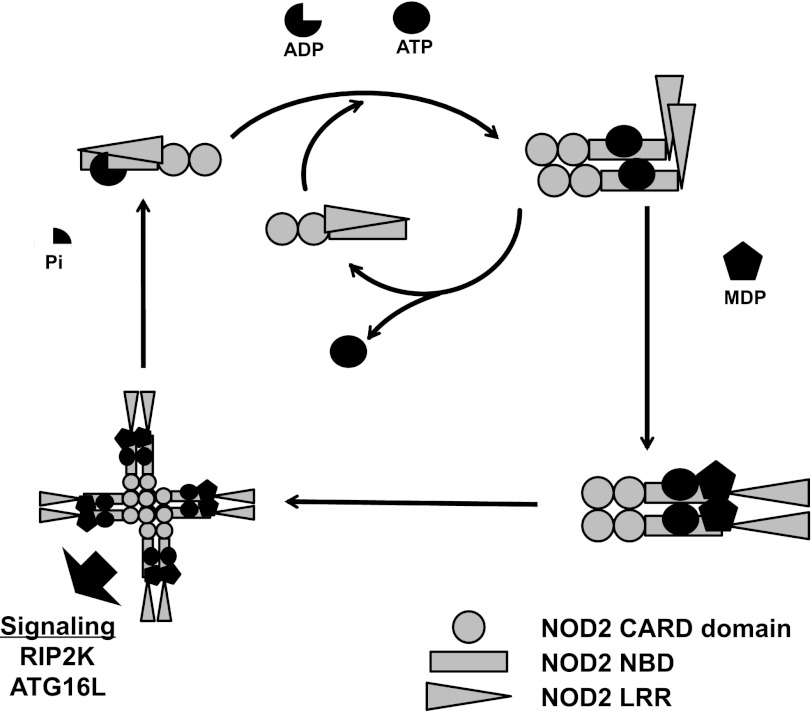
Based on the biochemical properties of purified NOD2 that we have presented and the analysis of mutant NOD2 molecules in cellular assays published by others (20), we propose a model as a working hypothesis for the role of nucleotide binding in NOD2 signaling (Fig. 8). In this model, NOD2 can rapidly cycle between the ATP-bound and open state while signaling is inactive. However, binding to ATP leads to the formation of a state that is competent for homo-oligomerization and MDP binding. Homo-oligomerization is facilitated probably by both CARD-CARD and NBD-NBD interactions. ATP-binding by NOD2 probably induces conformational changes that are dependent on intact Walker A and Walker B motifs that facilitate association with MDP and assembly of a signaling-competent complex. Our data do not currently distinguish binding of MDP by ATP-NOD2 monomers or oligomers. Data from the literature suggest that the binding of MDP then leads to formation of an active signaling complex that contains both oligomeric ATP-NOD2 and other adapter and effector molecules, particularly binding of RIP2K through CARD-CARD interactions. It is likely that NOD2 CARD domains can support multiple protein protein interactions through distinct surface patches, as has been described for other CARD domains (54). The signaling complex is shut off by hydrolysis of ATP by NOD2 and disassembly of the oligomer. Although the data presented herein support the NOD2 interactions with ATP and MDP and the proposed changes in oligomerization state, further experiments are required to refine this working model and determine the sites in the ATP hydrolysis cycle that are regulated by (or regulate) interactions between NOD2 and other cellular proteins involved in this signaling axis.
FIGURE 8.
Hypothetical model for NOD2 signaling activation. In the basal, autoinhibited state, NOD2 is monomeric either bound to ADP (three-quarters circle) or free of nucleotide. Binding of ATP (full circle) promotes an open conformation that allows for homo-oligomerization and the ability to respond to MDP (pentagon). Binding of MDP to ATP-NOD2 promotes higher order oligomerization and/or stabilizes ATP-induced oligomerization to promote induction of signaling. Hydrolysis of ATP to ADP restores the basal state.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA131645 (to B. K. D. and J. A. D.) and AI088255 (to J. A. D.). This work was supported by the Burroughs Wellcome Fund Career Award for Medical Scientists (to J. A. D.), the Crohn's and Colitis Foundation Career Development Award and Franklin and Marshall College (to B. K. D.), Wellcome Trust Grant WT0805090MA (to T. P. M.), and Biotechnology and Biological Sciences Research Council Grant RG52820 (to T. P. M.).

This article contains supplemental Table 1 and Fig. 1.
- NLR
- nucleotide-binding domain and leucine-rich repeat-containing protein
- CARD
- caspase activation and recruitment domain
- MDP
- muramyl dipeptide
- CIITA
- class II transcriptional activator
- TEV
- tobacco etch virus
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- tri-DAP
- l-alanyl-γ-d-glutamyl-meso-diaminopimelic acid
- NBD
- nucleotide-binding domain
- LRR
- leucine-rich repeat.
REFERENCES
- 1. Ting J. P., Lovering R. C., Alnemri E. S., Bertin J., Boss J. M., Davis B. K., Flavell R. A., Girardin S. E., Godzik A., Harton J. A., Hoffman H. M., Hugot J. P., Inohara N., Mackenzie A., Maltais L. J., Nunez G., Ogura Y., Otten L. A., Philpott D., Reed J. C., Reith W., Schreiber S., Steimle V., Ward P. A. (2008) The NLR gene family. A standard nomenclature. Immunity 28, 285–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harton J. A., Linhoff M. W., Zhang J., Ting J. P. (2002) Cutting edge. CATERPILLER. a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J. Immunol. 169, 4088–4093 [DOI] [PubMed] [Google Scholar]
- 3. Inohara N., Ogura Y., Nuñez G. (2002) Nods. A family of cytosolic proteins that regulate the host response to pathogens. Curr. Opin. Microbiol. 5, 76–80 [DOI] [PubMed] [Google Scholar]
- 4. Ting J. P., Kastner D. L., Hoffman H. M. (2006) CATERPILLERs, pyrin, and hereditary immunological disorders. Nat. Rev. Immunol. 6, 183–195 [DOI] [PubMed] [Google Scholar]
- 5. Hampe J., Cuthbert A., Croucher P. J., Mirza M. M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A. J., Bridger S., van Deventer S., Forbes A., Nikolaus S., Lennard-Jones J. E., Foelsch U. R., Krawczak M., Lewis C., Schreiber S., Mathew C. G. (2001) Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet 357, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 6. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 7. Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 8. Miceli-Richard C., Lesage S., Rybojad M., Prieur A. M., Manouvrier-Hanu S., Häfner R., Chamaillard M., Zouali H., Thomas G., Hugot J. P. (2001) CARD15 mutations in Blau syndrome. Nat. Genet. 29, 19–20 [DOI] [PubMed] [Google Scholar]
- 9. Wang X., Kuivaniemi H., Bonavita G., Mutkus L., Mau U., Blau E., Inohara N., Nunez G., Tromp G., Williams C. J. (2002) CARD15 mutations in familial granulomatosis syndromes. A study of the original Blau syndrome kindred and other families with large-vessel arteritis and cranial neuropathy. Arthritis Rheum. 46, 3041–3045 [DOI] [PubMed] [Google Scholar]
- 10. Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 11. Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S. J., Moran A. P., Fernandez-Luna J. L., Nuñez G. (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 [DOI] [PubMed] [Google Scholar]
- 12. Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Núñez G., Janeway C. A., Medzhitov R., Flavell R. A. (2002) RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 [DOI] [PubMed] [Google Scholar]
- 13. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 14. Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D. J., Campbell B. J., Jewell D., Simmons A. (2010) NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 [DOI] [PubMed] [Google Scholar]
- 15. Homer C. R., Richmond A. L., Rebert N. A., Achkar J. P., McDonald C. (2010) ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology 139, 1630–1641, 1641.e1–1641.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Travassos L. H., Carneiro L. A., Ramjeet M., Hussey S., Kim Y. G., Magalhães J. G., Yuan L., Soares F., Chea E., Le Bourhis L., Boneca I. G., Allaoui A., Jones N. L., Nuñez G., Girardin S. E., Philpott D. J. (2010) Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 [DOI] [PubMed] [Google Scholar]
- 17. Chamaillard M., Philpott D., Girardin S. E., Zouali H., Lesage S., Chareyre F., Bui T. H., Giovannini M., Zaehringer U., Penard-Lacronique V., Sansonetti P. J., Hugot J. P., Thomas G. (2003) Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc. Natl. Acad. Sci. U.S.A. 100, 3455–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonen D. K., Ogura Y., Nicolae D. L., Inohara N., Saab L., Tanabe T., Chen F. F., Foster S. J., Duerr R. H., Brant S. R., Cho J. H., Nuñez G. (2003) Crohn's disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology 124, 140–146 [DOI] [PubMed] [Google Scholar]
- 19. Tanabe T., Chamaillard M., Ogura Y., Zhu L., Qiu S., Masumoto J., Ghosh P., Moran A., Predergast M. M., Tromp G., Williams C. J., Inohara N., Núñez G. (2004) Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 23, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zurek B., Proell M., Wagner R. N., Schwarzenbacher R., Kufer T. A. (2012) Mutational analysis of human NOD1 and NOD2 NACHT domains reveals different modes of activation. Innate Immunity 18, 100–111 [DOI] [PubMed] [Google Scholar]
- 21. Albrecht M., Domingues F. S., Schreiber S., Lengauer T. (2003) Structural localization of disease-associated sequence variations in the NACHT and LRR domains of PYPAF1 and NOD2. FEBS Lett. 554, 520–528 [DOI] [PubMed] [Google Scholar]
- 22. Albrecht M., Lengauer T., Schreiber S. (2003) Disease-associated variants in PYPAF1 and NOD2 result in similar alterations of conserved sequence. Bioinformatics 19, 2171–2175 [DOI] [PubMed] [Google Scholar]
- 23. Tameling W. I., Elzinga S. D., Darmin P. S., Vossen J. H., Takken F. L., Haring M. A., Cornelissen B. J. (2002) The tomato R gene products I-2 and MI-1 are functional ATP-binding proteins with ATPase activity. Plant Cell 14, 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tameling W. I., Vossen J. H., Albrecht M., Lengauer T., Berden J. A., Haring M. A., Cornelissen B. J., Takken F. L. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Askari N., Correa R. G., Zhai D., Reed J. C. (2012) Expression, purification, and characterization of recombinant NOD1 (NLRC1). A NLR family member. J. Biotechnol. 157, 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duncan J. A., Bergstralh D. T., Wang Y., Willingham S. B., Ye Z., Zimmermann A. G., Ting J. P. (2007) Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 8041–8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ye Z., Lich J. D., Moore C. B., Duncan J. A., Williams K. L., Ting J. P. (2008) ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol. Cell. Biol. 28, 1841–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu C., Wang A., Wang L., Dorsch M., Ocain T. D., Xu Y. (2005) Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem. Biophys. Res. Commun. 331, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 29. Harton J. A., Cressman D. E., Chin K. C., Der C. J., Ting J. P. (1999) GTP binding by class II transactivator. Role in nuclear import. Science 285, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 30. Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., Reed J. C. (2007) Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 25, 713–724 [DOI] [PubMed] [Google Scholar]
- 31. Kim H. E., Du F., Fang M., Wang X. (2005) Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. U.S.A. 102, 17545–17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bewry N. N., Bolick S. C., Wright K. L., Harton J. A. (2007) GTP-dependent recruitment of CIITA to the class II major histocompatibility complex promoter. J. Biol. Chem. 282, 26178–26184 [DOI] [PubMed] [Google Scholar]
- 33. Silverman N., Maniatis T. (2001) NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342 [DOI] [PubMed] [Google Scholar]
- 34. Northup J. K., Smigel M. D., Gilman A. G. (1982) The guanine nucleotide-activating site of the regulatory component of adenylate cyclase. Identification by ligand binding. J. Biol. Chem. 257, 11416–11423 [PubMed] [Google Scholar]
- 35. Brandt D. R., Asano T., Pedersen S. E., Ross E. M. (1983) Reconstitution of catecholamine-stimulated guanosinetriphosphatase activity. Biochemistry 22, 4357–4362 [DOI] [PubMed] [Google Scholar]
- 36. Traut T. W. (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem. 222, 9–19 [DOI] [PubMed] [Google Scholar]
- 37. Girardin S. E., Jéhanno M., Mengin-Lecreulx D., Sansonetti P. J., Alzari P. M., Philpott D. J. (2005) Identification of the critical residues involved in peptidoglycan detection by Nod1. J. Biol. Chem. 280, 38648–38656 [DOI] [PubMed] [Google Scholar]
- 38. Wagner R. N., Proell M., Kufer T. A., Schwarzenbacher R. (2009) Evaluation of Nod-like receptor (NLR) effector domain interactions. PLoS One 4, e4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sansom F. M., Robson S. C., Hartland E. L. (2008) Possible effects of microbial ecto-nucleoside triphosphate diphosphohydrolases on host-pathogen interactions. Microbiol. Mol. Biol. Rev. 72, 765–781, Table of Contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao Y., Alonso C., Ballester I., Song J. H., Chang S. Y., Guleng B., Arihiro S., Murray P. J., Xavier R., Kobayashi K. S., Reinecker H. C. (2012) Control of NOD2 and Rip2-dependent innate immune activation by GEF-H1. Inflamm. Bowel Dis. 18, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kufer T. A., Kremmer E., Banks D. J., Philpott D. J. (2006) Role for erbin in bacterial activation of Nod2. Infect. Immun. 74, 3115–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macdonald T. T., Monteleone G. (2005) Immunity, inflammation, and allergy in the gut. Science 307, 1920–1925 [DOI] [PubMed] [Google Scholar]
- 43. von Kampen O., Lipinski S., Till A., Martin S. J., Nietfeld W., Lehrach H., Schreiber S., Rosenstiel P. (2010) Caspase recruitment domain-containing protein 8 (CARD8) negatively regulates NOD2-mediated signaling. J. Biol. Chem. 285, 19921–19926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. LeBlanc P. M., Yeretssian G., Rutherford N., Doiron K., Nadiri A., Zhu L., Green D. R., Gruenheid S., Saleh M. (2008) Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe 3, 146–157 [DOI] [PubMed] [Google Scholar]
- 45. Mayor A., Martinon F., De Smedt T., Pétrilli V., Tschopp J. (2007) A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 8, 497–503 [DOI] [PubMed] [Google Scholar]
- 46. Marinis J. M., Homer C. R., McDonald C., Abbott D. W. (2011) A novel motif in the Crohn's disease susceptibility protein, NOD2, allows TRAF4 to down-regulate innate immune responses. J. Biol. Chem. 286, 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hitotsumatsu O., Ahmad R. C., Tavares R., Wang M., Philpott D., Turer E. E., Lee B. L., Shiffin N., Advincula R., Malynn B. A., Werts C., Ma A. (2008) The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity 28, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inohara N., Ogura Y., Chen F. F., Muto A., Nuñez G. (2001) Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J. Biol. Chem. 276, 2551–2554 [DOI] [PubMed] [Google Scholar]
- 49. Chamaillard M., Hashimoto M., Horie Y., Masumoto J., Qiu S., Saab L., Ogura Y., Kawasaki A., Fukase K., Kusumoto S., Valvano M. A., Foster S. J., Mak T. W., Nuñez G., Inohara N. (2003) An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4, 702–707 [DOI] [PubMed] [Google Scholar]
- 50. Girardin S. E., Boneca I. G., Carneiro L. A., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M. K., Labigne A., Zähringer U., Coyle A. J., DiStefano P. S., Bertin J., Sansonetti P. J., Philpott D. J. (2003) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300, 1584–1587 [DOI] [PubMed] [Google Scholar]
- 51. Chen D., Duggan C., Reden T. B., Kooragayala L. M., Texada D. E., Langford M. P. (2004) Calreticulin is a binding protein for muramyl dipeptide and peptidoglycan in RK13 cells. Biochemistry 43, 11796–11801 [DOI] [PubMed] [Google Scholar]
- 52. Chen D., Texada D. E., Duggan C., Liang C., Reden T. B., Kooragayala L. M., Langford M. P. (2005) Surface calreticulin mediates muramyl dipeptide-induced apoptosis in RK13 cells. J. Biol. Chem. 280, 22425–22436 [DOI] [PubMed] [Google Scholar]
- 53. Laroui H., Yan Y., Narui Y., Ingersoll S. A., Ayyadurai S., Charania M. A., Zhou F., Wang B., Salaita K., Sitaraman S. V., Merlin D. (2011) l-Ala-γ-d-Glu-meso-diaminopimelic acid (DAP) interacts directly with leucine-rich region domain of nucleotide-binding oligomerization domain 1, increasing phosphorylation activity of receptor-interacting serine/threonine-protein kinase 2 and its interaction with nucleotide-binding oligomerization domain 1. J. Biol. Chem. 286, 31003–31013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kersse K., Lamkanfi M., Bertrand M. J., Vanden Berghe T., Vandenabeele P. (2011) Interaction patches of procaspase-1 caspase recruitment domains (CARDs) are differently involved in procaspase-1 activation and receptor-interacting protein 2 (RIP2)-dependent nuclear factor κB signaling. J. Biol. Chem. 286, 35874–35882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.